Abstract
UV photofootprinting and repair of pyrimidine dimers by photolyase was used to investigate chromatin structure, protein-DNA interactions, and DNA repair in the spacer and promoter of Saccharomyces cerevisiae rRNA genes. Saccharomyces cerevisiae contains about 150 copies of rRNA genes separated by nontranscribed spacers. Under exponential growth conditions about half of the genes are transcribed by RNA polymerase I (RNAP-I). Initiation of transcription requires the assembly of the upstream activating factor (UAF), the core factor (CF), TATA binding protein, and RNAP-I with Rrn3p on the upstream element and core promoter. We show that UV irradiation of wild-type cells and transcription factor mutants generates photofootprints in the promoter elements. The core footprint depends on UAF, while the UAF footprint was also detected in absence of the CFs. Fractionation of active and inactive promoters showed the core footprint mainly in the active fraction and similar UAF footprints in both fractions. DNA repair by photolyase was strongly inhibited in active promoters but efficient in inactive promoters. The data suggest that UAF is present in vivo in active and inactive promoters and that recruitment of CF and RNAP-I to active promoters generates a stable complex which inhibits repair.
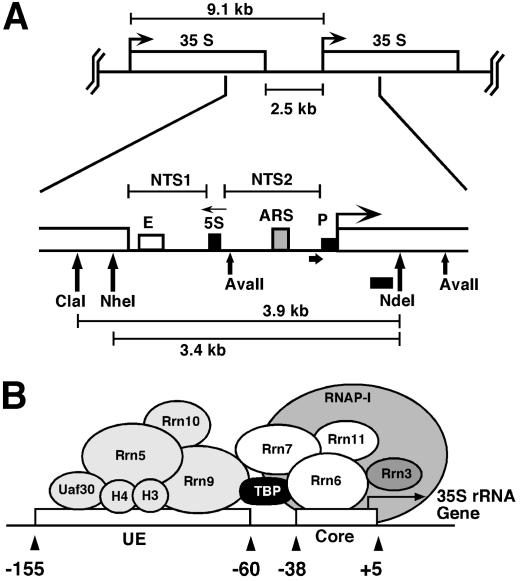
In Saccharomyces cerevisiae, about 150 copies of rRNA genes are clustered as tandem repeats in the nucleolus. Each repeat contains a gene for the 35S rRNA transcribed by RNA polymerase I (RNAP-I) and a gene for the 5S rRNA transcribed by RNA polymerase III. A nontranscribed spacer 1 (NTS1) extends from the transcription termination site to the 5S gene and includes the enhancer. A nontranscribed spacer 2 (NTS2) spans the region between the 5S gene and the start of the 35S gene and contains a ribosomal origin of replication (rARS) and the promoter with the core and the upstream elements (Fig. 1) (35, 36).
FIG. 1.
The intergenic rRNA gene spacer of the yeast S. cerevisiae. (A) One unit of the rRNA gene repeat consists of 9.1 kb of DNA. It contains the following elements: the 35S-rRNA gene transcribed by RNAP-I (white box; 35S); a 5S-rRNA gene transcribed by RNAP-III (black box; 5S); a ribosomal origin of replication (gray box; ARS); an enhancer (white box; E); and two nontranscribed spacers between the 35S and 5S genes (NTS1 and NTS2). Relevant restriction sites and fragments (AvaII, ClaI, NheI, NdeI, 3.9 kb, and 3.4 kb), the probe used for indirect end labeling from the NdeI site (black bar), and the primer used for high resolution footprinting (fat horizontal arrow) are indicated. (B) Schematic illustration of the promoter elements and transcription factors: upstream element (UE), core element (core), UAF containing Rrn5, Rrn9, Rrn10, Uaf30, histone H3, and histone H4, the CF containing Rrn6, Rrn7, Rrn11, the TATA binding protein (TBP), and the RNA-polymerase I (RNAP-I) with the associated Rrn3 (36).
rRNA genes are transcribed with high efficiency to keep up with the cell's metabolic activity and demand for ribosomes (19). The rate of rRNA synthesis is regulated as a response to the cellular environment, either by a change of the initiation rate in transcriptionally competent genes (3, 15) or by activation and inactivation of additional gene copies (11, 41). The transcribed genes are free of nucleosomes (“open”), while the inactive genes are packaged in nucleosomes (“closed”). The spacer region is always nucleosomal (11). Nucleosomes are positioned in NTS2 between the promoter and the 5S gene (18, 28, 52). A nucleosome with multiple positions was reported previously for the 5S gene (7).
Transcription initiation involves coordinated interactions of at least four transcription factors with promoter elements and RNAP-I: the upstream activating factor (UAF) containing Rrn5, Rrn9, Rrn10, the H3 and H4 histones and Uaf30p; the core factor (CF) containing Rrn6, Rrn7, and Rrn11; TBP, the TATA binding protein; and Rrn3p, a factor that binds RNAP-I (reviewed in references 35 and 36). TFIIH and CSB are additional factors that are involved in RNAP-I transcription in mammalian cells (6, 20, 21). In vitro studies revealed that UAF strongly binds the upstream element and recruits CF with the help of TBP and, finally, the Rrn3p-RNAP-I complex to initiate transcription (23, 24, 44). Upon transcription initiation, RNAP-I-Rrn3p and CF dissociate from the promoter, while UAF remains behind. These findings support a model in which the RNAP-I basal machinery cycles on and off the promoter with each round of transcription (2). Nuclease digestions provided an indication of genomic footprints attributable to CF and UAF (52). Moreover, DNase I footprints in CF-deficient and UAF-deficient cells suggest that UAF is necessary for CF binding (5). Chromatin immunoprecipitation (ChIP) assays revealed that the association of RNAP-I with the promoter and the coding region of rRNA genes was decreased in stationary phase, where the rate of rRNA synthesis is reduced, but association of transcription factor UAF with the promoter is unchanged (9). Since neither the nuclease nor the chromatin immunoprecipitation assays could discriminate between active and inactive promoters in the rRNA gene cluster, it remains unknown which of the factors bind to active and inactive promoters in living cells and how stable the interactions are.
Cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4) pyrimidone photoproducts (6-4PPs) are the two major classes of DNA lesions generated by UV light (16). Since the formation directly depends on the DNA structure, the sequence context, and protein-DNA interaction, UV light can be used to monitor protein-DNA contacts (termed “UV photofootprinting”) (4). Both classes of photolesions are repaired by nucleotide excision repair (NER), a multienzyme pathway that involves damage recognition, excision of the lesion, and DNA repair synthesis (12, 37). Alternatively, many organisms, including yeast, have photolyase, an enzyme which binds to CPDs and reverses the damage with the energy of light (photoreactivation [PR]) (40). NER and PR are modulated by protein-DNA interactions, positioned nucleosomes, and heterochromatin (26, 50). Moreover, NER rapidly repairs the transcribed strand of genes transcribed by RNAP-II (referred to as transcription-coupled repair) (49). Photolyase, however, is inhibited on the transcribed strand (TS), because RNAP-II is stalled at DNA lesions (27, 46).
