Abstract
The evolutionary conserved SET domain is present in many eukaryotic chromatin-associated proteins, including some members of the trithorax (TrxG) group and the polycomb (PcG) group of epigenetic transcriptional regulators and modifiers of position effect variegation. All SET domains examined exhibited histone lysine methyltransferase activity, implicating these proteins in the generation of epigenetic marks. However, the mode of the initial recruitment of SET proteins to target genes and the way that their association with the genes is maintained after replication are not known. We found that SET-containing proteins of the SET1 and SET2 families contain motifs in the pre-SET region or at the pre-SET-SET and SET-post-SET boundaries which very tightly bind single-stranded DNA (ssDNA) and RNA. These motifs also bind stretches of ssDNA generated by superhelical tension or during the in vitro transcription of duplex DNA. Importantly, such binding withstands nucleosome assembly, interfering with the formation of regular nucleosomal arrays. Two representatives of the SUV39 SET family, SU(VAR)3-9 and G9a, did not bind ssDNA. The trxZ11 homeotic point mutation, which is located within TRX SET and disrupts embryonic development, impairs the ssDNA binding capacity of the protein. We suggest that the motifs described here may be directly involved in the biological function(s) of SET-containing proteins. The binding of single-stranded nucleic acids might play a role in the initial recruitment of the proteins to target genes, in the maintenance of their association after DNA replication, or in sustaining DNA stretches in a single-stranded configuration to allow for continuous transcription.
The SET domain is an ∼130-amino-acid (aa) sequence which was initially identified in the protein products of three regulatory genes in Drosophila, whose names account for the name SET, i.e, SU(VAR)3-9, enhancer of Zeste [E(Z)], and trithorax (Trx). SET domains are usually located at the carboxyl termini of proteins, although in some proteins, such as ASH1 (another regulator belonging to the TrxG family), the motif is located in the middle of the protein (Fig. 1A), and in the proteins RIZ and BLIMP this motif maps to the N termini. In addition to the highly conserved SET motif, less conserved ∼50- to 80-aa pre-SET and post-SET regions may be present at the amino and carboxyl boundaries, respectively, of the SET domain (Fig. 1A) (reviewed in reference 12). Genes encoding SET domains are widely represented in eukaryotic genomes (>300 database entries). Based on the homology of their SET motifs, the most characterized human proteins were classified into four major families, SET1, SET2, SUV39, and RIZ (15). A recently discovered function of the SET domain is the methylation of lysine residues in nucleosomal histones. SET domains of different proteins target specific lysines, and these modifications have different consequences on gene expression. For example, the methylation of Lys 4 of histone H3 is usually associated with activation, while the methylation of H3 Lys 9 or Lys 27 is associated with silencing (reviewed in reference 28).
FIG. 1.
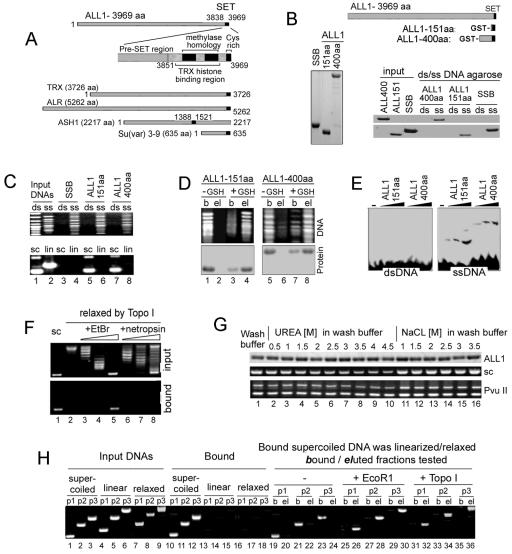
SET-domain-containing polypeptides bind single-stranded and supercoiled DNAs and RNA. (A) Schemes of SET domains in some of the proteins studied here and motifs within TRX SET. (B) Binding of GST-tagged ALL-1 C-terminal polypeptides and the E. coli SSB protein to ss- and dsDNAs immobilized on Sepharose. Binding was performed in a buffer containing 100 mM NaCl and 0.05% NP-40, and washing was done with a buffer containing 0.5 M NaCl and 0.1% NP-40. Bound proteins were resolved in SDS-10% polyacrylamide gels and stained with Coomassie blue. The relative migration of the GST polypeptides in the 10% gel is shown on the left. (C) Binding of GST-tagged ALL-1 C-terminal polypeptides and the E. coli SSB protein, immobilized on glutathione-Sepharose, to a single-stranded but not double-stranded 100-bp DNA ladder and to supercoiled but not linear 6-kb DNA. Binding and washing were done as described above. Retained DNAs were analyzed in 1% agarose gels. (D) ssDNAs complexed with immobilized GST-SET were incubated in the presence or absence of 10 mM reduced glutathione. DNAs (top) and proteins (bottom) in the bound (b) and eluted (el) fractions were resolved in gels. (E) Single- and double-stranded DNA templates (63 bp) were incubated with either no protein or with increasing concentrations of ALL-1 C-terminal fragments (151 and 400 aa) and resolved by 4% native polyacrylamide gel electrophoresis. (F) The 220-aa ALL-1 C-terminal fragment binds to negatively supercoiled but not to relaxed or positively supercoiled 4.5-kb plasmid. Negative and positive plasmids were produced by Topo I relaxation in the presence of ethidium bromide or netropsin, respectively. The input corresponds to 25% of the material used for binding. (G) Complexes between the ALL-1 C-terminal 350-aa polypeptide and a 6-kb supercoiled plasmid or the temperature-denatured PvuII fragments of the same plasmid are stable at high concentrations of chaotropic agents. The top panel shows the relative amounts of the ALL-1 polypeptide retained on the beads after washings. (H) Stable complexes between the ALL-1 220-aa C-terminal polypeptide and supercoiled DNAs are disrupted upon DNA relaxation. The supercoiled, relaxed, and linear forms of plasmids P1 (3 kb), P2 (4.5 kb), and P3 (6 kb) were tested for binding (lanes 10 to 18). The bound supercoiled plasmids were washed with 0.5 M NaCl and 0.1% NP-40 and were either linearized with EcoRI (lanes 25 to 30), relaxed with Topo I (lanes 31 to 36), or left untreated (lanes 19 to 24). The DNAs were washed as described for panel B, and the eluted (el) and bound (b) species were resolved by electrophoresis.
