Abstract
Escherichia coli ribonuclease III (RNase III; EC 3.1.24) is a double-stranded(ds)-RNA-specific endonuclease with key roles in diverse RNA maturation and decay pathways. E.coli RNase III is a member of a structurally distinct superfamily that includes Dicer, a central enzyme in the mechanism of RNA interference. E.coli RNase III requires a divalent metal ion for activity, with Mg2+ as the preferred species. However, neither the function(s) nor the number of metal ions involved in catalysis is known. To gain information on metal ion involvement in catalysis, the rate of cleavage of the model substrate R1.1 RNA was determined as a function of Mg2+ concentration. Single-turnover conditions were applied, wherein phosphodiester cleavage was the rate-limiting event. The measured Hill coefficient (nH) is 2.0 ± 0.1, indicative of the involvement of two Mg2+ ions in phosphodiester hydrolysis. It is also shown that 2-hydroxy-4H-isoquinoline-1,3-dione—an inhibitor of ribonucleases that employ two divalent metal ions in their catalytic sites—inhibits E.coli RNase III cleavage of R1.1 RNA. The IC50 for the compound is 14 μM for the Mg2+-supported reaction, and 8 μM for the Mn2+-supported reaction. The compound exhibits noncompetitive inhibitory kinetics, indicating that it does not perturb substrate binding. Neither the O-methylated version of the compound nor the unsubstituted imide inhibit substrate cleavage, which is consistent with a specific interaction of the N-hydroxyimide with two closely positioned divalent metal ions. A preliminary model is presented for functional roles of two divalent metal ions in the RNase III catalytic mechanism.
INTRODUCTION
The processing of double-stranded (ds) RNA is an essential step in the expression and regulation of many eukaryotic and bacterial genes (1–3). Members of the ribonuclease III (RNase III) superfamily are primarily responsible for cleavage of dsRNA, and participate in the maturation and degradation of coding and noncoding RNAs (1–3). The eukaryotic RNase III ortholog Dicer initiates RNA interference by processing dsRNAs to 21–23 bp short interfering (si) RNAs (4–6). The siRNAs in turn are incorporated into a macromolecular complex that identifies and degrades target RNAs containing sequences complementary to the siRNAs (7–9). Mammalian Dicer, in conjunction with the RNase III ortholog Drosha, cleaves precursors to microRNAs, which regulate translation in a cistron-specific manner (10–12).
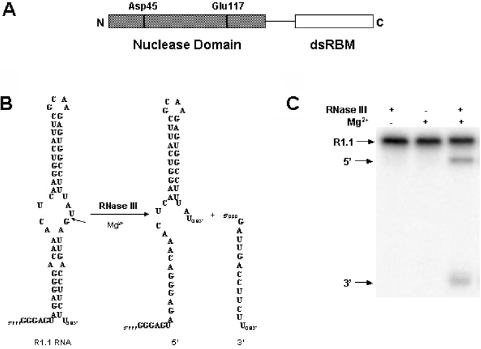
Much of what is currently known of the mechanisms of substrate recognition and cleavage by RNase III orthologs has been provided by studies on the Escherichia coli enzyme [reviewed in (1,3,13–15)]. Cellular and viral substrates of E.coli RNase III typically possess a dsRNA element of ∼20 bp and are cleaved in a site-specific manner which is required for the proper function of the mature species, or for efficient degradation. E.coli RNase III functions as a homodimer, with the subunit polypeptide (226 amino acids) containing an N-terminal nuclease domain and a C-terminal dsRNA-binding domain (dsRBD) (Figure 1A). The latter domain consists of a single copy of the dsRNA-binding motif (dsRBM), which is present in many other dsRNA-binding proteins (16–17). The dsRBD is important for RNase III activity in vivo (18) and under standard reaction conditions in vitro (19). However, a truncated version of RNase III containing only the nuclease domain retains the ability to cleave substrate in vitro in the presence of low salt and Mn2+ ion (19). The nuclease domain contains highly conserved residues important for activity, including an aspartic acid at position 45 and a glutamic acid at position 117 (Figure 1A). A crystallographic analysis of the nuclease domain of Aquifex aeolicus RNase III reveals a homodimeric structure, with the subunit polypeptide exhibiting a novel, all-alpha fold (20).
Figure 1.
E.coli ribonuclease III and the phage T7 substrate, R1.1 RNA. (A) Domain structure of the E.coli RNase III polypeptide. The positions of several conserved residues in the nuclease domain are indicated. (B) Sequence and structure of R1.1 RNA. The secondary structure is that originally proposed by Dunn and Studier (31). The cleavage site is indicated by the arrow, with the two products of cleavage 47 and 13 nt in size. (C) Gel electrophoretic pattern of R1.1 RNA cleavage, showing the requirement for Mg2+. Experimental conditions involved incubation of internally 32P-labeled R1.1 RNA (synthesized using [α-32P]UTP) with RNase III for 5 min at 37°C (see Materials and Methods). The reaction products were separated by electrophoresis in a 15% polyacrylamide gel and then visualized by phosphorimaging. The partial reaction displayed is similar to the gel electrophoretic patterns analyzed in the kinetic analyses (see Results).
