Abstract
The ability of herpes simplex virus (HSV) to establish lifelong latency in neurons suggests that HSV-derived vectors hold promise for gene delivery to the nervous system. However, vector toxicity and transgene silencing have created significant barriers to vector applications to the brain. Recently, we described a vector defective for all immediate-early gene expression and deleted for the joint region between the two unique genome segments that proved capable of extended transgene expression in non-neuronal cells. Sustained expression required the proximity of boundary elements from the latency locus. As confirmed here, we have also found that a transgene cassette introduced into the ICP4 locus is highly active in neurons but silent in primary fibroblasts. Remarkably, we observed that removal of the virion host shutoff (vhs) gene further improved transgene expression in neurons without inducing expression of viral genes. In rat hippocampus, the vhs-deleted vector showed robust transgene expression exclusively in neurons for at least 1 month without evidence of toxicity or inflammation. This HSV vector design holds promise for gene delivery to the brain, including durable expression of large or complex transgene cassettes.
Keywords: HSV, gene therapy, CNS, virion host shutoff gene, ICP4, ICP0
Introduction
Replication-defective herpes simplex virus (HSV)-based vectors have the potential to provide an invaluable tool to both studies of brain function and treatment of CNS disorders. HSV genomes persisting in neurons do not integrate and can serve as a platform for therapeutic gene expression. Replication-defective HSV vectors have been tested for gene therapy approaches in the CNS1, 2, 3 and peripheral nervous system.4, 5, 6, 7 However, CNS applications have been plagued by residual neuronal toxicity and progressive loss of transgene expression.8, 9, 10 Viral toxicity results from expression of the viral immediate early (IE) protein ICP0, a protein with functions related to viral chromatin modification and resistance to innate anti-viral responses.11 In the absence of ICP0, rapid genome silencing ensues, resulting in low transgene expression, while, in the presence of ICP0, significant toxicity results in loss of neuronal cell function and viability.
In sensory neurons, ICP0 fails to accumulate in the nucleus,12 and ICP0-mediated cytotoxicity is further avoided by silencing of the viral genome and the establishment of latency. The viral latency-associated transcript (LAT) locus remains transcriptionally active during latency, and the LAT promoter elements have been used to express transgenes in sensory neurons.4, 5, 13, 14 However, expression was typically short-term or low,4, 15 and promoter activity was not seen in all latently infected neurons.16 We have recently reported that, in the absence of IE gene activity, transgene expression from a non-viral promoter can be maintained in non-neuronal cells when the expression cassette is placed between two clusters of CTCF binding motifs referred to as CTRL1 and CTRL217 flanking the LAT promoter.18 This arrangement remained active when relocated to different intergenic regions unrelated to the latency locus. Interestingly, primary neurons isolated from fetal rat dorsal root ganglia (rDRG) also displayed expression of a second reporter cassette located in the deleted ICP4 locus. This finding prompted further examination of the ability of the ICP4 locus to support transgene expression in neurons in the brain.
Preliminary observations in our lab suggested that elimination of the virion host shut-off function (vhs) may further reduce the already minimal toxicity of IE gene-depleted vectors. Vhs functions as a virulence factor that reduces immune recognition of infected cells in vivo and helps evade the innate anti-viral interferon (IFN) responses;19, 20, 21 vhs-deficient viruses are attenuated for neurovirulence. Since vhs is an mRNA-specific RNase, we reasoned that it could potentially reduce transgene mRNA and corresponding protein expression. Thus we deleted the vhs (UL41) gene from our most recent vector, JΔNI5,18 generating a new backbone that was named JΔNI8, and compared the 2 vectors for transgene expression from the deleted ICP4 locus in rat brain.
The results of our analyses show that the human ubiquitin C (UbC) promoter placed between the end points of the ICP4 gene deletion allows expression of the linked reporter gene in vivo in rat hippocampal neurons. Furthermore, expression was enhanced by deletion of the vhs gene. No vector-associated toxicity was observed following rat brain injections and expression persisted in neurons of the CNS for at least 1 month, providing an effective platform for the delivery of therapeutic transgenes to the CNS, including multiple transgenes or transgenes that are too large for accommodation by other vector systems.
Results
Vector Construction and Characterization in Non-complementing Fibroblasts
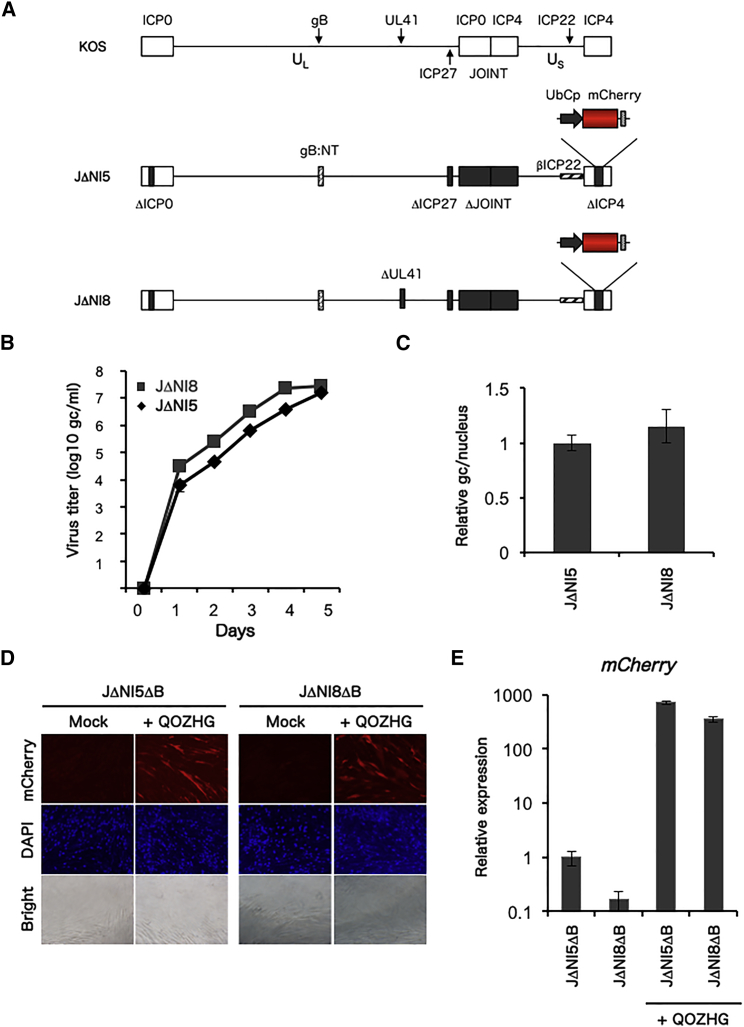
JΔNI8 was derived from JΔNI5 by deletion of the vhs locus (Figure 1A). JΔNI5 and JΔNI8 grew at comparable rates on ICP4- and ICP27-complementing U2OS cells [U2OS-ICP4/27]18 (Figure 1B), showing that vhs did not clearly contribute to virus yield in cultured cells. Infection of human dermal fibroblasts (HDFs) with JΔNI5 and JΔNI8 at 5,000 genome copies (gc)/cell resulted in comparable levels of viral DNA in the nuclei of infected cells at 2 hr post-infection (hpi) (Figure 1C), indicating that deletion of vhs did not alter virus entry or transport of viral DNA into the nucleus.
Figure 1.
Structure and Analysis of the JΔNI8 Vector
(A) Genomic structure of KOS-37 BAC and the derivative JΔNI5 and JΔNI8 vectors loxP-flanked (BAC regions between UL37 and UL38 not shown). UL, unique long segment; US, unique short segment. The terminal and internal inverted repeats are represented by open boxes. Deletions in JΔNI518 and JΔNI8 are indicated by black boxes (Δ). Both vectors contain the entry-enhancing N/T mutations in the gB gene (gB:NT)45 and a ubiquitin C promoter (UbCp)-mCherry cassette in the ICP4 locus; the SV40 poly(A) region of the mCherry cassette is represented by a small patterned box. The cross-hatched horizontal box (βICP22) indicates conversion of the ICP22 IE gene to early-expression kinetics by promoter TAATGARAT deletion. (B) Growth curves of JΔNI5 and JΔNI8 in complementing cells. U2OS-ICP4/27 cells were infected with JΔNI5 and JΔNI8 vectors at 1 gc/cell, and supernatant virus titers were determined in gc per milliliter every day for 5 days (averages of duplicate wells). (C) Relative nuclear viral DNA levels at 2 hr post-infection of HDFs with 5,000 gc of JΔNI5 and JΔNI8. Viral gc numbers were determined by qPCR for the gD gene normalized to the cellular 18S rRNA gene content (averages of duplicate wells). (D) mCherry protein and (E) mRNA expression in vector-infected HDFs without and with ICP0 complementation. Cells were infected at 25,000 gc/cell with JΔNI5ΔB or JΔNI8ΔB and superinfected 6 days later with mock or QOZHG vector (108 gc/well). One day later, microscopy fields were imaged for mCherry fluorescence, DAPI-stained nuclei, and bright field. Separately, mRNA was collected 1 day after superinfection, and mCherry mRNA levels in duplicate samples were measured in triplicate, averaged, and normalized to viral gc in the same samples. Normalized values ±SD are presented relative to JΔNI5ΔB infected cells at 7 dpi.
