Abstract
DR5 (also called TRAIL receptor 2 and KILLER) is an apoptosis-inducing membrane receptor for tumor necrosis factor-related apoptosis-inducing ligand (also called TRAIL and Apo2 ligand). DR5 is a transcriptional target of p53, and its overexpression induces cell death in vitro. However, the in vivo biology of DR5 has remained largely unexplored. To better understand the role of DR5 in development and in adult tissues, we have created a knockout mouse lacking DR5. This mouse is viable and develops normally with the exception of having an enlarged thymus. We show that DR5 is not expressed in developing embryos but is present in the decidua and chorion early in development. DR5-null mouse embryo fibroblasts expressing E1A are resistant to treatment with TRAIL, suggesting that DR5 may be the primary proapoptotic receptor for TRAIL in the mouse. When exposed to ionizing radiation, DR5-null tissues exhibit reduced amounts of apoptosis compared to wild-type thymus, spleen, Peyer's patches, and the white matter of the brain. In the ileum, colon, and stomach, DR5 deficiency was associated with a subtle phenotype of radiation-induced cell death. These results indicate that DR5 has a limited role during embryogenesis and early stages of development but plays an organ-specific role in the response to DNA-damaging stimuli.
DR5 (also called TRAIL receptor 2, KILLER, and TRICK2) is a member of the tumor necrosis factor receptor superfamily (22). The ligands of this family include tumor necrosis factor, FasL, and TRAIL (tumor necrosis factor-related apoptosis-inducting ligand; also called Apo2L). DR5 is a proapoptotic receptor for TRAIL (4, 18, 23, 26, 28, 32), as is the related receptor DR4 (24). When trimerized by TRAIL, these receptors signal through an intracellular death domain. Association of death domains induces the formation of a death-inducing signaling complex, which includes the Fas-associated death domain and pro-caspase 8 or pro-caspase 10. Death-inducing signaling complex-activated autocleavage of pro-caspase 8 to active caspase 8 allows downstream cleavage of caspases 3, 7, and 6 and BID, leading to the execution of apoptosis (22). Two members of the TRAIL receptor family, TRID (also called DcR1 and TRAIL receptor 3) and TRUNDD (also called DcR2 and TRAIL receptor 4), lack functional death domains and thus do not signal apoptosis (16). TRAIL has been shown to kill tumor cells but not normal cells, and is currently being investigated as an anticancer agent (16).
The tumor suppressor gene TP53 is mutated in roughly half of human cancers (30). p53 functions as a tumor suppressor by signaling a number of responses to oncogenic stressors that may include oncogenes, DNA damage, and hypoxia (31). p53 signals apoptosis through transactivation of various target genes that can be grouped as inducing apoptosis either intrinsically (via the mitochondria) or extrinsically (via death receptors). p53 targets that function through the mitochondria include Bax (6), Puma (39), Noxa (20), and E124/PIG8 (10). p53 death receptor targets include Fas (also called Apo and CD95) (1, 21) and DR5 (also called TRAIL receptor 2 and KILLER) (35).
In mice, only one death domain containing a TRAIL receptor has been identified, DR5 (mouse KILLER) (36). This receptor is a homologue of human DR5 and DR4 (79 and 76% amino acid homology, respectively) and binds TRAIL with an affinity similar to that of human DR4 and DR5 (36). Baseline expression of murine DR5 predominates in the heart, lung, and kidney, whereas in the human DR4 and DR5 are expressed in almost all tissues (22). Human DR5 is also expressed in the fetal kidney, liver, and lung but not the brain (28). It is currently unknown whether other death-inducing receptors for TRAIL exist in the mouse.
In recent years, knockout mice have been generated to explore the in vivo roles of various genes instrumental in death receptor-mediated apoptosis. Homozygous caspase 8 deletion is embryo lethal, highlighting an important role for this death receptor-activated caspase in hematopoiesis and development of the heart (29). Bid-null mice develop normally but are protected from hepatocellular apoptosis and hemorrhagic necrosis when injected with an antibody directed against Fas (38). TRAIL has also been targeted in mice, producing a phenotype that develops normally but shows increased susceptibility to tumor burden and autoimmune diseases (15, 27).
In order to better understand the in vivo biology of DR5, we have generated a knockout mouse for this gene. In response to ionizing radiation, these mice are partially protected from apoptosis in the brain, spleen, thymus, and Peyer's patches.
(This work was presented at the 95th American Association for Cancer Research meeting in Orlando, Fla., March 2004.)
MATERIALS AND METHODS
Generation of DR5-deficient mice.
Phage clones containing the murine DR5 gene were isolated by screening a 129/J genomic library with the full-length cDNA of mouse DR5 (36). A targeting vector was designed to replace exons 3 and 4 and part of exon 5 with a phosphoglycerol kinase-neomycin resistance cassette in the antisense orientation, leading to replacement of the extracellular domain of the DR5 receptor encoding the cysteine-rich repeats and a downstream frameshift. A total of 30 μg of the linearized targeting vector was used to electroporate 5 × 106 E14K embryonic stem (ES) cells (Bio-Rad Gene Pulser, 0.34 kV, 0.25 mF), which were subsequently cultured for 10 days in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, glutamine, pyruvate, β-mercaptoethanol, and leukemia inhibitory factor, containing 300 μg of G418 (Sigma) per ml.
Recombinants were identified by PCR and confirmed by Southern blot analysis with EcoRI-digested genomic DNA hybridized to an external flanking probe, which detects an 8-kb band for the wild-type locus and a 5-kb band for the mutant allele. Single integration was confirmed with a probe corresponding to the neo gene; 13 correctly targeted ES clones were identified. Three of those were injected into C57BL/6 blastocysts. Germ line chimeras were successfully derived for two of these ES cell clones.
Genotyping of mice was performed by PCR with tail genomic DNA with the following primers: forward, 5′-GACAACCGATCACATGTACGTGATTC-3′, reverse (wild-type), 5′-ATGGCTATCACAACTGAGGGACTGCC-3′, and reverse (transgene), 5′-GGGTGGGATTAGATAAATGCCTGCTC-3′. These primers give rise to a 607-bp PCR product for the wild-type locus and 440-bp PCR product for the mutant allele. The following PCR conditions were used (35 cycles): 94°C for 45 s followed by 63°C for 30 s followed by 72°C for 45 s.
Animals and treatments.
Four- to six-week-old DR5−/− mice and age- and sex-matched C57BL/6 wild-type controls (Jackson Laboratories) were subject to 5 Gy of total body irradiation with a 137Cs gamma source at a rate of 1.4 Gy/min. Fifteen TRAIL−/− mice were subjected to the same regimen. In dexamethasone-treated animals, 0.5 mg/animal was delivered by single-bolus intraperitoneal injection. At 6 or 8 h, animals were euthanized with an approved Institutional Animal Care and Use Committee Protocol, which followed the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association. Tissues were harvested and fixed in freshly prepared 4% paraformaldehyde overnight at 4°C. Tissues were washed in phosphate-buffered saline and then 70% ethanol and embedded in paraffin for sectioning. For intraperitoneal injections, soluble mouse TRAIL (50 μg/mouse) was obtained from BioMol.
Reverse transcription-PCR.
Total RNA was isolated from individual tissues as previously described (3). Total RNA samples were electrophoresed on a morpholinepropanesulfonic acid (MOPS)-formaldehyde gel, and the patterns of the 28S and 18S ribosomal bands were observed to verify the quality of the total RNA preparation. Oligonucleotide primers for mouse DR5 were designed as follows: forward, 5′-AAAACGGCTTGGGCATCTTGGC-3′, and reverse, 5′-AGACGGTTCCAGGAGTCAAAGG-3′. Glyceraldehyde-3-phosphate dehydrogenase primers were obtained from PE Applied Biosystems. Comparative quantitative reverse transcription-PCR analysis was performed on RNA isolated from brain with the 1 QuantumRNA Classic II 18S internal standard kit (Ambion). Two micrograms of total RNA was used for reverse transcription (Advantage RT-for-PCR; BD Biosciences) and generation of cDNA. Multiplex PCR (HotStartTaq; QIAGEN) was performed on the cDNA and PCR products separated on a 2% agarose gel. Mean band density was determined with NIH Image 1.62 and normalized to the band density of 18S rRNA.
In situ hybridization.