Some of the most studied SET proteins are the products of members of the TrxG and PcG gene families and are modifiers of position effect variegation that are involved in maintaining stable and heritable states of gene expression during the development of higher eukaryotes (reviewed in references 24 and 32). For example, the initiation of expression of the best-studied targets of the TrxG and PcG proteins, the homeotic (HOX) genes, is achieved by the early-acting segmentation genes of Drosophila. The TrxG and PcG proteins are required later during development to maintain the active and repressed states of HOX transcription, respectively (24, 32). Some of the TrxG and PcG proteins are found in multiprotein complexes that contain enzymes capable of remodeling nucleosomes or marking histones at the target loci by several specific histone modifications. These markings serve as signals for proteins which change the architecture of nucleosomes and subsequently bring about transcriptional activation or silencing. It is thought that the combination of different modifications at a particular histone constitutes a cellular memory propagated to the progeny cells (35, 37; reviewed in reference 7).
Although the methylation of histone residues is widely believed to be the central function of the SET domains, important aspects of this process, such as how SET proteins are recruited in the first place and how the histone modifications survive replication, are not understood. Here we describe another biochemical feature associated with SET domain proteins which might play a role in the aforementioned processes and in other processes. A motif present within the general area of the SET domains of several proteins binds single-stranded and supercoiled DNAs as well as RNA. This binding is very tight so that it will last and interfere with nucleosome assembly.
MATERIALS AND METHODS
DNA procedures.
PCR fragments encoding the desired polypeptides were cloned in frame into the NdeI and EcoR1 sites of the pGEX-2TX-derived plasmid pGEX-2TKN (kindly provided by K. Katsani and P. Verrijzer) or into the BamHI and EcoRI sites of pGEX-4T-3 (Pharmacia). Residues encoded by the various deletion constructs are indicated in the figures. The trxZ11 mutation in TRX SET was generated by PCR-based site-directed mutagenesis. Positively and negatively supercoiled DNA templates were prepared by relaxing covalently closed plasmids with topoisomerase I in the presence of ethidium bromide or netropsin (9). Single-stranded DNAs (ssDNAs) were prepared by boiling DNA templates (usually a 100-bp DNA ladder [Fisher]) for 2 to 3 min followed by an immediate transfer to ice for 5 min. For preparations of immobilized double- and single-stranded DNAs, enzyme-quality calf thymus DNA was biotinylated and immobilized on avidin-Sepharose either directly or after denaturing by boiling for 5 min. A 63-base random sequence oligonucleotide containing two SacI sites at its terminus and the same DNA sequence amplified from the SacI site of pERSVΔCAT (22) were used as single- and double-stranded DNA probes, respectively, for gel retardation assays.
Protein procedures.
Recombinant glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli strain Origami B (Novagen), purified by standard procedures (including immobilization on glutathione-Sepharose), and analyzed in sodium dodecyl sulfate (SDS)-13% polyacrylamide gels. In some cases, to remove bound endogenous nucleic acids, we additionally treated the immobilized GST-SET polypeptides with DNase I and RNase A and washed them with HEMG buffer (50 mM HEPES-KOH [pH 7.6], 0.2 mM EDTA, 25 mM MgCl2, 20% glycerol [vol/vol], 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride) containing 500 mM NaCl, 0.1% NP-40, and 2 to 3 M urea. GST pull-down experiments were performed in EX protein buffer (17) (10 mM HEPES-KOH [pH 7.6], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM EGTA, 10% glycerol [vol/vol], 1 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride) containing the desired amounts of NaCl (indicated in the figure legends) and 0.05% NP-40. Loading was followed by a series of washes with HEMG buffer containing 500 mM NaCl and 0.1% NP-40. Bound DNAs were isolated by phenol extraction of the glutathione-Sepharose beads and were analyzed in 1% agarose gels.
Coupled in vitro transcription and GST pull-down assay.
The transcription of pGEM Express positive control template DNA (Promega) was performed directly in the buffer for the GST pull-down assay, supplemented with a complete or partial set of transcription components (a 0.1 mM concentration of each ribonucleoside triphosphate, 1 U each of SP6, T3, and T7 RNA polymerases [Promega], 1 U of RNasin [Boehringer], 3 U each of RNases A and H [Boehringer], and 3 U of DNase I [Boehringer]) as indicated in Fig. 3.
FIG. 3.
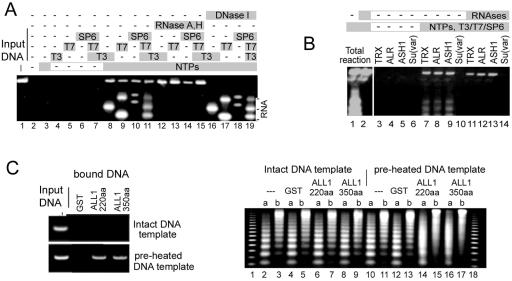
SET regions bind transcribed DNA and nascent RNA (A and B) and prevent the assembly of regular nucleosome arrays on locally melted DNA (C). The C-terminal 220-aa ALL-1 polypeptide was used for panel A, and a description of the other polypeptides is given in Fig. 1. Immobilized GST-SET polypeptides were incubated with in vitro transcription mixtures containing the pGEM Express positive control template DNA, a mixture of all four ribonucleoside triphosphates (NTPs; lanes 3 and 8 to 19), polymerases T3, T7, and SP6 (as indicated in the figure), RNases A and H (lanes 12 to 15), and DNase I (lanes 16 to 19). After being washed with 0.5 M NaCl-0.1% NP-40, bound DNAs were resolved in 1% agarose gels. (C) The assembly of nucleosomes was performed with the linearized 6-kb plasmid DNA, either intact as double-stranded DNA (lanes 3 to 9) or preheated to 82 to 87°C to create local regions of ssDNA (lanes 10 to 17). GST alone or GST-ALL-1 fusions (220 and 350 aa of ALL-1) were preincubated with the DNA template for 10 min before the addition of a Drosophila extract, ATP, and an energy regeneration mixture (control binding experiments with immobilized GST-ALL-1 fusions to double-stranded and partially melted DNAs are shown on the left). After completion of the assembly, samples of reconstituted chromatin were digested with micrococcal nuclease for 30 s (lanes a) or 3 min (lanes b). Isolated DNAs were resolved in a 1.3% agarose gel. Lanes 1 and 18 show the migration of a 123-bp DNA ladder.