We previously demonstrated that removal of bacterial artificial chromosome (BAC) sequences from BAC-based viral vectors reduces vector toxicity for non-complementing cells.18 Cre recombination was used to remove the bacterial sequences from JΔNI5 and JΔNI8, creating JΔNI5ΔB and JΔNI8ΔB, respectively. Following transduction of HDFs with JΔNI5ΔB and JΔNI8ΔB, neither vector showed noticeable mCherry transgene expression at 7 dpi (Figure 1D), consistent with our previous results for JΔNI5.18 The presence of viral DNA in the nuclei of infected HDFs (Figure 1C) with little corresponding protein expression suggested that both viral genomes were epigenetically silenced. Restoration of the IE gene ICP0 to ICP0-deficient vectors has been shown to counteract epigenetic repression.22, 23 As anticipated, when JΔNI5ΔB- and JΔNI8ΔB-infected HDFs were superinfected with QOZHG virus12 to supply ICP0 expression, mCherry expression was restored (Figures 1D and 1E). While the qRT-PCR data (Figure 1E) suggested that the vhs deletion caused a decrease in mCherry mRNA levels (∼6-fold), these levels were extremely low compared to those in the presence of ICP0 (Figure 1E).
Characterization of Gene Expression in Neuronal Cells
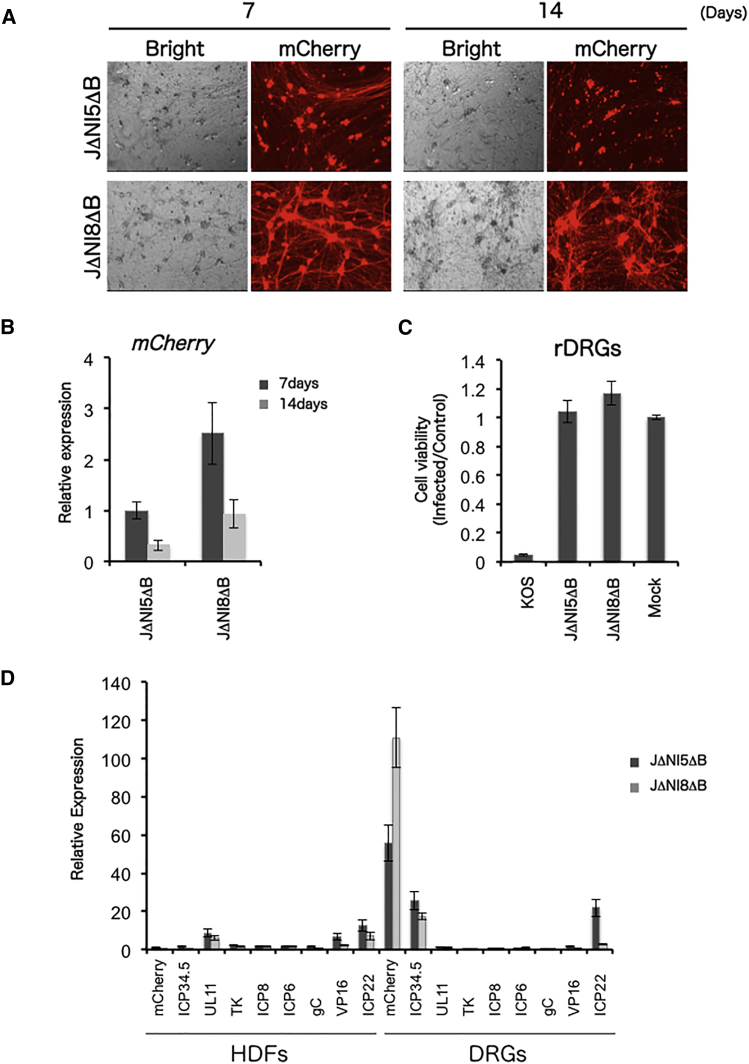
We previously reported that mCherry expression from JΔNI5-based vectors was markedly higher in rDRG neurons than in fibroblasts in vitro at 3 dpi. We therefore infected rDRG cultures with JΔNI5ΔB and JΔNI8ΔB to assess the effect of vhs deletion on transgene expression in these cells. At 7 and 14 dpi, rDRGs infected with either virus showed abundant mCherry fluorescence, but the signals were stronger in JΔNI8ΔB- than in JΔNI5ΔB-infected cultures (Figures 2A and S1). Analysis of reverse transcribed mRNA by qPCR revealed that mCherry gene expression in JΔNI8ΔB-infected rDRGs was approximately 2.5- and 3-fold higher than in JΔNI5ΔB-infected rDRGs at 7 and 14 dpi, respectively (Figure 2B). To eliminate the possibility that mCherry expression was affected by cytotoxicity, we assessed cell viability after transduction of rDRGs with JΔNI5ΔB or JΔNI8ΔB. No difference in cell viability was observed between vector- and mock-infected cells, indicating that neither vector displayed noticeable cytotoxicity (Figure 2C). Together, these results showed that in neuronal cells, both JΔNI5 and JΔNI8 can mediate prolonged transgene expression from the UbC promoter at the ICP4 locus without apparent cytotoxicity and that increased transgene expression correlates with the vhs gene deletion in JΔNI8.
Figure 2.
JΔNI5ΔB and JΔNI8ΔB Transgene Expression in HDFs and rDRGs
(A) mCherry transgene expression in rDRG cultures. Cells were infected with 3,000 gc/cell of JΔNI5ΔB (top) or JΔNI8ΔB (bottom). Bright-field (left) and mCherry fluorescence images (right) were taken at 7 (left panels) and 14 dpi (right panels). (B) mCherry mRNA levels in JΔNI5ΔB- and JΔNI8ΔB-infected rDRGs at 7 and 14 dpi. mCherry mRNA levels were normalized to viral gc in the same samples, and the normalized values are presented relative to JΔNI5ΔB-infected cells at 7 dpi. (C) Cytotoxicity for rDRGs in culture. Cells were infected with KOS, JΔNI5ΔB, or JΔNI8ΔB virus at 3,000 gc/cell or mock-infected, and cell viability was measured in triplicate by MTT assay at 5 dpi. Plotted values represent the mean ratios of virus-infected to mock-infected cells. (D) Transgene and viral gene expression in HDFs (left) and rDRGs (right). HDF and rDRG cultures were infected with JΔNI5ΔB or JΔNI8ΔB vectors at 25,000 gc/cell or 3,000 gc/cell, respectively. At 7 dpi, mRNA levels of selected genes were measured by qRT-PCR analysis. Data conversion was performed using JΔNI5 BAC DNA standard curves generated with the same gene-specific primers and normalization to viral gc in each sample. Values are presented relative to JΔNI5ΔB-infected HDF at 7 dpi. Data in (B) and (D) represent averages ±SD of two independent experiments.
To determine whether the vhs gene influences global viral gene expression, we measured the mRNA levels of the mCherry transgene and selected viral genes representing different kinetic classes and regions of the viral genome in both HDFs and rDRGs (Figure 2D). In HDFs, expression of most viral genes from JΔNI5ΔB and JΔNI8ΔB was comparable, regardless of kinetic class or genomic location, indicating that removal of vhs did not dramatically alter viral gene expression (Figure 2D, left side). In DRGs, expression of individual genes was also similar between JΔNI5ΔB and JΔNI8ΔB, but with a few notable exceptions. mCherry expression per genome copy was 50- to 100-fold higher than in HDFs, considerably higher than expression of any of the viral genes, and approximately 2-fold higher from JΔNI8ΔB than from JΔNI5ΔB (Figure 2D, right side). Of interest, the expression of ICP34.5 in DRGs, while similar between the two vectors, was also elevated compared to HDFs (∼25-fold for JΔNI8ΔB, ∼17-fold for JΔNI5ΔB). The ICP34.5 gene (γ134.5) and the mCherry transgene are both located in a terminal repeat, relatively close to one another in the circular genome. The kinetically modified promoter of the ICP22 gene is also located in a terminal repeat in our JΔNI viruses and ICP22 was expressed more abundantly than the majority of viral genes in JΔNI5ΔB-infected DRGs. However, this was not seen in JΔNI8ΔB-infected DRGs. Together, these results indicated that the vhs deletion caused selective upregulation of mCherry expression in a cell-restricted manner without considerable changes in the global pattern of HSV gene expression.