The catalytic mechanism of E.coli RNase III is only partly defined. A divalent metal ion is a required cofactor, with Mg2+ most likely the physiologically relevant species (21–23). Mn2+, Co2+ and Ni2+ also support catalysis, while Ca2+, Zn2+ and Sr2+ are inactive (23–25). The nuclease domain of A.aeolicus RNase III, crystallized in the presence of Mg2+ or Mn2+, reveals a single metal ion bound to each of the two active sites (20,26). However, the structural data do not shed light on the number of metal ions needed for catalysis. Moreover, the probable occurrence of two active sites in the holoenzyme prompts the question whether one or more metal ions must bind to both sites to support activity. To address these questions, we present kinetic- and inhibitor-based analyses of the divalent metal ion dependence of E.coli RNase III. The data are used to develop a preliminary model for divalent metal ion function in the RNase III catalytic mechanism.
MATERIALS AND METHODS
Materials
Water was deionized and distilled. Chemicals and reagents were of molecular biology grade and were obtained from Sigma (St Louis, MO) or Fisher Scientific (Chicago, IL). Standardized 1 M solutions of MgCl2 and MnCl2 were obtained from Sigma. The radiolabeled ribonucleoside 5′-triphosphates [α-32P]CTP (3000 Ci/mmol) and [α-32P]UTP (3000 Ci/mmol) were from Perkin-Elmer (Boston, MA), while unlabeled ribonucleoside 5′-triphosphates were from Amersham-Pharmacia Biotech (Piscataway, NJ). T7 RNA polymerase was purified in-house as described in (27). 2-hydroxy-4H-isoquinoline-1,3-dione was synthesized according to Edafiogho and coworkers (28) using homophthalic anhydride as starting material. 2-Methoxy-1,3(2H,4H)-isoquinolinedione was synthesized according to Bouzide (29) using 2-hydroxy-4H-isoquinoline-1,3-dione as starting material. 1,3(2H,4H)-Isoquinolinedione was synthesized according to Crockett and coworkers (30). Stock solutions of the compounds were prepared in ethanol and stored at −20°C.
Protein overexpression and purification
Purification of N-terminal (His)6-tagged RNase III is described elsewhere (25). Briefly, enzyme was overproduced in IPTG-treated E.coli BL21(DE3)rnc105, recA cells containing plasmid pET-15b(rnc) (25), and purified from the supernatant fraction of sonicated cell lysates using an immobilized Ni2+ affinity column (HisBind resin, Novagen). Protein was extensively dialyzed against 1 M NaCl, 60 mM Tris–HCl (pH 7.9) and stored at −20°C in a 50% glycerol-containing buffer, consisting of 0.5 M NaCl, 30 mM Tris–HCl (pH 7.9), 0.5 mM DTT and 0.5 mM EDTA. Protein concentrations were determined by the Bradford assay, using bovine serum albumin as the standard. The (His)6 tag was not removed, as it has only a minor effect on activity (25). For convenience, (His)6-RNase III will be referred to in this report as RNase III.
Substrate synthesis and purification
R1.1 RNA (Figure 1B) is a small substrate that corresponds to the R1.1 processing signal within the phage T7 polycistronic early mRNA precursor (31,32). R1.1 RNA was enzymatically synthesized and purified as described in (25,33). Internally, 32P-labeled R1.1 RNA was prepared by including [α-32P]CTP or [α-32P]UTP in the transcription reaction. The specific activity of 32P-labeled R1.1 RNA was 100 Ci/mol, and was stored at −20°C in 10 mM Tris–HCl (pH 7.5), 1 mM EDTA.
Substrate cleavage assay
Cleavage assays were performed essentially as described (19,25,34) using internally 32P-labeled R1.1 RNA, purified RNase III (see above) and buffer consisting of 160 mM NaCl, 30 mM Tris–HCl (either pH 6.8 or 7.9, as indicated), 5 mM spermidine, 0.1 μg/ml tRNA, 0.1 mM DTT and 0.1 mM EDTA. The salt concentration was identical in all assays. Reactions were initiated by addition of either RNase III or divalent metal ion to the mixture, which was pre-incubated at 37°C for 5 min. Essentially, the same cleavage rates were obtained by either method. Reactions were allowed to proceed for 10 s at 37°C and stopped by adding EDTA (20 mM final concentration). Other specific experimental conditions are provided in the appropriate figure legends. Reactions were electrophoresed as described previously (19,25,34) in a 15% polyacrylamide gel containing TBE buffer and 7 M urea, and were visualized by phosphorimaging (Amersham Biosciences Storm 860 system or Typhoon 9400 system). Substrate cleavage rates were measured using ImageQuant software. Kinetic parameters were determined by curve fitting using KaleidaGraph, v3.5 (Synergy Software).
RESULTS
Kinetic analysis of the Mg2+ concentration dependence of RNase III cleavage of R1.1 RNA
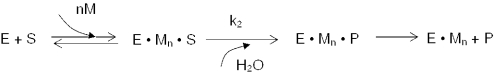
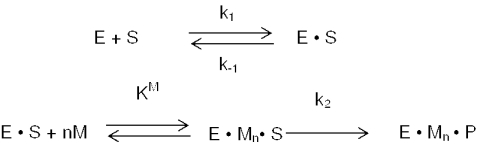
E.coli RNase III cleavage of substrate follows Michaelis–Menten kinetics (Scheme 1) (23,24). In this scheme, E is RNase III; S, substrate; M, Mg2+, n, the number of Mg2+ ions and P, products.
Scheme 1.
The rate of substrate cleavage exhibits an apparent hyperbolic dependence on the Mg2+ concentration, with maximal activity attained by ∼5 mM Mg2+ and a KMg (pH 7.5) of ∼0.5 mM (34). Scheme 2 (see above) incorporates the observations that RNase III can bind substrate in the absence of divalent metal ion, and that substrate binding may promote metal ion binding (24,34,35).
