Abstract
Transcription of the HIV-1 genome yields a single primary transcript, which is alternatively spliced to >30 mRNAs. Productive infection depends on inefficient and regulated splicing and appears to proceed in a tight 5′ to 3′ order. To analyse whether sequential splicing is mediated by the quality of splice sites or by the position of an intron, we inserted the efficient β-globin intron (BGI) into the 3′ region or 5′UTR of a subgenomic expression vector or an infectious proviral plasmid. RNA analysis revealed splicing of the 3′ BGI only if all upstream introns were removed, while splicing of the same intron in the 5′UTR was efficient and independent of further splicing. Furthermore, mutation of the upstream splice signal in the subgenomic vector did not eliminate the inhibition of 3′ splicing, although the BGI sequence was the only intron in this case. These results suggest that downstream splicing of HIV-1 RNAs is completely dependent on prior splicing of all upstream intron(s). This hypothesis was supported by the mutation of the major 5′ splice site in the HIV-1 genome, which completely abolished all splicing. It appears likely that the tight order of splicing is important for HIV-1 replication, which requires the stable production of intron containing RNAs, while splicing of 3′ introns on incompletely spliced RNAs would be likely to render them subject to nonsense-mediated decay.
INTRODUCTION
The hallmark of retroviral replication is the reverse transcription of the genomic RNA into dsDNA and the subsequent integration into a host cell chromosome. The proviral DNA then functions as a single expression unit with the 5′ long terminal repeat (LTR) serving as promoter and the 3′ LTR carrying the signals for 3′ processing, respectively (1). In the case of most retroviruses, a single primary transcript is produced by cellular RNA polymerase II. This polycistronic pre-mRNA is used as genomic RNA and mRNA and, in addition, serves as precursor for all retroviral mRNAs. Successful infection and production of new infectious viruses requires the balanced expression of all viral genes. This is accomplished by a combination of alternative splicing, intron retention and regulated nuclear export of the primary transcript (reviewed in 1–4). Accordingly, retroviral splicing has to be incomplete and intron-containing RNAs need to be exported from the nucleus, where they are normally retained in the case of cellular RNA (5–7).
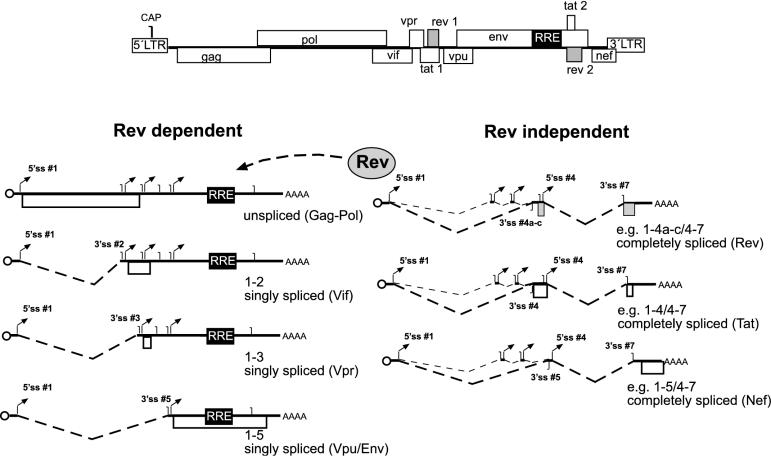
The genomic organization of the primary transcript of all retroviruses is similar. The gag and pol open reading frames (ORF) encoding the inner structural proteins (Gag) and the replication enzymes (Pol) are located in the 5′ half of the transcript (Figure 1) and are expressed from the unspliced primary transcript as polyproteins (Pr55 and Pr160 in the case of human immunodeficiency virus type 1, HIV-1). All primary retroviral transcripts contain a major 5′ splice site (5′ss) upstream of the gag ORF in the 5′ untranslated region (UTR) and a 3′ss at the end of the pol ORF, thus defining gag–pol as an intron. The viral envelope glycoproteins (Env) are encoded in the 3′ half of the genome and are translated from a spliced RNA lacking the gag–pol intron (1–4). In the case of simple retroviruses, this is the only splice event, while complex retroviruses contain additional genes with regulatory functions in the 3′ part of their genome. In the case of HIV-1, there are 6 accessory genes besides gag, pol and env (Figure 1). Production of the mRNAs for the Tat, Rev, Vif, Vpr and Nef proteins involves alternative splicing at four 5′ss (5′ss # 1–4) and at least seven 3′ss (3′ss #1–7; Figure 1). More than 30 different mRNAs have been observed in HIV-1 infected cells (8,9) and have been grouped into three different classes. The unspliced primary transcript (∼9 kb), a class of singly spliced RNAs (∼4 kb) lacking the gag–pol coding region and a class of completely spliced RNAs (∼2 kb) lacking the env coding region in addition are shown in Figure 1 (8). The singly spliced mRNAs encode the Env proteins and the viral regulatory proteins Vif, Vpr and Vpu, while Tat, Rev and Nef are produced from RNAs spliced at multiple sites. In the early phase of HIV-1 gene expression, only completely spliced mRNAs are exported to the cytoplasm, giving rise to the Tat, Rev and Nef proteins. Subsequently, Rev binds to its target sequence on incompletely spliced HIV-1 RNAs [termed Rev response element, RRE, (10)] and mediates their nuclear export by delivering them into the Crm1-dependent export pathway (11,12). HIV-1 RNAs can thus be divided into Rev-independent RNAs lacking an intron (Figure 1, right) and Rev-dependent RNAs containing at least one functional intron and requiring an active nuclear export pathway (Figure 1, left).
Figure 1.
Splicing pattern of HIV-1. The genomic organization of HIV-1 is depicted on top with open boxes representing ORFs and 5′ and 3′ LTRs, respectively. Classes of Rev-dependent RNAs are shown below on the left side, Rev-independent RNAs on the right side. A circle represents the 5′ cap, translated reading frames are depicted as open boxes, the RRE as a black box, and 5′ and 3′ss as arrows and vertical lines, respectively. The nomenclature of splice sites was adapted from Purcell and Martin, (8). Below each RNA, the splice sites used for its generation and the translation product are identified.
The requirement for cytoplasmic accumulation of unspliced, partially spliced and completely spliced RNAs indicates that splicing of the primary HIV-1 transcript must be inefficient, allowing Rev to access the RRE-containing RNAs and promote their nuclear export prior to splicing (13–15). This is in contrast to cellular gene expression, where pre-mRNAs are immediately spliced to completion and only intronless RNAs are exported (16). Most 5′ss in the HIV-1 genome match the consensus sequence (17) and the delayed and inefficient splice reaction has mostly been attributed to non-consensus 3′ss with short polypyrimidine tracts interrupted by purines and non-canonical branchpoint sequences (8,15,18–21). The recognition of splice signals and removal of introns is further modulated by intronic and exonic splice enhancers surrounding the 3′ss sites, which are recognized by splice-enhancing factors of the SR-protein family (e.g. SF2/ASF, 22–24). Splice silencers have also been observed, which recruit members of the heterogeneous ribonucleoprotein family (hnRNPs) to suppress the splice site recognition (e.g. hnRNP A/B, 25–27).
In general, splice signals are recognized co-transcriptionally in the order of their appearance, suggesting a sequential removal of introns from the 5′ end [28; reviewed in (29)]. However, the order of splicing can vary significantly and there are many cases where a downstream intron is removed before the upstream intron (30). Examples are the adenine phospho-ribosyltransferase (aprt) gene, where the third of the four introns is removed first (31) and the thymidine kinase mRNA (32). In case of the insulin pre-mRNA, even the metabolic status of the cells seems to influence the order of splicing (33). It should also be noted that the removal of an intron can have different effects, depending on its relative position with respect to the ORF. Splicing of an intron in the 5′ UTR promotes nuclear export and expression by splice-dependent recruitment of factors to the mRNA (34–36). In contrast, splicing of introns in the 3′ UTR can be detrimental due to the induction of nonsense-mediated mRNA decay (NMD) and, accordingly, most cellular mRNAs lack introns in the 3′ untranslated region (37,38; reviewed in 39,40). Interestingly, the splicing of retroviral RNAs appears to have a very tight directionality from the 5′ to the 3′ end (Figure 1). In this report, we show that the 5′ to 3′ directionality of splicing is preserved even when efficient heterologous introns are inserted into different positions of the HIV-1 genome. Splicing of a 3′ intron is tightly inhibited unless the 5′ intron is removed. Accordingly, mutating a single splice site in the HIV-1 genome (5′ss #1) blocks all downstream splice events and leads to the accumulation of the unspliced primary transcript. This tight control of the order of splicing may have important implications for viral gene expression.