Transgene Expression from the LAT Region in Neuronal and Non-neuronal Cells
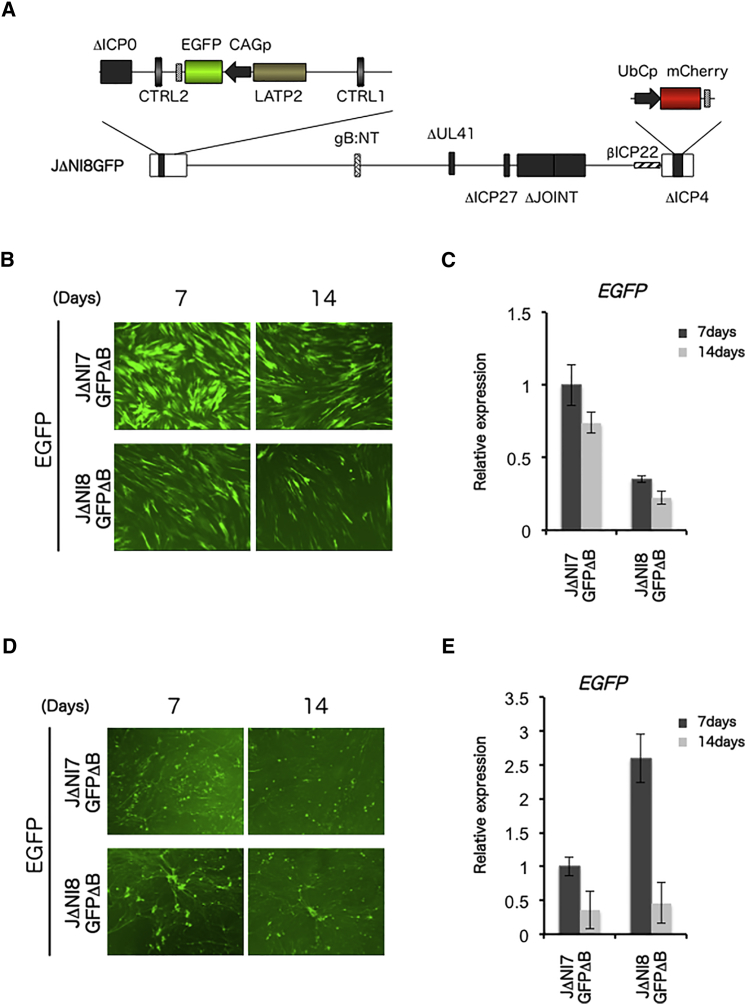
We recently reported that GFP transgene expression from the CAG promoter (CMV immediate-early enhancer/chicken β-actin promoter/chimeric intron) was maintained in non-neuronal cells when the expression cassette was placed between two clusters of CTCF binding motifs flanking the LAT promoter in JΔNI5; this vector was referred to as JΔNI7GFP.18 We deleted the vhs gene from JΔNI7GFP, creating JΔNI8GFP (Figure 3A), to assess the effect on GFP expression. HDFs and rDRGs were infected with BAC-deleted (ΔB) JΔNI7GFP or JΔNI8GFP, GFP fluorescence from infected cells was visualized at 7 and 14 dpi, and GFP mRNA levels were measured in the same cells. In HDFs, GFP fluorescence in JΔNI7GFPΔB-infected cells was higher at both time points than in JΔNI8GFPΔB-infected cells (Figure 3B), and mRNA levels were approximately 3-fold higher in JΔNI7GFPΔB- than in JΔNI8GFPΔB-infected cells at both 7 and 14 dpi, decreasing by ∼25% (JΔNI7GFPΔB) and ∼35% (JΔNI8GFPΔB) between the two time points (Figure 3C). In contrast, GFP fluorescence in rDRGs was higher at 7 and 14 days post-infection with JΔNI8GFPΔB than with JΔNI7GFPΔB (Figure 3D), and GFP mRNA abundance was 2.5-fold higher in JΔNI8GFPΔB- than in JΔNI7GFPΔB-infected cultures at 7 dpi, although a difference was no longer observed at 14 dpi (Figure 3E). While the increase in GFP expression in rDRGs caused by the vhs deletion was thus transient compared to the increase in mCherry expression in rDRGs noted earlier, these results indicated that the vhs deletion promoted transgene expression in neuronal cells independent of the promoter or location of the expression cassette within the viral genome. We confirmed that the disruption of the LAT locus and insertion of the GFP cassette did not noticeably alter the effect of the vhs deletion on mCherry expression (Figure S2). The reduced expression of GFP observed in HDFs highlights the cell-type dependence of the effect of vhs removal on transgene expression.
Figure 3.
GFP Expression from JΔNI7GFPΔB and JΔNI8GFPΔB in Infected HDFs and DRGs
(A) Schematic diagram of the genomic structure of the JΔNI8GFPΔB vector derived from JΔNI7GFP18 by deletion of the vhs (UL41) gene. Like JΔNI7GFP, JΔNI8GFP contains a CAG promoter-eGFP expression cassette in the LAT intron between an enhancer-like LAT region (LAP2 or LATP2)4, 46, 47 and CTRL2. (B) GFP fluorescence and (C) relative GFP mRNA levels in JΔNI7GFPΔB- and JΔNI8GFPΔB-infected HDFs (25,000 gc/cell) at 7 and 14 dpi. GFP qRT-PCR data were normalized to viral gc in the same samples and are presented as expression relative to that in JΔNI7GFPΔB infected cells at 7 dpi. (D) GFP fluorescence and (E) relative GFP mRNA levels in JΔNI7GFPΔB- and JΔNI8GFPΔB-infected rDRGs (3,000 gc/cell) at 7 and 14 dpi. Data in (C) and (E) represent averages ±SD of two independent experiments.