Scheme 2.
In this scheme, KM is the metal ion dissociation constant for the enzyme–substrate complex. The two schemes provide a kinetic approach to determine the number of Mg2+ ions required for catalysis. Since the rate constant (k1) for substrate binding to E.coli RNase III is ∼1 × 108 M−1 s−1 (24), and since the apparent dissociation constant (K′d = k−1/k1) of the RNase III–substrate complex is ∼2 nM (34), the calculated dissociation rate constant (k−1) is 0.18 s−1. A pulse-chase kinetic study of RNase III cleavage of R1.1 RNA (24) revealed that the rate constant for the cleavage step (k2) is >10-fold slower than the dissociation rate constant (k−1). Based on these observations, experimental conditions were chosen such that the pseudo first-order rate constant for substrate binding is substantially greater than the cleavage rate constant. These conditions would provide kobs values that reflect only the rate of the chemical step. Also, use of single-turnover reaction conditions served to eliminate any contribution to the rate of events subsequent to cleavage, product release in particular. Based on Scheme 1, and assuming rapid equilibration of substrate and metal ion binding to enzyme, Equation 1 describes the dependence of the observed rate constant on the divalent metal ion concentration:
| 1 |
Here, kobs (min−1) is the initial rate, normalized to the total enzyme concentration; k2 is the rate constant for phosphodiester bond cleavage; [M] is the divalent metal ion concentration; n is the number of divalent metal ions needed for hydrolysis; and KM the apparent metal ion dissociation constant for the enzyme–divalent metal ion–substrate complex. The best-fit curve of data to Equation 1 would indicate the number of divalent metal ions needed for catalysis, and would also provide the apparent metal dissociation constant as well as the rate constant for phosphodiester cleavage. Finally, the assumption in the kinetic schemes is that RNase III is catalytically active only when divalent metal ion occupies all functionally essential binding sites (i.e. an ‘all-or-none’ functional cooperativity of metal ion).
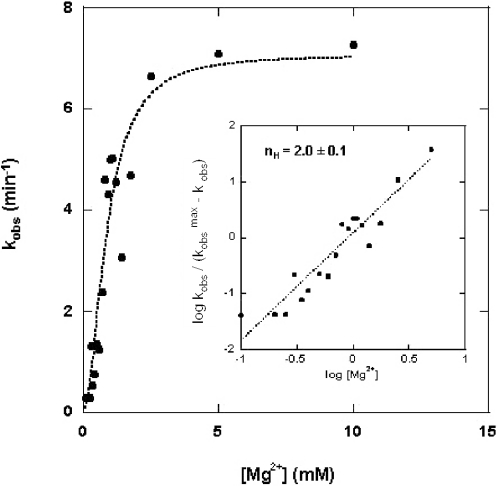
The rate of RNase III cleavage of an internally 32P-labeled model substrate was measured as a function of Mg2+ concentration at pH 6.8, and using conditions described above and in Materials and Methods. R1.1 RNA (Figure 1B) was used as substrate, since it undergoes cleavage at a single phosphodiester bond (31,32), and provides an easily quantifiable gel electrophoretic pattern (Figure 1C). Figure 2 shows the dependence of kobs on the Mg2+ concentration. The best-fit curve of the kinetic data to Equation 1 used a value of n = 2. The data were also subjected to a Hill analysis, according to Equation 2.
| 2 |
Here, nH (Hill coefficient) corresponds to the slope of the curve in the region of half-maximal reaction velocity. Assuming an ‘all-or-none’ functional cooperativity (see above), the Hill coefficient would indicate the number of divalent metal ions needed for the catalytic step (36,37). The inset diagram in Figure 2 is a plot of the kinetic data according to Equation 2. The best-fit line has a slope (nH value) of 2.0 ± 0.1 (Table 1). A similar kinetic analysis carried out at pH 7.9 provided an nH of 2.0 ± 0.1 (Table 1). In summary, the measured Hill coefficients indicate the involvement of two Mg2+ ions in the hydrolysis of the single scissile bond of R1.1 RNA.
Figure 2.
Mg2+ concentration dependence of the rate of substrate cleavage. Cleavage reactions involved internally 32P-labeled R1.1 RNA (1.1 μM) and RNase III (790 nM) (see Materials and Methods). Cleavage rates were based on the production of the 47 nt product (see Figure 1B and C). The kobs values (min−1) were determined and plotted as a function of the Mg2+ concentration according to Equation 1 (see Results). The best-fit curve used an n value of 2. The inset displays the Hill analysis of the kinetic data. Here, the kobs values were plotted as a function of the log of the Mg2+ concentration according to Equation 2 (see Results). The slope of the best-fit line is 2.0, with a standard error of ±0.1.
Table 1.
Hill coefficients and apparent Mg2+ dissociation constantsa
| pH 6.8 | pH 7.9 | |
|---|---|---|
| nH (Mg2+) | 2.0 ± 0.1 | 2.0 ± 0.1 |
| KMg (mM2) | 0.78 ± 0.18 | 0.16 ± 0.01 |
| KdMg(mM)b | 0.88 ± 0.04 | 0.40 ± 0.03 |
aCleavage assays were performed as described in Materials and Methods.
bThe apparent dissociation constant for Mg2+ (KdMg) is calculated as the square root of the KMg (see Equation 1) and assumes that the two Mg2+ ions that are required for cleavage bind with comparable affinities. The apparently stronger binding of Mg2+ at higher pH is consistent with the observed pH dependence of the cleavage rate at subsaturating Mg2+ concentrations (24).