DNA repair of UV lesions has not been extensively studied in rRNA genes. Recent work revealed transcription-coupled NER in yeast RNAP-I genes (10, 30) but not in mammals (8, 17). Active genes were repaired faster by photolyase than inactive genes, providing evidence for an open chromatin structure facilitating repair (30). Here, we show UV photofootprinting and repair data in active and inactive promoters, suggesting that UAF is present in vivo in active and inactive promoters and that recruitment of CF and RNAP-I to active promoters generates a stable initiation complex which enhances DNA damage formation and inhibits repair.
MATERIALS AND METHODS
Yeast strains.
The following yeast strains were used: W303.1a (mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100) (provided by R. Sternglanz); UCC510 (mata ade2-101 his3-Δ200 leu2-Δ1 lys2-801 trp1-Δ1 ura3-53 URA3, flanking telomere of chromosome V) (38) (provided by D. Gottschling); AMY3 (W303.1a, but rad1Δ::URA3) (30), and NOY556 (mata ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100), NOY604 (matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn3Δ::HIS3), NOY699 (matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn5Δ::LEU2), NOY567 (matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn6Δ::HIS3), NOY558 (matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn7Δ::LEU2), NOY703 (mata ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn9Δ::HIS3), NOY704 (mata ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn10Δ::LEU2), NOY730 (mata ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rrn11Δ::LEU2), and NOY408-1a (matα ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 rpa135Δ::LEU2) (provided by M. Nomura). The “NOY” strains except NOY556 are defective in a transcription factor for the RNAP-I machinery. They grow in galactose medium due to a helper plasmid (pNOY102 or pNOY103) carrying the 35S gene under the control of the GAL7 promoter (33, 34). Cultures of NOY strains were grown in full medium containing galactose (1% yeast extract, 2% peptone, 2% galactose); all other strains were grown in full medium containing glucose (1% yeast extract, 2% peptone, 2% dextrose [YPD]) (43).
UV irradiation and repair of yeast cultures was done as described previously (30). Briefly, yeast cultures were grown in YPD at 30°C to a density of about 0.5 × 107 cells/ml, resuspended in minimal medium (2% dextrose or galactose, 0.67% yeast nitrogen base without amino acids) to about 3 × 107 cells/ml. Suspensions 4 mm deep were irradiated with UV light by use of Sylvania G15T8 germicidal lamps (predominantly 254 nm) at a dose of 150 J/m2 (measured by an UVX radiometer; UVP Inc., Upland, Calif.). For repair, the irradiated cultures were supplemented with the appropriate amino acids and uracil. For photoreactivation, the cell suspensions were exposed to photoreactivating light (Sylvania type F15 T8/BLB bulbs) (peak emission at 375 nm) at ∼1.3 mW/cm2 (measured by a UVX radiometer with a 365-nm photocell) at 24 to 26°C. Samples were collected and chilled on ice, and genomic DNA was extracted following yeast DNA isolation protocols (QIAGEN genomic DNA handbook, 1999). All steps until lysis of cells were performed in yellow safety light.
UV irradiation and photoreactivation of DNA in vitro.
A total of 500 ng of DNA, isolated from nonirradiated cells, was irradiated with 80 J/m2 at 254 nm in 10 mM Tris-1 mM EDTA, pH 8.0. For photoreactivation in vitro, about 300 ng of DNA was mixed with 0.5 μg of Escherichia coli photolyase (Becton Dickinson) and exposed on ice to photoreactivating light for 60 min at a flux of about 24 J/m2 sec. DNA was purified using the QIAEX II protocol (QIAGEN, 1997).
Fractionation of active and inactive 35S promoters.
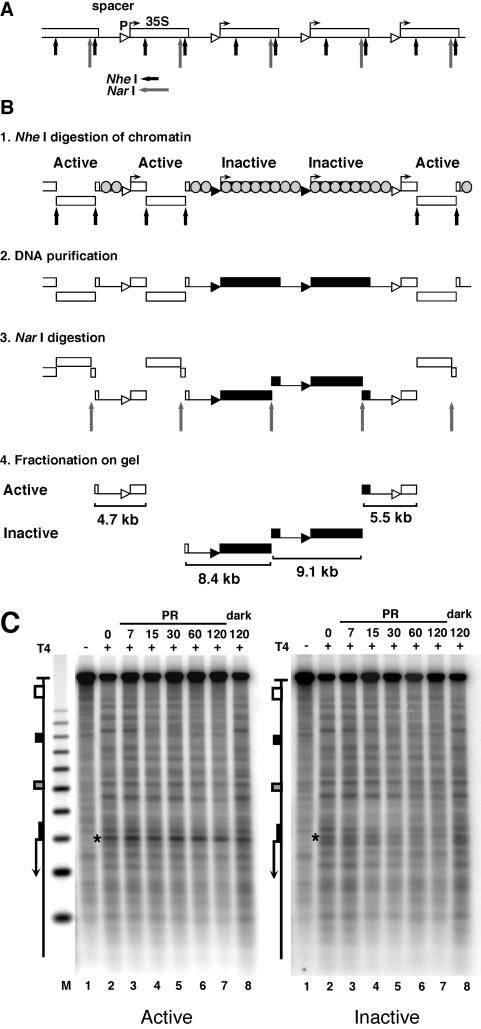
Yeast nuclei were purified according to a method described previously (32). Briefly, 100 ml of cells (3 × 107 cells/ml) were harvested, resuspended in cold water, and resuspended in 2 ml of cold nuclear isolation buffer (17% glycerol, 50 mM MOPS [morpholinepropanesulfonic acid], 150 mM potassium acetate, 2 mM MgCl2, 0.5 mM spermidine, 0.15 mM spermine, pH 7.2). The suspension was vortexed with 2 ml of glass beads (SIGMA) (diameter of 0.5 mm; acid washed and equilibrated in nuclear isolation buffer) until about 90% of the cells were broken (as checked by microscopy). The broken cells were centrifuged (10 min, 4°C, 4,500 × g), and the pellet containing nuclei was resuspended in 2 ml of restriction buffer (33 mM Tris-acetate, 10 mM MgAc, 66 mM KAc, 100 μg of bovine serum albumin/ml, pH 7.9). Aliquots corresponding to 1.5 × 109 cells were digested with 160 U of NheI (Roche Diagnostics) at 37°C for 1 h to release the active ribosomal genes. Genomic DNA was extracted following the yeast DNA isolation protocol (QIAGEN genomic DNA handbook, 1999), digested with NarI (New England Biolabs), and electrophoresed at 4°C in 0.8% low-melting agarose gels (SeaPlaque agarose; FMC BioProducts) in 1× Tris-borate-EDTA. The DNA fragments (see Fig. 3B) were purified according to the AgarACE protocol (Promega) without exposure to UV light. The fractions were redigested with appropriate restriction enzymes and purified by phenol extraction.
FIG. 3.