Analysis of the JΔNI8ΔB Vector in Neurons In Vivo
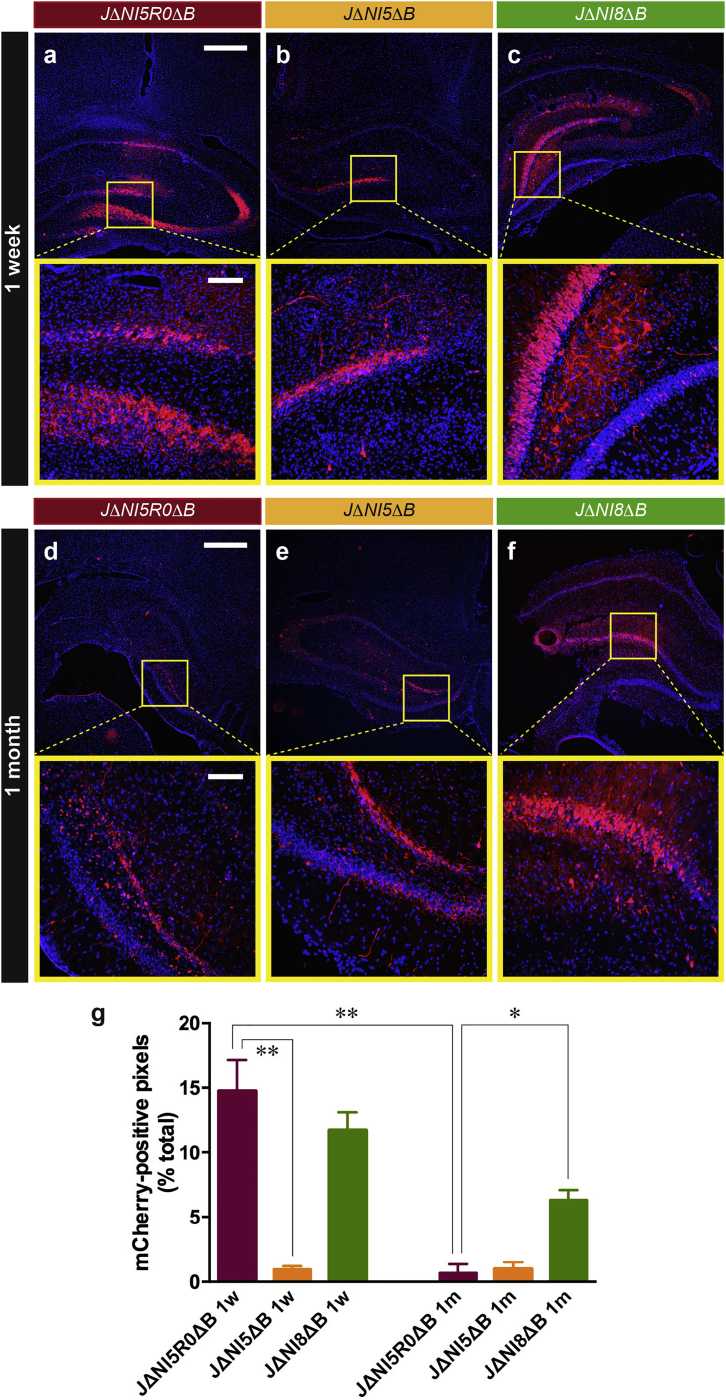
To determine whether our vectors were capable of non-cytotoxic transgene expression in neuronal cells in vivo, we injected JΔNI5ΔB and JΔNI8ΔB into rat hippocampus. We used JΔNI5R0ΔB, an ICP0-rescued derivative of JΔNI5 (Figure S3), as a positive control for mCherry expression and vector-associated toxicity. At 7 days post-vector administration, JΔNI5ΔB showed relatively low mCherry expression and normal neuronal morphology (Figure 4B) that were maintained at 1 month (Figure 4E). In comparison, more robust mCherry expression was observed in JΔNI5R0ΔB-injected animals at 7 days (Figure 4A), but a significant loss of mCherry signal was observed after 1 month, and the morphology of the mCherry positive cells indicated substantial cytotoxicity (Figure 4D). JΔNI8ΔB-injected animals displayed enhanced mCherry expression in the hippocampus at 7 days compared to JΔNI5ΔB (Figure 4C), and only a modest reduction in mCherry signal was apparent at 1 month without overt change in cell morphology (Figure 4F); as opposed to JΔNI5R0ΔB-injected hippocampi, cells in JΔNI8ΔB-injected hippocampi maintained a clear neuronal morphology with many dendritic and axonal elongations. We confirmed that JΔNI8ΔB transgene expression was restricted to neurons by double-label immunofluorescence for mCherry and markers of neurons (NissI), astrocytes (GFAP), and oligodendrocytes (O4) (Figure S4). Quantification of mCherry expression in the hippocampus (Figure 4G) demonstrated that, in JΔNI5ΔB-injected animals, only approximately 2% of the pixels were mCherry positive at both time points, whereas some 12% were positive in JΔNI8ΔB-injected animals at 7 days with only a modest reduction to approximately 7% by 1 month. The JΔNI5R0ΔB injection yielded about 15% mCherry positive pixels at 7 days, but this fell to less than 2% by 1 month. Both the loss of mCherry positive cells and the changes in cell morphology in the JΔNI5R0ΔB-injected animals were consistent with vector cytotoxicity that was not observed in animals injected with either of the ICP0-deficient vectors.
Figure 4.
Rat Hippocampal Transduction by JΔNI5R0ΔB, JΔNI5ΔB, and JΔNI8ΔB
2 × 109 gc of either vector was injected in the rat hippocampus, and DAPI (blue) and mCherry fluorescence (red) were imaged at 7 days (A–C) or 1 month (D–F) after injection. Upper images for each time point show a section of the injected hippocampus; bottom images are enlargements of the boxed areas. Scale bars in (A) and (D) upper panels (for all upper panels), 500 μm; scale bars in (A) and (D) lower panels (for all lower panels), 100 μm. (G) Quantification of mCherry expression in the hippocampus of rats injected with JΔNI5R0ΔB (red), JΔNI5ΔB (orange), or JΔNI8ΔB (green) at 1 week (1w) or 1 month (1m) after vector inoculation. Data are means ± SEM (n = three to five animals/group, five sections/animal). *p < 0.05; **p < 0.01 (ANOVA and post hoc Kruskal-Wallis test).
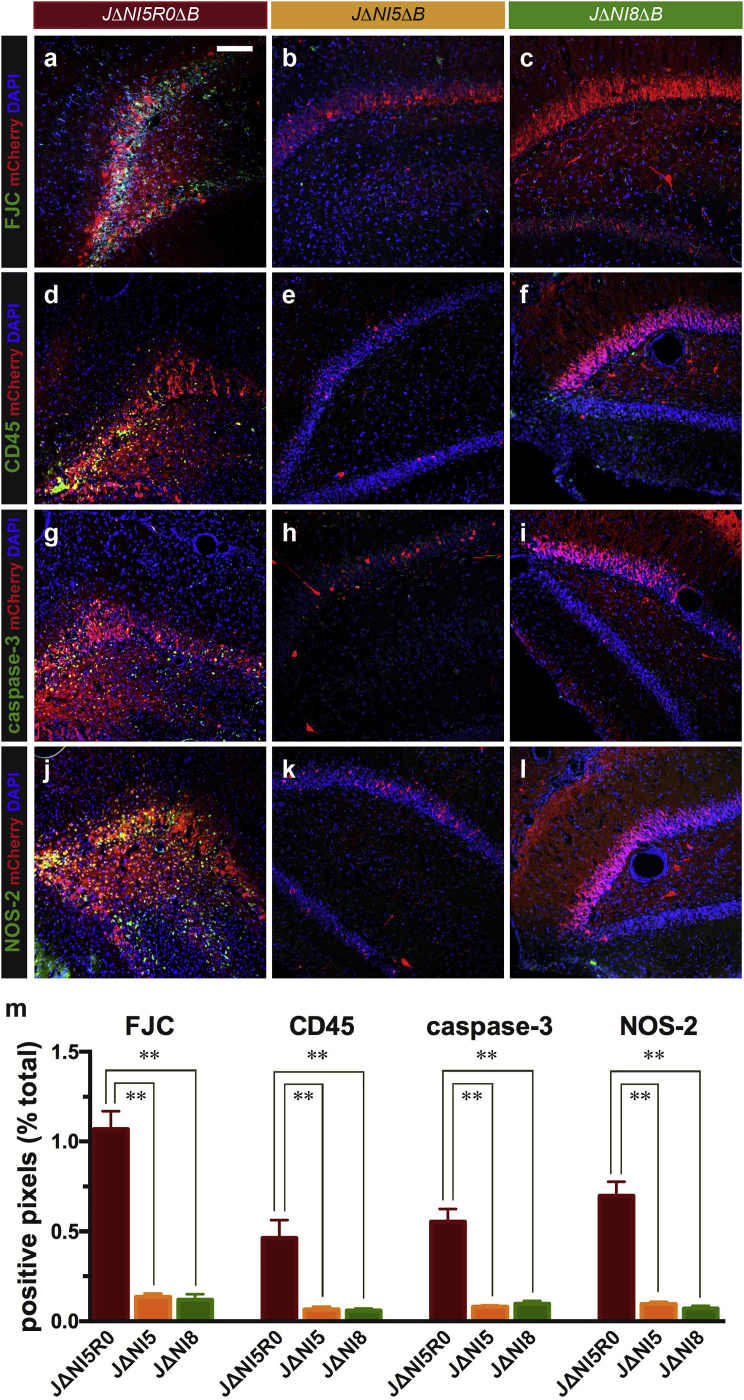
To further assess toxicity in rat brain, Fluoro-Jade C staining was performed on brain sections at 7 days post-vector administration. Fluoro-Jade C stains degenerating neurons, regardless of the cause of degeneration, and provides a clear indication of cytotoxicity. In JΔNI5R0ΔB-injected animals, many green, Fluoro-Jade C-positive cells were observed, whereas no signal was detected in either JΔNI5ΔB- or JΔNI8ΔB-injected animals (Figures 5A–5C). We also examined the expression of CD45 as a marker for the accumulation of inflammatory cells in the injected area. Many CD45 positive cells accumulated near the mCherry positive cells in JΔNI5R0ΔB-injected animals, while only a few CD45 positive cells were observed in either JΔNI5ΔB- or JΔNI8ΔB-injected animals (Figures 5D–5F). Similarly, both caspase-3, an apoptotic marker (Figures 5G–5I), and NOS-2, an inducible nitric oxide synthase involved in the immune response (Figures 5J–5L), were induced by JΔNI5R0ΔB injection, but not by either JΔNI5ΔB or JΔNI8ΔB injection. Quantification of these data is shown in Figure 5M. Taken together, these observations confirmed that the presence of ICP0 results in neuronal cytotoxicity and apoptosis in the hippocampus, along with activation of inflammatory cell recruitment. Deletion of ICP0 reduced these negative consequences although transgene expression was also reduced. Importantly, removal of the vhs gene from the ICP0-deficient vector resulted in enhanced transgene expression in the hippocampus without toxicity.