In situ hybridization was performed by Phylogeny Inc. as described (17). Briefly, fetuses and tissues were fixed in paraformaldehyde and mounted with paraffin. Serial sections were deparaffinized and digested with proteinase K, postfixed, treated with triethanolamine-acetic anhydride, washed, and dehydrated. cRNAs were prepared from linearized cDNA templates to generate antisense and sense probes. The cRNA transcripts were synthesized according to the manufacturer's conditions (Ambion) and labeled with [35S]UTP (>1,000 Ci/mmol; Amersham). Sections were hybridized overnight at 52°C in 50% deionized formamide-0.3 M NaCl-20 mM Tris-HCl (pH 7.4)-5 mM EDTA-10 mM NaPO4-dextran sulfate-1× Denhardt's-50 μg of total yeast RNA per ml-5 to 75,000 cpm of 35S-labeled cRNA probe/μl. The tissue was subjected to stringent washing at 65°C in 50% formamide-2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-10 mM dithiothreitol and washed in phosphate-buffered saline before treatment with 20 μg of RNase A per ml at 37°C for 30 min. Following washes in 2× SSC and 0.1× SSC for 10 min at 37°C, the slides were dehydrated and dipped in Kodak NTB-2 nuclear track emulsion and exposed for 2 to 3 weeks in light-tight boxes with desiccant at 4°C. Photographic development was carried out in Kodak D-19. Slides were counterstained lightly with toluidine blue and analyzed with both light- and dark-field optics. Sense control cRNA probes indicate background levels of the hybridization signal.
Generation of recombinant retrovirus.
The pCEP4 vector containing murine DR5 (MK) cDNA (36) was digested with XhoI and HindIII (New England Biolabs). The 1.5-kb insert was extracted from a 1.5% agarose gel and incubated with 1 U of Klenow enzyme (New England Biolabs) and subsequently blunt-end cloned into the EcoRI-digested retroviral vector pBabepuro. Positive clones were screened for by analyzing restriction enzyme (PstI) fragments with the Vector NTI software. Ecotropic Phoenix cells were transfected with pBabepuro-DR5, pWZL-hygromycin-E1A, LPC-E1A, and corresponding empty vectors. The supernatant containing recombinant retrovirus was harvested 48 h following transfection. Target cells (mouse embryo fibroblasts) were seeded at a density of 105 and incubated 1 h with the retroviral supernatant. Cells stably expressing DR5 or E1A or containing empty vector were selected by incubation with either 2 μg of puromycin per ml or 100 μg of hygromycin per ml for 3 days.
Cell death assays.
Mouse embryo fibroblasts (MEFs) were infected with a retroviral vector expressing either a long terminal repeat-puromycin-cytomegalovirus (LPC)-E1A cassette, LPC alone (vector), pWZL-hygromycin-E1A, pBabepuro-MK (murine DR5), or pBabepuro alone. Infected cells were selected in puromycin (2 mg/ml) for 48 h and then replated overnight at 5 × 104 cells per 1-ml well. The next day MEFs were treated as described for 24 h, and cell death was quantified by trypan blue exclusion.
Immunohistochemistry.
Immunohistochemistry was performed as described (9). Briefly, 5-μm paraffin sections were dewaxed and immunostained with the ABC peroxidase method (Vectastain) followed by counterstaining with hematoxylin or methyl green. Primary anti-active caspase 3 antibody was from Novacastra. For MEFs grown in vitro, caspase 3 was visualized by labeling with tetramethylrhodamine isothiocyanate-conjugated avidin (Pierce). Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays were performed with the Apoptag peroxidase kit per the manufacturer's instructions (Chemicon International). TRAIL was detected by incubating sections with rabbit anti-TRAIL antibodies (1:100, antibody 2435, AbCam) following dewaxing and rehydration. Subsequently, sections were incubated with goat anti-rabbit Fc (Jackson Immunochemicals) and donkey anti-goat Cy2-conjugated antibodies (1:200, Jackson Immunochemicals). Sections were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and evaluated under a fluorescence microscope.
Analysis of stained sections.
Sections stained for anti-active caspase 3 and TUNEL were analyzed in detail from three age- and sex-matched animals. Cells in the thymus and spleen staining positive for caspase 3 were counted. Five fields with an area of 0.0185 mm2 (containing approximately 1,000 cells) were counted, and an average was calculated per field and animal. Significant differences were determined with Student's t test. Sections stained with the TUNEL technique were analyzed with the ImageJ (Java-based version of NIH Image 1.62) software with the commands “density slicing” and “count particles” on binary images. The accuracy and appropriate cutoff level were determined by comparing counted images to the original. The average area (in square pixels for each section) positive for “apoptotic clusters” of cells was calculated for three animals per genotype and treatment. The results were analyzed with Student's t test.
Western blot.
Tissues were homogenized in radioimmunoprecipitation assay (RIPA) buffer (1x phosphate-buffered saline, 1% Nonidet P-40 or Igepal CA-630, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) and snap-frozen in liquid nitrogen. Homogenates were centrifuged at 14,000 rpm for 20 min, and the protein concentration was determined by the Bradford method (Bio-Rad). Proteins were separated on sodium dodecyl sulfate-12.5% polyacrylamide gels and transferred to polyvinylidene difluoride membranes. Mouse DR5 was detected with AF837 (1:200, R&D systems), and Ran was detected with a mouse monoclonal (1:1,000, BD Transduction Laboratories). Membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (1:4,000) and detected by the enhanced chemiluminescence method (Amersham).
RESULTS
Generation of DR5-null mice.
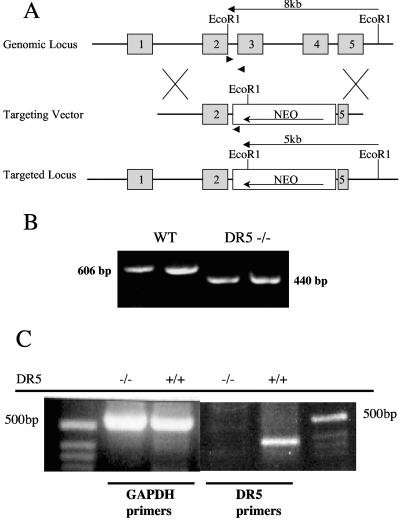
We abolished TRAIL-DR5 signaling by targeted disruption of the extracellular cysteine-rich domain of DR5 by homologous recombination. The targeting vector replaced exons 3 and 4 and part of 5 with a neomycin resistance cassette in the antisense orientation (Fig. 1A). Targeted E14K ES cells were used to generate DR5-null mice. PCR on genomic DNA isolated from thymocytes confirmed the absence of the deleted sequence (Fig. 1B). Lack of functional gene transcription was verified by reverse transcription-PCR of RNA derived from the small bowel (Fig. 1C).
FIG. 1.
Targeting the DR5 gene. (A) Schematic of the gene targeting strategy. Exons 3, 4, and part of 5 were targeted for deletion by homologous recombination with a vector encoding a neomycin resistance cassette. Arrowheads show the locations of forward and reverse primers used to genotype the mice. EcoRI restriction fragments are shown. (B) Multiplex PCR for wild-type and recombinant alleles. DNA was isolated from thymocytes originating from wild-type (WT) and DR5−/− female mice and amplified with three different primers. The PCR products were 606 bp and 440 bp for the wild-type and recombinant alleles, respectively. (C) DR5 is not expressed in somatic tissue of knockout mice. Reverse transcription-PCR for DR5 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was performed on RNA extracts from the small intestine of DR5−/− and DR5+/+ animals. The predicted size of the DR5 PCR product is 343 bp.