MATERIALS AND METHODS
Plasmids
To construct the subgenomic Env expression vector pNLenv, the sequence between the 5′ss #1 [nucleotide 743, numbering according to Adachi et al. (41)] and the 3′ss #5 (nucleotide 5976) was deleted from the infectious HIV-1 proviral plasmid pNL4-3. To this end, the 5′ half of the env gene was PCR-amplified using primers 5′MSD env (GAA GCG CGC ACG GCA AGA GGC GAG GGG CGG CGA CTG GAA GAA GCG GAG ACA GCG ACG AAG AGC; introducing a BssHII site shown underlined) and 3′BstEII env (AGA CGG TGA CCC ACA ATT TTT CTG TAG C; introducing a BstEII site shown underlined). The PCR product was cut with BssHII and BstEII and cloned into the molecular clone pNL4-3 BstEII (42), which had been cleaved with BssHII (nucleotide 743) and BstEII (nucleotide 6331). The second intron from the rabbit β-globin gene (BGI) was amplified using primers 5′BGintron (GGC ATC GAT TG AGA ACT TCA GGG TGA GTT TGG GG) and 3′BGintron (GGC TAG CTA TCT TTG CCA AAA TGA TGA GAC AGC) introducing ClaI sites (underlined) at both ends. The BGI sequence was inserted in sense and antisense orientation between the env and nef coding regions of a pNLenv derivative, carrying a ClaI site (underlined) between the stop codon of the env (bold) and the initiation codon of the nef (bold) ORF [TAATCGATAGCACCATG; (43)] to give pNLenvBGIs and pNLenvBGIas. To generate pNLenvBGB, the BGI sequence was amplified with primers introducing a BssHII site at each end and cloned into the BssHII site of pNLenv. Plasmid pNLenvM3 was generated by overlap PCR and contains three point mutations in the 5′ss #4 (Figure 5; details of the cloning procedure available upon request). The proviral plasmid pNLC4-3 has been described previously (44). The BGI sequence was cloned into the 3′region of a tat/rev negative derivative of pNLC4-3 by exchanging the BamHI with XhoI fragment (nucleotide 8465–8887) for that from pNLenvBGIs and pNLenvBGIas, respectively. To insert the BGI sequence into the 5′UTR of the proviral plasmid, a PCR fragment with flanking BssHII sites was cloned into a tat/rev negative derivative of pNL4-3 (derived from a subgenomic plasmid kindly provided by H. Schaal, Düsseldorf). Mutations in the major 5′ss were also introduced into a tat/rev-negative derivative of pNL4-3 containing a newly introduced NgoM I site immediately adjacent to the major 5′ss. Introducing the NgoM I site into wild-type pNL4-3 did not influence the titer and replication kinetics of the resulting virus (data not shown). The sequence of the major 5′ss was changed by PCR-mutagenesis using a primer, including the newly introduced NgoM I site to give pNL4-3tr−M3. The plasmids pTat and pRev (gifts of P. Krammer, Heidelberg (45) and S. Modrow, Regensburg, respectively) are pcDNA3 (Invitrogen) based expression vectors containing the respective ORFs under control of the CMV promoter.
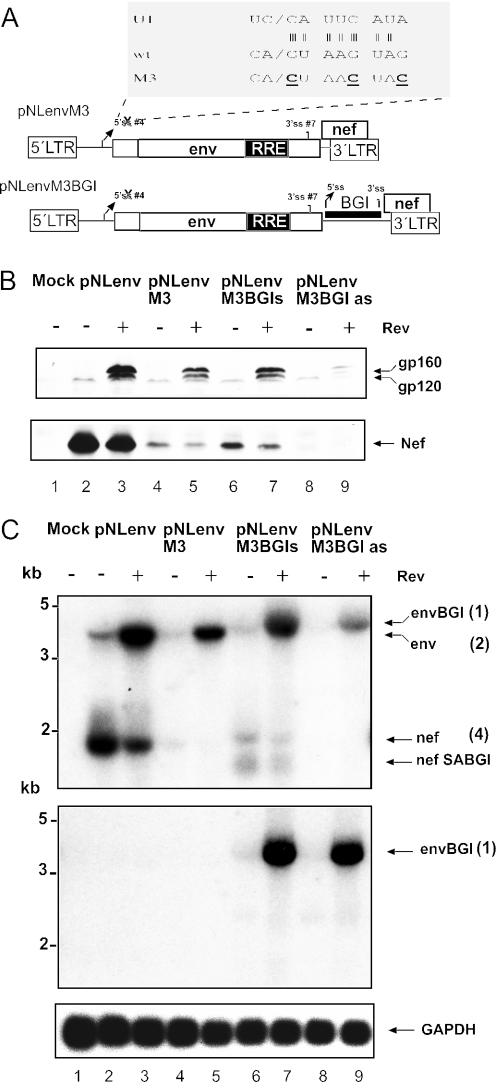
Figure 5.
Mutation of the HIV-1 5′ss in pNLenv does not relieve the 3′ splice inhibition. (A) Schematic depiction of plasmids pNLenvM3 and pNLenvM3BGI. The sequence at the HIV-1 5′ss #4 (wt) and the complementary sequence of the U1 RNA is expanded on top. The three mutations introduced into the 5′ss are depicted below in bold and underlined (M3). (B) Western blot analysis of HeLa P4 cells transfected with the constructs indicated above each lane. A Tat expression plasmid was cotransfected in all cases. HIV-1 proteins are identified on the right as in Figure 2B. (C) Northern blot analysis of 10 μg of RNA obtained from the same transfection as in panel B. The upper panel was probed with an LTR-specific probe, the middle panel with a BGI-specific probe, and the bottom panel with a GAPDH-specific probe. The observed RNA species are identified as in Figure 3C; numbering is according to the drawing in Figure 3A.
Cells, transfection and protein detection
HeLa P4 cells were maintained in DMEM growth medium with 10% heat inactivated fetal calf serum and antibiotics. For transfection, 8 × 105 cells were seeded and transfected with 10 μg of the respective plasmid, 5 μg of Tat and Rev expression plasmids as indicated and 1 μg of a reporter construct encoding green fluorescent protein (eGFP) using the modified calcium phosphate coprecipitation technique (46). For immunoblot analysis, cells were harvested 48 h post transfection (60 h post transfection in the experiment shown in Figure 8). Cell lysates were normalized for transfection efficiency, according to the number of GFP positive cells and analysed by SDS–PAGE and immunoblotting using antisera against HIV-1 capsid (CA) (1:10 000), Env (1:5000, provided by V. Bosch, Heidelberg) or Nef proteins (1:5000) and peroxidase conjugated second antibody (Dianova, Hamburg). Enhanced chemiluminescence (Amersham) was used for detection, according to the manufacturer's protocol. Indirect immunofluorescence was performed on cells fixed with 4% paraformaldehyde, using monoclonal antibody 902 directed against HIV-1 Env (obtained through the NIH AIDS Research and Reference reagent programme) or a polyclonal rabbit anti-Nef serum and phycoerythrin-conjugated secondary antibody (Dianova, Hamburg).
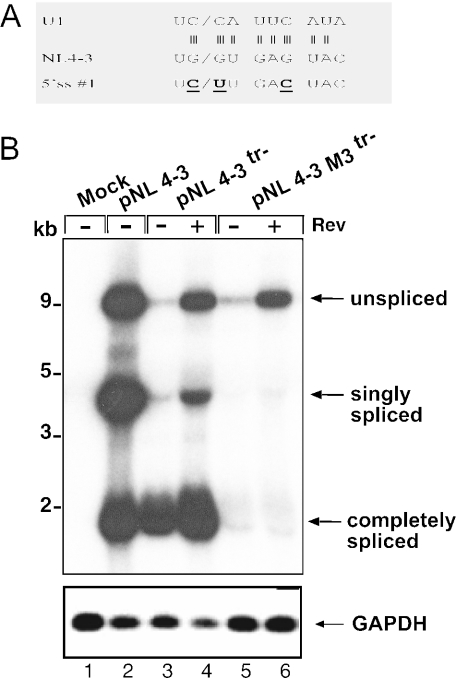
Figure 8.