N-hydroxyimide inhibition of Mg2+-supported cleavage of R1.1 RNA by RNase III
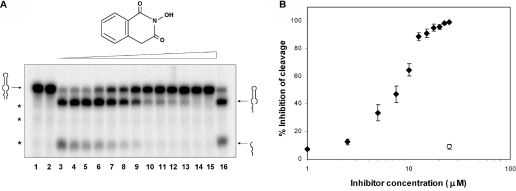
Specific N-hydroxyimides, including 2-hydroxy,4H-isoquinoline-1,3-dione (compound 1, see Figure 3A) are inhibitors of several ribonucleases that employ two divalent metal ions in their catalytic sites (38–40). It has been proposed that inhibition derives from a stereochemically compatible interaction of the N-hydroxy and keto functional groups with two divalent metal ions separated by ∼4 Å in the active site (38–40). There is evidence that the enol form of the compound interacts with the divalent metal ion (38). Given the mode of interaction of the compound with metal ion, 2-hydroxy,4H-isoquinoline-1,3-dione is a potentially informative probe of metal ion occupancy of enzyme active sites. We determined whether the N-hydroxyimide could inhibit RNase III cleavage of R1.1 RNA. A cleavage assay (Figure 3A) reveals a strong inhibitory effect of the N-hydroxyimide, with a concentration of 14 ± 2 μM providing half-maximal inhibition (Figure 3C). This value can be compared to the IC50 values for the influenza virus cap-dependent endoribonuclease (15 μM) (38), and for the HIV RNase H (0.4–0.6 μM) (39,40). Methylation of the hydroxyl group abolishes the inhibitory action of the N-hydroxyimide towards the other ribonucleases, presumably due to steric interference of metal ion binding (38,39). Here also, O-methylation relieves the inhibitory action of the N-hydroxyimide (Figure 3B and C). Finally, the corresponding imide (i.e. 1,3[2H,4H]-isoquinolinedione), which lacks the N-hydroxyl group, and which is expected to possess a weaker affinity for metal, also is an ineffective inhibitor (data not shown).
Figure 3.
N-hydroxyimide inhibition of R1.1 RNA cleavage by RNase III in the presence of Mg2+ ion. (A) Cleavage assay carried out in the presence of increasing concentrations of 2-hydroxy-4H-isoquinoline-1,3-dione (structure shown above gel image). The amount of internally 32P-labeled R1.1 RNA (synthesized using [α-32P]CTP) was 10–40 nmol, and the amount of RNase III was ∼100 fmol. MgCl2 (10 mM final concentration) was added to initiate the reaction, with a reaction time of 2 min at 37°C. Reactions were electrophoresed in a 15% polyacrylamide gel and visualized by phosphorimaging (see Materials and Methods for additional information). Lane 1 displays a reaction where substrate was incubated with RNase III in the absence of Mg2+. Lane 2 displays a reaction where substrate was incubated with Mg2+ in the absence of RNase III. Lane 3 is the complete reaction, but without added inhibitor. Lanes 4–13 display reactions carried out in the presence of 2.5, 5, 7.5, 10, 12.5, 15, 17.5, 20, 22.5 and 25 μM inhibitor, respectively. Lane 14 is a control reaction lacking inhibitor, but containing the same amount of ethanol as in the reaction in lane 13. The position of R1.1 RNA is shown on the left and the positions of the two product fragments are on the right. The asterisks indicate positions of small amounts of nonenzymatic breakdown products. (B) Cleavage assay carried out in the presence of increasing concentrations of 2-methoxy-1,3(2H,4H)-isoquinolinedione (structure shown above the gel image). The reactions in lanes 1–3 correspond to those of lanes 1–3 in the experiment shown in (A) (see above), and the reactions in lanes 4–8 contain 5, 10, 25, 100 and 250 μM of the compound, respectively. Lane 9 is a control reaction lacking the compound, but containing the same amount of ethanol as in the reaction in lane 8. (C) Graphic representation of the inhibitory action of the N-hydroxyimide. The solid triangles represent the percentage inhibition of cleavage by the N-hydroxyimide. The solid squares indicate inhibition by the O-methylated N-hydroxyimide. The open circle represents the effect of ethanol alone on the cleavage reaction.
Studies suggest that 2-hydroxy,4H-isoquinoline-1,3-dione inhibits ribonuclease activity in a reversible manner (38–40). To gain additional information on the mode of inhibition of RNase III, the dependence of the rate of cleavage of R1.1 RNA was determined as a function of the N-hydroxyimide concentration and substrate concentration. A double-reciprocal analysis (Figure 4) reveals noncompetitive inhibitory behavior. This mode of inhibition indicates that substrate recognition by RNase III under catalytic conditions is not affected by the N-hydroxymide. A gel mobility shift assay (data not shown) reveals that the N-hydroxyimide also does not cause dissociation of the R1.1 RNA–RNase III complex, formed under noncatalytic conditions. These lines of evidence indicate that the N-hydroxyimide inhibits substrate cleavage by a mechanism that does not involve inhibition of substrate binding (see also Discussion).
Figure 4.