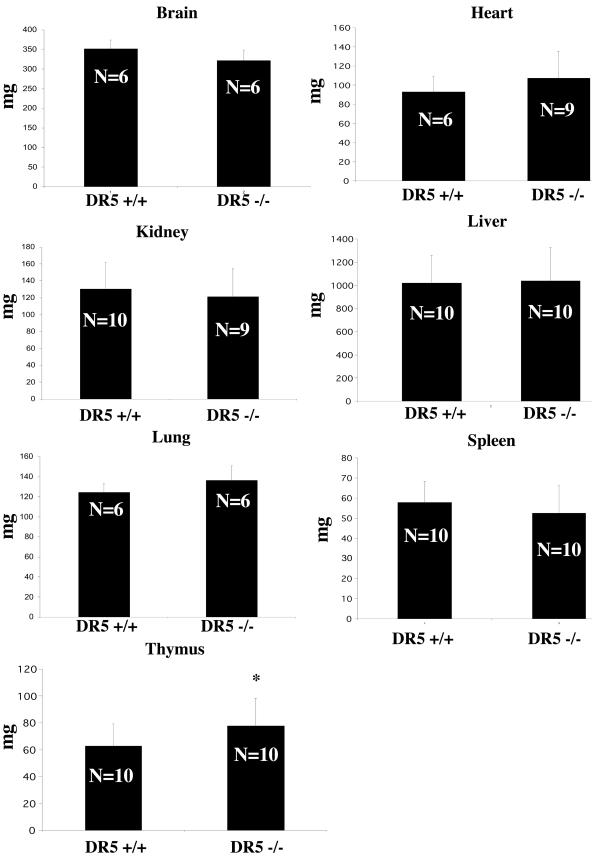
DR5-null mice show no developmental abnormalities and are fertile. We found no histological differences in the tissues examined (spleen, thymus, liver, lung, brain, intestine, colon, skin, esophagus, and kidney) from 4-week-old mice. When the organ mass of 4-to 6-week-old knockout mice was compared to that of wild-type mice, we found a statistically significant increase in the mass of the thymus in DR5-null mice (Fig. 2). No particular histological compartment of the thymus appeared to be enlarged. There was no statistically significant difference in the masses of brain, heart, kidney, liver, lung, or spleen from DR5-null mice compared to wild-type mice.
FIG. 2.
Mass of the DR5 null thymus is increased compared to controls. Organs from 4- to 6-week-old mice were weighed. Statistical significance was determined by Student’s t test. *, P < 0.05.
DR5 and TRAIL are expressed in the decidua and chorion at E8.5.
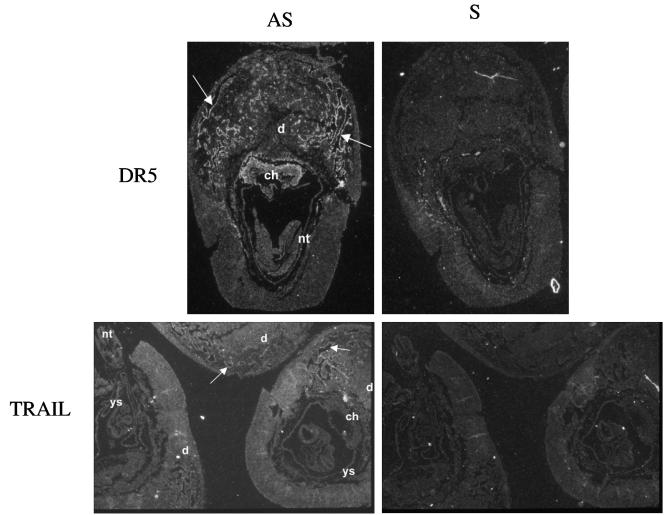
To determine the function of DR5 signaling during development, we examined gene expression patterning of DR5 and TRAIL during embryogenesis by in situ hybridization. Antisense probes labeled with [35S]UTP specific for DR5 and TRAIL mRNAs were hybridized on sections of embryos at embryonic day 8.5 (E8.5), E10.5, E12.5, E13.5, E16.5, and E17.5. Sense probes for DR5 and TRAIL were used as controls. Expression of DR5 and TRAIL was close to the background in the embryo proper at all time points. However, we found DR5 expressed strongly in the vessels of the decidua and in the chorion at E8.5 (Fig. 3). TRAIL expression was similar at E8.5, although the signal was weaker. TRAIL and DR5 expression was not significant in the extraembryonic tissues at other time points. Despite expression of DR5 and TRAIL in placental tissues, we have observed that DR5-null mice reproduce at a normal rate and with normal litter sizes (data not shown). Thus, the absence of TRAIL and DR5 in the placenta does not affect the reproductive process.
FIG. 3.
DR5 and TRAIL are expressed at E8.5 in the placenta of wild-type mice. In situ hybridization shows staining in the decidua (d) and chorion (ch). Arrows indicate staining in the vessels of the decidua. Antisense (AS) mRNA were used on the left, and sense (S) controls were used on the right. nt, notochord; ys, yolk sac.
Tissue-specific expression of DR5.
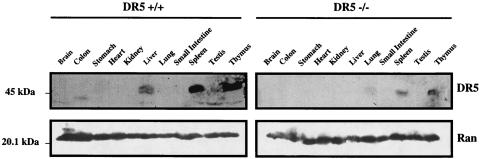
Previous analysis of baseline expression of murine DR5 by Northern blotting suggested that DR5 expression predominates in the heart, lung, and kidney (22). Interestingly, Western blotting suggests that baseline expression of DR5 protein predominates in the thymus, spleen, and somewhat in the liver (Fig. 4). By Western immunoblotting, tissues from DR5−/− animals did not produce a band of the size predicted for wild-type DR5. The discrepancy between the levels of mRNA and protein suggests that posttranslational mechanisms may modulate DR5 protein levels in (some) tissues during unstressed conditions in vivo.
FIG. 4.
Tissue-specific expression of DR5. DR5 protein is most prevalent in the thymus and spleen, as detected by Western immunoblotting. Weak expression of DR5 is also observed in the liver. A nonspecific band of lower molecular weight can be seen in tissue homogenates from DR5−/− mice.
In vitro, lack of DR5 diminishes the apoptotic response to TRAIL.
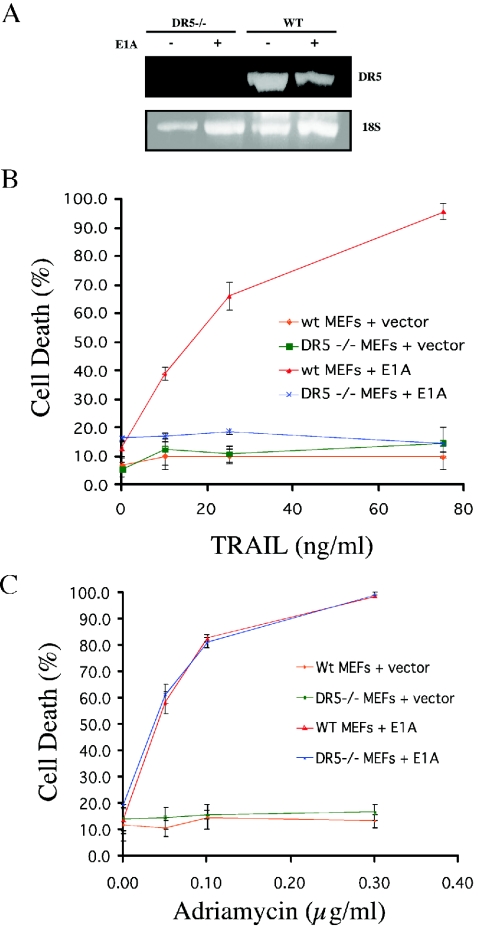
Thus far, DR5 is the only death-signaling TRAIL receptor isolated from the mouse. In order to establish whether other death-transducing TRAIL receptors exist in the mouse, we isolated DR5-null MEFs and transfected them with either E1A or empty vector. E1A transfection is associated with nuclear p53 stabilization in part through the p19ARF pathway (7). Recent reports have also uncovered decreased proteasomal degradation of p53 following E1A transfection (40). Surprisingly, E1A transfection did not significantly alter the levels of DR5 mRNA present in MEFs (Fig. 5A), indicating that E1A did not increase transcription of the DR5 gene. Interestingly, E1A sensitized wild-type MEFs in a dose-dependent fashion to TRAIL, whereas DR5-null E1A MEFs remained resistant to TRAIL (Fig. 5B).
FIG. 5.