Inhibition of photoreactivation in active promoters. (A) Schematic illustration of five rRNA gene repeats with the 35S rRNA genes (boxes), promoters (triangles), and spacer and restriction sites for NheI (short arrows) and NarI (long arrows). (B) Fractionation procedure for active and inactive 35S promoters in rRNA gene chromatin. White and black triangles and boxes depict active and inactive promoters and genes, respectively. Spacers and inactive genes are packaged in nucleosomes (grey-shaded circles); active genes are free of nucleosomes (11). Panel 1: nuclear chromatin was digested with NheI to release transcriptionally active genes (30, 32) and a fragment containing an rRNA gene spacer flanked by active genes. All other promoters remain in long, uncut chromatin fragments. Panel 2: DNA was purified. Panel 3: DNA was digested with NarI, which generates spacers containing fragments of 4.7, 8.4, 9.1, and 5.5 kb. Panel 4: the DNA was fractionated on an agarose gel, and fragments containing active promoters (open triangles; 4.7 and 5.5 kb) and inactive promoters (black triangles; 8.4 and 9.1 kb) were excised and purified. (C) AMY3cells were irradiated as described for Fig. 2. DNA fragments containing promoters of active and inactive genes were purified (as depicted in panel B), digested with NheI and NdeI, cut at CPDs with T4-endoV (lanes 2 to 8), fractionated on alkaline agarose gels, and blotted and hybridized with strand-specific probes for the bottom strand (see Fig. 1A). Lanes 1, same as lanes 2 but with no T4-endoV treatment; M, size marker as described for Fig. 2. The 35S promoter is marked with a star.
CPD analysis by indirect end labeling.
DNA was cut with NdeI, ClaI, or NheI (Fig. 1) and incubated for 2 h at 37°C with T4-endonuclease V (T4-endoV) (Epicentre) in 50 mM Tris-5 mM EDTA (pH 7.5) or mock treated with the same buffer. The DNA was electrophoresed in 1.5% alkaline agarose gels, blotted to Zeta GT nylon membranes, and hybridized with radioactively labeled strand-specific DNA probes (Fig. 1). The signals were analyzed with a PhosphorImager (Amersham Biosciences) using ImageQuant (Amersham Biosciences). The CPD content (CPDs/top strand and CPDs/bottom strand) was calculated using the Poisson expression [−ln(RFa/RFb)], where RFa and RFb represent the signal intensity of the intact restriction fragment of the T4-endoV and mock-treated DNA, respectively (31). Region-specific damage was calculated as the signal of that region in the T4-endoV DNA divided by the signal for the whole lane. The corresponding signal of the mock-treated DNA was subtracted as background. To generate repair curves, the values were normalized with respect to the initial damage (0 min = 100% damage).
Chromatin mapping by micrococcal nuclease.
Yeast cells of strain UCC510 were grown at 30°C in YPD to an optical density at 600 nm of about 1.0, and genomic chromatin was prepared as described previously (51). Half of it was used for DNA purification. Purified DNA and the chromatin were digested with micrococcal nuclease (MNase; Roche Diagnostics). DNA was purified by phenol extractions, cut with ClaI and NdeI, and analyzed by indirect end labeling as described above for CPDs.
Radioactive probes.
DNA fragments for generation of radioactive probes (Fig. 1A) were generated by whole-cell PCR. The oligonucleotides used for the 268-bp rRNA genes probe were 5′-GTGCTATGGTATGGTGACG-3′ (top strand) and 5′-ACTACTGGCAGGATCAACC-3′ (bottom strand). Strand-specific probes were generated by separate primer extensions with one oligonucleotide for each strand by use of QIAGEN Taq polymerase.
Primer extension.
Primer extension was done as described previously (53) with minor modifications. The primer used for analysis of the bottom strand of the 35S promoter was 5′-GTATGTTTTGTATGTTCCCGCG-3′. The 3′ end of the oligonucleotide hybridizes 266 bp upstream of the transcription start site of the 35S rRNA gene (Fig. 1A). A total of 10 pmol of the primer was labeled at the 5′ end by use of 10 U of T4-polynucleotide kinase (New England Biolabs) and 15 pmol of [γ-32P]ATP (Hartmann Analytics) (5,000 Ci/mmol, 10 mCi/ml) at 37°C for 30 to 60 min. Nonincorporated nucleotides were removed using Sephadex G-50 quick spin columns (Roche Diagnostics). For primer extension, 30 to 40 ng of DNA was mixed with 18 μl of end-labeled primer (0.5 to 0.7 pmol), 2 μl of dimethyl sulfoxide (100%), 4 μl of 10× QIAGEN Taq buffer, and 1.2 μl of dNTPs (Pharmacia ultrapure dNTPs) (5 mM each) and adjusted to a final volume of 35 μl. Samples were heated at 95°C for 10 min and then chilled on ice. A total of 5 μl of Taq polymerase (QIAGEN) (1 U) was added, and the samples were subjected to 30 cycles of repeated denaturation (94°C for 45 s), annealing (58°C for 4 min 30 s), and extension (72°C for 3 min). The reaction products were ethanol precipitated and analyzed on a 5% acrylamide-42% urea sequencing gel (29). The gel was dried on Whatman DE81 paper and analyzed with a PhosphorImager (Amersham Biosciences). DNA sequencing was done in parallel by the chain termination method using the same primer and the same conditions as described above. To obtain the fraction of molecules that contained a dimer at a defined site, the signal of individual bands or clusters of bands was measured and divided by the signal of the whole lane. The corresponding signal in the lane with photoreactivated DNA was subtracted as non-CPD background. The damage at each repair time was normalized with respect to the initial damage (no repair = 100% damage).
RESULTS
Chromatin structure and repair in the spacer region.
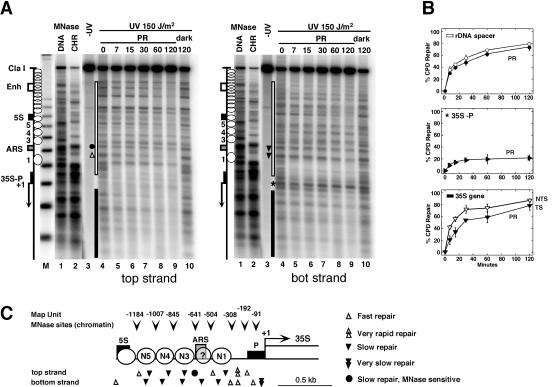
DNA repair by photolyase (photoreactivation) was investigated in AMY3 with inactivated NER (rad1Δ) (Fig. 2). Cells were irradiated in suspension with 150 J/m2 and exposed at room temperature to light for photoreactivation or in the dark for testing NER activity. DNA was isolated and cut with ClaI and NdeI, which generates a 3.9-kb fragment containing the rRNA gene spacer (Fig. 1). The DNA was cut at CPDs with T4-endonucleaseV (T4-endoV), fractionated on an alkaline gel, blotted to a nylon membrane, and hybridized to strand-specific probes abutting the NdeI site.
FIG. 2.