Figure 5.
Vector Toxicity in Rat Hippocampus
2 × 109 genome copies of JΔNI5R0ΔB, JΔNI5ΔB, or JΔNI8ΔB were injected into rat hippocampi, and brain sections were collected and processed 7 days later. (A–C) Fluoro-Jade C (FJC)-stained hippocampal sections from JΔNI-injected animals. Representative merged images of FJC staining (green), DAPI staining (blue), and mCherry fluorescence (red) are shown. (D–L) Immunohistochemical staining of brain sections for CD45 (D–F), caspase-3 (G–I), and NOS-2 (J–L) (all in green) is shown as merged images with DAPI staining (blue) and mCherry fluorescence (red). (M) Quantification of FJC, CD45, caspase-3, and NOS-2 in the hippocampus of rats 1 week after injection of JΔNI5R0ΔB, JΔNI5ΔB, or JΔNI8ΔB. Data are means ± SEM (n = three to five animals/group, five sections/animal). *p < 0.05; **p < 0.01 (ANOVA and post hoc Kruskal-Wallis test).
Discussion
HSV vectors offer the distinct advantage that they can accommodate large payloads; for example, our current replication-defective backbones offer approximately 30 kb of usable space. This makes HSV an ideal vector for gene therapy requiring the expression of large or multiple transgenes. Given HSV’s ability to enter and establish latency in sensory ganglia, current gene therapy studies using HSV have focused on the peripheral nervous system, treating conditions such as chronic pain and neuropathy.24 Establishing a platform for replication-defective gene delivery to the CNS has been hampered by problems including early silencing of CMV-promoter driven transgenes and limited, low-level transgene expression from other promoters such as the viral latency promoter.2, 9 The level of transgene expression is intricately tied to expression of the viral ICP0 protein: while ICP0 increases transgene expression, it also increases cytotoxicity.25
We recently generated an ICP0-deficient vector, JΔNI5, that is nontoxic and allowed transgene expression from an ectopic promoter in multiple non-neuronal cell lines and rDRGs in vitro when the transgene cassette was inserted between CTRLs associated with the viral latency locus. Consistent with an overall silencing of expression from the viral genome, a UbC-promoter-driven second transgene present in the deleted ICP4 locus was not expressed in non-neuronal cells. Surprisingly, however, it was expressed in DRGs. This observation prompted our current efforts to examine the potential of this vector and its vhs-deficient counterpart, JΔNI8, for CNS applications. The results indicated that both vectors are safe in the brain and express their transgene at detectable levels from the ICP4 locus for at least 1 month but also that JΔNI8 provides the more robust expression profile of the two.
Vhs plays a key role in attenuating the host anti-viral response by mediating mRNA degradation and inhibiting host protein synthesis. After virus entry into the cell through membrane fusion, vhs present in the virion tegument is released into the cytoplasm where it causes the shutoff of host protein synthesis. Available evidence suggests that vhs-induced host shutoff action contributes to the regulation of viral gene expression, in part by removing competition with cellular mRNAs for translation factors and sharpening the transition between the kinetic classes of viral genes, thereby supporting efficient virus replication during the lytic cycle.19, 20, 21 However, the functional significance of vhs in cell culture appears to vary between host cells. For example, U2OS cells were reported to be permissive for replication of viruses carrying a 588-nt deletion in the vhs gene26; these are the parental cells used to generate the U2OS-ICP4/27 cells in which we propagate our JΔNI viruses, and indeed we observed little difference in the growth rates of JΔNI5 and JΔNI8 in these cells (Figure 1B). In other reports, a nonsense mutant of vhs, UL41NHB, showed a growth delay in mouse embryonic fibroblasts27 but grew normally in Vero and C3H10T1/2 mouse embryo cells,28 while a vhs mutant that contained a lacZ gene insertion in the vhs open reading frame (ORF) exhibited a severe growth defect in human fibrosarcoma HT1080 cells.29 It has further been shown that vhs enhances the accumulation of viral late gene products in HeLa cells but is dispensable for late gene product accumulation in Vero and other permissive cells.26 In a similar vein, while treatment of human embryonic fibroblasts with IFN-α or -β significantly inhibited plaque formation by a vhs-deficient mutant,30 IFN-α treatment did not reduce the plaquing efficiency of a vhs mutant on Vero and U2OS cells.31 These observations clearly showed that the role of vhs is variable between cell types and indicated that vhs function is not critical in certain cell types. The host-cell-dependent mechanism of vhs action is further supported by evidence that primary sympathetic and sensory neuronal cultures are resistant to vhs-mediated shut off of protein synthesis,32 an effect that could be overcome by increasing the multiplicity of infection (MOI) and therefore the amount of vhs delivered to the cell during infection.21
The effect of vhs deletion has typically been assessed in vectors that still retained expression of ICP0. The ICP0 protein globally stimulates the expression of HSV genes, thereby enabling the production of viral gene products that may act to compensate for vhs deficiency, including ICP0 itself. ICP0 also has E3 ubiquitin ligase activity targeting the degradation of host-cell proteins involved in the antiviral response, some of which are also targets of vhs-mediated mRNA degradation.33, 34 Thus, any effects of vhs disruption may be masked by effects of ICP0. The vhs gene has been a favored target in replication-defective HSV vectors for replacement or insertion of therapeutic transgene cassettes because vhs is dispensable in Vero cells typically used for HSV growth.35 These vectors generally also expressed ICP0, potentially obscuring any remaining effects of vhs disruption. Using a vector that is deficient in all IE gene activity, our study reveals an unexpected consequence of deletion of the entire vhs ORF. While virus growth in complementing U2OS-ICP4/27 cells, transduction efficiency of fibroblasts, viral gene expression in fibroblasts, and toxicity for sensory neurons in culture and hippocampal neurons in vivo were largely unaffected by the deletion, reporter gene expression from the UbC promoter in a deleted ICP4 locus was specifically enhanced in neurons. In vivo, expression persisted at an elevated level for at least 1 month.
Both the cellular promoter and genomic location are of interest when considering our results. The UbC promoter has been tested for transgene expression both in lentivirally transduced cultured neurons in vitro and in transgenic mouse lines in vivo, revealing that it is active and drives consistent levels of transgene expression in neuronal cells.36, 37 With respect to the location of the transgene in the HSV genome, a recent report by Harkness et al. evaluating global HSV genome expression in non-neuronal MRC5 cells and trigeminal ganglion (TG) neurons infected with an IE gene-deficient HSV mutant, d109, made several observations of note. d109 was generally more transcriptionally active in TG neurons than in MRC5 cells (approximately 10-fold), and, in particular, the expression of LAT and other genes within, or adjacent to, the terminal/internal repeat regions was enhanced compared to the remainder of the genome in neurons.38 These findings are consistent with our observations that both JΔNI5 and JΔNI8 showed enhanced expression specifically in DRGs, but not in HDFs, of the UbC-promoter driven mCherry construct that is positioned at the deleted ICP4 locus in the terminal repeat region of the genome. Our findings indicate that additional deletion of vhs from the viral genome may further enhance the transcriptional activity of this genomic location in neuronal cells. While other repeat-associated genes were also expressed at higher levels in DRGs than in HDFs, and than genes within the unique regions of the viral genome in DRGs, vhs deletion did not further increase their expression (γ134.5) and in fact reduced ICP22 mRNA abundance. The effect of vhs removal on transgene expression from the terminal repeat-based LAT locus was also not dramatic. Expression in neurons was modestly enhanced by vhs deletion, but this effect was not as robust, nor was it maintained as long as at the ICP4 locus. In HDFs, vhs removal led to reduced reporter gene expression from the LAT locus and the already low levels of viral mRNAs appeared to trend toward further reduction. Together, these observations suggest distinctly separate utilities for the JΔNI5 and JΔNI8 vectors with respect to the target cell for transgene expression; the ICP4 locus of JΔNI8 is favored for expression in neurons, while the LAT locus of JΔNI5 is preferable for expression in non-neuronal cell types.