Noncompetitive inhibitory behavior of 2-hydroxy-4H-isoquinoline-1,3-dione. The initial rate of cleavage of 5′-32P-labeled R1.1 RNA was measured in the presence of several concentrations of the N-hydroxyimide. The RNase III concentration was 10 nM and the buffer consisted of 160 mM NaCl, 10 mM MgCl2, 30 mM Tris–HCl (pH 7.9), 0.1 mM EDTA, 0.1 mM DTT, 5% glycerol and 5 μg/ml tRNA. The reaction was initiated by adding MgCl2 and quenched by adding EDTA (20 mM) final concentration. Reaction times were 0.5 and 1 min (37°C), and the N-hydroxyimide concentrations were 0, 10 and 20 μM (designated by the filled circles, open circles and filled triangles, respectively). The best-fit lines in the double-reciprocal plot exhibit different y-intercepts but share the same x-intercept, consistent with a noncompetitive (mixed) mode of inhibition (36,37) (see also Discussion). The Km and kcat values for the reaction in the absence of inhibitor are 14 nM and 2.5 min−1, respectively. These can be compared with values of 42 nM and 1.16 min−1, as determined in a separate study of R1.1 RNA cleavage kinetics (24).
N-hydroxyimide inhibition of Mn2+-supported cleavage of R1.1 RNA by RNase III
E.coli RNase III can use Mn2+ as a cofactor, with a concentration of ∼1–2 mM conferring optimal activity (21,34). The question arises whether a two metal ion mechanism is applicable to an alternative species. Since the determination of the Hill coefficient for the Mn2+-dependent cleavage reaction is precluded by the inhibitory effect of higher Mn2+ concentrations (21,34), we examined instead the effect of the N-hydroxyimide on cleavage of R1.1 RNA by RNase III in the presence of 2 mM Mn2+. A representative cleavage assay (Figure 5A) shows that the compound also inhibits the Mn2+-supported reaction, with an IC50 of 8 ± 1 μM (Figure 5B). Neither the O-methylated form of the N-hydroxyimide nor the unsubstituted imide inhibits cleavage to a significant extent (data not shown). The similarity of N-hydroxyimide inhibition of the Mn2+- and Mg2+-supported substrate cleavage reactions indicates that RNase III can also employ two Mn2+ ions in its catalytic mechanism.
Figure 5.
2-hydroxy-4H-isoquinoline-1,3-dione inhibition of R1.1 RNA cleavage by RNase III in the presence of Mn2+ ion. (A) Cleavage assay carried out in the presence of increasing concentrations of 2-hydroxy-4H-isoquinoline-1,3-dione (structure shown above gel image). The amount of internally 32P-labeled R1.1 RNA (synthesized using [α-32P]UTP) was 10–40 nmol and the amount of RNase III was 100 fmol. MnCl2 (2 mM final concentration) was added to initiate the reaction, with a reaction time of 2 min. Reactions were electrophoresed in a 15% polyacrylamide gel and visualized by phosphorimaging (see Materials and Methods for additional information). Lane 1 displays a reaction where substrate was incubated with RNase III in the absence of Mn2+. Lane 2 displays a reaction where substrate was incubated with Mn2+ in the absence of RNase III. Lane 3 is the complete reaction, lacking inhibitor. Lanes 4–15 display reactions carried out in the presence of 0.5, 1, 2.5, 5, 7.5, 10, 12.5, 15, 17.5, 20, 22.5 and 25 μM inhibitor, respectively. Lane 16 displays a control reaction lacking inhibitor, but containing the same amount of ethanol as in the reaction in lane 15. The position of R1.1 RNA is shown on the left and the positions of the two product fragments are indicated on the right. (B) Graphic representation of the inhibitory action of the N-hydroxyimide on the Mn2+-supported cleavage reaction. The solid rhombuses (average of three experiments) represent inhibition by the N-hydroxyimide. The open circle represents the effect of ethanol alone on cleavage of substrate (reaction in lane 16).
DISCUSSION
This study has provided evidence for the involvement of two divalent metal ions in the hydrolysis of an RNA phosphodiester by E.coli RNase III. The dependence of the cleavage rate on the Mg2+ ion concentration and the potent inhibition by an N-hydroxyimide provide independent experimental approaches that yield congruent results. It may be argued that an additional metal ion(s) is involved in catalysis, which would not be detected in the kinetic assays if it were tightly bound to RNase III. However, the protein purification procedure involves extensive dialysis, and the crystal structure of the Aquifex aeolicus RNase III nuclease domain, determined in the absence of added divalent metal ion, is devoid of an intrinsic (i.e. tightly bound) metal (20).
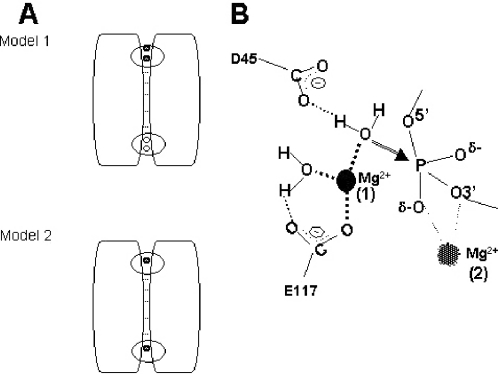
The N-hydroxyimide inhibition not only indicates that two metal ions are involved in catalysis, but that they are closely positioned. The structure of the A.aeolicus RNase III nuclease domain reveals a binding site for a single metal ion at each end of the subunit interface (20,26) (Figure 6A and B). However, since the distance between the two sites is considerably greater than ∼4 Å, the two metal ions seen in the crystal structure do not represent a pair consistent with the N-hydroxyimide inhibition. A more consistent model would be one in which catalytic activity requires binding of an additional Mg2+ to one or both sites (Model 1, Figure 6A). This model would include the possibility that binding of the second metal is promoted by substrate binding, as is seen with other phosphotransferases (41,42). Other experimental results are consistent with this model. Thus, while the inactivity of the RNase III[E117K] mutant (34,35) probably is due to disruption of metal binding to each active site (20), catalytic activity is retained by a mutant heterodimer in which one subunit (and therefore only one active site) carries the E117K mutation (44) or the E117A mutation (W. Meng and A.W. Nicholson, unpublished experiments). The retained activity of the heterodimer is incompatible with the model in which catalytic activity requires the binding of a single Mg2+ to each active site (Model 2, Figure 6A).