Chromatin structure modulates UV damage formation and repair in the rRNA gene spacer. (A) Chromatin footprinting by micrococcal nuclease (MNase) was compared with CPD repair by photolyase in AMY3 (rad1Δ). Cells were irradiated with UV light (150 J/m2) and exposed to photoreactivating light (PR). DNA was purified, digested with ClaI and NdeI, and cut at CPDs with T4-endoV. Chromatin (CHR) and genomic DNA (DNA) were isolated from UCC510, partially digested with micrococcal nuclease (MNase), and cut with ClaI and NdeI. The DNA was fractionated on alkaline agarose gels and blotted and hybridized with probes for the bottom strand (right panel) and top strand (left panel), respectively. The positions of the DNA elements (as described for Fig. 1A), positioned nucleosomes (white circles), and nucleosomes not positioned (overlapping circles) are indicated. M, a size marker with multiples of 256 bp; lanes 1, DNA digested with MNase; lanes 2, chromatin digested with MNase; lanes 3, DNA of unirradiated cells; lanes 4, DNA of irradiated cells with no repair (initial damage); lanes 5 to 9, DNA of cells after incubation in photoreactivating light for 7 to 120 min; lanes 10, DNA of cells after incubation in the dark for 120 min. Sites of differential repair in the ARS region (dot, triangles) and in the promoter (star) are indicated. (B) Repair curves of the whole spacer region (white bar in panel A), the promoter region (35S-P; star in panel A), and the transcribed region (black bar in panel A). Black symbols, bottom stand; white symbols, top strand. The bottom strand is the transcribed strand (TS); the top strand is the nontranscribed strands (NTS) of the 35S gene. Data are given as averages with standard deviations for at least three gels. The modulation of repair in the spacer, promoter, and coding region was reproduced in duplicate experiments. Strand-specific repair in the coding region confirmed previous observations (30). (C) Schematic summary and structural interpretation of DNA accessibility to MNase (arrowheads) and photolyase (triangles). Circles 1, 3, 4, and 5 represent positions of nucleosomes. Whether ARS is included in a nucleosome is unclear (circled question mark; see text).
Nonirradiated DNA showed the intact restriction fragment (Fig. 2A, lanes 3). Treatment of damaged DNA with T4-endoV generated bands which represent the yields and distribution of CPDs in pyrimidine clusters (lanes 4). The CPD patterns were different in both strands, demonstrating the strand specificity of the assay. The initial damage was 0.21 CPDs/kb (top strand) and 0.27 CPDs/kb (bottom strand).
Repair of CPDs was detected as a time-dependent decrease of the CPD bands and an increase of the intact restriction fragment. The CPDs were efficiently removed by photoreactivation (Fig. 2A, lanes 4 to 9). No NER activity was detected when cells were incubated in the dark (Fig. 2A, lanes 10). Repair was heterogenous, showing slow and fast repair at different sites.
To allow a side-by-side comparison of repair and chromatin structures (Fig. 2C), MNase footprinting lanes were included in the gels (Fig. 2A, lanes 1 and 2). For that purpose, chromatin and naked DNA were partially digested with MNase and the cutting sites were displayed on the same gels as the CPDs. MNase revealed four clear footprints between the promoter and the 5S gene, a result which is consistent with positioned nucleosomes (Fig. 2A) and supports previous observations (52). The absence of strong footprints between the 5S gene and the enhancer indicates that the nucleosomes were more randomly arranged (Fig. 2A). Photolyase removed about 80% of the lesions from both strands of the whole spacer region (Fig. 2B) in 2 h. The repair efficiency was similar to that observed previously in the inactive nucleosomal rRNA genes (30). Thus, the repair results are consistent with the presence of nucleosomes in the spacer region and indicate that the spacer chromatin was as compact as the inactive coding region.
The chromatin structure around the rARS element is unclear. Vogelauer et al. reported a positioned nucleosome (52), while psoralen cross-linking data argued against the presence of a nucleosome (28). Our MNase profile showed partial protection (Fig. 2A, lanes 1 and 2). A CPD cluster in the bottom strand of rARS was as slowly repaired as CPDs in flanking nucleosomes (bottom strand) (Fig. 2A), while a CPD site in the top strand was more rapidly removed (top strand). Surprisingly, CPDs in an MNase-sensitive site of rARS were slowly repaired (top strand) (Fig. 2A). The inhibition of repair and the partial nuclease footprint are consistent with the presence of a nucleosome or a specialized protein DNA complex such as the origin of replication complex. Since only a fraction of rARSs are active (32) and since yeast replication origins exist in two chromatin states during the cell cycle (13), the repair data are also consistent with different populations of complexes.
MNase digestion of the transcribed region (Fig. 2A) produced no footprints of nucleosomes, a result which is consistent with the absence of nucleosomes in active genes (11) and a lack of positioning in the inactive fraction. As observed previously (30), there was strand-specific repair in the transcribed region (35S gene) (Fig. 2B). Photolyase was slow on the transcribed strand, which is the bottom strand of rRNA genes, and reflects inhibition by stalled RNAP-I (30).
Inefficient repair in the 35S promoter.
The promoter region between the initiation site (+1) and nucleosome 1 was sensitive to MNase (Fig. 2A, lanes 2), but a pronounced heterogeneity in DNA repair was obvious. CPDs in the upstream region were rapidly repaired by photolyase (less than 7 min), which is consistent with the absence of nucleosomes (46, 47). However, CPDs located in the 35S promoter were resistant to photoreactivation (Fig. 2A). Promoter repair reached significantly lower levels than repair in the flanking spacer and coding regions (Fig. 2A and B). Moreover, repair reached a plateau after 30 min, indicating that two populations of promoters might exist, one that is accessible to repair and a second that is not accessible. This strong inhibition was confirmed in additional independent experiments (data not shown, but see Fig. 3 and 6). Moreover, very similar results were obtained with photolyase in presence of NER (not shown). Those “repair footprints” suggest that transcription factors and/or RNAP-I inhibits access of photolyase to the lesions.
FIG. 6.
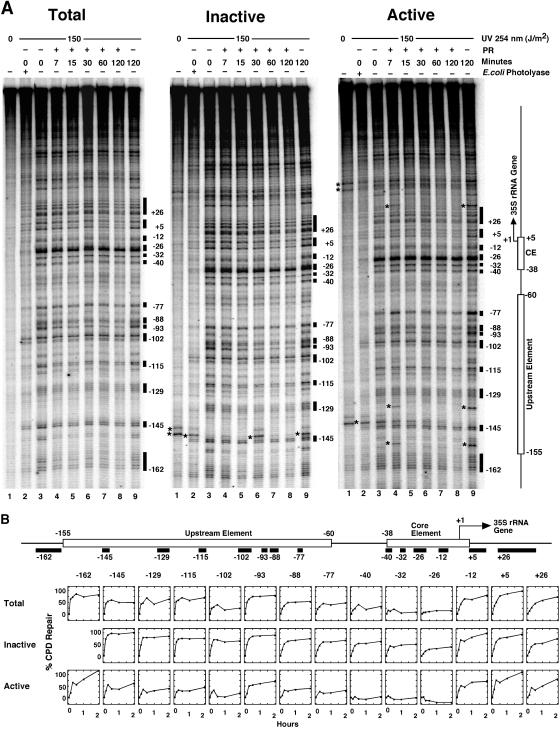
Modulation of photoreactivation in active and inactive promoters. (A) Irradiation of AMY3 and photoreactivation and fractionation of active and inactive 35S promoters were done as described for Fig. 2 and 3. The 35S promoter was analyzed by primer extension as described for Fig. 4. A set of data are shown for total, inactive, and active 35S promoters. The lanes in each set represent DNA of nonirradiated cells (lanes 1); DNA of cells irradiated with 150 J/m2 (lanes 2 to 9) and photoreactivated for 7 to 120 min (lanes 4 to 8) or incubated in the dark for 120 min (lanes 9); and damaged DNA as described for lane 3 but treated with E. coli photolyase (lanes 2). Damage sites used for quantification of repair and their position with respect to the transcription initiation site (+1) are indicated as black boxes. (B) Repair curves are shown for total 35S promoters (Total) and purified inactive and active 35S promoters (Inactive and Active). Repair curves represent the averages of results obtained with two gels (Total and Inactive) or three gels (Active) of the same repair experiment. Differential repair was confirmed in an independent experiment (data not shown).