The key event differentiating between JΔNI5 and JΔNI8 infection would appear to be the initial introduction of vhs protein into non-complementing cells as a component of the JΔNI5 virion as (1) vhs protein is synthesized and incorporated into infectious particles during JΔNI5, but not JΔNI8 virus production by ICP4/ICP27-complementing U2OS cells, and (2) de novo expression of viral genes is extremely low in neurons infected with our vectors. However, we observed expression differences between the two vectors out to at least 14 days in vitro and 1 month in vivo, long after the vhs protein is degraded. As the incoming HSV genome is not epigenetically modified and its subsequent silencing is controlled by cellular and viral factors upon entry into the host cell,39 it is conceivable that the initial exposure of JΔNI5-infected neurons to vhs indirectly affects the epigenetic status of the mCherry locus, causing persistent downregulation of the gene. While further studies are required to test this and other possibilities, our current results indicate that vhs deletion may be an attractive feature for highly defective HSV gene therapy vectors for nervous system disorders.
Materials and Methods
Cells
HDFs (PCS-201-010; ATCC) were cultured in DMEM (Lonza) with 10% (v/v) fetal bovine serum (FBS; Sigma) and penicillin-streptomycin (P/S). Vero-based ICP4/ICP27-complementing 7b cells (Vero-7b)40 were cultured in DMEM with 5% FBS and P/S. Primary fetal rDRGs were isolated and cultured as described previously.18 U2OS-ICP4/27 cells were grown in DMEM with 10% FBS and P/S in the presence of puromycin (2 μg/mL) and blasticidin (10 μg/mL), and U2OS-ICP4/27/Cre cells were maintained in DMEM with 10% FBS and P/S in the presence of puromycin, blasticidin, and hygromycin (200 μg/mL), as described previously.18
HSV-BAC Engineering
All BAC engineering was performed in E. coli strain GS178341 as described.18 JΔNI8 and JΔNI8GFP BAC constructs were derived, respectively, from JΔNI5 and JΔNI7GFP BACs.18 To delete the vhs (UL41) coding sequence (GenBank JQ673480, positions 91,088–92,557), the kanamycin selection marker, I-SceI-aphAI, was amplified from pEPkan-S2 by PCR with primers 5′-tatcaattgtggtcgttgttgtggaaaagcaccagctggatgatgttgtacacgcgcgaaggatgacgacgataagtagggata-3′ and 5′-aatctgcagggcgcgccctacaaccaattaaccaattctgattag-3′. The PCR product was gel-purified and recombined with the vhs gene of JΔNI5 BAC or JΔNI7GFP BAC, followed by removal of the aphA1 gene.
JΔNI5R0 was constructed as follows: KOS-BAC DNA42 was digested with DraI and PsiI to isolate a fragment containing the complete ICP0 gene. The ICP0 fragment was cloned into the pCR-Blunt vector using the Zero Blunt PCR cloning kit (Thermo Fisher Scientific), creating pCRBlunt-ICP0. The I-SceI-aphAI fragment was amplified by PCR with primers (5′-tatcaattgcgcaacacctgcccgctgtgcaacgccaagctggtgtacctgatagtgggaggatgacgacgataagtagggata-3′ and 5′-aatctgcagcaattgctacaaccaattaaccaattctgattag-3′), and the product was inserted into the MfeI site of pCRBlunt-ICP0, creating pCRBlunt-ICP0-KAN. The pCRBlunt-ICP0-KAN plasmid was digested with EcoRI and PstI, and the isolated ICP0-KAN fragment was used for recombination with JΔNI5 BAC DNA. All HSV-BAC modifications were confirmed by field inversion gel electrophoresis (FIGE) analysis of restriction enzyme digests (FIGE mapper; Bio-Rad), PCR analysis, and targeted DNA sequencing.
Viruses
Production of infectious virus from BAC recombinants, Cre-mediated BAC excision, and virus titration were performed as described previously.18 JΔNI viruses were grown in U2OS-ICP4/27 cells, the KOS virus stock was produced on Vero cells, and QOZHG virus12 was grown in Vero-7b cells.
Virus Growth Curves
Duplicate wells of 4 × 105 U2OS-ICP4/27 cells were infected with JΔNI viruses at 1 gc/cell for 2 hr at 37°C, treated with 0.1 M glycine (pH 3.0) for 1 min to inactivate extracellular virus, and incubated at 37°C and 5% CO2. The supernatants were collected daily for viral DNA extraction and titration by qPCR for the gD gene, as described.18
Cytotoxicity Assay
Cell viability was measured essentially as described.43 Briefly, dissociated rDRGs were plated in a 24-well plate (9 × 104 cells/well) and infected with KOS or JΔNI viruses at 3,000 gc/cell. Cell viability was determined 5 days later by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay.
qRT-PCR and Genomic qPCR
HDF and rDRG cells were plated in 24-well plates, and duplicate wells were infected 24 hr later with JΔNI viruses at MOIs of 25,000 gc/cell or 3,000 gc/cell, respectively. Total RNA was extracted, and RT was carried out by Cells-to-cDNA II Kit (Ambion). Real-time PCR for each sample was performed in triplicate using the StepOnePlus Real-Time PCR System (Applied Biosystems). The data were normalized to viral gc in the same samples determined by qPCR for the gD gene or (Figure 2D) using separate JΔNI5 BAC DNA standard curves generated for each gene with the gene-specific primer pairs used for cDNA qPCR to allow for mRNA/gc comparisons between genes and across host cells. All primers for qPCR were as previously reported.18 The gc titers of virus stocks and nuclear viral DNA copy numbers were determined as described.18
In Vivo Experiments
Under ketamine (90 mg/kg i.p.) and xylazine (13 mg/kg i.p.) anesthesia, 2 × 109 gc of JΔNI vectors were inoculated into the right dorsal hippocampus of male rats (Harlan Sprague Dawley, 300 g). The viral vectors (JΔNI5R0, JΔNI5, and JΔNI8) were injected by stereotactic implantation of a borosilicate glass needle linked to a microperfusion pump in a volume of 3 μL at a flow rate of 200 nL/min. In order to facilitate the needle entry into the brain tissue and to reduce the mechanical damage, the needle tip had been laser-chamfered (inner diameter at tip = 60 μm) using Leica Laser Microdissector CTR6000 (Leica Microsystems). The stereotactic coordinates for the hippocampal inoculation, based on the Paxinos atlas,44 were 2.1 mm lateral and 3 mm posterior to bregma, 3.5 mm deep from dura. Experiments were performed at the University of Ferrara in compliance with the university’s guidelines for the ethical treatment of experimental animals.
Tissue Preparation
Animals were anesthetized with pentobarbital at 7 or 30 days post-virus injection and perfused with 0.1 M PBS followed by 4% paraformaldehyde. The brains were quickly removed and post-fixed in 4% paraformaldehyde for 1 hr, cryoprotected in 30% sucrose at 4°C until the tissue sank, and snap frozen in isopentane at −80°C.
Immunofluorescence
20-μm coronal cryostat sections were cut at −20°C on a freezing microtome, rinsed in 0.1 M PBS for free-floating immunostaining, and blocked with 10% normal goat serum/0.3% Triton X-100. Sections were then incubated overnight at 4°C with primary antibodies dissolved in blocking solution as follows: CD45 (Santa Cruz Biotechnology, sc-53047), 1:100; cleaved caspase-3 (Cell Signaling Technology, 9664), 1:100; nitric oxide synthase-2 (Santa Cruz, sc-651), 1:100; mCherry (Thermo Fisher Scientific), 1:100; GFAP (Sigma), 1:200; O4 (Sigma), 1:100. After washing in PBS, sections were incubated in blocking solution with 10 μM DAPI (Molecular Probes, D3571) and either Alexa Fluor 488 goat anti-rabbit (Molecular Probes, A11008, 1:1,000) or Alexa Fluor 488 goat anti-mouse (Molecular Probes, A11029, 1:1,000) for 1 hr at room temperature.
Fluoro-Jade C Staining
Brain sections were mounted on frosted microscope slides (Thermo Fisher Scientific) and pretreated for 5 min in 80% EtOH/1% NaOH, 2 min in 70% EtOH, and 2 min in distilled water. Sections were then incubated for 10 min in a 0.06% KMnO4 solution and rinsed in distilled water for 3 min before incubation in a 0.0001% solution of Fluoro-Jade C (Immunological Sciences, IS-0012) in 0.1% acetic acid for 10 min. After three washes in distilled water for 1 min, slides were dried, dehydrated with xylene, and coverslipped with DPX mountant (Sigma) for microscopy.