Figure 6.
Model for a two-metal-ion-dependent catalytic mechanism for RNase III. (A) Alternative binding modes for two Mg2+ ions to the nuclease domain. The diagram depicts the homodimeric structure of the nuclease domain and the location of the binding sites for Mg2+ (or Mn2+) at each end of the subunit interface (20,26). The dsRBDs of each subunit are not shown. Metal ions are indicated by the filled circles. A rectangle indicates a functional active site; and a circle indicates a nonfunctional active site, with site functionality defined here by metal ion occupancy. The assumption in the two models is that, in the absence of substrate, RNase III already carries a single Mg2+ ion at each site, as observed in the crystal structures (20,26). A catalytic requirement for two Mg2+ ions either would indicate a requirement for double occupancy of at least one of the two sites (Model 1, Mg2+ ions denoted by filled circles), or single occupancy of both sites (Model 2). Note that Model 2 would not require the binding of additional metal ions. The experimental data support Model 1 (see Results and Discussion). A modified Model 1 would include the binding of a second Mg2+ ion to the second active site and would be invoked if a substrate is destined to be cleaved in a concomitant manner at two target sites. (B) Proposed involvement of two Mg2+ ions in the catalytic mechanism. The diagram derives from Model 1 (A) and is based on structural (20) and enzymological (24,34,56) data. The Mg2+ ion in binding site 1 (observed in the crystal structure) would activate the water nucleophile (see Discussion). For simplicity, the figure shows only two (D45 and E117) of the four (E41, D45, D114, E117) carboxyl side chains that have been shown in A.aeolicus RNase III to coordinate the Mg2+ ion in binding site 1 (20,26). Moreover, a third metal-bound water molecule is not shown. The Mg2+ ion in binding site 2 (proposed placement) could (i) enhance the electrophilicity of the phosphorus and neutralize the negative charge of the pentacoordinate intermediate, and (ii) protonate or otherwise increase the acidity of the 3′-oxygen leaving group. Note that binding of substrate would be involved in the creation of binding site 2. The identity(ies) of the protein moieties that coordinate the second Mg2+ ion are not known.
The noncompetitive inhibitory behavior of the N-hydroxyimide indicates that the compound does not disrupt enzyme–substrate binding. However, this behavior does not necessarily suggest that substrate and inhibitor can simultaneously bind to the active site. It has been shown elsewhere (19) that the dsRBD is the primary determinant of substrate recognition by E.coli RNase III. It is therefore possible that the inhibitor disrupts engagement of substrate by the active site in the nuclease domain, without inhibiting substrate binding by the dsRBD. The N-hydroxyimide inhibition shares a formal similarity to the effect of ethidium bromide, which reversibly inhibits cleavage without disrupting substrate binding by the dsRBD (45). In the latter situation, ethidium binds substrate to form a complex that is resistant to cleavage, yet is still competent to bind RNase III. It is possible that the structure of the enzyme–substrate complex in the presence of either inhibitor may be similar to structure of A.aeolicus RNase III bound to dsRNA in a catalytically unreactive conformation (26). Further structural studies will be needed to determine the precise mode of binding of the N-hydroxyimide to RNase III and characterize the specific features of the substrate–inhibitor–enzyme complex.
A proposed catalytic mechanism for E.coli RNase III
Figure 6B shows the features of a possible catalytic mechanism involving two magnesium ions. This mechanism assumes an SN2(P) reaction pathway that involves inversion of configuration at phosphorus, for which there is preliminary experimental evidence (24). One of the metal ions may serve to activate the water nucleophile. This function is based on the presence of several water molecules directly coordinated to the bound metal (20), and that the rate of the hydrolysis step is dependent on the pKa of the metal (24). Water activation also may involve participation of the D45 side chain, which is within hydrogen-bonding distance of one of the metal-bound waters (Figure 6B) (20). The functional role(s) as well as the specific location of the second Mg2+ ion are more speculative, since the crystallographic study showed only one metal ion in each active site (20). However, two functions can be proposed, based on other phosphodiesterases that employ two metal ions [e.g., see (46,47)] and the chemical requirements for efficient catalysis of phosphotransfer (48–50). One role in the presumed SN2(P) reaction path would be enhancement of phosphorus electrophilicity and stabilization of a transition state, both of which can be achieved through neutralization of negative charge on the non-bridging phosphodiester oxygens. A second role would be enhancement of 3′-oxygen leaving group ability, either by water-mediated protonation or through direct coordination (Figure 6B). As mentioned above, the binding site for the second metal ion may be dependent upon substrate binding. It is also possible that binding of the N-hydroxyimide could promote the binding of the second metal ion. If so, this pathway would obviate the need for substrate binding, if this event is a necessary prerequisite for creating the second metal binding site, and if substrate binding and inhibitor binding to the active site are mutually exclusive (see above).