Enhanced damage formation and inhibition of repair in active promoters.
To investigate whether the transcription factors were bound in active and inactive promoters, the two populations were fractionated as depicted in Fig. 3B and the CPDs were analyzed by indirect end labeling (Fig. 3C). The most striking observation was that of enhanced damage formation and inhibition of photoreactivation in the active promoter, while CPD levels were lower in the inactive fraction and CPDs were more efficiently repaired (Fig. 3C). These results support several conclusions. First, fractionation of active and inactive promoters was successful. Second, both fractions had distinct chromatin structures. Active promoters appear to be preferentially associated with transcription factors that alter the structure of DNA and enhance the formation of CPDs. Third, some proteins remained bound after the irradiation and inhibited repair by photolyase.
UV photofootprints in the core and upstream element: CF binding requires UAF.
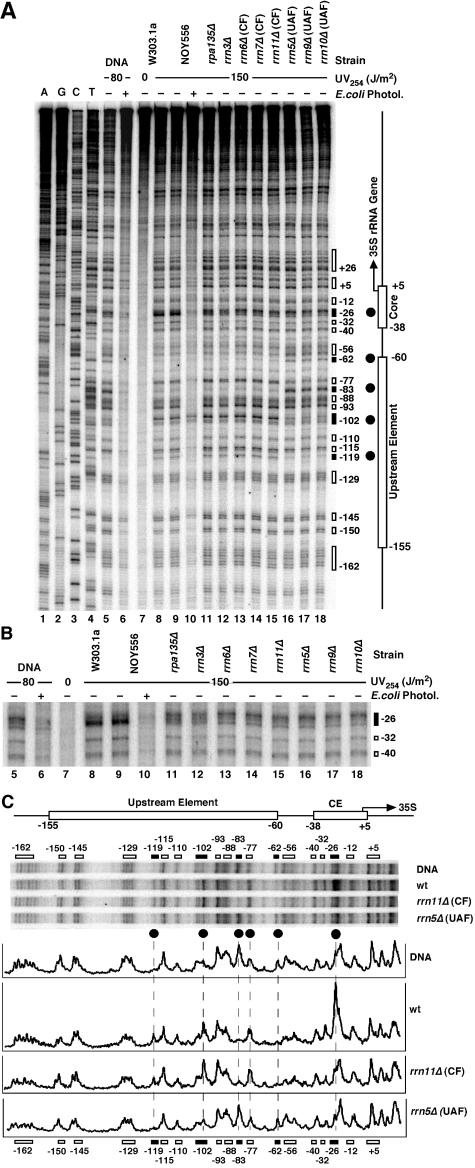
To identify the transcription factors that were responsible for the UV photofootprint, high-resolution UV photofootprinting experiments were done with yeast strains mutated in individual transcription factors or RNAP-I (Fig. 4). Those strains survive in galactose medium, since they carry the 35S rRNA gene on a plasmid under the control of the GAL7 promoter (36). After irradiation with 150 J/m2, the DNA was purified and subjected to primer extension analysis using Taq polymerase, which is efficiently blocked at CPDs and 6-4PPs (53). Primer extension products were separated on sequencing gels (Fig. 4). The intensity of the UV-induced bands represents the yields of lesions. The contribution of non-CPD lesions (presumably 6-4PPs) was low, as demonstrated by removal of CPDs with E. coli photolyase in vitro (Fig. 4A, lanes 6 and 10).
FIG. 4.
UV photofootprinting in 35S promoter of yeast strains defective in RNAP-I transcription factors. (A) Yeast strains were irradiated with 150 J/m2 in selective medium containing galactose. DNA was purified and digested with AvaII. An end-labeled primer was annealed to the bottom strand 280 bp upstream of the transcription initiation site (fat horizontal arrow in Fig. 1A) and was extended by use of Taq polymerase towards the transcription start site of the 35S gene. Products were separated on a 5% acrylamide-42% urea sequencing gel. Lanes 1 to 4, dideoxy sequencing reactions. Lane 5, DNA damaged in vitro with 80 J/m2. Lane 6, damaged DNA as described for lane 5treated with E. coli photolyase. Lane 7, DNA of nonirradiated W303.1a cells. Lanes 8 to 18, DNA of different yeast strains irradiated with 150 J/m2 (chromatin). Lane 8, W303.1a. Lane 9 and 10, NOY556. Lane 10, treatment with E. coli photolyase in vitro. Lane 11, NOY408-1a (rpa135Δ; defective in RNAP-I). Lane 12, NOY604 (rrn3Δ). Lane 13 to 15, strains with mutations in the CF: NOY567 (rrn6Δ), NOY558 (rrn7Δ), and NOY730 (rrn11Δ). Lane 16 to 18, strains with mutations in the UAF: NOY699 (rrn5Δ), NOY703 (rrn9Δ), and NOY704 (rrn10Δ). The elements of the promoter region (as described for Fig. 1) and damage clusters and their positions with respect to the transcription initiation site (+1) (black and white boxes) are indicated. (B) A magnification of the core element region. (C) Pyrimidine dimer patterns in the promoter region of irradiated DNA and chromatin of strain NOY556 (wt), the CF mutant NOY730 (rrn11Δ), and the UAF mutant NOY699 (rrn5Δ). A comparison of relative pyrimidine dimer yields is shown by PhosphorImager scanning results. Black circles and dashed lanes depict dimer positions with marked differences in damage formation (UV photofootprint; see text for details).
Comparison of naked DNA irradiated in vitro and DNA of irradiated wild-type cells revealed differences in the core element (core photofootprint) and the upstream element (UAF photofootprint) (Fig. 4A and C). The most pronounced effect in the core was enhanced damage formation in chromatin at position −26 (Fig. 4B, lanes 5, 8, and 9; gels in Fig. 4C). The UAF photofootprint is characterized by suppressed damage yields at −62 and −83 as well as by enhanced damage formation in chromatin at positions −77, −102, and −119. Most of those lesions were CPDs, since they could be removed by E. coli photolyase in vitro (lane 10). Position −102, however, included a substantial fraction of a non-CPD lesion (Fig. 4A, lane 10) generated in chromatin but not in free DNA (lane 6).