Chromatin reconstitution and analysis.
Nucleosome reconstitutions were performed as described previously (17) by the use of Drosophila extracts prepared from 0- to 3-h-old embryos without the addition of extra histones. Micrococcal analyses of reconstituted chromatin were performed as described previously (17).
Modeling of secondary structures.
The utilization of Expasy modeling software allows analyses of two proteins side-by-side or of several proteins simultaneously. The modeling templates, which were all SET-domain proteins with known three-dimensional (3D) structures, included CLR4 (NCBI no. mvxA and lmvhA), DIM5 (NCBI no. 1pegA, 1pegB, and 1m19A) and SET7 (1MUF, 1NGA, and 1NGC). For simultaneous threading of an ssDNA binding sequence into the three templates, we used SWISS-PdbViewer or the automated homology modeling servers SWISS-Model and Geno 3D (5). Protein structures were visualized with the RasMol viewer in the color mode “chain” or “structure,” which showed chains of proteins in different colors or structural elements in different colors, respectively.
RESULTS
The SET region of SET-domain proteins tightly binds ssDNA substrates.
To study DNA binding, we used GST-tagged polypeptides corresponding to segments of SET regions (we define a SET region as including the SET, pre-SET, and post-SET domains) derived from distinct SET-domain proteins. As a control, we used the 178-aa ssDNA-binding protein (SSB; protein no. NP290692.1) from E. coli, which binds tightly ssDNA under a wide range of environmental conditions.
We commonly used a commercial 100-bp DNA ladder (Fisher), either intact or denatured by heating at 98°C in Tris-EDTA buffer, as a dsDNA and ssDNA probe. The choice of DNA source was not critical, since the testing of several other ss- or dsDNA substrates yielded similar results (not shown). We also compared SET binding to the negatively supercoiled versus linear forms of DNA. DNA helixes of circular closed DNA molecules can have a deficiency or excess of coils compared to a linear DNA of the same size. These under- or overwound DNA states are referred to as negatively or positively supercoiled DNA, respectively. The typical plasmid DNA isolated from a bacterial source is negatively supercoiled at a rate of one supercoil per ∼200 bp of DNA. Negatively or positively supercoiled DNA molecules tend to relieve arising torsional tensions either by changing their helical twist (decreasing or increasing it, respectively) and/or by writhing of the axis. Thus, negative, in contrast to positive, superhelical stress is known to destabilize the DNA duplex, resulting in locally unwounded regions of DNA (reviewed in reference 6). As a control, we used relaxed forms of plasmids, in which superhelical stress was removed by a treatment with topoisomerase I (Topo I).
ALL-1, also termed MLL, HRX, or HTRX, is the human homologue of Drosophila TRX and acts as a positive regulator of HOX genes (26, 40). SET fragments of the ALL-1 and E. coli SSB proteins were expressed as GST fusions in bacteria (Fig. 1B), and their binding with various DNA substrates was assessed in several assays under stringent washing conditions for DNA-protein complexes by use of a buffer containing 500 mM NaCl and 0.1% NP-40.
Like the case for the E. coli SSB protein, the 151- and 400-aa C-terminal fragments of ALL-1 associated tightly with immobilized single-stranded, but not double-stranded, DNA (Fig. 1B). When immobilized on a glutathione-Sepharose matrix via a GST moiety (Fig. 1C), the SET region of ALL-1 efficiently retained ssDNA fragments, whereas its binding to dsDNA was barely detectable (the input amounts of the ds- and ssDNA were equalized for DNA content, but since ethidium bromide binds ssDNA less efficiently than dsDNA, lanes with ssDNA appear to be underloaded). In contrast to the case for the GST-SSB protein of E. coli, ALL-1 C-terminal tails bound the negatively supercoiled form of the plasmid (Fig. 1C, bottom panel, lanes 5 to 8 versus lanes 3 and 4). Complexes of GST-SET with ssDNA could be further eluted with reduced glutathione (Fig. 1D), indicating that the physical association was not due to a nonspecific aggregation on the surface of the Sepharose matrix. Gel retardation assays performed with physiological ionic strengths (140 to 150 mM NaCl) also revealed a higher affinity of the ALL-1 SET region for ss- versus dsDNA (Fig. 1E).
We noted that at low ionic strengths (below 100 mM NaCl), ALL-1 SET fragments and some preparations of EcSSB bound small amounts of dsDNA (not shown), indicating that the difference in the affinities of SET fragments and EcSSB for dsDNA was quantitative rather than qualitative. The glutathione-Sepharose matrix or immobilized GST retained neither ss- nor dsDNA (not shown).
The SET regions of several other SET-containing proteins, including Drosophila E(Z), TRX, and ASH1, human ALR, the E(Z)-related Arabidopsis CLF, and yeast SET1 and SET2, also bound single-stranded and supercoiled DNAs (not shown; see Fig. 2 for detailed analyses of some of these), suggesting that such DNA binding may be a general feature of SET regions. Altogether, the tested proteins encompassed members of the SET domain families SET1 and SET2 (15). Interestingly, the corresponding polypeptides of two representatives of the SUV39 family, Drosophila SU(VAR)3-9 and human G9a, did not bind to supercoiled (not shown) and ssDNA (Fig. 2).
FIG. 2.