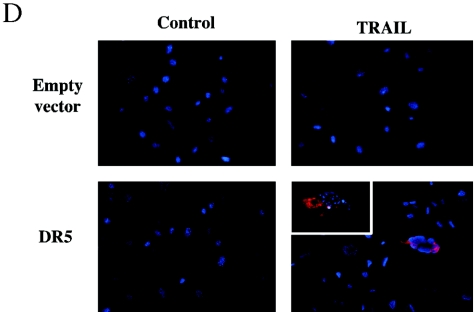
E1A immortalization sensitizes wild-type MEFs but not DR5−/− MEFs to TRAIL. MEFs were infected with a retroviral vector expressing E1A or empty vector. (A) E1A immortalization does not significantly alter DR5 mRNA expression as determined by reverse transcription-PCR analysis of RNA from DR5−/− and wild-type MEFs with and without E1A. MEFs were exposed to increasing concentrations of (B) TRAIL or (C) adriamycin. Cell death was quantitated after 24 h by trypan blue exclusion. (D) E1A-DR5−/− MEFs were infected with empty vector (pBabe Puro) and vector containing murine DR5 (MK) and subsequently treated with soluble mouse TRAIL (0.5 μg). Nuclei are visualized in blue (DAPI), whereas cytoplasmic red fluorescence (tetramethylrhodamine isothiocyanate) indicates activated caspase 3.
We also examined the effect of adriamycin on both wild-type and DR5-null E1A MEFs. We found no significant difference in the apoptotic response to adriamycin between DR5-null and wild-type E1A MEFs (Fig. 5C). In order to confirm that the DR5-null MEFs lack of sensitivity to TRAIL was related to the lack of DR5 expression, we transfected murine DR5 (Killer/MK) into the DR5−/− MEFs. Following infection with a DR5-expressing retrovirus, most MEFs died during the selection process (data not shown). Surviving DR5-infected clones showed increased active caspase 3 labeling in comparison to clones containing the empty vector when challenged with TRAIL (Fig. 5D).
DR5-null mice are compromised in radiation-induced apoptosis.
We have previously shown that DR5 is a major target gene of p53 upregulated in response to DNA damage in the spleen, thymus, and small bowel (3). Also, in response to ionizing radiation, p53-null mice exhibit decreased apoptosis in these tissues as well as in the colon (9). Therefore, we hypothesized that DR5-null mice may be resistant to ionizing radiation-induced apoptosis in the spleen, thymus, small bowel, and colon. To test this idea, we exposed 4- to 6-week-old female DR5-null mice and age- and sex-matched wild-type controls to 5 Gy of gamma radiation. A separate group of DR5-null and wild-type mice were also treated with dexamethasone as a positive control. Mice were sacrificed between 6 and 8 h after irradiation, and tissues were fixed in paraformaldehyde. The time points used were known from previous studies to elicit a p53-dependent apoptotic response associated with DR5 induction in several tissues (3, 9).
Apoptosis was examined in sectioned tissues by immunohistochemistry for activated (cleaved) caspase 3 or TUNEL. Detailed analysis of the stained sections was performed by either counting the number of cells/field (activated caspase 3) or determining the area fraction that stained positive by image analysis with ImageJ software (TUNEL). For active caspase 3, single cells could be discriminated, whereas TUNEL positivity appeared mainly in the form of “apoptotic clusters” in the lymphoid organs following ionizing irradiation. Thus, the TUNEL-positive area fraction reflects the size and number of such clusters.
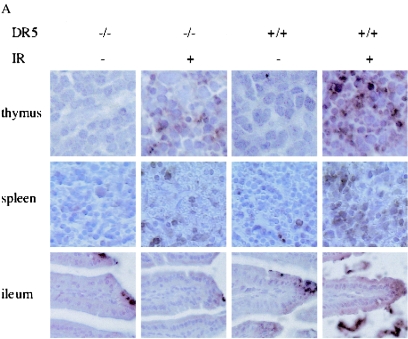
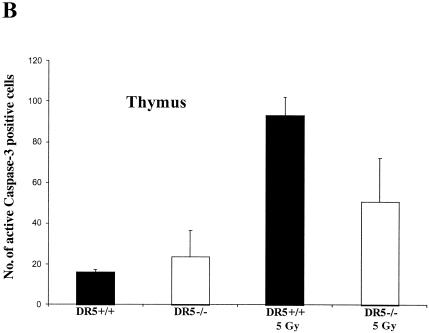
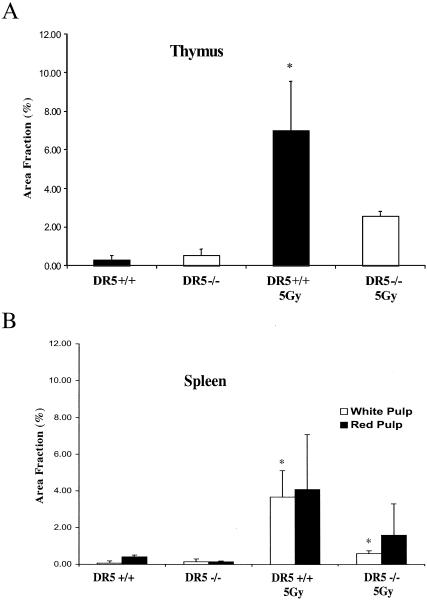
In the thymus, irradiated wild-type tissue showed approximately two times more apoptotic cells than DR5-null mice on average (93 ± 9 cells/field and 51 ± 21 cells/fields, respectively) as determined by counting caspase-3-positive cells (Fig. 6A and B). Approximately the same difference was observed when sections were TUNEL-stained, i.e., irradiated DR5+/+ mice had an average area fraction that was 7.03 ± 2.50 cells/field and the corresponding value for DR5−/− mice was 2.57 ± 0.25 cells/field (Fig. 7 and 8A). Thus, irradiated thymus from DR5+/+ mice shows significantly increased death (P < 0.05, Student's t test) in comparison to DR5-null tissue by both active caspase 3 and TUNEL staining. Apoptosis was observed most extensively in the thymic cortex but not in the medulla in both wild-type and DR5-null mice. These results indicate that DR5 is an important but not exclusive regulator of DNA damage-induced apoptosis in the thymic cortex.
FIG. 6.
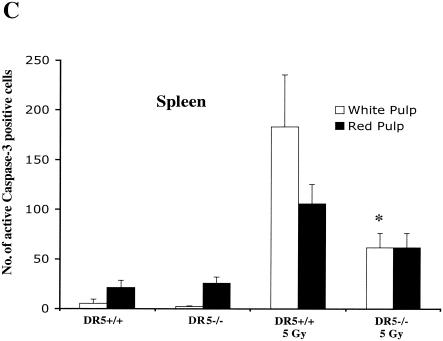
Radiation-induced death is attenuated in some tissues of DR5−/− and TRAIL−/− animals. (A) DR5+/+ and DR5−/− animals were treated with 5 Gy of ionizing radiation, and tissues were harvested after 8 h. DR5−/− animals show a reduced number of cells with activated caspase 3 in the thymus (B) and in the white pulp of the spleen (C) compared to DR5+/+ animals (P < 0.05, Student’s t test).
FIG. 7.
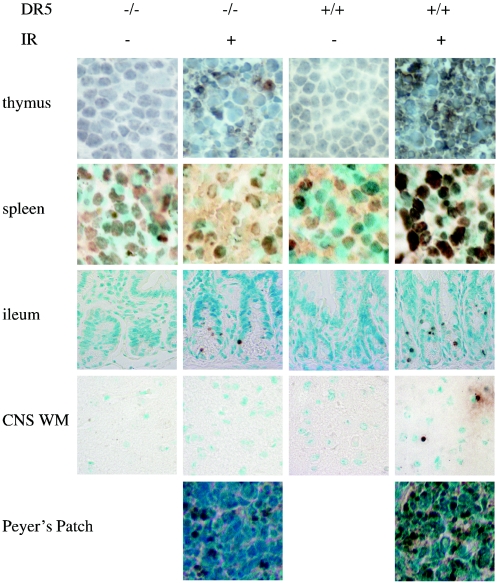
Radiation-induced death is attenuated in some tissues of DR5−/− animals. DR5+/+ and DR5−/− animals were treated with 5 Gy of ionizing radiation, and tissues were harvested after 8 h. Apoptosis was detected by TUNEL assay.
FIG. 8.
Image analysis of histological sections stained with TUNEL. (A) Sections from the thymus of DR5−/− animals show a smaller TUNEL-positive area fraction in comparison to thymus sections taken from DR5+/+ animals. (B) The white pulp of the spleen from DR5−/− animals displays a smaller area fraction than that of DR5+/+ animals. Images were analyzed with ImageJ software. Asterisks indicate P < 0.05, Student's t test.