Several CF mutants were examined (rrn6Δ, rrn7Δ, and rrn11Δ). All of them revealed a loss of the enhanced damage formation in the core element (−26), while the footprint in the upstream element was not altered (Fig. 4B, lanes 13 to 15, and Fig. 4C). A similar observation was made for mutants of RNAP-I (rpa135Δ) and RRN3 (rrn3Δ; Fig. 4A and B, lanes 11 and 12). Thus, the core footprint required not only a functional CF but also Rrn3p and RNAP-I. If only one factor was absent, the DNA-protein complex was altered and the footprint was lost.
In addition, several mutants of UAF proteins were tested (rrn5Δ, rrn9Δ, and rrn10Δ; Fig. 4, lanes 16 to 18). Damage levels were very similar in DNA and chromatin. Thus, all UAF mutants lost not only the upstream footprint but also the core footprint. Those in vivo results strongly suggest that UAF binding was required for subsequent recruitment of the other factors, as was proposed on the basis of in vitro transcription studies (44, 45) and DNase I footprinting experiments (5).
Different UV footprints in the core and upstream element of active and inactive promoters.
A central issue is how the transcription factors are distributed among active and inactive promoters. We therefore analyzed UV photofootprints in both promoter fractions (Fig. 5). This experiment was done with AMY3 (rad1Δ). Unfractionated promoters (total 35S-P) (Fig. 5) revealed the same footprints as observed in W303.1a and NOY556 (Fig. 4C), demonstrating that the footprint was not strain dependent.
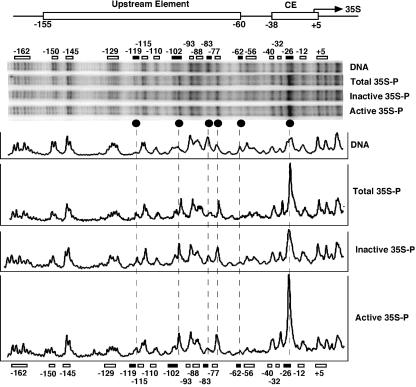
FIG. 5.
UV photofootprints in active and inactive promoters. DNA was irradiated with 80 J/m2. AMY3 (rad1Δ) cells were irradiated with 150 J/m2. Active and inactive 35S promoters were isolated as described for Fig. 3. Pyrimidine dimers were detected by primer extension as described for Fig. 4. Pyrimidine dimer distribution in irradiated naked DNA (DNA), in total DNA of irradiated cells (Total 35S-P), and in purified fragments containing active and inactive promoters (Inactive 35S-P; Active 35S-P) are shown. A comparison of relative pyrimidine dimer yields is shown (PhosphorImager scans). Symbols are as defined for Fig. 4C.
In active promoters, a very high yield of lesions was observed at position −26 (core footprint). In inactive promoters, however, this peak was reduced compared with the active fraction results. Thus, the enhanced damage yield in active promoters is consistent with the strong signal detected in the promoter region of active promoters at low resolution (Fig. 3C) and demonstrates that the CF, RNAP-I, and Rrn3 were complexed with the core element of active promoters.
While the core footprints differed in active and inactive promoters, we found that the upstream footprints were remarkably similar in both fractions and clearly different from irradiated naked DNA results (Fig. 5). This is strong evidence that UAF was present in a substantial fraction of promoters irrespective of the transcriptional activity of the downstream gene.
Inhibition of photoreactivation in the core and upstream element of active 35S promoters.
Having observed slow photoreactivation in active core promoters (Fig. 3) and differential binding of UAF and CF in active and inactive promoters (Fig. 5), we investigated photoreactivation in the upstream and the core element of fractionated promoters by primer extension. Photoreactivation of total rRNA genes and inactive and active 35S promoters is shown in Fig. 6. In all sites, repair of total promoters was faster than in the active fraction but slower than in the inactive fraction, demonstrating that the mixed population of promoters was successfully fractionated (Fig. 6B).
In active promoters, the core element (Fig. 6A, positions −26, −32, and −40) was not repaired and repair was slow in the upstream element (positions −77, −102, −115, and −129). Less than 50% of the lesions were removed in 2 h. Thus, the CF and UAF remained bound to the active promoters after damage formation and inhibited access of photolyase to CPDs. Moreover, the relatively rapid repair at position −93 indicates that not all DNA in the upstream element was covered by proteins and inaccessible to repair enzymes. In the inactive promoters, all DNA lesions of the core element and the upstream element were more rapidly repaired than in active promoters. Fast repair of the core element correlates with the lack of a UV footprint and the absence of CFs. However, enhanced repair of the upstream element occurred despite the presence of a UV footprint. Thus, the UV photofootprinting and repair experiments demonstrate that UAF, together with CF and RNAP-I, forms a stable complex in active 35S promoters.
ChIP analysis of UAF binding to the promoter, however, did not reveal any significant difference between exponentially growing cells with about 50% active genes and stationary cells with reduced rates of RNA synthesis (9). Judging on the basis of the UV photofootprinting and repair experiments, it seems possible that UAF in the inactive promoter is less tightly bound than in the active promoter. Alternatively, an altered conformation or composition might facilitate access to photolyase in inactive promoters.
DISCUSSION
The UV photofootprinting and repair data provide novel information with respect to the protein complexes bound in active and inactive promoters and repair in the rRNA gene spacer.
Since only a fraction of all rRNA gene copies is transcribed, a central issue is that of which factors are associated with active and inactive promoters of living cells. ChIP approaches were used with genetically modified yeast cells but could not discriminate between the two populations in wild-type cells (9). Compared with conventional nuclease footprinting (5), the UV photofootprinting and repair approach has two major advantages: First, UV irradiation and repair is done in living cells. Second, it leaves the DNA intact and the transcribed genes remain nucleosome free, which allows the fractionation of active and inactive genes (10, 17, 30) and promoters (Fig. 3). Indeed, the UV photofootprint of the CF (enhanced CPD formation) and the repair inhibition were found in the active promoters (Fig. 3, 5, and 6) and argue for a successful fractionation. On the other hand, the inactive fraction might contain some active promoters if the restriction digest in nuclei were incomplete or if in some gene transcription were initiated but elongation did not proceed far enough to disrupt the nucleosomes or if a fraction of genes with active promoters were to fold into nucleosomes after damage induction and blockage of RNAP-I (30). This may explain the mild UV photofootprint in the core region of the inactive fraction (Fig. 5).
RNAP-I transcription is initiated by UAF interaction with the upstream element of the 35S promoter, which leads to the recruitment of the CF and Rrn3-RNAP-I (23, 24, 44). Upon transcription initiation, RNAP-I-Rrn3p and CF dissociate from the promoter, while UAF remains behind (2). We observed characteristic UV photofootprints in the core and upstream element. The core footprint was lost when CFs, Rrn3, RNAP-I, or UAF factors were mutated. The UAF footprint, however, was independent of CF, Rrn3, and RNAP-I mutations. Thus, our in vivo results stand in general agreement with the results of DNase I footprinting studies (5) and in vitro transcription studies (2, 23, 24, 44) that support a binding hierarchy of the RNAP-I transcription factors in living cells.