Mutation of the 5′ss #1 of HIV-1 abolishes splicing. (A) Schematic depiction of the wild-type sequence at the 5′ss #1 of pNL4-3 (middle), the complementary sequence of U1 (top) and the M3 mutations (bold and underlined; bottom). Vertical lines indicate hydrogen bonds between the 5′ss and U1. (B) Northern blot analysis of RNA from HeLa P4 cells transfected with the constructs indicated above each lane and hybridized with a HIV-1 LTR-specific probe. pNL4-3tr− is a derivative of pNL4-3 with mutations in the tat and rev genes. A Tat expression vector was cotransfected in the experiments shown in lanes 3–6. Unspliced, singly spliced and completely spliced RNAs are identified on the right, molecular mass standards are shown on the left. The bottom panel shows re-hybridization of the same blot with a GAPDH-specific probe.
RNA and DNA preparation and analysis
Preparation of total RNA, gel electrophoresis, blotting and detection with a radiolabeled probe were performed as described (47). Nuclear RNA was prepared according to Weil et al. using a mild lysis step in the cold (48). In order to detect HIV-1 specific RNAs, pNLenv was cleaved with BamHI and HindIII, thereby generating a specific probe for the 3′ LTR that is present on all mRNAs. The BGI-sequence was detected with a probe corresponding to an NcoI restriction fragment from pNLenvBGI. The GAPDH sequence was detected with a probe corresponding to a 1.2 kb EcoRI fragment from the GAPDH expression plasmid (gift of K. Harbers, Hamburg). Blots were washed, sealed and exposed to X-ray films (Kodak X-omat-AR). For RT–PCR, 5 μg of total RNA were digested with 5 U of DNase (Promega) for 1 h at 37°C and purified using RNeasy columns (Qiagen) to remove plasmid contaminations. Subsequently, 1 μg of RNA was used for reverse transcription using 100 pmol of the reverse primer (GAA TTC CGC CAG GCA CAA GCG CAT TGT TAG CTG C; Figure 3A) and Superscript II (GIBCO) reverse transcriptase. The reaction product was used in a standard PCR reaction using Taq polymerase (Qiagen), the described reverse primer and forward primer (GGA ATT CCG GCG ACT GGA AGA AGC GGA GAG ACA GCG; Figure 3A).
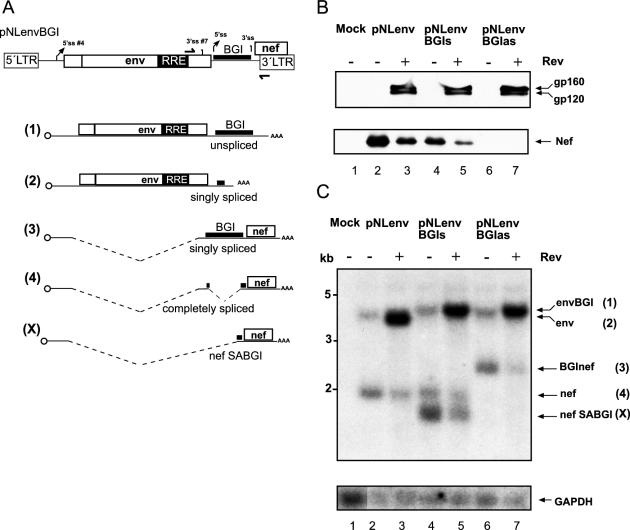
Figure 3.
Splicing of a heterologous intron in the 3′ region of HIV-1. (A) Schematic depiction of the construct (top) and the RNA products. The second intron of the rabbit BGI was cloned in sense or antisense orientation between the reading frames of env and nef of pNLenv. Drawings (1–4) show the potential RNA species plus the shorter product (X). Identification of ORFs, cap and splice signals is as shown in Figure 4; the BGI sequence and the remaining heterologous sequence after splicing of this intron are shown as thick lines. Primers for RT–PCR analysis (see Figure 5) are also indicated in the top panel. (B) Western blot analysis of HeLa P4 cells transfected with the plasmids identified above each lane. A Tat expression plasmid was cotransfected in all cases. The blot was stained and labeled as described in Figure 2B. (C) Northern blot analysis of cell extracts from the same transfection as in panel B. RNA separation and hybridization was performed as described in Figure 2C. The respective RNA species and their numbering in panel A is shown on the right. The shortest RNA species labeled nefSABGI (X) corresponds to the product of splicing from the HIV-1 5′ss #4 to the 3′ss of the BGI sequence and therefore, yields an RNA shorter than authentic nef mRNA. The blot was re-hybridized with a GAPDH-specific probe (bottom).
RESULTS
A subgenomic HIV-1 expression system for the analysis of splicing
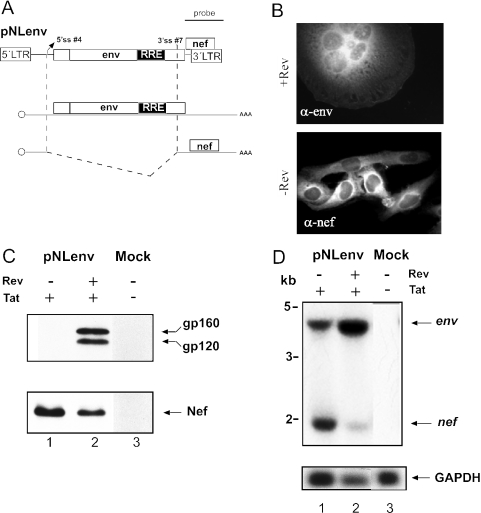
As outlined in Figure 1, splicing of the genomic RNA of HIV-1 leads to at least 30 differentially spliced RNAs, making the analysis of splicing regulation very complicated. We therefore developed a subgenomic expression system closely matching the situation in virus infected cells and not containing heterologous splice sites. Most previous reports analysed HIV-1 splicing using heterologous expression systems (e.g. 25,49,50), which did not produce authentic HIV-1 RNAs. We selected the 3′ part of the HIV-1 genome for our expression system, which should give rise to an unspliced Rev-dependent RNA for Vpu and Env and a singly spliced Rev-independent mRNA for Nef (Figure 2A). Plasmid pNLenv was based on the proviral HIV-1 plasmid pNL4-3 (41) and contains the viral 5′ and 3′ LTRs. The sequence between the major 5′ splice site (5′ss #1) and the 3′ss upstream of the vpu/env coding region [3′ss #5; nomenclature according to Purcell and Martin, (8)] was removed. Transcription from the 5′ LTR of pNLenv is dependent on the viral Tat protein and yields a primary transcript with a sequence identical to that of the authentic env mRNA produced in HIV-1 infected cells (Figure 1, bottom left). This RNA can undergo a single splice event from 5′ss #4 to 3′ss #7, which leads to RNA identical in sequence to that of the genuine nef mRNA, in HIV-1 infected cells (Figure 1, bottom right).
Figure 2.
The subgenomic pNLenv expression system. (A) Schematic depiction of pNLenv (top). The vpu, env and nef ORFs as well as the 5′ and 3′ LTRs and the localization of the probe used for northern blot analysis are shown. The middle and bottom drawings depict the unspliced (middle) and spliced RNA produced from pNLenv. (B) Indirect immunofluorescence analysis of HeLa P4 cells transfected with pNLenv with (top) or without (bottom) a Rev expression plasmid. A Tat expression plasmid was cotransfected to induce LTR-dependent transcription. Cells were stained with antiserum against Env (top) or Nef (bottom). (C) Western blot analysis of transfected HeLa P4 cells. Cells were mock-transfected (lane 3) or transfected with pNLenv and a Tat expression plasmid either with (lane 2) or without (lane 1) a Rev expression plasmid. Blots were stained with antiserum against Env (top) or Nef (bottom). The viral glycoprotein precursor gp160 and the surface glycoprotein gp120 (which contains the epitope for the antibody) as well as the Nef protein are identified on the right. (D) Northern blot analysis of RNA from cell extracts from the same transfection as in panel C. Ten μg of RNA was separated on a denaturing gel. The blot was hybridized with a 3′ LTR-specific probe as indicated in panel A. Re-hybridization with a GAPDH-specific probe served as the loading control (bottom panel). The unspliced and spliced RNAs are identified on the right and molecular mass standards are shown on the left.