In the spleen, apoptosis was strongly induced in irradiated wild-type tissues as detected by active caspase 3 and TUNEL staining (Fig. 6A and C and 8B). Irradiated DR5-null white pulp had approximately a threefold lower apoptotic response in the white pulp on average (i.e., 182 ± 52.3 and 61.4 ± 14.4 positive cells/field for DR5+/+ and DR5−/−, respectively) of the spleen as detected by staining for active caspase 3 (Fig. 6C). The corresponding value for the irradiated white pulp with TUNEL-staining and image analysis was 3.7 ± 1.5 positive cells/field for DR5+/+ mice compared to 0.60 ± 0.15 DR5+/+for DR5−/− mice on average, i.e., a sixfold decrease of the TUNEL-positive area (Fig. 8B). Thus, analysis of caspase 3 and TUNEL-stained sections of irradiated white pulp from DR5+/+ and DR5−/− mice shows that DR5+/+ mice have significantly (P < 0.05, Student's t test) increased levels of apoptosis in the white pulp.
Irradiated red pulp showed a less than twofold increase in the number of cells with activated caspase 3 (105.4 ± 19.4 and 61.4 ± 14.4 positive cells/field for DR5+/+ and DR5−/−, respectively) (Fig. 6C), whereas analysis of the TUNEL-positive area fraction showed an approximately twofold increase (4.1 ± 3.0 positive cells/field for DR5+/+ animals and 1.6 ± 1.7 positive cells/field for DR5−/− animals) (Fig. 8B) in DR5+/+ mice compared to DR5−/− mice. Although there was a trend towards increased apoptosis in the irradiated red pulp of DR5+/+ mice, this was not statistically significant. Thus, as with the immune cells of the thymus, DR5 plays a role in the DNA damage response of the spleen and in particular in lymphocytes of the white pulp. However, since apoptotic cells are still present in both irradiated red and white pulp of DR5−/− mice, DR5 is not the sole effector of apoptosis in this tissue.
In the ileum, irradiated wild-type tissue retained a cytoplasmic active caspase 3 stain along the sides of the villi that was not evident in irradiated DR5-null ileum (Fig. 6A). TUNEL staining showed rare instances of apoptosis in the villi but consistent amounts of apoptosis in the crypts that was of a very slightly reduced level between the irradiated DR5-null and wild-type tissues (Fig. 7). In the Peyer's patches of the ileum, the percentage of apoptotic cells was significantly greater in irradiated wild-type than DR5-null tissues (Fig. 7).
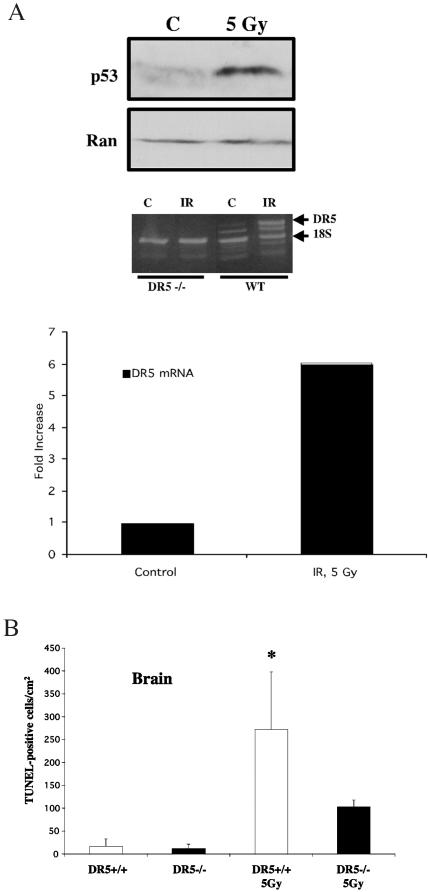
In the brain, more apoptotic cells were evident by TUNEL in the white matter of irradiated wild-type compared to DR5-null mice (Fig. 7). Interestingly, no radiation-induced cell death was observed in the grey matter of the DR5−/− or wild-type brain (data not shown). Since it was unclear if DR5 was upregulated in the brain of mice following irradiation, we performed comparative quantitative reverse transcription-PCR on RNA isolated from irradiated brains of wild-type and DR5−/− mice (Fig. 9A). By analyzing band density, we were able to document a sixfold upregulation of DR5 mRNA in the brain at 8 h following 5 Gy of whole-body irradiation (Fig. 9A). In order to further quantify the differences in cell death in the white matter, we counted the number of apoptotic cells per section and normalized it over the surface area of the section (Fig. 9B). From this analysis we conclude that there are more than twice as many TUNEL-positive cells in the wild-type white matter as in the DR5-null white matter (272 ± 126 and 104 ± 14 per mm2, respectively) following irradiation. This suggests that DR5 is critical to DNA damage-induced apoptosis in neural tissue.
FIG. 9.
Radiation-induced cell death in the brains of DR5-null animals. (A) Western blot analysis shows that p53 protein levels are increased in the brain of wild-type mice (WT) 8 h following 5 Gy of gamma irradiation. Also, at this time point DR5 mRNA is upregulated in the brain of DR5+/+ mice but not DR5−/− mice. Comparative quantitative analysis of the upregulation was done with NIH Image 1.62 software and 18S RNA as a housekeeping gene. (B) Quantitation of radiation-induced cell death in the brain of DR5-null and wild-type animals by TUNEL staining.
Apoptosis was examined in other irradiated tissues, including the stomach, proximal colon, kidney, lung, and esophagus. In these tissues, the level of cell death was similar between wild-type and DR5-null mice (data not shown). In the colon, apoptosis was primarily observed basally in the crypts and became more sporadic toward the apical layer. Radiation did not induce a significant amount of cell death in the kidney, lung, or esophagus.
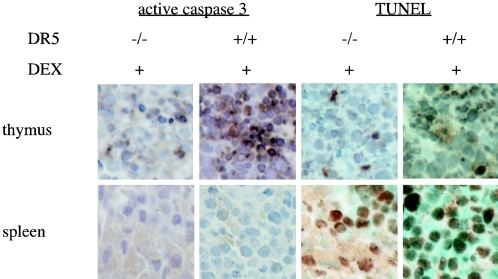
Dexamethasone is a steroid known to induce apoptosis in the spleen and thymus (37). We have previously shown both in vitro (19) and in vivo (3) that dexamethasone treatment causes upregulation of DR5. We observed that in dexamethasone-treated mice, apoptosis was more pervasive in the thymus of wild-type mice compared to DR5-null mice (Fig. 10). This difference was not observed in the spleen. These results indicate that DR5 contributes to the mechanism of dexamethasone-induced cell death in the thymus.
FIG. 10.
Apoptosis in immune tissues of dexamethasone-treated animals. DR5+/+ and DR5−/− animals were injected intraperitoneally with 0.5 mg of dexamethasone/animal, and tissues were harvested after 8 h. Apoptosis was assayed by staining for active caspase 3 and TUNEL.
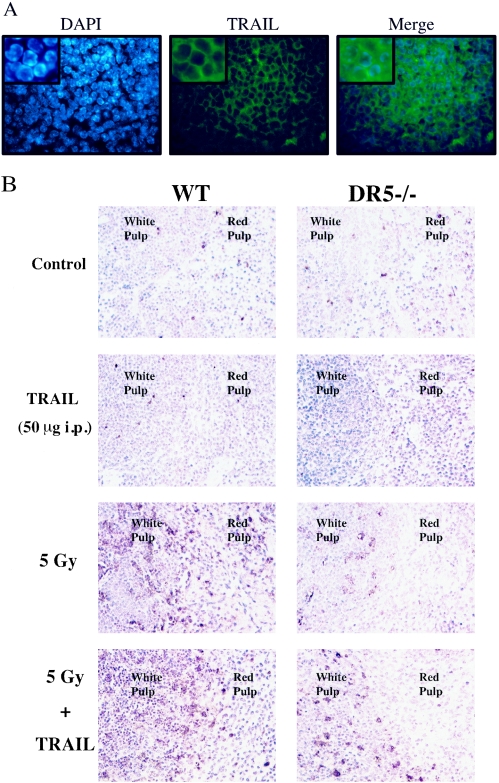
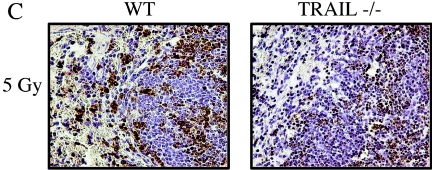
We hypothesized that either membrane-bound or soluble TRAIL might mediate cell death through the DR5 receptor in the lymphoid organs following irradiation. Thus, TRAIL deficiency could contribute to an attenuated apoptotic response. By performing immunofluorescence, we could confirm that TRAIL was expressed on the membrane of lymphocytes in the white pulp of the spleen (Fig. 11A). We also irradiated TRAIL−/− mice with 5 Gy of ionizing radiation and analyzed the amount of apoptosis in the spleen by TUNEL staining (Fig. 11C). There was a marked decrease in the number of TUNEL-positive cells in the white pulp of the spleen from TRAIL−/− animals analogous to that observed in the spleen of DR5−/− animals. We also observed a slight decrease in the number of apoptotic cells in the small intestine (data not shown).