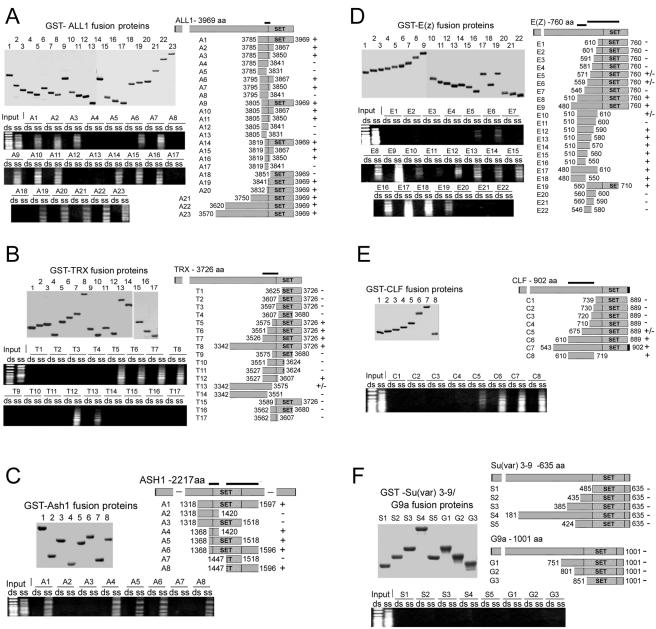
Mapping of ssDNA-binding motifs within the related proteins ALL-1 (A) and TRX (B), E(Z) (D) and CLF (E), and ASHI (C). Ranges of the C-terminal polypeptides of the five proteins were expressed in E. coli as GST fusion proteins, immobilized on glutathione-Sepharose beads, and incubated with a single-stranded or double-stranded 100-bp DNA ladder. Binding and washing conditions were as described in the legend to Fig. 1. Dark horizontal lines above the schemes of the polypeptides indicate the smallest polypeptide which bound ssDNA with an efficiency comparable to that of the largest positive polypeptide. For an evaluation of the results, see the text. (F) C-terminal fragments containing the SET domains of SU(VAR)3-9 and G9a proteins did not bind DNA.
To examine in more detail the effect of superhelicity on the binding of DNA to the SET region, we prepared several DNA templates with different levels of superhelical tension. Negative or positive supercoiling was introduced into DNAs by relaxing covalently closed plasmids with Topo I in the presence of ethidium bromide or netropsin, respectively (Fig. 1F, top panel, lanes 3 to 5 or 6 to 8) (9). The level of the induced superhelicity was monitored by gel electrophoresis, as the relative migration of plasmids in agarose gel increases proportionally to the increase in their supercoiling level. At high concentrations of ethidium bromide (Fig. 1F, lane 5) or netropsin (Fig. 1F, lane 8), the level of induced supercoiling was similar to that of the native supercoiled plasmids (Fig. 1F, lane 1). Only the highly negatively supercoiled plasmids were retained on the matrix containing ALL-1 GST-SET fragments (Fig. 1F, bottom panel, lanes 1 and 5). These results imply that the local untwisting of DNA driven by negative superhelicity, and not the alteration in DNA tertiary structure, is the primary factor responsible for the strong interaction of DNA with the SET region.
We wondered how strong the binding was between the SET-containing polypeptides and supercoiled or ssDNAs. The ALL-1 SET region was bound to both templates and subjected to washings at increasing stringencies (Fig. 1G, middle and bottom panels). We found that the polypeptide was still bound in the presence of 4.5 M urea and 3.5 M NaCl (or even 5 M NaCl [not shown]), suggesting a strong association.
We next demonstrated that even though SET binding to supercoiled DNA was very stable, it was reversible: DNA supercoiling was essential not only for establishing but also for maintaining binding to the ALL-1 SET domain (Fig. 1H). To this end, the GST-tagged 220-aa ALL-1 C-terminal SET domain was immobilized on glutathione-Sepharose and assayed for binding to the negatively supercoiled, linear, and relaxed forms of three plasmids (3, 4.5, and 6 kb). Only the torsionally stressed plasmids remained bound to ALL-1 SET after washing with 0.5 M NaCl-0.1% NP-40 buffer (Fig. 1H, lanes 10 to 12 versus lanes 13 to 18). When the bound supercoiled plasmids were subsequently linearized or relaxed by treatment with a restriction enzyme or with Topo I, most of the plasmid DNA was eluted in the wash buffer, whereas unmodified supercoiled plasmids remained bound to immobilized ALL-1 SET (Fig. 1H, compare lanes 19, 21, and 23 to lanes 25, 27, 29, 31, 33, and 35). Thus, the DNA-protein complexes dissociate upon relief from torsional tension.
Mapping the ssDNA-binding motifs within SET regions.
We applied deletion analysis to map the ssDNA recognition motifs within several representative SET-domain proteins. These included ALL-1 and its Drosophila homologue TRX (Fig. 2A and B), Drosophila E(Z) and the related plant protein CLF (Fig. 2D and E), and Drosophila ASH1 (Fig. 2C). The black bars at the tops of the schemes indicate the smallest polypeptides capable of binding to ssDNA with an affinity comparable to that of the full-length SET regions. The data in Fig. 2 show that the ALL-1 and TRX ssDNA-binding domains are located at the pre-SET-SET boundaries. The corresponding domains of E(Z) and CLF were mapped to the pre-SET region, and E(Z) had a second binding domain within pre-SET-SET. The ASH1 protein, in which the SET domain is in the middle of the protein, contains two ssDNA-binding domains, at the pre-SET-SET and SET-post-SET boundaries (Fig. 2C). In some instances, polypeptides spanning the minimal binding sequences together with some additional sequences did not bind DNA [T11 of TRX, E7 of E(Z), and A2,3 of ASH1]. This was likely due to the polypeptides assuming steric conformations which prevented DNA binding. No binding of ssDNA was observed for the SET regions of SU(VAR) 3-9 and G9a (Fig. 2F) with polypeptides spanning SET and extending upstream.
The ssDNA-binding motif associates with in vitro-transcribed DNA and with nascent RNA.
A major physiological process that causes an opening of the DNA duplex is the traversing of DNA by RNA polymerases. To assess if transcription of a DNA template is sufficient for making it available for an interaction with the SET region, we performed a coupled in vitro transcription-GST pull-down assay by using phage RNA polymerases and an appropriate DNA template. The latter, the pGEM Express positive control template (Promega), contains two promoters for each of the T3, T7, and SP6 polymerases which generate RNAs of 250 and 1,525, 1,065 and 2,346, and 1,787 and 2,566 bases, respectively. In the absence of ongoing transcription, the interaction of the DNA template and the 220-aa ALL-1 C-terminal SET fragment occurred only at background levels (Fig. 3A, lanes 2 to 7). However, if a full set of reaction components was supplied to allow transcription, the DNA template was efficiently associated with the ALL-1 SET region, as was the nascent RNA species (lanes 8 to 11). Transcribed DNA and the RNA product interacted with the polypeptide independently of each other, as indicated by the selective removal of DNA (lanes 16 to 19) or RNA (lanes 12 to 15) after a treatment with DNase I or a mixture of RNases A and H, respectively (we took advantage of RNase H's ability to destroy RNA within RNA-DNA hybrids). We did not test whether processing by eukaryotic RNA polymerases can also induce an association of DNA with the polypeptide. However, the basic principles of eukaryotic and prokaryotic transcription are well conserved, and it can be presumed that the SET region is associated with transcribing eukaryotic genes as well. We tested whether SET regions of some other proteins could also bind transcribed DNA structures. Indeed, SET-containing fragments of TRX, ALR, and ASH1, but not SU(VAR)3-9, also exhibited binding to in vitro-transcribed DNA (Fig. 3B). Note that SET polypeptides isolated from bacteria are found associated with various amounts of nucleic acids which can be removed by incubation with DNase and RNase (not shown). The associated nucleic acids do not appear to impair histone lysine methyltransferase activity (26).