Cotransfection of HeLa P4 cells with pNLenv and expression vectors for the HIV-1 Tat and Rev proteins led to the efficient production of viral Env protein as detected by indirect immunofluorescence (Figure 2B) and immunoblot analysis (Figure 2C). Nef protein, which is synthesized from a spliced RNA was also observed (Figure 2B and C). Surface expression of functional HIV-1 Env proteins was confirmed by syncytia formation of transfected cells with neighbouring HeLaP4 cells, which express the HIV receptor CD4 and the co-receptor CXCR4 [Figure 2B; (51)]. Env production was dependent on the HIV-1 Tat (data not shown) and Rev proteins. No Env proteins were detected in the absence of Rev and no syncytia were observed, while Nef expression was increased (Figure 2B and C). Thus, pNLenv reproduces the Rev dependence of the authentic viral RNAs as expected. Northern blot analysis was performed to analyse splicing. RNA was extracted from transfected cells and detected with a 3′ UTR-specific probe that recognizes all HIV-1 specific RNAs (indicated in Figure 2A). The unspliced transcript encoding Env (∼ 4 kb) and the spliced nef mRNA (∼1.6 kb) were detected both in the presence and absence of Rev (Figure 2D), but there was a much stronger signal for the unspliced RNA in the presence of Rev as expected (Figure 2D, lanes 2 and 3). No other HIV-1 specific RNAs were observed.
Splicing of a heterologous intron in the 3′ region of pNLenv requires removal of the upstream intron
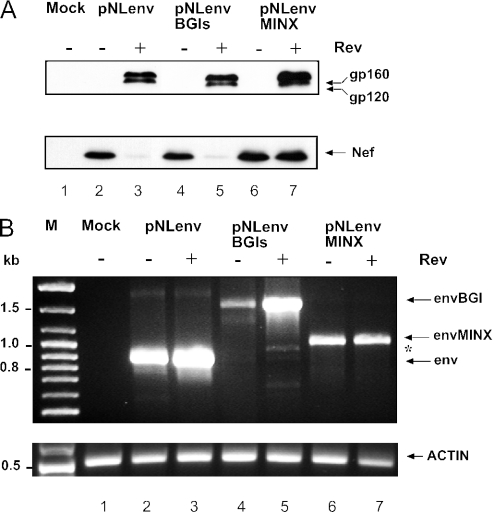
To analyse whether the order of splicing is dependent on the specific splice signals or on the position of the intron, heterologous introns were inserted into pNLenv. Intron 2 from the rabbit BGI was selected as an efficient heterologous intron, which had been used in previous studies (52,53). The BGI sequence was cloned into the 3′ region of pNLenv between the termination codon of env and the initiation codon of nef (pNLenvBGIs; Figure 3A). The same sequence was inserted in antisense orientation as a control (pNLenvBGIas). HeLaP4 cells were transfected with pNLenv, pNLenvBGIs and pNLenvBGIas, respectively, and cell extracts were analysed by immunoblotting using antisera against Env and Nef and by northern blot. Efficient Rev-dependent Env expression was observed in all cases and was not altered by the presence of the BGI intron (Figure 3B, upper panel). In contrast, Nef expression was reduced in the case of pNLenvBGIs and completely lost for pNLenvBGIas (Figure 3B, lower panel). The latter result is probably due to the 700 nt long BGI sequence in the 5′ untranslated region [5′ UTR, (54)] of the nef mRNA, which should not be spliced in the antisense orientation.
To analyse the splicing pattern, RNA from the same transfection was subjected to northern blot hybridization. The unspliced 4 kb RNA and the spliced 1.6 kb RNA was again observed in pNLenv-transfected cells (Figure 3C, lanes 2 and 3), while a longer unspliced RNA, containing the BGI sequence, was detected in cells transfected with pNLenvBGIs and pNLenvBGIas, respectively (Figure 3C, lanes 4–7). In the case of pNLenvBGIas, there was only one more specific RNA product, which lacks the env intron and contains the BGI sequence in antisense orientation (Figure 3C, lanes 6 and 7). Transfection of pNLenvBGIs, on the other hand, yielded a doubly spliced RNA co-migrating with genuine nef mRNA and a shorter product (Figure 3C, lanes 4 and 5). There was no detectable signal corresponding to singly spliced RNAs, where either the BGI sequence (Figure 3A; RNA 2) or the HIV-1 intron (Figure 3A; RNA 3) had been removed. The shorter product was characterized by RT–PCR and sequence analysis and shown to correspond to an RNA that was spliced from the HIV-1 derived 5′ss #4 to the 3′ss in the BGI sequence (termed SABGI). Thus, it appears that the transcript from pNLenvBGIs is either not spliced at all or both introns are removed efficiently. Accordingly, the very efficient BGI sequence is retained unless the relatively inefficient HIV-1 intron is also removed. To further characterize the order of intron removal, nuclear RNA was isolated from cells transfected as above and hybridized with HIV-1 LTR- and BGI-specific probes. Analysis with the BGI-probe confirmed the identity of the unspliced RNA in cells transfected with pNLenvBGIs and pNLenvBGIas and the singly spliced product in cells transfected with pNLenvBGIas (data not shown). Importantly, a BGI-reactive product corresponding to the singly spliced RNA lacking the HIV-1 intron was also detected in cells transfected with pNLenvBGIs, while no product for the singly spliced RNA lacking the BGI intron was observed (data not shown). Taken together, these results indicate that BGI splicing is dependent on splicing of the upstream HIV-1 intron, both in the presence and absence of Rev.
Inefficient splicing of the heterologous intron in the 3′ region was also observed when the cryptic splice sites for the Tev protein (55) in pNLenv were removed together with most of the intronic sequence between 5′ss # 4 and the RRE (data not shown). To analyse whether this inefficient splicing is specific for the BGI sequence, the very efficient MINX intron (56) was cloned into the same position of pNLenv to give pNLenvMINX. The MINX intron is a short synthetic intron containing adenoviral splice sites with perfect consensus sequences and a very strong polypyrimidine tract. Western blot analysis of HeLaP4 cells transfected with pNLenvMINX and controls is shown in Figure 4A. Again, Rev-dependent Env expression and Rev-independent Nef expression was observed as in the case of pNLenv and pNLenvBGIs, respectively. However, higher levels of Nef were found in pNLenvMINX transfected cells in the presence of Rev (Figure 4A, lower panel, lane 7). To determine whether there is a retention of the MINX intron, similar to what had been observed for the BGI sequence, RNA from transfected cells was analysed by RT–PCR using primers that recognize HIV-1 sequences upstream of the 3′ss #7 and in the 3′ LTR. These primers recognize only unspliced and singly spliced RNA containing the env sequences (Figure 3A; RNAs 1 and 2), but not RNAs lacking the HIV-1 intron. Analysis of RNA from pNLenv-transfected cells showed a single product corresponding to the unspliced env mRNA (Figure 4B, lanes 2 and 3). Products with the expected sizes of the respective unspliced RNA were also observed in pNLenvBGIs (Figure 4B, lanes 4 and 5) and pNLenvMINX transfected cells (Figure 4B, lanes 6 and 7). However, only a very weak signal was detected for the spliced mRNA lacking the BGI sequence (Figure 4B, asterisk). This result indicates that retention of the 3′ intron is tight with virtually no partially spliced RNA lacking this intron, despite the presence of very efficient splice sites and a short intronic sequence in case of the MINX intron. Thus, inefficient splicing of an intron in the 3′ region of pNLenv is not restricted to the BGI sequence.
Figure 4.
Analysis of the MINX intron in the 3′ region of pNLenv. The synthetic MINX intron was cloned into the 3′ region of pNLenv in the same position as described for the BGI sequence (pNLenvMINX). (A) Western blot analysis of HeLaP4 cells transfected with the constructs indicated above each lane. A Tat expression plasmid was cotransfected in all cases. HIV-1 specific proteins were identified as described in Figure 2B. (B) RT–PCR analysis of RNA isolated from the same transfection as in panel A. RNA was reverse transcribed and PCR amplified with primers hybridizing to the 3′ end of the env ORF and to the 3′ LTR (indicated in Figure 3A), and PCR products were separated on an agarose gel and stained with ethidium bromide. Specific products are identified on the right, an asterisk marks the position of the spliced product where the BGI sequence had been removed and the env intron had been retained. Molecular mass standards are given on the left. A parallel RT–PCR reaction with actin-specific primers (bottom panel) served as the RNA control. Control reactions without RT enzyme were negative (data not shown).