A dual catalytic mechanism for RNase III has been proposed in which the two phosphodiesters at a dsRNA target site are cleaved by separate chemical reactions, involving different groups of residues and/or metal ion(s) in the active site (20). The single-site reactivity of R1.1 RNA would necessarily involve only one of the two proposed chemistries. Alternatively, it is possible that a single catalytic mechanism is responsible for cleavage of both phosphodiesters at a given dsRNA target site. Here, each phosphodiester would be cleaved in a random initial order manner, and a structural change in the enzyme–substrate complex following cleavage of the first phosphodiester would allow cleavage of the second bond (3).
The N-hydroxyimide inhibition of the Mn2+-supported reaction indicates that the catalytic mechanism also can employ two Mn2+ ions, positioned ∼4 Å apart. Furthermore, the observation that Mg2+ and Mn2+ at their respective optimal concentrations confer similar catalytic efficiencies (34) provides additional evidence that RNase III can effectively use an alternative metal. Moreover, with the evidence indicating a two-metal-ion catalytic mechanism, the observation that higher Mn2+ concentrations inhibit RNase III provides further evidence for a third binding site on the enzyme that is specific for Mn2+ (34).
It is likely that most if not all RNase III orthologs employ the same active site chemistry. Zhang and coworkers determined that mammalian Dicer is active in monomeric form, and that the tandem nuclease domain sequences associate in an intramolecular fashion, forming an internal dimeric structure formally equivalent to the homodimeric structure of bacterial RNase III (51). The pattern of cleavage of miRNA precursors suggests that both catalytic sites are employed in the processing reaction, at least for this class of substrate. It therefore would be anticipated that four metal ions (i.e. two in each site) would be needed for miRNA production, if the two cleavage events derive from a single binding event. The identification of RNase III-like polypeptides in the Leishmania tarentolae mitochondrial RNA editing complex (52) suggests that the cleavage events associated with uridine insertion/deletion editing have a similar divalent metal ion requirement. A formal similarity of E.coli RNase III and E.coli RNase HI has been noted (19,34), in particular with respect to inhibition by Mn2+. However, E.coli RNase HI apparently requires only a single Mg2+ ion (53,54), which may function to bind a water molecule that protonates the 3′-oxygen leaving group (55). Also, the water nucleophile may be activated by a histidine side chain acting as a general base, rather than by a divalent metal ion (55).
Acknowledgments
The authors thank Peiling Chen (Temple University) for synthesis of compounds, and Scott Sieburth (Temple University) for important advice on the synthetic strategies. The authors also thank Xinhua Ji (National Cancer Institute at Frederick) for insightful comments on the study, especially with regard to modes of metal ion binding to RNase III. We are indebted to Bruce Palfey (University of Michigan) and Rhonda Nicholson (Temple University) for a critique of the manuscript, and also are grateful for Wenzhao Meng and other members of the laboratory for their advice and encouragement. This work was supported by NIH Grant GM56457. Funding to pay the open access publication charges for this article was provided by Temple University.
REFERENCES
- 1.Court D.L. RNA processing and degradation by RNase III. In: Belasco J.G., Brawerman G., editors. Control of Messenger RNA Stability. New York: Academic Press; 1993. pp. 71–116. [Google Scholar]
- 2.LaMontagne B., Larose S., Boulanger J., AbouElela S. The RNase III family: a conserved structure and expanding functions in eukaryotic dsRNA metabolism. Curr. Issues Mol. Biol. 2001;3:71–78. [PubMed] [Google Scholar]
- 3.Nicholson A.W. The ribonuclease superfamily: forms and functions in RNA maturation, decay, and gene silencing. In: Hannon G.J., editor. RNAi: A Guide to Gene Silencing. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2003. pp. 149–174. [Google Scholar]
- 4.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate nuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 5.Provost P., Dishart D., Doucet J., Frendewey D., Samuelsson B., Radmark O. Ribonuclease activity and RNA binding of recombinant human dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Kolb F.A., Brondani V., Billy E., Filipowicz W. Human dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 8.Martinez J., Patkaniowska A., Urlaub H., Luhrmann R., Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 9.Chiu Y.L., Rana T.M. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutvagner G., McLachlan J., Pasquinelli A.E., Balint E., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 11.Ketting R.F., Fischer S.E.J., Bernstein E., Sijen T., Hannon G.J., Plasterk R.H.A. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y., Ahn C., Han J., Choi H., Kim J., Lee J., Provost P., Radmark O., Kim S., Kim V.N. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 13.Robertson H.D. Escherichia coli ribonuclease III cleavage sites. Cell. 1982;30:669–672. doi: 10.1016/0092-8674(82)90270-7. [DOI] [PubMed] [Google Scholar]
- 14.Dunn J.J. Ribonuclease III. In: Boyer P.D., editor. The Enzymes. New York: Academic Press; 1982. pp. 485–499. [Google Scholar]