The ssDNA-binding motif is not displaced during in vitro assembly of nucleosomes and prevents the formation of regular nucleosome arrays.
Within the cell nucleus, DNA is exposed to interactions with numerous factors, some of which result in very stable associations. For example, most eukaryotic DNA is found in complex with histones, forming chromatin structures (10) that can be destroyed only by treatments with high concentrations of salt or ionic detergents. We tested whether nucleosomes would exclude or remove bound ALL-1 SET regions from the template DNA during assembly (Fig. 3C). The template DNA used for this experiment contained single-stranded regions generated by heating of the DNA to temperatures sufficient to open the easily melting sequences but not the entire DNA duplex (21). Such transiently melted DNA regions usually renature quickly and do not significantly affect nucleosome assembly, but they appear to be sufficient to provide strong binding of the ALL-1 SET region (Fig. 3C, left panel). The GST-tagged 220- and 350-aa ALL-1 SET fragments were incubated with such partially melted DNA and, as a control, intact double-stranded DNAs, and subsequent nucleosome assembly was performed as previously described (18) by use of a Drosophila embryo extract and an ATP regeneration system (Fig. 3C, right panel). Under these conditions, the ATP-dependent spacing activities of the extract generate continuous arrays of dynamic nucleosomes with regular spacing (19). Upon the completion of assembly, the integrity of the nucleosomal arrays was assayed by the use of staphylococcal nuclease, which cleaves regularly spaced nucleosome arrays to produce a typical ladder of the oligonucleosomal DNA fragments. The digestion of chromatin assembled on the intact dsDNA resulted in a set of well-resolved DNA fragments (Fig. 3C, right panel, lanes 2 to 9), implying that preincubation with GST-SET or GST polypeptides did not interfere with nucleosome assembly. However, when the DNA template was partially melted and preincubated with GST-ALL-1 SET fragments (but not with GST alone) prior to the assembly reaction, the nucleosome ladder was significantly less pronounced (compare lanes 11 to 13 with lanes 14 to 17). This result suggests that the association between the SET fragments and ssDNA survives assembly, preventing the formation of regular nucleosome arrays. Thus, the binding of SET-domain proteins to ssDNA efficiently competes with the extremely strong affinity between nucleosomal proteins and DNA.
The homeotic mutation trxZ11 impairs binding of the TRX SET region to ssDNA.
The TRX SET domain spans the well-studied developmental mutation trxZ11, in which a highly conserved glycine (G3601) is replaced with serine (34). This mutation results in homeotic transformations in flies that are heterozygous for trxZ11 and causes lethality at the pupal stages in hemizygous animals (4). To assess whether this mutation affects the binding of the TRX SET region to ssDNA, we applied a pull-down assay to compare the activities of the GST-tagged TRX SET fragments derived from the trxZ11 and wild-type proteins (Fig. 4). trxZ11 polypeptides, but not their wild-type counterparts, showed progressively reduced abilities to bind ssDNA when the ionic strength of the binding buffer was increased from 125 to 175 mM. This effect was apparent with the 385- and 151-aa TRX SET regions and became striking (Fig. 4, bottom panel) when an 81-residue TRX polypeptide which was the smallest to bind ssDNA was used (composed of pre-SET and a small region of SET).
FIG. 4.
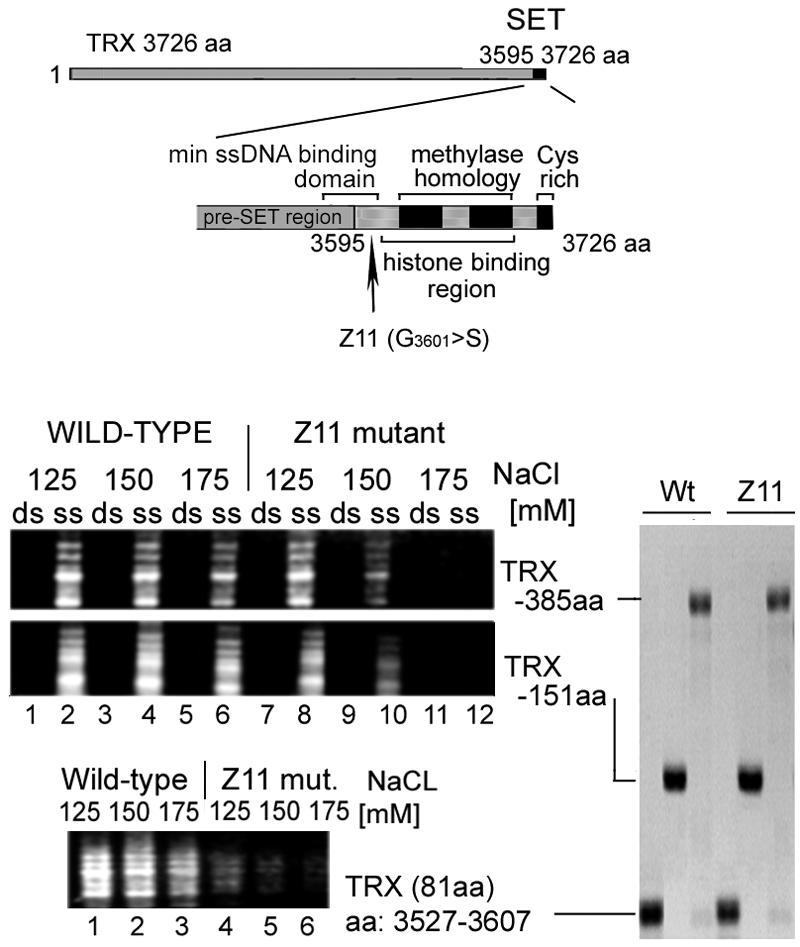
The trxZ11 point mutation in the TRX SET region impedes the association with ssDNA. The ssDNA-binding abilities of equal amounts of the wild type and of trxZ11 GST-TRX C-terminal fragments were compared under different ionic strength conditions. Immobilized GST-TRX C-terminal tails (151 and 385 aa) and the GST-TRX 81-aa pre-SET-SET boundary fragment corresponding to the smallest binding protein (aa 3527 to 3607) were incubated with a single-stranded or double-stranded 100-bp DNA ladder in a buffer containing 0.05% NP-40 and 125 to 175 mM NaC1 (as indicated). Unbound DNAs were washed out with a buffer containing 0.1% NP-40-0.5 M NaCl. Bound DNAs were resolved in 1% agarose gels. The relative migration of the GST fusion proteins by SDS-10% polyacrylamide gel electrophoresis is shown on the right.