We next analysed whether the heterologous intron in the 3′ region of pNLenv can be spliced if the 5′ss of the upstream HIV-1 intron is destroyed and the BGI sequence is the only functional intron. To this end, three nucleotides in the 5′ss #4 were mutated, thereby preventing recognition by U1snRNP (Figure 5A). This should completely block the splicing of the HIV-1 intron. The resulting plasmids were termed pNLenvM3, pNLenvM3BGIs and pNLenvM3BGIas, respectively (Figure 5A). Figures 5B and C show western and northern blot analyses of transfected HeLa P4 cells. Mutation of the 5′ss led to a slight reduction of Env expression and to an almost complete loss of Nef expression compared with pNLenv (Figure 5B, lanes 3 and 5; it is noted that the Nef blot was overexposed to visualize residual Nef expression). Northern blot analysis of pNLenvM3-transfected cells revealed an almost complete loss of the spliced RNA in the presence and absence of Rev (Figure 5C, upper panel, lanes 4 and 5), indicating that the M3 mutation efficiently blocked splicing. The residual Nef expression may be explained by a cryptic 5′ss immediately downstream of 5′ss #4. No significant difference in the expression pattern was observed for pNLenvM3BGIs compared with pNLenvM3 (Figure 5B, lanes 5 and 7), while the BGI sequence in antisense orientation abolished HIV-1 protein expression almost completely (Figure 5B, lanes 8 and 9). The latter result was unexpected because the BGI sequence in antisense orientation had not affected Env expression in the case of pNLenvBGIa. The reason for this difference is currently not clear. Importantly, however, northern blot analysis of both, pNLenvM3BGIs and pNLenvM3BGIas transfected cells, revealed a strong signal for the unspliced primary transcript containing the BGI sequence (Figure 5C, upper panel, lanes 6–8). This was confirmed by hybridization of the blot with a BGI-specific probe (Figure 5C, middle panel). In addition, weak signals corresponding to nef mRNA and the previously described shorter product were seen in pNLenvM3BGIs transfected cells (especially in the absence of Rev, Figure 5C, middle panel, lane 6) and are probably due to the cryptic 5′ss mentioned above. The signal for the unspliced RNA was strongly decreased in the absence of Rev in all cases (Figure 5C, upper panel), indicating degradation if this RNA is not exported. The results show that there was no splicing of the efficient heterologous intron in the 3′ region of pNLenv when splicing of the upstream HIV-1 intron was prevented by a 5′ss mutation. Thus, splicing of an intron in this position appears to be inhibited until the upstream splicing event has occurred and this inhibition does not seem to depend on the quality of the splice signals.
The BGI is efficiently spliced when placed into the 5′ UTR of pNLenv
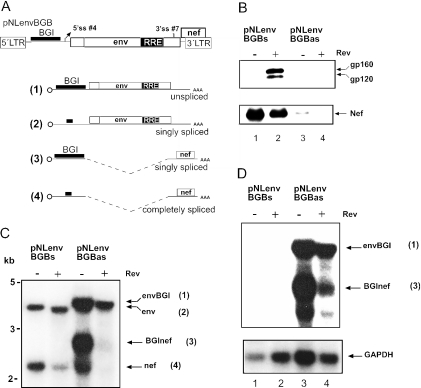
To analyse whether the retention of the BGI sequence is dependent on the position of the intron, the same sequence was inserted in sense or antisense orientation upstream of 5′ss #4 into the 5′ UTR of pNLenv (Figure 6A). The resulting plasmids were named pNLenvBGBs and pNLenvBGBas, respectively. Western blot and northern blot analysis of HeLaP4 cells transfected with these constructs is shown in Figure 6. Efficient Rev-dependent expression of Env and Rev-independent expression of Nef was observed in the case of pNLenvBGBs (Figure 6B, lanes 1 and 2), which was indistinguishable from pNLenv-transfected cells. In contrast, there was very little HIV-1 specific protein expression in pNLenvBGBas transfected cells (Figure 6B, lanes 3 and 4). This is most probably due to the presence of the long heterologous sequence in the 5′ UTR, which cannot be spliced, and may prevent translation of the mRNA (54). Accordingly, the normal production of Env and Nef proteins in pNLenvBGBs transfected cells indicated that the BGI sequence was efficiently removed in this case. This was confirmed by northern blot hybridization of nuclear RNA from transfected cells with HIV-1 (Figure 6C) and BGI-specific (Figure 6D) probes. The HIV-1 specific RNA pattern in pNLenvBGBs transfected cells in the presence or absence of Rev (Figure 6C, lanes 1 and 2) was identical to that observed for pNLenv (Figure 2C, lanes 2 and 3). Importantly, no signal was detected with the BGI-specific probe (Figure 6D, lanes 1 and 2), indicating that the 4 kb RNA in this case lacks the BGI sequence. Thus, insertion of the efficient heterologous intron into the 5′ UTR of pNLenv led to its rapid splicing and degradation of the heterologous intron, while the (inefficient) HIV-1 intron was only partially spliced. In the case of pNLenvBGBas transfected cells (Figure 6C and D, lanes 3 and 4), signals were observed for the primary transcript containing the BGI sequence in antisense orientation [Figure 6A, RNA (1)] and the spliced RNA lacking the HIV-1 intron but containing the BGI sequence (Figure 6A, RNA 3). The weak signal for nef mRNA may be due to cryptic splicing from a 5′ss within the BGI sequence in antisense orientation to 3′ss #7 and may thus explain the weak Nef expression observed (Figure 6B, lane 3).
Figure 6.
Splicing of a heterologous intron in the 5′ region of HIV-1. (A) Schematic depiction of pNLenvBGB (top). The BGI sequence was cloned into the BssHII site located upstream of 5′ss #1. ORFs, splice signals and cap are identified as described in Figure 1. The possible RNA products are shown below. The thick line identifies the BGI sequence or the residual sequence after splicing of this intron. (B) Western blot analysis of HeLa P4 cells transfected with the plasmids indicated above each lane. A Tat expression vector was cotransfected in all cases. HIV-1 specific proteins were detected and labelled as described in Figure 2B. (C) Northern blot analysis of 10 μg of nuclear RNA derived from the same transfection as in panel B. Cells were fractionated and nuclear RNA was separated and hybridized with a probe specific for the HIV-1 LTR as described in Figure 2C. The respective RNA species and their numbering in panel A is shown on the right. (D) Re-hybridization of the same blot as in panel B with a BGI-specific probe (upper panel) and with a GAPDH-specific probe (lower panel).
Insertion of the BGI into different positions of an infectious HIV-1 plasmid recapitulates the results obtained with pNLenv
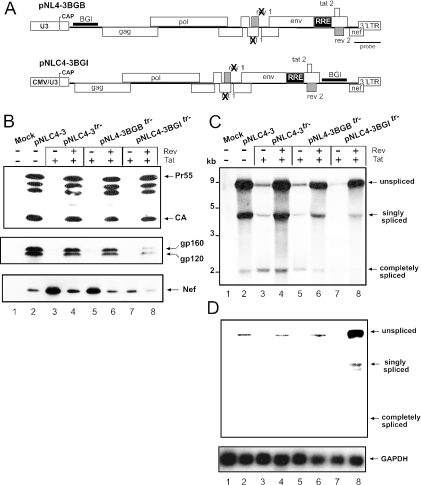
To analyse whether the position-dependent retention of a heterologous intron also occurs in the context of the complete HIV-1 genome, we inserted the BGI sequence into an infectious proviral plasmid. The heterologous intron was placed into the same positions in the 5′ UTR and upstream of the nef gene as described for pNLenv. A variant of pNL4-3 containing mutations in the tat and rev genes (pNL4-3tr−) was used for insertion of the BGI sequence into the 5′ UTR (Figure 7A), and Tat and Rev expression vectors were cotransfected as indicated. For insertion into the 3′ region, a pNL4-3 derivative containing a CMV promoter driving HIV-1 transcription [pNLC4-3; (44)] and with mutations in the tat and rev genes was used (Figure 7A). Transfection of pNLC4-3 was previously shown to cause production of infectious virus with a similar efficiency as pNL4-3 (44). Figure 7B indicates that similar amounts of Gag-derived products were observed when transfection of pNLC4-3 was compared with transfection of pNLC4-3 tr− and pNL4-3BGBtr− in the presence of Tat and Rev (Figure 7B, upper panel, compare lane 2 with lanes 4 and 6). No Gag-specific proteins were observed in the absence of Rev (Figure 7B, upper panel, lanes 3 and 5). This result suggested that the BGI sequence in the 5′ UTR of the HIV-1 genomic RNA was efficiently removed; similar to what had been observed for pNLenvBGB. Immunoblot analysis with antisera against HIV-1 Env (Figure 7B, middle panel) and Nef (Figure 7B, lower panel) confirmed that protein expression from singly spliced RNAs and RNAs that were spliced at multiple sites was unaffected by insertion of the BGI sequence. Northern blot analysis of transfected cells revealed the expected pattern of three classes of RNA, corresponding to the genomic RNA, singly spliced RNAs and RNAs that were spliced at multiple sites in the case of pNLC4-3 (Figure 7C, lane 2). A similar pattern was observed for pNLC4-3tr− and for pNL4-3BGBtr− in the presence of Tat and Rev (Figure 7C, lanes 4 and 6). The genomic RNA in the case of pNL4-3BGBtr− was observed in the same position as for pNLC4-3tr− and hybridization of the blot with a BGI-specific probe detected no signal for a BGI-containing RNA in this case (Figure 7D, lane 6). The background signal in lanes 2, 4 and 6 is derived from the HIV-specific probe used in the first hybridization. This result shows that the BGI-sequence in the 5′UTR was rapidly and completely spliced as had been observed for pNLenvBGB.