Interestingly, one of the sites with enhanced UV damage formation (position −77) coincides with a hypersensitive site detected by DNase I footprinting in vitro (described in reference 5). However, the most pronounced UV photofootprint was the enhanced induction of CPDs in the core element of active promoters (position −26). Note that enhanced damage formation was also observed in the TATA box of the SNR6 gene which is transcribed by RNAP-III and in the active GAL1 and GAL10 promoters (1, 42). In both the GAL10 and SNR6 promoters, a substantial fraction of the enhanced lesions consisted of 6-4PPs generated by binding of the TATA binding protein (TBP) (1). The enhanced damage in the rRNA genes core element (position −26), however, represents preferentially CPDs (Fig. 4). Thus, the structural deformations in DNA generated by the initiation complexes are remarkably different in the rRNA gene promoter compared with the results seen with RNAP-II- and RNAP-III promoters. The in vivo photofootprinting results support the conclusion derived from in vitro transcription assays that the role of TBP in RNAP-I transcription is fundamentally different from its role in RNAP-II or RNAP-III transcription (23).
The inhibition of repair by photolyase provides insight into the stability of protein DNA interactions after UV damage formation (repair footprint). The strong inhibition of promoter repair seen in the bottom strand (Fig. 2 and 3) in active promoters, but not in inactive promoters (Fig. 3), was most pronounced. The strong inhibition is restricted to the core element, while some repair was detected in the upstream element (Fig. 6). Thus, the CF and UAF remain bound to the DNA after damage formation and inhibit repair. In vitro experiments showed that the CF, Rrn3, and RNAP-I dissociate from the template after each transcription initiation event, supporting a model in which the RNAP-I basal machinery cycles on and off the promoter with each round of transcription (2). Reloading of RNAP-I in vivo was estimated to occur in about 1 s in heavily transcribed genes (14, 15). Thus, rapid reloading of the factors could explain the tight inhibition of repair. Similar experiments done with the SNR6 and GAL10 genes revealed inhibition of repair by photolyase in the SNR6 promoter and efficient repair in the active GAL10 promoter, respectively (1). We therefore take the repair observations as an indication of the different stability or different initiation frequencies of the initiation complexes.
The UV photofootprints of UAF in the upstream element were surprisingly similar in active and inactive promoters, suggesting that UAF is constitutively present irrespective of the transcriptional activity of the downstream gene (Fig. 5). The same conclusion was emphasized on the basis of ChIP experiments with growing cultures containing about 50% active genes and stationary cultures containing reduced rates of rRNA gene transcription (9). A tight interaction of UAF with the upstream element in vitro led to the prediction that UAF assembles after replication on all promoters (22). The UAF photofootprints and repair footprints detected in inactive promoters and the ChIP data are consistent with this hypothesis. This leads to the following question: why are CF, Rrn3, and RNAP-I not recruited to inactive promoters? One reason could be the presence of limited amounts of at least one of the factors RNAP-I, Rrn3, TBP, and CF. Alternatively, UAF might be incomplete or modified in a way that does not allow recruitment of the CFs and yet is sufficient to provide a footprint. The relative rapid repair of the upstream element in inactive promoters (Fig. 6) supports the idea of an instability of the complex.
Complementary to UV photofootprinting, repair by photolyase directly probes for the accessibility of CPDs in chromatin in vivo and provides an indication of the dynamic properties of chromatin components. Under our conditions, nucleosome-free DNA (e.g., promoters and linker DNA) is repaired in about 15 min and the DNA in nucleosomes is repaired in about 2 h (46, 48). Here, we identified rapidly repaired sites in the nuclease-sensitive promoter region (top strand) (Fig. 2A and C), as well as slowly repaired sites in the spacer region. Moreover, repair of the spacer overall was similar to repair of the inactive nucleosomal rRNA genes (30). Thus, the modulation of repair is consistent with a nucleosomal organization of the spacer region. It is interesting that in addition to regulation of transcription by RNA-polymerase I, the rRNA genes locus exhibits a silencing effect on recombination and expression of reporter genes inserted into rRNA genes (25, 39). Thus, photoreactivation might be used to investigate how chromatin structures affect silencing in the rRNA genes.
Acknowledgments
We thank U. Suter for continuous support, M. Nomura, D. Gottschling, and R. Sternglanz for providing the yeast strains, and M. Nomura, J. Sogo, and the reviewers for comments on the manuscript.
This work was supported by grants from the Swiss National Science Foundation, the ETH Zürich, the Roche Research Foundation, and the Janggen-Pöhn-Stiftung.
REFERENCES
- 1.Aboussekhra, A., and F. Thoma. 1999. TATA-binding protein promotes the selective formation of UV-induced (6-4)-photoproducts and modulates DNA repair in the TATA box. EMBO J. 18:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aprikian, P., B. Moorefield, and R. H. Reeder. 2001. New model for the yeast RNA polymerase I transcription cycle. Mol. Cell. Biol. 21:4847-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banditt, M., T. Koller, and J. M. Sogo. 1999. Transcriptional activity and chromatin structure of enhancer-deleted rRNA genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4953-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, M. M., and J. C. Wang. 1984. Use of light for footprinting DNA in vivo. Nature 309:682-687. [DOI] [PubMed] [Google Scholar]
- 5.Bordi, L., F. Cioci, and G. Camilloni. 2001. In vivo binding and hierarchy of assembly of the yeast RNA polymerase I transcription factors. Mol. Biol. Cell 12:753-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradsher, J., J. Auriol, L. Proietti de Santis, S. Iben, J. L. Vonesch, I. Grummt, and J. M. Egly. 2002. CSB is a component of RNA pol I transcription. Mol. Cell 10:819-829. [DOI] [PubMed] [Google Scholar]
- 7.Buttinelli, M., E. Di Mauro, and R. Negri. 1993. Multiple nucleosome positioning with unique rotational setting for the Saccharomyces cerevisiae 5S rRNA gene in vitro and in vivo. Proc. Natl. Acad. Sci. USA 90:9315-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christians, F. C., and P. C. Hanawalt. 1993. Lack of transcription-coupled repair in mammalian ribosomal RNA genes. Biochemistry 32:10512-10518. [DOI] [PubMed] [Google Scholar]
- 9.Claypool, J. A., S. L. French, K. Johzuka, K. Eliason, L. Vu, J. A. Dodd, A. L. Beyer, and M. Nomura. 2004. Tor pathway regulates Rrn3p-dependent recruitment of yeast RNA polymerase I to the promoter but does not participate in alteration of the number of active genes. Mol. Biol. Cell 15:946-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conconi, A., V. A. Bespalov, and M. J. Smerdon. 2002. Transcription-coupled repair in RNA polymerase I-transcribed genes of yeast. Proc. Natl. Acad. Sci. USA 99:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1993. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 21:2331-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Laat, W. L., N. G. Jaspers, and J. H. Hoeijmakers. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13:768-785. [DOI] [PubMed] [Google Scholar]