Hippocampal Image Quantifications
Brain sections were imaged using a DMRA2 Leica microscope (Leica Microsystems) and Hamamatsu C11440 camera. Confocal images were captured using a HAL 100 camera (Zeiss) mounted on a Zeiss LSM510 confocal microscope. Imaged mCherry expression was quantified with MetaMorph (Universal Imaging). The entire hippocampus was selected as the region of interest (ROI). mCherry-positive pixels were identified by thresholding at the gray level corresponding to the mean plus the difference between average and minimum. Using this approach, only those pixels that were significantly above background (i.e., mCherry-positive) were selected. The percentage of mCherry-positive pixels in the hippocampus was calculated as the ratio of pixels above threshold to total pixels in the ROI.
A similar procedure was employed for quantification of FJC, CD45, caspase-3, and NOS-2 signals. An outline was drawn around five positive cells, and mean gray values were measured and averaged; the resulting value was used to set the image threshold. Pixels above the threshold were identified, as described above, and the number of positive pixels was expressed as the percentage of total pixels.
For each injected animal, the number of mCherry-, FJC-, CD45-, caspase-3-, and NOS-2-positive pixels was quantified from five regularly interspaced sections spanning across the injection site (one of every five 20-μm sections cut across the injection site, i.e., one per 100 μm with the third at the site of injection), and the average per animal (three to five animals/group) was determined. These averages were used for statistical analysis.
Statistical Analyses
Data from in vitro experiments are presented as the mean ± SD. Data from in vivo experiments were analyzed by the Kruskal-Wallis test to determine significance.
Author Contributions
Conceptualization, Y.M.; Investigation, Y.M., G.V., F.H., H.U., S.Z., and W.F.G.; Formal Analysis, Y.M., G.V., B.R., M.S., J.B.C., and J.C.G.; Supervision, M.S., J.B.C., and J.C.G.; Writing – Original Draft, Y.M.; Writing – Review & Editing, B.R., J.B.C., and J.C.G.; Funding Acquisition, W.F.G., M.S., J.B.C., and J.C.G.
Conflicts of Interest
G.V., M.S., and J.C.G. are founders of NuvoVec srl. Y.M., J.B.C., and J.C.G. are co-inventors of intellectual property licensed to SwitchBio, Inc. H.U., J.B.C., and J.C.G. are co-inventors of intellectual property licensed to Oncorus, Inc. J.C.G. is a founder and consultant of SwitchBio, Inc. and Oncorus, Inc. B.R. and W.F.G. are consultants of Oncorus, Inc.
Acknowledgments
We are grateful to Klaus Osterrieder (Free University of Berlin, Germany) for plasmid pEPkan-S2, Greg Smith (Northwestern University) for E. coli strain GS1783, David Leib (Dartmouth Medical School) for KOS-37 BAC DNA, Mingdi Zhang for rat DRG cultures, and Jim Smiley (University of Alberta, Canada) for helpful comments on the manuscript. This work was supported by grants to J.C.G. from the NIH (NS064988 and DK044935), the CHDI Foundation (A3777 and A8790), and the Commonwealth of Pennsylvania (SAP #4100061184) and to M.S. from the European Community (FP7-PEOPLE-2011-IAPP project 285827 [EPIXCHANGE]) and the Italian Ministry for Education, University and Research (PRIN project 2010N8PBAA [INBDNF]).
Footnotes
Supplemental Information includes four figures and can be found with this article online at http://dx.doi.org/10.1016/j.omtm.2017.06.001.
Supplemental Information
References
- 1.Puskovic V., Wolfe D., Goss J., Huang S., Mata M., Glorioso J.C., Fink D.J. Prolonged biologically active transgene expression driven by HSV LAP2 in brain in vivo. Mol. Ther. 2004;10:67–75. doi: 10.1016/j.ymthe.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Smith C., Lachmann R.H., Efstathiou S. Expression from the herpes simplex virus type 1 latency-associated promoter in the murine central nervous system. J. Gen. Virol. 2000;81:649–662. doi: 10.1099/0022-1317-81-3-649. [DOI] [PubMed] [Google Scholar]
- 3.Zhu J., Kang W., Wolfe J.H., Fraser N.W. Significantly increased expression of beta-glucuronidase in the central nervous system of mucopolysaccharidosis type VII mice from the latency-associated transcript promoter in a nonpathogenic herpes simplex virus type 1 vector. Mol. Ther. 2000;2:82–94. doi: 10.1006/mthe.2000.0093. [DOI] [PubMed] [Google Scholar]
- 4.Palmer J.A., Branston R.H., Lilley C.E., Robinson M.J., Groutsi F., Smith J., Latchman D.S., Coffin R.S. Development and optimization of herpes simplex virus vectors for multiple long-term gene delivery to the peripheral nervous system. J. Virol. 2000;74:5604–5618. doi: 10.1128/jvi.74.12.5604-5618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez M.C., Hunt S.P., Coffin R.S., Palmer J.A. Comparative analysis of genomic HSV vectors for gene delivery to motor neurons following peripheral inoculation in vivo. Gene Ther. 2004;11:1023–1032. doi: 10.1038/sj.gt.3302258. [DOI] [PubMed] [Google Scholar]
- 6.Goss J.R., Cascio M., Goins W.F., Huang S., Krisky D.M., Clarke R.J., Johnson J.W., Yokoyama H., Yoshimura N., Gold M. HSV delivery of a ligand-regulated endogenous ion channel gene to sensory neurons results in pain control following channel activation. Mol. Ther. 2011;19:500–506. doi: 10.1038/mt.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goss J.R., Mata M., Goins W.F., Wu H.H., Glorioso J.C., Fink D.J. Antinociceptive effect of a genomic herpes simplex virus-based vector expressing human proenkephalin in rat dorsal root ganglion. Gene Ther. 2001;8:551–556. doi: 10.1038/sj.gt.3301430. [DOI] [PubMed] [Google Scholar]
- 8.McMenamin M.M., Byrnes A.P., Charlton H.M., Coffin R.S., Latchman D.S., Wood M.J. A gamma34.5 mutant of herpes simplex 1 causes severe inflammation in the brain. Neuroscience. 1998;83:1225–1237. doi: 10.1016/s0306-4522(97)00513-7. [DOI] [PubMed] [Google Scholar]
- 9.Scarpini C.G., May J., Lachmann R.H., Preston C.M., Dunnett S.B., Torres E.M., Efstathiou S. Latency associated promoter transgene expression in the central nervous system after stereotaxic delivery of replication-defective HSV-1-based vectors. Gene Ther. 2001;8:1057–1071. doi: 10.1038/sj.gt.3301497. [DOI] [PubMed] [Google Scholar]
- 10.Bloom D.C., Maidment N.T., Tan A., Dissette V.B., Feldman L.T., Stevens J.G. Long-term expression of a reporter gene from latent herpes simplex virus in the rat hippocampus. Brain Res. Mol. Brain Res. 1995;31:48–60. doi: 10.1016/0169-328x(95)00031-m. [DOI] [PubMed] [Google Scholar]
- 11.Boutell C., Everett R.D. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J. Gen. Virol. 2013;94:465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- 12.Chen X., Li J., Mata M., Goss J., Wolfe D., Glorioso J.C., Fink D.J. Herpes simplex virus type 1 ICP0 protein does not accumulate in the nucleus of primary neurons in culture. J. Virol. 2000;74:10132–10141. doi: 10.1128/jvi.74.21.10132-10141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goins W.F., Lee K.A., Cavalcoli J.D., O’Malley M.E., DeKosky S.T., Fink D.J., Glorioso J.C. Herpes simplex virus type 1 vector-mediated expression of nerve growth factor protects dorsal root ganglion neurons from peroxide toxicity. J. Virol. 1999;73:519–532. doi: 10.1128/jvi.73.1.519-532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chattopadhyay M., Wolfe D., Mata M., Huang S., Glorioso J.C., Fink D.J. Long-term neuroprotection achieved with latency-associated promoter-driven herpes simplex virus gene transfer to the peripheral nervous system. Mol. Ther. 2005;12:307–313. doi: 10.1016/j.ymthe.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Margolis T.P., Bloom D.C., Dobson A.T., Feldman L.T., Stevens J.G. Decreased reporter gene expression during latent infection with HSV LAT promoter constructs. Virology. 1993;197:585–592. doi: 10.1006/viro.1993.1632. [DOI] [PubMed] [Google Scholar]
- 16.Labetoulle M., Maillet S., Efstathiou S., Dezelee S., Frau E., Lafay F. HSV1 latency sites after inoculation in the lip: Assessment of their localization and connections to the eye. Invest. Ophthalmol. Vis. Sci. 2003;44:217–225. doi: 10.1167/iovs.02-0464. [DOI] [PubMed] [Google Scholar]