Figure 7.
Analysis of the splicing of a heterologous intron in the complete HIV-1 genome. (A) Schematic depiction of the proviral constructs containing the BGI sequence in the 5′ region (pNL4–3BGBtr−; top) or in the 3′ LTR (pNLC4–3BGItr−, bottom). The BGI sequence was inserted into the same positions as described for pNLenv. It is noted that the proviral clones with mutations in the tat and rev genes were used and Tat and Rev expression plasmids were cotransfected as indicated. In the case of pNLC4–3BGItr−, a derivative of pNL4-3 containing a cytomegalovirus promoter instead of the U3 region of the 5′ LTR was used (pNLC4-3). This construct had been shown to produce an infectious RNA with a titer comparable to pNL4-3 (44). (B) Hela P4 cells were transfected with the plasmids indicated above each lane. Cell extracts were analysed by western blot using an antiserum against the HIV-1 Gag derived capsid protein (CA). This antiserum detects the Gag precursor Pr55, several cleavage intermediates and the completely cleaved CA protein. Antisera against Env and Nef were used in the middle and lower panels, respectively. Viral proteins are marked on the right as in Figure 2B. (C) Northern blot analysis of RNA isolated from the same transfections as in panel B. The blot was hybridized with a LTR-specific probe as indicated in panel A. (D) Re-hybridization of the same blot as in panel C with a BGI (upper panel) and a GAPDH-specific (lower panel) probe. The positions of unspliced, singly spliced and completely spliced RNAs are shown on the right.
The insertion of the BGI sequence into the 3′ region of the HIV-1 genome also did not lead to altered Gag expression (Figure 7B, upper panel, lane 8), but led to reduced levels of Env and Nef compared with wild type and pNLC4-3tr− (Figure 7B, middle and lower panels, lanes 2, 4, 7 and 8). Northern blot analysis with a HIV-1 specific probe revealed a similar pattern as observed for pNLC4-3tr− but hybridization of the blot with a BGI-specific probe showed that the heterologous intron in the 3′ region of the HIV-1 genome was retained (Figure 7D, lane 8). The BGI sequence was detected in the unspliced genomic, as well as in the singly spliced RNA, while no signal was observed for the completely spliced RNAs. The presence of the BGI in the 3′ region also leads to reduced levels of singly and completely spliced RNAs (Figure 7C, lane 8) and consequently to lower expression levels for Env and Nef (Figure 7B, middle and lower panels, lanes 8). The reason for this difference, which was not observed for the construct with intact tat and rev genes (data not shown), is currently not clear. Retention of the 3′ intron was further supported by analysing proviral DNA after infection of a T cell line with an infectious virus carrying the BGI sequence. PCR analysis of cells infected for 6 days revealed a strong signal for genomes that retained the BGI sequence in the 3′ region and a very weak signal for genomes that had lost this sequence by splicing (supplementary Figure S1). Taken together, these data indicate that splicing of the heterologous intron in the 3′ region of HIV-1 genomic RNA depends on removal of all upstream introns as had been observed for pNLenv.
Mutation of 5′ss #1 in an infectious proviral HIV-1 plasmid abolishes downstream splicing
If the observation that the splicing of a downstream intron is dependent on removal of upstream introns holds true for the HIV-1 splice signals, it can be predicted that mutation of the first 5′ss should completely abolish HIV-1 splicing. To test this hypothesis, three point mutations were introduced into the 5′ss #1 in the HIV-1 proviral clone pNL4-3 (M3; Figure 8A). As before, a derivative of pNL4-3 with mutations in the tat and rev genes was used for these experiments and Tat and Rev expression vectors were cotransfected as indicated. The M3 mutation introduces three alterations into the binding site for U1snRNP (Figure 8A) and should thus abolish U1 binding and splicing. A cryptic 5′ss 4 nt downstream of the actual splice site was mutated in addition [Figure 8A, (8)]. Figure 8B shows northern blot analysis of HeLaP4 cells transfected with the indicated plasmids. Transfection of pNL4-3 yielded the described pattern of genomic RNA (∼9 kb), singly spliced RNA (∼4 kb) and completely spliced RNA (∼2 kb) (Figure 8B, lane 2). A very similar pattern was observed for pNL4-3tr− in the presence of Rev (Figure 8B, lane 4), while completely spliced RNA was the main product in the absence of Rev (Figure 8B, lane 3) as previously reported (14). In contrast, only the unspliced genomic RNA was detected in the case of pNL4-3 M3tr− (Figure 8B, lanes 5 and 6). There was a strong signal for the genomic RNA in the presence of Rev (Figure 8B, lane 6), which was significantly reduced if Rev was lacking (Figure 8B, lane 5), consistent with Rev stabilizing this RNA by facilitating its nuclear export (14,57). No signals corresponding to singly spliced RNAs or RNAs that were spliced at multiple sites, were observed for pNL4-3M3tr− in the presence or absence of Rev, indicating that all downstream splice events of HIV-1 are dependent on the removal of upstream introns as predicted from the results with heterologous introns.
DISCUSSION
In this report, we provide evidence that splicing of HIV-1 involves the ordered removal of introns from the 5′ end of the genomic RNA, and that this process is mediated by 3′ splice inhibition. This inhibition was also seen for heterologous introns independent of the quality of their splice signals. We observed the removal of native and heterologous introns from HIV-1 RNAs only when the respective intron was located in the 5′ proximal position on this RNA. Splicing of 3′ located introns was completely dependent on the removal of all upstream introns. Accordingly, mutation of the 5′ss #1 in the 5′ UTR of the HIV-1 genome completely abolished splicing despite the presence of three additional 5′ss and seven 3′ss on this RNA, which are normally used in HIV-1 splicing.
Given that splicing is a co-transcriptional event and splice sites are recognized as they are transcribed (58,59), it may not appear surprising that intron removal proceeds in a 5′ to 3′ direction. Several reports indicated, however, that splicing of cellular pre-mRNAs does not follow a strict 5′ to 3′ order and 3′ introns may be removed before the 5′ proximal introns. Thus, splice site recognition and intron-removal do not necessarily occur in the same order (30–33). This also holds true for intron 2 of the β-globin gene, which was used as a heterologous intron in the current study. This intron has been shown to be spliced prior to the upstream intron 1 in its native context (60), while splicing was inhibited when it was placed into the 3′ region, but not the 5′ region of the HIV-1 genome. It should also be considered that splicing of cellular pre-mRNAs is normally a very rapid event and very little, if any, intron containing RNAs are observed under steady-state conditions (16). In contrast, HIV-1 splicing is necessarily inefficient although the same machinery is used (3). This leads to the nuclear accumulation of unspliced and singly spliced RNAs, which can either be further spliced, exported via the Rev/Crm1-dependent pathway or degraded. Thus, in the case of HIV-1, there is a sustained nuclear pool of intron-containing RNAs, which could undergo further splicing. This would make it more likely that splicing deviates from a strict 5′ to 3′ order than in the case of rapidly spliced cellular mRNAs, while the opposite is in fact observed. This may be important for viral gene expression (see below).
Splicing of the heterologous BGI sequence in the 3′ region of HIV-1 was dependent on the removal of all upstream introns, both in a subgenomic expression vector and in the HIV-1 genome. This required a single splice event in the case of pNLenv, while removal of two introns—the gag–pol and the env intron—was required in case of the complete genome. Northern blot analysis revealed that the BGI sequence was retained on the unspliced genomic and on the singly spliced RNA, while it was completely removed from all RNAs that were spliced at multiple sites. Thus, splicing of the efficient heterologous intron was apparently suppressed on the sustained pool of nuclear incompletely spliced RNAs, while it appeared to be activated upon removal of all upstream introns. This inhibition could not be relieved by mutation of the upstream splice signals, and splicing of the BGI in the 3′ region was still suppressed when the 5′ ss of the single upstream intron was mutated. Thus, activation of downstream splicing in the case of HIV-1 appears to require prior splicing of all upstream introns. Accordingly, the BGI-sequence was efficiently and completely spliced when placed into the 5′ UTR of the subgenomic vector or the proviral genome and no trace of BGI-containing sequence was detected.