- 13.Diffley, J. F. X., J. H. Cocker, S. J. Dowell, and A. Rowley. 1994. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78:303-316. [DOI] [PubMed] [Google Scholar]
- 14.Dundr, M., U. Hoffmann-Rohrer, Q. Hu, I. Grummt, L. I. Rothblum, R. D. Phair, and T. Misteli. 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298:1623-1626. [DOI] [PubMed] [Google Scholar]
- 15.French, S. L., Y. N. Osheim, F. Cioci, M. Nomura, and A. L. Beyer. 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol 23:1558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 17.Fritz, L. K., and M. J. Smerdon. 1995. Repair of UV damage in actively transcribed ribosomal genes. Biochemistry 34:13117-13124. [DOI] [PubMed] [Google Scholar]
- 18.Fritze, C. E., K. Verschueren, R. Strich, and E. Easton, R. 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16:6495-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grummt, I. 2003. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17:1691-1702. [DOI] [PubMed] [Google Scholar]
- 20.Hoogstraten, D., A. L. Nigg, H. Heath, L. H. Mullenders, R. van Driel, J. H. Hoeijmakers, W. Vermeulen, and A. B. Houtsmuller. 2002. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell 10:1163-1174. [DOI] [PubMed] [Google Scholar]
- 21.Iben, S., H. Tschochner, M. Bier, D. Hoogstraten, P. Hozak, J. M. Egly, and I. Grummt. 2002. TFIIH plays an essential role in RNA polymerase I transcription. Cell 109:297-306. [DOI] [PubMed] [Google Scholar]
- 22.Keener, J., J. A. Dodd, D. Lalo, and M. Nomura. 1997. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl. Acad. Sci. USA 94:13458-13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keener, J., C. A. Josaitis, J. A. Dodd, and M. Nomura. 1998. Reconstitution of yeast RNA polymerase I transcription in vitro from purified components. TATA-binding protein is not required for basal transcription. J. Biol. Chem. 273:33795-33802. [DOI] [PubMed] [Google Scholar]
- 24.Keys, D. A., B. S. Lee, J. A. Dodd, T. T. Nguyen, L. Vu, E. Fantino, L. M. Burson, Y. Nogi, and M. Nomura. 1996. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes Dev. 10:887-903. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, T., T. Horiuchi, P. Tongaonkar, L. Vu, and M. Nomura. 2004. SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117:441-453. [DOI] [PubMed] [Google Scholar]
- 26.Livingstone-Zatchej, M., R. Marcionelli, K. Moller, R. De Pril, and F. Thoma. 2003. Repair of UV lesions in silenced chromatin provides in vivo evidence for a compact chromatin structure. J. Biol. Chem. 278:37471-37479. [DOI] [PubMed] [Google Scholar]
- 27.Livingstone-Zatchej, M., A. Meier, B. Suter, and F. Thoma. 1997. RNA polymerase II transcription inhibits DNA repair by photolyase in the transcribed strand of active yeast genes. Nucleic Acids Res. 25:3795-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucchini, R., R. E. Wellinger, and J. M. Sogo. 2001. Nucleosome positioning at the replication fork. EMBO J. 20:7294-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Meier, A., M. Livingstone-Zatchej, and F. Thoma. 2002. Repair of active and silenced rDNA in yeast: the contributions of photolyase and transcription-coupled nucleotide excision repair. J. Biol. Chem. 277:11845-11852. [DOI] [PubMed] [Google Scholar]
- 31.Mellon, I., G. Spivak, and P. C. Hanawalt. 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51:241-249. [DOI] [PubMed] [Google Scholar]
- 32.Muller, M., R. Lucchini, and J. M. Sogo. 2000. Replication of yeast rDNA initiates downstream of transcriptionally active genes. Mol. Cell 5:767-777. [DOI] [PubMed] [Google Scholar]
- 33.Nogi, Y., L. Vu, and M. Nomura. 1991. An approach for isolation of mutants defective in 35S ribosomal RNA synthesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:7026-7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nogi, Y., R. Yano, and M. Nomura. 1991. Synthesis of large rRNAs by RNA polymerase II in mutants of Saccharomyces cerevisiae defective in RNA polymerase I. Proc. Natl. Acad. Sci. USA 88:3962-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nomura, M. 2001. Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harbor Symp. Quant. Biol. 66:555-565. [DOI] [PubMed] [Google Scholar]
- 36.Nomura, M. 1998. Transcription factors used by Saccharomyces cerevisiae RNA polymerase I and the mechanism of initiation, p. 155-172. In M. R. Paule (ed.), Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I. Springer-Verlag, Berlin, Germany.
- 37.Prakash, S., and L. Prakash. 2000. Nucleotide excision repair in yeast. Mutat. Res. 451:13-24. [DOI] [PubMed] [Google Scholar]
- 38.Renauld, H., O. M. Aparicio, P. D. Zierath, B. L. Billington, S. K. Chhablani, and D. E. Gottschling. 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7:1133-1145. [DOI] [PubMed] [Google Scholar]
- 39.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 40.Sancar, A. 2003. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103:2203-2238. [DOI] [PubMed] [Google Scholar]
- 41.Sandmeier, J. J., S. French, Y. Osheim, W. L. Cheung, C. M. Gallo, A. L. Beyer, and J. S. Smith. 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21:4959-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selleck, S. B., and J. Majors. 1987. Photofootprinting in vivo detects transcription-dependent changes in yeast TATA boxes. Nature 325:173-177. [DOI] [PubMed] [Google Scholar]
- 43.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 44.Steffan, J. S., D. A. Keys, J. A. Dodd, and M. Nomura. 1996. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes Dev. 10:2551-2563. [DOI] [PubMed] [Google Scholar]
- 45.Steffan, J. S., D. A. Keys, L. Vu, and M. Nomura. 1998. Interaction of TATA-binding protein with upstream activation factor is required for activated transcription of ribosomal DNA by RNA polymerase I in Saccharomyces cerevisiae in vivo. Mol. Cell. Biol. 18:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suter, B., Z. M. Livingstone, and F. Thoma. 1997. Chromatin structure modulates DNA repair by photolyase in vivo. EMBO J. 16:2150-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suter, B., G. Schnappauf, and F. Thoma. 2000. Poly(dA.dT) sequences exist as rigid DNA structures in nucleosome-free yeast promoters in vivo. Nucleic Acids Res. 28:4083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suter, B., and F. Thoma. 2002. DNA-repair by photolyase reveals dynamic properties of nucleosome positioning in vivo. J. Mol. Biol. 319:395-406. [DOI] [PubMed] [Google Scholar]
- 49.Svejstrup, J. Q. 2003. Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 116:447-451. [DOI] [PubMed] [Google Scholar]
- 50.Thoma, F. 1999. Light and dark in chromatin repair: repair of UV-induced DNA lesions by photolyase and nucleotide excision repair. EMBO J. 18:6585-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thoma, F. 1996. Mapping of nucleosome positions. Methods Enzymol. 274:197-214. [DOI] [PubMed] [Google Scholar]
- 52.Vogelauer, M., F. Cioci, and G. Camilloni. 1998. DNA protein-interactions at the Saccharomyces cerevisiae 35 S rRNA promoter and in its surrounding region. J. Mol. Biol. 275:197-209. [DOI] [PubMed] [Google Scholar]
- 53.Wellinger, R. E., and F. Thoma. 1996. Taq DNA polymerase blockage at pyrimidine dimers. Nucleic Acids Res. 24:1578-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]