- 17.Amelio A.L., McAnany P.K., Bloom D.C. A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J. Virol. 2006;80:2358–2368. doi: 10.1128/JVI.80.5.2358-2368.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyagawa Y., Marino P., Verlengia G., Uchida H., Goins W.F., Yokota S., Geller D.A., Yoshida O., Mester J., Cohen J.B. Herpes simplex viral-vector design for efficient transduction of nonneuronal cells without cytotoxicity. Proc. Natl. Acad. Sci. USA. 2015;112:E1632–E1641. doi: 10.1073/pnas.1423556112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivas H.G., Schmaling S.K., Gaglia M.M. Shutoff of host gene expression in influenza A virus and herpesviruses: Similar mechanisms and common themes. Viruses. 2016;8:102. doi: 10.3390/v8040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smiley J.R. Herpes simplex virus virion host shutoff protein: Immune evasion mediated by a viral RNase? J. Virol. 2004;78:1063–1068. doi: 10.1128/JVI.78.3.1063-1068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strand S.S., Vanheyningen T.K., Leib D.A. The virion host shutoff protein of herpes simplex virus type 1 has RNA degradation activity in primary neurons. J. Virol. 2004;78:8400–8403. doi: 10.1128/JVI.78.15.8400-8403.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferenczy M.W., DeLuca N.A. Epigenetic modulation of gene expression from quiescent herpes simplex virus genomes. J. Virol. 2009;83:8514–8524. doi: 10.1128/JVI.00785-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hobbs W.E., Brough D.E., Kovesdi I., DeLuca N.A. Efficient activation of viral genomes by levels of herpes simplex virus ICP0 insufficient to affect cellular gene expression or cell survival. J. Virol. 2001;75:3391–3403. doi: 10.1128/JVI.75.7.3391-3403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glorioso J.C., Fink D.J. Herpes vector-mediated gene transfer in the treatment of chronic pain. Mol. Ther. 2009;17:13–18. doi: 10.1038/mt.2008.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samaniego L.A., Neiderhiser L., DeLuca N.A. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 1998;72:3307–3320. doi: 10.1128/jvi.72.4.3307-3320.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dauber B., Pelletier J., Smiley J.R. The herpes simplex virus 1 vhs protein enhances translation of viral true late mRNAs and virus production in a cell type-dependent manner. J. Virol. 2011;85:5363–5373. doi: 10.1128/JVI.00115-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasieka T.J., Lu B., Crosby S.D., Wylie K.M., Morrison L.A., Alexander D.E., Menachery V.D., Leib D.A. Herpes simplex virus virion host shutoff attenuates establishment of the antiviral state. J. Virol. 2008;82:5527–5535. doi: 10.1128/JVI.02047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strelow L.I., Leib D.A. Role of the virion host shutoff (vhs) of herpes simplex virus type 1 in latency and pathogenesis. J. Virol. 1995;69:6779–6786. doi: 10.1128/jvi.69.11.6779-6786.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sciortino M.T., Parisi T., Siracusano G., Mastino A., Taddeo B., Roizman B. The virion host shutoff RNase plays a key role in blocking the activation of protein kinase R in cells infected with herpes simplex virus 1. J. Virol. 2013;87:3271–3276. doi: 10.1128/JVI.03049-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzutani T., Nagamine M., Shibaki T., Ogasawara M., Yoshida I., Daikoku T., Nishiyama Y., Azuma M. The role of the UL41 gene of herpes simplex virus type 1 in evasion of non-specific host defence mechanisms during primary infection. J. Gen. Virol. 2000;81:1763–1771. doi: 10.1099/0022-1317-81-7-1763. [DOI] [PubMed] [Google Scholar]
- 31.Mossman K.L., Saffran H.A., Smiley J.R. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 2000;74:2052–2056. doi: 10.1128/jvi.74.4.2052-2056.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nichol P.F., Chang J.Y., Johnson E.M., Jr., Olivo P.D. Infection of sympathetic and sensory neurones with herpes simplex virus does not elicit a shut-off of cellular protein synthesis: Implications for viral latency and herpes vectors. Neurobiol. Dis. 1994;1:83–94. doi: 10.1006/nbdi.1994.0011. [DOI] [PubMed] [Google Scholar]
- 33.Lin R., Noyce R.S., Collins S.E., Everett R.D., Mossman K.L. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 2004;78:1675–1684. doi: 10.1128/JVI.78.4.1675-1684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orzalli M.H., Broekema N.M., Knipe D.M. Relative contributions of herpes simplex virus 1 ICP0 and vhs to loss of cellular IFI16 vary in different human cell types. J. Virol. 2016;90:8351–8359. doi: 10.1128/JVI.00939-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goins W.F., Krisky D.M., Wechuck J.B., Wolfe D., Huang S., Glorioso J.C. Generation of replication-competent and -defective HSV vectors. Cold Spring Harb. Protoc. 2011 doi: 10.1101/pdb.prot5615. Published online May 1, 2011. [DOI] [PubMed] [Google Scholar]
- 36.Li M., Husic N., Lin Y., Christensen H., Malik I., McIver S., LaPash Daniels C.M., Harris D.A., Kotzbauer P.T., Goldberg M.P. Optimal promoter usage for lentiviral vector-mediated transduction of cultured central nervous system cells. J. Neurosci. Methods. 2010;189:56–64. doi: 10.1016/j.jneumeth.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilhelm F., Winkler U., Morawski M., Jager C., Reinecke L., Rossner M.J., Hirrlinger P.G., Hirrlinger J. The human ubiquitin C promoter drives selective expression in principal neurons in the brain of a transgenic mouse line. Neurochem. Int. 2011;59:976–980. doi: 10.1016/j.neuint.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 38.Harkness J.M., Kader M., DeLuca N.A. Transcription of the herpes simplex virus 1 genome during productive and quiescent infection of neuronal and nonneuronal cells. J. Virol. 2014;88:6847–6861. doi: 10.1128/JVI.00516-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knipe D.M. Nuclear sensing of viral DNA, epigenetic regulation of herpes simplex virus infection, and innate immunity. Virology. 2015;479-480:153–159. doi: 10.1016/j.virol.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krisky D.M., Wolfe D., Goins W.F., Marconi P.C., Ramakrishnan R., Mata M., Rouse R.J., Fink D.J., Glorioso J.C. Deletion of multiple immediate-early genes from herpes simplex virus reduces cytotoxicity and permits long-term gene expression in neurons. Gene Ther. 1998;5:1593–1603. doi: 10.1038/sj.gt.3300766. [DOI] [PubMed] [Google Scholar]
- 41.Tischer B.K., Smith G.A., Osterrieder N. En passant mutagenesis: A two step markerless red recombination system. Methods Mol. Biol. 2010;634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 42.Gierasch W.W., Zimmerman D.L., Ward S.L., Vanheyningen T.K., Romine J.D., Leib D.A. Construction and characterization of bacterial artificial chromosomes containing HSV-1 strains 17 and KOS. J. Virol. Methods. 2006;135:197–206. doi: 10.1016/j.jviromet.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Uchida H., Marzulli M., Nakano K., Goins W.F., Chan J., Hong C.S., Mazzacurati L., Yoo J.Y., Haseley A., Nakashima H. Effective treatment of an orthotopic xenograft model of human glioblastoma using an EGFR-retargeted oncolytic herpes simplex virus. Mol. Ther. 2013;21:561–569. doi: 10.1038/mt.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paxinos G., Watson C. Academic Press; 1982. The Rat Brain in Stereotaxic Coordinates. [DOI] [PubMed] [Google Scholar]
- 45.Uchida H., Chan J., Goins W.F., Grandi P., Kumagai I., Cohen J.B., Glorioso J.C. A double mutation in glycoprotein gB compensates for ineffective gD-dependent initiation of herpes simplex virus type 1 infection. J. Virol. 2010;84:12200–12209. doi: 10.1128/JVI.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berthomme H., Thomas J., Texier P., Epstein A., Feldman L.T. Enhancer and long-term expression functions of herpes simplex virus type 1 latency-associated promoter are both located in the same region. J. Virol. 2001;75:4386–4393. doi: 10.1128/JVI.75.9.4386-4393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goins W.F., Sternberg L.R., Croen K.D., Krause P.R., Hendricks R.L., Fink D.J., Straus S.E., Levine M., Glorioso J.C. A novel latency-active promoter is contained within the herpes simplex virus type 1 UL flanking repeats. J. Virol. 1994;68:2239–2252. doi: 10.1128/jvi.68.4.2239-2252.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.