Inhibition of 3′ BGI splicing was not dependent on the cryptic tev splices sites present in the env-intron (55,61) or on the length of the intron. Removal of a large part of the env region, including the cryptic splice signals did not alter the splicing pattern, confirming previous results from Cochrane et al. that intronic sequences do not influence the inefficiency of 3′ss #7 (15). Furthermore, placing the very efficient synthetic MINX intron (56) into the same position of the subgenomic vector yielded a very similar result. This intron is short (120 nt) with optimal consensus splice signals and is normally rapidly and autonomously spliced even in a heterologous context (62). These results suggest that cis-acting elements in the HIV-1 genome override efficient splice signals, thereby suppressing the fast removal of the intron in a position-dependent manner. This inhibition is not a property of the respective intron and can be overcome if the upstream intron is spliced. The same effect is also observed in the case of the authentic HIV-1 introns, where splicing of all downstream introns requires prior removal of the gag–pol intron. Based on these results, we reasoned that a mutation affecting only the major 5′ss in the HIV-1 genome should abolish all splicing. This was indeed the case and was also observed when the BGI-sequence was placed into the 3′ region of a HIV-1 genome with the M3 mutation in 5′ss #1 (data not shown). Thus, splicing of all downstream HIV-1 (or heterologous) introns is completely dependent on removal of the 5′ gag–pol intron and functional downstream splice sites are not utilized unless they are placed into the 5′ proximal position.
One indication of how HIV-1 splicing is regulated can be deduced from the observation of an additional spliced product when the BGI sequence was placed into the 3′ region of HIV-1. This product is derived from splicing at 5′ss #4 and the 3′ss of the globin intron, thus deleting the sequences between 3′ss #7 of HIV-1 and the 3′ss of the BGI. This corresponds to the skipping of a newly introduced exon (3′ss #7 to BGI 5′ss), which may be due to weak exon definition (63) because of inefficient recognition of 3′ss #7. This 3′ss exhibits a weak polypyrimidine tract (15), contains a U-residue instead of A as a branchpoint (19) and is further regulated through adjacent splice enhancers and silencers (22,26,50,64,65). According to a mechanism proposed by Branlant (65) and Krainer (22), splice enhancer sequences help to define 3′ss #7 through recruitment of splice factors (e.g. ASF/SF2) and subsequent binding of U2AF to the 3′ss. Alternatively, binding of hnRNPA1 to splice silencers, prevents the use of 3′ss #7 (65) and possibly of downstream splice signals. In this way, 3′ss #7 is only used on the part of the transcripts giving rise to separate pools of singly spliced and completely spliced RNAs, while the BGI sequence is only spliced when the exon is defined through recognition of 3′ss #7 or when this exon is skipped. However, exon definition involving 3′ss #7 cannot explain suppression of all 3′ introns in the HIV-1 genome because mutation of 5′ss #1 abolished all splice events despite the presence of functional downstream introns. It appears likely; therefore, that additional crosstalk across the introns occurs.
A simple explanation for sequential splicing would be that the splice signals on the genomic RNA are sorted by their efficiency, having the most efficient splice sites at the 5′ end. However, this is not the case, since all 5′ss on the HIV-1 genome closely match the consensus sequence, while 3′ss are weak (8,17). Furthermore, insertion of introns with optimal splice signals into the 3′ region cannot overcome the inhibition. Another interesting observation is that all HIV 5′ and 3′ ss are arranged in an alternating order where an inefficient 3′ss is paired with an efficient 5′ss. If and how this contributes to regulation of sequential splicing is the subject of ongoing research.
From the pattern of viral gene expression, it appears likely that the sequential splicing of HIV-1 genomic RNA is biologically significant. As stated above, viral protein synthesis and production of infectious particles critically depends on inefficient splicing, which yields stable pools of unspliced, singly spliced and completely spliced RNAs. Removal of the 5′ proximal intron(s) is required to produce mRNAs for the viral proteins Env, Vpr, Vif, Vpu, Tat, Rev and Nef. If the 3′ intron would be removed prior to the 5′ intron(s) in these cases, the respective RNAs would likely be subject to NMD (39). Thus, sequential removal of introns from the 5′ end of the HIV-1 genomic RNA may be critical to maintain a high and balanced viral gene expression and may be an intrinsic property of retroviral RNAs.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank P. Krammer, H. Schaal, K. Harbers and V. Bosch for reagents, and J. Tazi for helpful discussion. Jens Bohne was supported by the Boehringer Ingelheim Fond and DFG grant 1837/Ba4. Funding to pay the Open Access publication charges for this article was provided by institutional funding, University of Heidelberg.
REFERENCES
- 1.Coffin J.M., Hughes S.H., Varmus H.E. In: Retroviruses. Coffin J.M., editor. Cold Spring Harbor, NY: 1997. [PubMed] [Google Scholar]
- 2.Frankel A.D., Young J.A. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 1998;67:1–25. doi: 10.1146/annurev.biochem.67.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Pollard V.W., Malim M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 4.Wodrich H., Krausslich H.G. Nucleocytoplasmic RNA transport in retroviral replication. Results Probl. Cell Differ. 2001;34:197–217. doi: 10.1007/978-3-540-40025-7_12. [DOI] [PubMed] [Google Scholar]
- 5.Chang D.D., Sharp P.A. Regulation by HIV Rev depends upon recognition of splice sites. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 6.Nakielny S., Fischer U., Michael W.M., Dreyfuss G. RNA transport. Annu. Rev. Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- 7.Legrain P., Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 8.Purcell D.F., Martin M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz S., Felber B.K., Benko D.M., Fenyo E.M., Pavlakis G.N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daly T.J., Cook K.S., Gray G.S., Maione T.E., Rusche J.R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature. 1989;342:816–819. doi: 10.1038/342816a0. [DOI] [PubMed] [Google Scholar]
- 11.Fischer U., Huber J., Boelens W.C., Mattaj I.W., Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 12.Fornerod M., Ohno M., Yoshida M., Mattaj I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 13.Malim M.H., Hauber J., Le S.Y., Maizel J.V., Cullen B.R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 14.Malim M.H., Cullen B.R. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RNA processing in eukaryotes. Mol. Cell. Biol. 1993;13:6180–6189. doi: 10.1128/mcb.13.10.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staffa A., Cochrane A. The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3′ splice site. J. Virol. 1994;68:3071–3079. doi: 10.1128/jvi.68.5.3071-3079.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Izaurralde E., Mattaj I.W. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 17.O'Reilly M.M., McNally M.T., Beemon K.L. Two strong 5′ splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology. 1995;213:373–385. doi: 10.1006/viro.1995.0010. [DOI] [PubMed] [Google Scholar]
- 18.Amendt B.A., Si Z.H., Stoltzfus C.M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dyhr-Mikkelsen H., Kjems J. Inefficient spliceosome assembly and abnormal branch site selection in splicing of an HIV-1 transcript in vitro. J. Biol. Chem. 1995;270:24060–25066. doi: 10.1074/jbc.270.41.24060. [DOI] [PubMed] [Google Scholar]
- 20.Si Z., Amendt B.A., Stoltzfus C.M. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 1997;25:861–867. doi: 10.1093/nar/25.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Si Z.H., Rauch D., Stoltzfus C.M. The exon splicing silencer in human immunodeficiency virus type 1 Tat exon 3 is bipartite and acts early in spliceosome assembly. Mol. Cell. Biol. 1998;18:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J., Mayeda A., Krainer A.R. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol. Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 23.Ropers D., Ayadi L., Gattoni R., Jacquenet S., Damier L., Branlant C., Stevenin J. Differential effects of the SR proteins 9G8, SC35, ASF/SF2, and SRp40 on the utilization of the A1 to A5 splicing sites of HIV-1 RNA. J. Biol. Chem. 2004;279:29963–29973. doi: 10.1074/jbc.M404452200. Epub 2004 Apr 27. [DOI] [PubMed] [Google Scholar]
- 24.Zahler A.M., Damgaard C.K., Kjems J., Caputi M. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J. Biol. Chem. 2004;279:10077–10084. doi: 10.1074/jbc.M312743200. Epub 2003 Dec 31. [DOI] [PubMed] [Google Scholar]