We concluded that the point mutation, which results in the strong obliteration of the TRX biological function, also brings about a reduction in the affinity of the TRX SET region for ssDNA. It should be noted that the trxZ11 mutation also impairs the ability of TRX SET to bind to histones H3 and H4 (13) and hampers H3 lysine 4 methylation activity (33). The residue that is mutated in trxZ11 maps to the very N terminus of the SET domain. It is included in the ssDNA-binding motif, but not within the histone binding and histone lysine methyltransferase domains. The deleterious effect of trxZ11 on all three functions, including the two located away from the affected residue, suggests an alteration in the tertiary structure of the entire SET region. Nevertheless, the correlation between the in vivo effect of trxZ11 and the diminution in the in vitro ssDNA-binding activity of the TRX SET region is consistent with our suggestion that this binding plays an important physiological role.
DISCUSSION
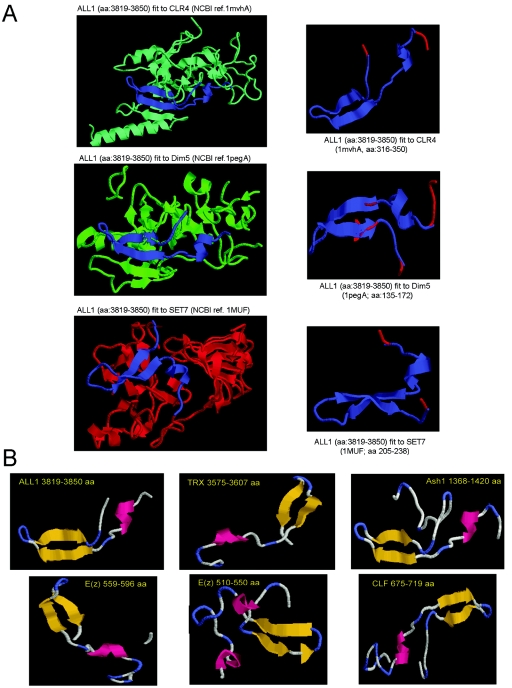
The motif identified here shares with the E. coli SSB protein the ability to tightly bind ssDNA, but unlike the latter it also binds supercoiled DNA. We named the motif SSBLS, for single-stranded nucleic acid binding linked to SET. The motif is present in SET proteins of unicellular yeast as well as of metazoans. This suggests involvement in a similar function associated with a fundamental feature of many or all SET domains. Since the motif was identified in both transcriptional activators (ALL-1, TRX, ALR, SET1, and ASH1) and repressors [E(Z), CLF, and SET2], the activity is likely to play a role in processes associated with both gene activation and silencing. It is interesting that we failed to identify SSBLS in Drosophila SU(VAR)3-9 and human G9a. Unless the failure was of a technical nature, it might reflect the classification of the two proteins in the SUV39 family correlated with H3 Lys9 methylation activity and the presence of a pre-SET domain necessary for the activity (15). Although the motif localizes to the general area of SET, its precise position shows some variation in different proteins, ranging from the pre-SET region [E(Z) and CLF] to the boundary of pre-SET and SET [ALL-1, TRX, E(Z), and ASH1] or to the boundary of the SET and post-SET regions (ASH1). It is interesting that the different positions of the motif fit with the classification of the relevant SET domains (2, 12, 15). The position of the motif relative to SET is conserved between ALL and its Drosophila homologue TRX and between Drosophila E(Z) and its plant homologue CLF. The homology in the primary sequence between the ALL-1 and TRX motifs and between the E(Z) and CLF motifs is high (not shown). In contrast, an overall primary sequence comparison of the different SSBLS motifs mapped in the five proteins has not revealed common sequences that can be attributed to a consensus. To inquire whether despite the lack of a consensus sequence the different motifs might share a common secondary configuration, we attempted a rough assessment of their structures. Structural predictions were performed by using Expasy modeling software (see Materials and Methods). Figure 5A shows the alignment of the ALL-1 ssDNA-binding sequence onto the published 3D structures, obtained by X-ray crystallography, of the SET-domain proteins CLR4, DIM5, and SET7. Each of the alignments returned a similar configuration, which also obtained when the DNA binding sequence was threaded into the three templates simultaneously. The same approach was used to predict the secondary structures of the ssDNA-binding regions of the other proteins studied here. In all cases (Fig. 5B), a configuration composed of two antiparallel beta sheets and an alpha helix was indicated. Despite the very rough structural assessment that such modeling can provide, it raises the possibility that the secondary conformations of the individual SSBLS motifs may be similar. This issue will be further investigated experimentally. In this context, we noted that several proteins that bind single-stranded and unusually structured nucleic acids, though lacking any sequence homology, share a common steric configuration designated the OB-fold (oligonucleotide-oligosaccharide binding). This is a highly variable 50- to 150-aa motif with a distinct three-dimensional barrel topology, assumed through the assembly of five antiparallel beta sheets, often accompanied by an alpha helix (reviewed in references 1, 14, and 36).
FIG. 5.