FIG. 11.
Cell death in the spleen of TRAIL−/− and TRAIL-treated animals. (A) Immunohistochemistry with Cy2-conjugated antibodies shows that membrane-bound TRAIL (green, middle panel) was expressed by splenocytes. Nuclei were visualized by counterstaining with DAPI (blue, left panel). (B) DR5+/+ and DR5−/− animals were treated with 50 μg of soluble mouse TRAIL, 5 Gy of whole-body irradiation, and 50 μg of soluble mouse TRAIL in combination with 5 Gy of whole-body irradiation. TRAIL was injected (intraperitoneally) 4 h after the irradiation, and the tissues were harvested 8 h following irradiation. Cell death was detected by TUNEL staining. (C) Wild-type (WT) and TRAIL−/− animals were irradiated with 5 Gy, and the spleen was isolated at 6 h following irradiation. The number of TUNEL-positive cells was increased in the spleen, in particular the white pulp, of wild-type animals.
In order to confirm the role of TRAIL in promoting cell death through DR5 following DNA damage, we treated nonirradiated mice and mice irradiated with 5 Gy of ionizing radiation with soluble mouse TRAIL (50 μg/mouse injected intraperitoneally) at 4 h following the time of irradiation (Fig. 11B). Surprisingly, injection of TRAIL alone did not alter the number of apoptotic cells, as detected by TUNEL staining, in the spleen in comparison to the control. However, the combination treatment with TRAIL and ionizing radiation resulted in a further increase in the number of TUNEL-positive cells in the white pulp of the spleen from wild-type mice in comparison to the white pulp of DR5−/− mice at 8 h following irradiation. In some cases the white pulp of wild-type animals seemed to be completely eradicated of nonapoptotic cells. This suggests that TRAIL can modulate the level of cell death of lymphocytes in the spleen through DR5 in vivo following DNA damage.
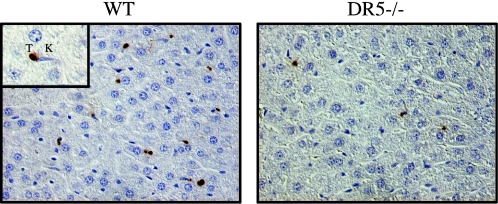
We detected an increased number of TUNEL-positive cells in the livers of wild-type mice in comparison to the livers of DR5-deficient mice (Fig. 12). TUNEL-positive cells were found primarily in the sinusoids in both wild-type and DR5-deficient mice. Kupffer cells were frequently observed in the vicinity of TUNEL-positive cells and contained TUNEL-positive particles, suggesting homing of Kupffer cells to apoptotic cells and subsequent phagocytosis. We did not find any morphological evidence for hepatocyte death in the liver, and hepatocytes were rarely TUNEL positive. We suggest that the TUNEL-positive cells found in the sinusoids of the liver may be lymphocytes based on their morphology and our previous data showing a connection between DR5 and apoptosis following irradiation in the thymus and spleen.
FIG. 12.
Cell death in the liver is reduced in DR5−/− mice. Eight hours following 5 Gy of ionizing radiation, TUNEL-positive (T) cells were detected in the liver. The cells were located primarily in the sinusoids and frequently located in the vicinity of Kupffer cells (K).
DISCUSSION
In order to better understand the in vivo biology of DR5, we have constructed a knockout mouse by targeting exons 3, 4, and 5 of the genomic locus. These knockout mice develop normally and can reproduce.
To explore the function of DR5 and its ligand TRAIL during development, we performed in situ hybridization with antisense mRNAs to these two genes. We found that both TRAIL and DR5 are expressed in the vessels of the decidua and the chorion early in development (E8.5). However, the fact that mice develop normally indicates that DR5 expression on day E8.5 is not essential for organogenesis in the embryo. It has been suggested that DR5 may play a role in limiting autoimmunity in the placenta, but we note that DR5-null females are able to have multiple litters without complications to the mother or pups (unpublished observations).
Because DR5 is the only known receptor for TRAIL in the mouse, we compared the histology and apoptotic responses to radiation in DR5-deficient and TRAIL-deficient mice. The TRAIL-deficient thymus contains more thymocytes than wild-type mice, suggesting deficient negative selection (15). The observation that DR5-deficient mice develop slightly larger thymuses suggests that negative selection might be controlled (in part) through the DR5 receptor. However, TRAIL-deficient mice also show enlarged spleens and lymph nodes in addition to an increased cell number in the thymus. We found no evidence for an altered spleen weight in DR5−/− animals at 4 to 6 weeks of age. This suggests that TRAIL may function independently of DR5 in regulating the turnover of cells in the spleen during physiological conditions or that TRAIL decoy receptors may modulate the response to TRAIL in conjunction with the proapoptotic DR5 receptor. Although DR5−/− mice are at this point less investigated in the context of autoimmunity, we have not found any evidence that DR5−/− mice are susceptible to spontaneous autoimmune diseases. We have documented four cases (out of 12 animals) of skin lesions (dermatitis) exclusively in female DR5−/− mice 8 to 14 months of age, whereas no case has been documented in wild-type female mice of the same age (eight animals) (data not shown).
In vitro studies of DR5-null MEFs.
DR5-null MEFs that express E1A do not undergo apoptosis after treatment with TRAIL. This suggests that DR5 may be the only death-inducing TRAIL receptor that is upregulated in the mouse in the setting of E1A. Although it is well established that E1A stabilizes p53 and may alter expression of its target genes, we detected no increase in the level of DR5 mRNA following E1A transfection. A recent report suggests that E1A activates p53 through inhibition of proteasomal regulatory subunit S2, preventing degradation of p53 (40). A possible explanation for our findings is that E1A stabilizes DR5 through inhibition of the proteasome. Indeed, several proteasome inhibitors have been shown to sensitize tumor cells to TRAIL-mediated killing (11, 14, 25a). We are currently investigating how proteasome inhibitors affect TRAIL sensitivity through DR5 (N. Finnberg and W. S. El-Deiry, unpublished data). The finding that E1A does sensitize DR5-null MEFs contrasts with the existence of two death-inducing TRAIL receptors, as in humans. However, we cannot rule out the existence of TRAIL receptors that may be activated by cytokines or other means.
Cell killing by toxic doses of adriamycin appears to be independent of DR5 status in MEFs. This may simply be due to the preponderance of proapoptotic signals induced by adriamycin or due to the paucity of TRAIL-DR5 signaling in vitro. This suggests a role for DR5 in the response of tumor cells to chemotherapy or immune cell killing in combination with additional TRAIL.
Irradiation of DR5-null mice.
The p53-dependent radiation response has been the subject of intense scrutiny. Experiments with p53-null mice have shown that the response to radiation differs based on dosage and tissue type. This is thought to be caused by transactivation of different p53 target genes within a tissue compartment. Differing expression of Waf1, Bax, mdm2, and Fas has been shown in the spleen, small intestine, liver, lung, heart, thymus, brain, and kidney in response to radiation (2). Similar studies in our laboratory have examined induction of p21, Bax, DR5, Fas, and PIG8. DR5 was specifically shown to be a dominant mRNA transcript upregulated in response to radiation in the spleen, thymus, and small intestine (3). In that study, DR5 upregulation following ionizing radiation was tightly correlated to the p53 status in the spleen, whereas upregulation of DR5 in the thymus in part was p53 independent. In a different study, we also showed radiation-dependent upregulation of DR5 in the colon (9). Based on these studies, we hypothesized there would be differential death due to radiation between DR5-null and wild-type mice in the spleen, thymus, small intestine, and colon.