- 25.Caputi M., Mayeda A., Krainer A.R., Zahler A.M. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tange T.O., Damgaard C.K., Guth S., Valcarcel J., Kjems J. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 2001;20:5748–5758. doi: 10.1093/emboj/20.20.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Domsic J.K., Wang Y., Mayeda A., Krainer A.R., Stoltzfus C.M. Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol. Cell. Biol. 2003;23:8762–8772. doi: 10.1128/MCB.23.23.8762-8772.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beyer A.L., Osheim Y.N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 29.Neugebauer K.M. On the importance of being co-transcriptional. J. Cell. Sci. 2002;115:3865–3871. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 30.Wetterberg I., Bauren G., Wieslander L. The intranuclear site of excision of each intron in Balbiani ring 3 pre-mRNA is influenced by the time remaining to transcription termination and different excision efficiencies for the various introns. RNA. 1996;2:641–651. [PMC free article] [PubMed] [Google Scholar]
- 31.Kessler O., Jiang Y., Chasin L.A. Order of intron removal during splicing of endogenous adenine phosphoribosyltransferase and dihydrofolate reductase pre-mRNA. Mol. Cell. Biol. 1993;13:6211–6222. doi: 10.1128/mcb.13.10.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gudas J.M., Knight G.B., Pardee A.B., Matsumoto K., Wassarman K.M., Wolffe A.P. Ordered splicing of thymidine kinase pre-mRNA during the S phase of the cell cycle. Mol. Cell. Biol. 1990;10:5591–5595. doi: 10.1128/mcb.10.10.5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Shen L., Najafi H., Kolberg J., Matschinsky F.M., Urdea M., German M., Luo Z.D., Camp S., Mutero A., Taylor P., Gudas J.M., Knight G.B., Pardee A.B., Matsumoto K., Wassarman K.M., Wolffe A.P., Schell T., Kulozik A.E., Hentze M.W., Maquat L.E., Dittmar M.T., Eichler S., Reinberger S., Henning L., Krausslich H.G., Lang K.M., Spritz R.A., Westendorp M.O., Li-Weber M., Frank R.W., Krammer P.H. Regulation of insulin preRNA splicing by glucose. Proc. Natl Acad. Sci. USA. 1997;94:4360–4365. doi: 10.1073/pnas.94.9.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo M.J., Reed R. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl Acad. Sci. USA. 1999;96:14937–14942. doi: 10.1073/pnas.96.26.14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nott A., Le Hir H., Moore M.J., Wiegand H.L., Lu S., Cullen B.R. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiegand H.L., Lu S., Cullen B.R. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl Acad. Sci. USA. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. Epub 2003 Sep 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumoto K., Wassarman K.M., Wolffe A.P. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J., Sun X., Qian Y., LaDuca J.P., Maquat L.E. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hentze M.W., Kulozik A.E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 40.Schell T., Kulozik A.E., Hentze M.W., Maquat L.E., Dittmar M.T., Eichler S., Reinberger S., Henning L., Krausslich H.G., Lang K.M., Spritz R.A., Westendorp M.O., Li-Weber M., Frank R.W., Krammer P.H. Integration of splicing, transport and translation to achieve mRNA quality control by the nonsense-mediated decay pathway. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-3-reviews1006. REVIEWS1006. Epub 2002 Feb 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adachi A., Gendelman H.E., Koenig S., Folks T., Willey R., Rabson A., Martin M.A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dittmar M.T., Eichler S., Reinberger S., Henning L., Krausslich H.G. A recombinant virus assay using full-length envelope sequences to detect changes in HIV-1 co-receptor usage. Virus Genes. 2001;23:281–290. doi: 10.1023/a:1012569206007. [DOI] [PubMed] [Google Scholar]
- 43.Welker R., Harris M., Cardel B., Krausslich H.G. Virion incorporation of human immunodeficiency virus type 1 Nef is mediated by a bipartite membrane-targeting signal: analysis of its role in enhancement of viral infectivity. J. Virol. 1998;72:8833–8840. doi: 10.1128/jvi.72.11.8833-8840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bohne J., Krausslich H.G. Mutation of the major 5′ splice site renders a CMV-driven HIV-1 proviral clone Tat-dependent: connections between transcription and splicing. FEBS Lett. 2004;563:113–118. doi: 10.1016/S0014-5793(04)00277-7. [DOI] [PubMed] [Google Scholar]
- 45.Westendorp M.O., Li-Weber M., Frank R.W., Krammer P.H. Human immunodeficiency virus type 1 Tat upregulates interleukin-2 secretion in activated T cells. J. Virol. 1994;68:4177–4185. doi: 10.1128/jvi.68.7.4177-4185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen C., Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wodrich H., Bohne J., Gumz E., Welker R., Krausslich H.G. A new RNA element located in the coding region of a murine endogenous retrovirus can functionally replace the Rev/Rev-responsive element system in human immunodeficiency virus type 1 Gag expression. J. Virol. 2001;75:10670–10682. doi: 10.1128/JVI.75.22.10670-10682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weil D., Boutain S., Audibert A., Dautry F. Mature mRNAs accumulated in the nucleus are neither the molecules in transit to the cytoplasm nor constitute a stockpile for gene expression. RNA. 2000;6:962–975. doi: 10.1017/s1355838200000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kammler S., Leurs C., Freund M., Krummheuer J., Seidel K., Tange T.O., Lund M.K., Kjems J., Scheid A., Schaal H. The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA. 2001;7:421–434. doi: 10.1017/s1355838201001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tange T.O., Kjems J. SF2/ASF binds to a splicing enhancer in the third HIV-1 tat exon and stimulates U2AF binding independently of the RS domain. J. Mol. Biol. 2001;312:649–662. doi: 10.1006/jmbi.2001.4971. [DOI] [PubMed] [Google Scholar]
- 51.Kimpton J., Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mader S., White J.H. A steroid-inducible promoter for the controlled overexpression of cloned genes in eukaryotic cells. Proc. Natl Acad. Sci. USA. 1993;90:5603–5607. doi: 10.1073/pnas.90.12.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buchman A.R., Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol. Cell. Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki Y., Ishihara D., Sasaki M., Nakagawa H., Hata H., Tsunoda T., Watanabe M., Komatsu T., Ota T., Isogai T., Suyama A., Sugano S. Statistical analysis of the 5′ untranslated region of human mRNA using ‘Oligo-Capped’ cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- 55.Caputi M., Zahler A.M. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zapp M.L., Berget S.M. Evidence for nuclear factors involved in recognition of 5′ splice sites. Nucleic Acids Res. 1989;17:2655–2674. doi: 10.1093/nar/17.7.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D'Agostino D.M., Felber B.K., Harrison J.E., Pavlakis G.N. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell. Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldstrohm A.C., Greenleaf A.L., Garcia-Blanco M.A. Co-transcriptional splicing of pre-messenger RNAs: considerations for the mechanism of alternative splicing. Gene. 2001;277:31–47. doi: 10.1016/s0378-1119(01)00695-3. [DOI] [PubMed] [Google Scholar]
- 59.Kotovic K.M., Lockshon D., Boric L., Neugebauer K.M. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol. Cell. Biol. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Audibert A., Weil D., Dautry F. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol. Cell. Biol. 2002;22:6706–6718. doi: 10.1128/MCB.22.19.6706-6718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hammarskjold M.L., Li H., Rekosh D., Prasad S. Human immunodeficiency virus env expression becomes Rev-independent if the env region is not defined as an intron. J. Virol. 1994;68:951–958. doi: 10.1128/jvi.68.2.951-958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thermann R., Neu-Yilik G., Deters A., Frede U., Wehr K., Hagemeier C., Hentze M.W., Kulozik A.E. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berget S.M. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 64.Suh D., Seguin B., Atkinson S., Ozdamar B., Staffa A., Emili A., Mouland A., Cochrane A. Mapping of determinants required for the function of the HIV-1 env nuclear retention sequence. Virology. 2003;310:85–99. doi: 10.1016/s0042-6822(03)00073-4. [DOI] [PubMed] [Google Scholar]
- 65.Marchand V., Mereau A., Jacquenet S., Thomas D., Mougin A., Gattoni R., Stevenin J., Branlant C. A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J. Mol. Biol. 2002;323:629–652. doi: 10.1016/s0022-2836(02)00967-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.