Structural assessment by modeling of ssDNA-binding motifs in SET-domain regions of ALL-1, TRX, E(Z), CLF, and ASH1. (A) Structural alignment of ALL-1 ssDNA-binding sequence onto the 3D structures of the SET7, CLR4, and DIM5 SET-domain histone lysine methyltransferases. Modeling was performed by using the stand-alone Swiss-PdbViewer with an installed protein loop database. For each of the three templates (1mvxA, 1pegA, and 1MUF), the modeling was performed first by using the complete template (larger window, shown at left) and was continued by using the relevant portion of the template (smaller window, shown at right). Protein structures were visualized with the RasMol viewer in color chain mode. (B) Results of 3D modeling of ssDNA-binding motifs of ALL-1, TRX, E(Z), CLF, and ASH1. For ALL-1, E(Z) 510-550, and ASH1 1368-1420, we used the smallest polypeptides that reacted with ssDNA. For analysis of the other three regions, the sizes of the modeled structures were minimized by selecting the overlapping domains of two interacting polypeptides [T5 and T12 for TRX, C5 and C8 for CLF, and E6 and E12 for E(Z) 559-596]. Protein structures were visualized with the RasMol viewer in color structure mode.
Considering that the in vitro association of the SSBLS motif with an opened DNA duplex is very strong so as to tolerate high concentrations of monovalent ions and urea and to be sustained during energetically efficient processes such as nucleosome assembly, it is very likely that such interactions also occur in vivo. Note that the SSB proteins, which also possess a high affinity for ssDNA, are involved in vivo in all processes examined associated with the generation or manipulation of single-stranded DNA. The physiological significance of the SSBLS motif was shown here by the demonstration that the trxZ11 point mutation, which exhibits dramatic biological effects in Drosophila, confers impaired binding to ssDNA. The critical function of the residue mutated in trxZ11 was also emphasized by studies in yeast, in which telomeric silencing and normal growth were disrupted upon the introduction of a similar mutation into the yeast SET1 protein (27).
When possible functions of SSBLS are considered, one potential role will involve the recruitment of SET proteins to their targets. The mechanism(s) associated with this process is largely unknown. Recent results with fission yeast, plants, flies, and vertebrates (8, 29, 31, 38, 39) suggest that short interfering RNAs (siRNAs) function to recruit H3 Lys9 methyltransferases to specific loci to direct heterochromatin assembly. It has been speculated that the protein complex RISC/RITS, which contains siRNA, associates with the H3 Lys9 methyltransferase and is targeted to specific chromosome regions through base pairing of the former to single-stranded DNA bubbles or nascent transcripts. In principle, this model can be extended to other histone methyltransferases that act in the local decondensation or condensation of euchromatin [e.g., ALL-1, ASH1, and E(Z)]. A motif within SET proteins which binds single-stranded nucleic acids may facilitate the targeting and anchoring of the RISC/RITS complex to specific chromosome regions. Moreover, since each RISC contains only one of the two strands of the siRNA duplex (25), the SET-domain proteins might physically link to RISC through the binding of SSBLS to complex-associated single-strand RNA.
A role for transcription of a target gene in the recruitment of a SET polypeptide was also deduced from our recent work, in which we found that the TRX SET region encoded by a transgene was recruited in vivo to the heat shock genes only during active transcription of the latter (33). Moreover, the TRX SET domain is indispensable for recruiting the TRX protein and the components of the TAC1 multiprotein complex to heat shock genes (33). Furthermore, several reports have indicated that initial transcription of the regulatory region of the Drosophila homeotic gene ultrabithorax is required for establishing the maintenance state (and presumably the corresponding covalent histone modifications) of this gene's transcriptional activity (3, 11, 30). These recent findings are consistent with the ability of SSBLS to bind transcribed DNA (and RNA) as well as with its ability to interact in a reversible way with torsionally stressed DNA, which is considered a hallmark of functionally active DNA structures (6, 16, 21). Another possible role for SSBLS is to prevent the formation of regular nucleosome arrays in transcribed regions so as to maintain the required level of transcription. Finally, the capacity of SET-domain proteins to bind ssDNA may play a mechanistic role in the inheritance of histone covalent modifications. One can imagine a scenario in which, during progression of the replication fork, SET-domain proteins dissociate from the parental DNA strands to be quickly recruited to the newly synthesized DNA due to the strong interaction between the SET regions and ssDNA. In this scenario, SET-domain proteins play an active role not only in marking histones for transcription and silencing states, but also in propagating these states in the progeny cells.
Acknowledgments
We are very grateful to Yaroslav Gursky for his generous help with molecular modeling, to V. Sobolev for useful discussions, and to P. Verrijzer, K. Katsani, J. Arredondo, and T. Rozovskaia for their generous gifts of plasmids.
This study was supported by NCI grant CA50507, grants from the Israeli Science Foundation, the Israel Cancer Research fund, and U.S.-Israel BSF, and grants to W.K. from the Institute of Developmental Biology (Russian Academy of Science) and the Leukemia Research Foundation (Evanston, Ill.).