Our results have confirmed the importance of DR5 in the radiation response of the spleen and thymus. Apoptosis is attenuated by three- to sixfold in the white pulp of the spleen and approximately twofold in the thymus of DR5−/− mice compared to wild-type mice following ionizing irradiation. Clearly DR5 has a role in the radiation response in the lymphoid cells of these tissues. The observation that TRAIL enhances killing in the white pulp of wild-type mice following irradiation but does not affect the white pulp of DR5−/− mice suggests that TRAIL is of central importance in mediating cell death in the white pulp of the spleen. It also suggests that DR5 is the main TRAIL-responsive receptor mediating cell death following ionizing radiation in the murine white pulp and supports the in vitro data on DR5−/− MEFs and TRAIL (see above). Although we cannot rule out the presence of other TRAIL-responsive receptors contributing to cell death upon treatment with different concentrations of TRAIL or at different time points and/or stimuli, the connection between TRAIL and DR5 is emphasized by the observation that TRAIL-deficient mice have an attenuated radiation response in their lymphoid tissues. However, because gene targeting of DR5 failed to eliminate all death in these organs, it is likely that DR5 is only one of several p53 target genes that are important in this response.
In the colon, our results differ from our hypothesis, as we observed equal amounts of radiation-induced death in DR5-null and wild-type mice. This may be explained by the fact that although DR5 was previously shown to be induced in the colon, it is not the dominant gene is this tissue that signals apoptosis. Although DR5 may not play a significant role in radiation-induced apoptosis in the colon, it may still play a role in suppressing tumor formation or progression. In this regard, we recently found that stable or inducible silencing of DR5 in human colon carcinoma cells significantly enhanced their tumorigenic growth in vivo (32a). In the ileum, we observed cytoplasmic staining along the villi by active caspase-3 immunostaining that was greater in the wild-type than in the DR5-null tissues. However, by TUNEL, only staining in the crypts was observed and no signal was apparent in the nuclei of the villi. Cytoplasmic active caspase 3 staining without fulminant DNA nick end-labeling may be the result of an apoptotic process that was initiated in the cytoplasmic compartment but was halted or has not yet progressed to the nuclei. Possibly, at later time points, TUNEL-positive nuclei might become positive in the villi of the ileum. We conclude that DR5 is not as important for apoptosis in the intestinal crypts but that it may play a role in apoptosis along the villi.
Surprisingly, cell death in response to ionizing radiation was significantly abolished in the central nervous system white matter of the DR5-null mice. This result supports the role of p53-dependent death in the nervous system, as has been explored in various mouse models. p53−/− mice and ATM−/− mice both show reduced apoptosis in various regions of the central nervous system in response to ionizing radiation (12, 34). Bax also plays a role in cell death in the brain as Bax-null mice have increased numbers of motor neurons (8) and are deficient in naturally occurring neural death during development (33). Additionally, mice nullizygous for Bax and ATM show reduced levels of radiation-induced apoptosis compared to ATM-null mice (5). The fact that we observed more than a 50% reduction of radiation-induced death in the white matter of DR5-null mice indicates that DR5 is an important gene induced by DNA damage in this tissue. However, at this point it is not clear if loss of DR5 leads only to a defective extrinsic proapoptotic pathway or if proapoptotic signal transduction pathways involving the mitochondria (intrinsic) are affected as well.
To address this question, we are currently performing expression profiling on irradiated DR5-null tissues to elucidate altered proapoptotic pathways (N. Finnberg and W. S. El-Deiry, unpublished data). In support of a model involving cross talk between a proapoptotic death receptor(s) and mitochondrial pathways, a previous report has shown that p53 does not upregulate Bax in response to radiation in cultured neurons (13). Also, Bid is cleaved in response to oxygen or glucose deprivation (a stimulus known to upregulate p53), and Bid-null neurons are resistant to oxygen and glucose deprivation (25). Bid is cleaved by activated caspase 8 in a death receptor-dependent manner. Thus, our data may support a model in the brain in which p53 upregulation of DR5 leads to cleavage of Bid and subsequent downstream activation of Bax instead of direct p53 transactivation of Bax. Although our data suggest that DR5 is an important initiating death receptor in the brain, we cannot exclude a role for other (p53-regulated) genes or death receptors, some of which clearly have an effect in the radiation response.
In conclusion, DR5-null mice are viable and develop normally with the exception of enlarged thymuses. We have established that DR5 and TRAIL are expressed early in development and only in the decidua and chorion. We have shown that loss of DR5 diminishes apoptosis in immortalized cells. We show that DR5-null mice are compromised in radiation-induced apoptosis in a tissue-specific manner and that this apoptotic response can be modulated in vivo by TRAIL in a DR5-dependent fashion. These results help to explain the differential toxicity of radiation to various tissues and tumor types and should pave the way for more rational use of DNA-damaging therapy in cancer.
Acknowledgments
This work was supported by NIH grants CA75454 and CA75138 and by funds from the Howard Hughes Medical Institute to W.S.E.-D.
REFERENCES
- 1.Bennett, M., K. Macdonald, S. W. Chan, J. P. Luzio, R. Simari, and P. Weissberg. 1998. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science 282:290-293. [DOI] [PubMed] [Google Scholar]
- 2.Bouvard, V., T. Zaitchouk, M. Vacher, A. Duthu, M. Canivet, C. Choisy-Rossi, M. Nieruchalski, and E. May. 2000. Tissue and cell-specific expression of the p53-target genes: bax, fas, mdm2 and waf1/p21, before and following ionising irradiation in mice. Oncogene 19:649-660. [DOI] [PubMed] [Google Scholar]
- 3.Burns, T. F., E. J. Bernhard, and W. S. El-Deiry. 2001. Tissue specific expression of p53 target genes suggests a key role for KILLER/DR5 in p53-dependent apoptosis in vivo. Oncogene 20:4601-4612. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhary, P. M., M. Eby, A. Jasmin, A. Bookwalter, J. Murray, and L. Hood. 1997. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kappaB pathway. Immunity 7:821-830. [DOI] [PubMed] [Google Scholar]
- 5.Chong, M. J., M. R. Murray, E. C. Gosink, H. R. Russell, A. Srinivasan, M. Kapsetaki, S. J. Korsmeyer, and P. J. McKinnon. 2000. Atm and Bax cooperate in ionizing radiation-induced apoptosis in the central nervous system. Proc. Natl. Acad. Sci. USA 97:889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cory, S., and J. M. Adams. 2002. The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2:647-656. [DOI] [PubMed] [Google Scholar]
- 8.Deckwerth, T. L., J. L. Elliott, C. M. Knudson, E. M. Johnson, Jr., W. D. Snider, and S. J. Korsmeyer. 1996. BAX is required for neuronal death after trophic factor deprivation and during development. Neuron 17:401-411. [DOI] [PubMed] [Google Scholar]
- 7.de Stanchina, E., M. E. McCurrach, F. Zindy, S. Y. Shieh, G. Ferbeyre, A. V. Samuelson, C. Prives, M. F. Roussel, C. J. Sherr, and S. W. Lowe. 1998. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 12:2434-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fei, P., E. J. Bernhard, and W. S. El-Deiry. 2002. Tissue-specific induction of p53 targets in vivo. Cancer Res. 62:7316-7327. [PubMed] [Google Scholar]
- 10.Gu, Z., C. Flemington, T. Chittenden, and G. P. Zambetti. 2000. ei24, a p53 response gene involved in growth suppression and apoptosis. Mol. Cell. Biol. 20:233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, Q., Y. Huang, and M. S. Sheikh. 2004. Proteasome inhibitor MG132 upregulates death receptor 5 and cooperates with Apo2L/TRAIL to induce apoptosis in Bax-proficient and -deficient cells. Oncogene 23:2554-2558. [DOI] [PubMed] [Google Scholar]
- 12.Herzog, K. H., M. J. Chong, M. Kapsetaki, J. I. Morgan, and P. J. McKinnon. 1998. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science 280:1089-1091. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, M. D., H. Xiang, S. London, Y. Kinoshita, M. Knudson, M. Mayberg, S. J. Korsmeyer, and R. S. Morrison. 1998. Evidence for involvement of Bax and p53, but not caspases, in radiation-induced cell death of cultured postnatal hippocampal neurons. J. Neurosci. Res. 54:721-733. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, T. R., K. Stone, M. Nikrad, T. Yeh, W. X. Zong, C. B. Thompson, A. Nesterov, A. S. Kraft. 2003. The proteasome inhibitor PS-341 overcomes TRAIL resistance in Bax and caspase 9-negative or Bcl-xL overexpressing cells. Oncogene 22:4953-4963. [DOI] [PubMed] [Google Scholar]
- 15.Lamhamedi-Cherradi, S. E., S. J. Zheng, K. A. Maguschak, J. Peschon, and Y. H. Chen. 2003. Defective thymocyte apoptosis and accelerated autoimmune diseases in TRAIL−/− mice. Nat. Immunol. 4:255-260. [DOI] [PubMed] [Google Scholar]
- 16.LeBlanc, H. N., and A. Ashkenazi. 2003. Apo2L/TRAIL and its death and decoy receptors. Cell Death Differ. 10:66-75. [DOI] [PubMed] [Google Scholar]
- 17.Lyons, G. E., S. Schiaffino, D. Sassoon, P. Barton, and M. Buckingham. 1990. Developmental regulation of myosin gene expression in mouse cardiac muscle. J. Cell Biol. 111:2427-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacFarlane, M., M. Ahmad, S. M. Srinivasula, T. Fernandes-Alnemri, G. M. Cohen, and E. S. Alnemri. 1997. Identification and molecular cloning of two novel receptors for the cytotoxic ligand TRAIL. J. Biol. Chem. 272:25417-25420. [DOI] [PubMed] [Google Scholar]
- 19.Meng, R. D., and W. S. El-Deiry. 2001. p53-independent upregulation of KILLER/DR5 TRAIL receptor expression by glucocorticoids and interferon-gamma. Exp. Cell Res. 262:154-169. [DOI] [PubMed] [Google Scholar]
- 20.Oda, E., R. Ohki, H. Murasawa, J. Nemoto, T. Shibue, T. Yamashita, T. Tokino, T. Taniguchi, and N. Tanaka. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288:1053-1058. [DOI] [PubMed] [Google Scholar]
- 21.Owen-Schaub, L. B., W. Zhang, J. C. Cusack, L. S. Angelo, S. M. Santee, T. Fujiwara, J. A. Roth, A. B. Deisseroth, W. W. Zhang, E. Kruzel, et al. 1995. Wild-type human p53 and a temperature-sensitive mutant induce Fas/APO-1 expression. Mol. Cell. Biol. 15:3032-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozoren, N., and W. S. El-Deiry. 2003. Cell surface Death Receptor signaling in normal and cancer cells. Semin. Cancer Biol. 13:135-147. [DOI] [PubMed] [Google Scholar]
- 23.Pan, G., J. Ni, Y. F. Wei, G. Yu, R. Gentz, and V. M. Dixit. 1997. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277:815-818. [DOI] [PubMed] [Google Scholar]
- 24.Pan, G., K. O'Rourke, A. M. Chinnaiyan, R. Gentz, R. Ebner, J. Ni, and V. M. Dixit. 1997. The receptor for the cytotoxic ligand TRAIL. Science 276:111-113. [DOI] [PubMed] [Google Scholar]
- 25.Plesnila, N., S. Zinkel, D. A. Le, S. Amin-Hanjani, Y. Wu, J. Qiu, A. Chiarugi, S. S. Thomas, D. S. Kohane, S. J. Korsmeyer, and M. A. Moskowitz. 2001. BID mediates neuronal cell death after oxygen/ glucose deprivation and focal cerebral ischemia. Proc. Natl. Acad. Sci. USA 98:15318-15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Sayers, T. J., A. D. Brooks, C. Y. Koh, W. Ma, N. Seki, A. Raziuddin, B. R. Blazar, X. Zhang, P. J. Elliott, and W. J. Murphy. 2003. The proteasome inhibitor PS-341 sensitizes neoplastic cells to TRAIL-mediated apoptosis by reducing levels of c-FLIP. Blood 102:303-310. [DOI] [PubMed] [Google Scholar]
- 26.Screaton, G. R., J. Mongkolsapaya, X. N. Xu, A. E. Cowper, A. J. McMichael, and J. I. Bell. 1997. TRICK2, a new alternatively spliced receptor that transduces the cytotoxic signal from TRAIL. Curr. Biol. 7:693-696. [DOI] [PubMed] [Google Scholar]
- 27.Sedger, L. M., M. B. Glaccum, J. C. Schuh, S. T. Kanaly, E. Williamson, N. Kayagaki, T. Yun, P. Smolak, T. Le, R. Goodwin, and B. Gliniak. 2002. Characterization of the in vivo function of tumor necrosis factor-alpha-related apoptosis-inducing ligand, TRAIL/Apo2L, using TRAIL/Apo2L gene-deficient mice. Eur. J. Immunol. 32:2246-2254. [DOI] [PubMed] [Google Scholar]
- 28.Sheridan, J. P., S. A. Marsters, R. M. Pitti, A. Gurney, M. Skubatch, D. Baldwin, L. Ramakrishnan, C. L. Gray, K. Baker, W. I. Wood, A. D. Goddard, P. Godowski, and A. Ashkenazi. 1997. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science 277:818-821. [DOI] [PubMed] [Google Scholar]
- 29.Varfolomeev, E. E., M. Schuchmann, V. Luria, N. Chiannilkulchai, J. S. Beckmann, I. L. Mett, D. Rebrikov, V. M. Brodianski, O. C. Kemper, O. Kollet, T. Lapidot, D. Soffer, T. Sobe, K. B. Avraham, T. Goncharov, H. Holtmann, P. Lonai, and D. Wallach. 1998. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 9:267-276. [DOI] [PubMed] [Google Scholar]
- 30.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 31.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 32.Walczak, H., M. A. Degli-Esposti, R. S. Johnson, P. J. Smolak, J. Y. Waugh, N. Boiani, M. S. Timour, M. J. Gerhart, K. A. Schooley, C. A. Smith, R. G. Goodwin, and C. T. Rauch. 1997. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J. 16:5386-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Wang, S., and W. S. El-Deiry. 2004. Inducible silencing of KILLER/DR5 in vivo promotes bioluminescent colon tumor xenograft growth and confers resistance to chemotherapeutic agent 5-fluorouracil. Cancer Res. 64:6666-6672. [DOI] [PubMed] [Google Scholar]
- 33.White, F. A., C. R. Keller-Peck, C. M. Knudson, S. J. Korsmeyer, and W. D. Snider. 1998. Widespread elimination of naturally occurring neuronal death in Bax-deficient mice. J. Neurosci. 18:1428-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood, K. A., and R. J. Youle. 1995. The role of free radicals and p53 in neuron apoptosis in vivo. J. Neurosci. 15:5851-5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, G. S., T. F. Burns, E. R. McDonald, 3rd, W. Jiang, R. Meng, I. D. Krantz, G. Kao, D. D. Gan, J. Y. Zhou, R. Muschel, S. R. Hamilton, N. B. Spinner, S. Markowitz, G. Wu, and W. S. el-Deiry. 1997. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet. 17:141-143. [DOI] [PubMed] [Google Scholar]
- 36.Wu, G. S., T. F. Burns, Y. Zhan, E. S. Alnemri, and W. S. El-Deiry. 1999. Molecular cloning and functional analysis of the mouse homologue of the KILLER/DR5 tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor. Cancer Res. 59:2770-2775. [PubMed] [Google Scholar]
- 37.Wyllie, A. H. 1980. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284:555-556. [DOI] [PubMed] [Google Scholar]
- 38.Yin, X. M., K. Wang, A. Gross, Y. Zhao, S. Zinkel, B. Klocke, K. A. Roth, and S. J. Korsmeyer. 1999. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature 400:886-891. [DOI] [PubMed] [Google Scholar]
- 39.Yu, J., Z. Wang, K. W. Kinzler, B. Vogelstein, and L. Zhang. 2003. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc. Natl. Acad. Sci. USA 100:1931-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, X., A. S. Turnell, C. Gorbea, J. S. Mymryk, P. H. Gallimore, and R. J. Grand. 2004. The targeting of the proteasomal regulatory subunit S2 by adenovirus E1A causes inhibition of proteasomal activity and increased p53 expression. J. Biol. Chem. 279:25122-25133. [DOI] [PubMed] [Google Scholar]