REFERENCES
- 1.Agrawal, V., and K. V. Kishan. 2003. OB-fold: growing bigger with functional consistency. Curr. Protein Pept. Sci. 4:195-206. [DOI] [PubMed] [Google Scholar]
- 2.Alvares-Venegas, R., and Z. Avramova. 2002. SET-domain proteins of the Su(var)3-9, E(z) and trithorax families. Gene 285:25-37. [DOI] [PubMed] [Google Scholar]
- 3.Bender, W., and D. P. Fitzgerald. 2002. Transcription activates repressed domains in the Drosophila birthorax complex. Development 129:4923-4930. [DOI] [PubMed] [Google Scholar]
- 4.Breen, T. R. 1999. Mutant alleles of the Drosophila trithorax gene produce common and unusual homeotic and other developmental phenotypes. Genetics 152:319-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Combet, C., M. Jambon, G. Deleage, and G. Geourjon. 2002. Geno3D: automatic comparative molecular modelling of protein. Bioinformatics 18:213-214. [DOI] [PubMed] [Google Scholar]
- 6.Esposito, F., and R. R. Sinden. 1988. DNA supercoiling and eukaryotic gene expression. Oxf. Surv. Eukaryot. Genes 5:1-50. [PubMed] [Google Scholar]
- 7.Francis, N. J., and R. E. Kingston. 2001. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2:409-421. [DOI] [PubMed] [Google Scholar]
- 8.Fugakawa, T., M. Nogami, M. Yoshikawa, M. Ikeno, T. Okazaki, T. Nakayama, and M. Oshimura. 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6:784-791. [DOI] [PubMed] [Google Scholar]
- 9.Hamiche, A., V. Carot, M. Alilat, F. De Lucia, M. F. O'Donohue, B. Revet, and A. Prunell. 1996. Interaction of the histone (H3-H4)2 tetramer of the nucleosome with positively supercoiled DNA minicircles: potential flipping of the protein from a left- to a right-handed superhelical form. Proc. Natl. Acad. Sci. USA 93:7588-7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes, J. J., and J. C. Hansen. 2001. Nucleosomes and the chromatin fiber. Curr. Opin. Genet. Dev. 11:124-129. [DOI] [PubMed] [Google Scholar]
- 11.Hogga, I., and F. Karch. 2002. Transcription through the Iab-7 cis-regulatory domain of the bithorax complex interferes with polycomb mediated silencing. Development 129:4915-4955. [DOI] [PubMed] [Google Scholar]
- 12.Jenuwein, T., G. Laible, R. Dorn, and G. Reuter. 1998. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell. Mol. Life Sci. 54:80-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kastani, K. R., J. J. Arredondo, A. J. Kal, and P. C. Verrijzer. 2001. A homeotic mutation in the trithorax SET domain implies histone binding. Genes Dev. 15:2197-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerr, I. D., R. I. Wadsworth, L. Cubeddu, W. Blankenfeldt, J. H. Naismith, and M. F. White. 2003. Insights into ssDNA recognition by the OB fold from a structural and thermodynamic study of SSB protein. EMBO J. 22:2561-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 16.Krajewski, W. A. 1996. Enhancement of transcription by short alternating C.G tracts incorporated within a Rous sarcoma virus-based chimeric promoter: in vivo studies. Mol. Gen. Genet. 252:249-254. [DOI] [PubMed] [Google Scholar]
- 17.Krajewski, W. A. 2000. Histone hyperacetylation facilitates chromatin remodeling in a Drosophila embryo cell-free system. Mol. Gen. Genet. 263:38-47. [DOI] [PubMed] [Google Scholar]
- 18.Krajewski, W. A. 2002. Histone acetylation status and DNA sequence modulate ATP-dependent nucleosome repositioning. J. Biol. Chem. 277:14509-14513. [DOI] [PubMed] [Google Scholar]
- 19.Krajewski, W. A., and P. B. Becker. 1999. Reconstitution and analysis of hyperacetylated chromatin. Methods Mol. Biol. 119:207-217. [DOI] [PubMed] [Google Scholar]
- 20.Krajewski, W. A., and S. V. Razin. 1993. DNA-protein interactions and spatial organization of DNA. Mol. Biol. Rep. 18:167-175. [DOI] [PubMed] [Google Scholar]
- 21.Krajewski, W. A., and S. V. Razin. 1992. Organization of specific DNA sequence elements in the region of the replication origin and matrix attachment site in the chicken alpha-globin gene domain. Mol. Gen. Genet. 235:381-388. [DOI] [PubMed] [Google Scholar]
- 22.Krajewski, W. A., and K. A. W. Lee. 1994. A monomeric derivative of the cellular transcription factor CREB functions as a constitutive activator. Mol. Cell. Biol. 14:7204-7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohman, T. M., and M. E. Ferrari. 1994. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63:527-570. [DOI] [PubMed] [Google Scholar]
- 24.Mahmoudi, T., and C. P. Verrijzer. 2001. Chromatin silencing and activation by polycomb and trithorax group proteins. Oncogene 20:3055-3066. [DOI] [PubMed] [Google Scholar]
- 25.Martinez, J., A. Patkaniowska, H. Urlaub, R. Lührmann, and T. Tuschl. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563-574. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura, T., T. Mori, S. Tada, W. Krajewski, T. Rozovskaia, R. Wassel, G. Dubois, A. Mazo, C. M. Croce, and E. Canaani. 2002. ALL-1 is histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10:1119-1128. [DOI] [PubMed] [Google Scholar]
- 27.Nislow, C., E. Ray, and L. Pillus. 1997. SET1, a yeast member of the trithorax family, functions in transcriptional silencing and diverse cellular processes. Mol. Biol. Cell 8:2421-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orlando, V. 2003. Polycomb, epigenomes and control of cell identity. Cell 112:599-606. [DOI] [PubMed] [Google Scholar]
- 29.Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra, J. A. Birchler, and S. C. Elgin. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303:669-672. [DOI] [PubMed] [Google Scholar]
- 30.Rank, G., M. Prestel, and R. Paro. 2002. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol. Cell. Biol. 22:8026-8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schramke, V., and R. Allshire. 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301:1069-1074. [DOI] [PubMed] [Google Scholar]
- 32.Simon, J. A., and J. W. Tamkun. 2002. Programming off and on states in chromatin: mechanisms of polycomb and trithorax group complexes. Curr. Opin. Genet. Dev. 12:210-218. [DOI] [PubMed] [Google Scholar]
- 33.Smith, S. T., S. Petruck, Y. Sedkov, E. Cho, S. Tillib, E. Canaani, and A. Mazo. 2004. Modulation of heat shock gene expression by the TAC1 chromatin modifying complex. Nat. Cell Biol. 6:162-167. [DOI] [PubMed] [Google Scholar]
- 34.Stassen, M. J., D. Bailey, S. Nelson, V. Chinwalla, and P. J. Harte. 1995. The Drosophila trithorax proteins contain a novel variant of the nuclear receptor type DNA binding domain and an ancient conserved motif found in other chromosomal proteins. Mech. Dev. 52:209-223. [DOI] [PubMed] [Google Scholar]
- 35.Strahl, B. D., and C. D. Allis. 2002. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 36.Theobald, D. L., R. M. Mitton-Fry, and D. S. Wuttke. 2003. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 32:115-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 38.Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. Gygi, S. I. Grewal, and D. Moazed. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303:672-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine 9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 40.Yu, B. D., R. D. Hanson, J. L. Hess, S. E. Horning, and S. J. Korsmeyer. 1998. MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc. Natl. Acad. Sci. USA 95:10632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]