- 15.Nicholson A.W. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 16.St Johnston D., Brown N.H., Gall J.G., Jantsch M. A conserved double-stranded RNA-binding domain. Proc. Natl Acad. Sci. USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiero-Monti I., Mathews M.B. Proteins binding to duplexed RNA: one motif, multiple functions. Trends Biochem. Sci. 2000;25:241–246. doi: 10.1016/s0968-0004(00)01580-2. [DOI] [PubMed] [Google Scholar]
- 18.Nashimoto H., Miura A., Saito H., Uchida H. Suppressors of temperature-sensitive mutations in a ribosomal protein gene, rpsL (S12), of Escherichia coli K12. Mol. Gen. Genet. 1985;199:381–387. doi: 10.1007/BF00330746. [DOI] [PubMed] [Google Scholar]
- 19.Sun W., Jun E., Nicholson A.W. Intrinsic double-stranded-RNA processing activity of Escherichia coli ribonuclease III lacking the dsRNA-binding domain. Biochemistry. 2001;40:14976–14984. doi: 10.1021/bi011570u. [DOI] [PubMed] [Google Scholar]
- 20.Blaszczyk J., Tropea J.E., Bubunenko M., Routzahn K.M., Waugh D.S., Court D.L., Ji X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001;9:1225–1236. doi: 10.1016/s0969-2126(01)00685-2. [DOI] [PubMed] [Google Scholar]
- 21.Robertson H.D., Webster R.E., Zinder N. Purification and properties of ribonuclease III from Escherichia coli. J. Biol. Chem. 1968;243:82–91. [PubMed] [Google Scholar]
- 22.Dunn J.J. RNase III cleavage of single-stranded RNA. Effect of ionic strength on the fidelity of cleavage. J. Biol. Chem. 1976;251:3807–3814. [PubMed] [Google Scholar]
- 23.Li H., Chelladurai B.S., Zhang K., Nicholson A.W. Ribonuclease III cleavage of a bacteriophage T7 processing signal. Divalent cation specificity, and specific anion effects. Nucleic Acids Res. 1993;21:1919–1925. doi: 10.1093/nar/21.8.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell F.E., Jr, Cassano A.G., Anderson V.E., Harris M.E. Pre-steady-state and stopped-flow fluorescence analysis of Escherichia coli ribonuclease III: insights into mechanism and conformational changes associated with binding and catalysis. J. Mol. Biol. 2002;317:21–40. doi: 10.1006/jmbi.2002.5413. [DOI] [PubMed] [Google Scholar]
- 25.Amarasinghe A.K., Calin-Jageman I., Harmouch A., Sun W., Nicholson A.W. Escherichia coli ribonuclease III: affinity purification of hexahistidine-tagged enzyme and assays for substrate binding and cleavage. Methods Enzymol. 2001;342:143–158. doi: 10.1016/s0076-6879(01)42542-0. [DOI] [PubMed] [Google Scholar]
- 26.Blaszczyk J., Gan J., Tropea J.E., Court D.L., Waugh D.S., Ji X. Noncatalytic assembly of ribonuclease III with double-stranded RNA. Structure (Camb.) 2004;12:457–466. doi: 10.1016/j.str.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 27.He B., Rong M., Lyakhov D., Gartenstein H., Diaz G., Castagna R., McAllister W.T., Durbin R.K. Rapid mutagenesis and purification of phage RNA polymerases. Protein Expr. Purif. 1997;9:142–151. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- 28.Edafiogho I.O., Scott K.R., Moore J.A., Farrar V.A., Nicholson J.M. Synthesis and anticonvulsant activity of imidooxy derivatives. J. Med. Chem. 1991;34:387–392. doi: 10.1021/jm00105a059. [DOI] [PubMed] [Google Scholar]
- 29.Bouzide A. Magnesium bromide mediated highly diastereoselective heterogeneous hydrogenation of olefins. Org. Lett. 2002;4:1347–1350. doi: 10.1021/ol020032m. [DOI] [PubMed] [Google Scholar]
- 30.Crockett G.C., Swanson B.J., Anderson D.R., Koch T.H. A preferred method for imide preparation. Synthetic Communications. 1981;11:447–454. [Google Scholar]
- 31.Dunn J.J., Studier F.W. Complete nucleotide of bacteriophage T7 and locations of T7 genetic elements. J. Mol. Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 32.Chelladurai B., Li H., Zhang K., Nicholson A.W. Mutational analysis of a ribonuclease III processing signal. Biochemistry. 1993;32:7549–7558. doi: 10.1021/bi00080a029. [DOI] [PubMed] [Google Scholar]
- 33.Milligan J.F., Groebe D.F., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun W., Nicholson A.W. Mechanism of action of Escherichia coli ribonuclease III. Stringent chemical requirement for the glutamic acid 117 side chain, and Mn2+ rescue of the Glu117Asp mutant. Biochemistry. 2001;40:5102–5110. doi: 10.1021/bi010022d. [DOI] [PubMed] [Google Scholar]
- 35.Li H., Nicholson A.W. Defining the enzyme binding domain of a ribonuclease III processing signal. Ethylation interference and hydroxyl radical footprinting using catalytically inactive RNase III mutants. EMBO J. 1996;15:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon M., Webb E.C. Enzymes. 3rd. New York: Academic Press; 1979. [Google Scholar]
- 37.Fersht A. Structure and Mechanism in Protein Science. New York: W.H. Freeman and Company; 1999. [Google Scholar]
- 38.Parkes E.B., Ermert P., Fassler J., Ives J., Martin J.A., Merrett J.H., Obrecht D., Williams G., Klumpp K. Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. J. Med. Chem. 2003;46:1153–1164. doi: 10.1021/jm020334u. [DOI] [PubMed] [Google Scholar]
- 39.Klumpp K., Hang J.Q., Rajendran S., Yang Y., Derosier A., Wong Kai In P., Overton H., Parkes K.E.B., Cammack N., Martin J.A. Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res. 2003;31:6852–6859. doi: 10.1093/nar/gkg881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hang J.Q., Rajendran S., Yang Y., Li Y., Wong Kai In P., Overton H., Parkes K.E.B., Cammack N., Martin J.A., Klumpp K. Activity of the isolated HIV RNase H domain and specific inhibition by N-hydroxyimides. Biochem. Biophys. Res. Commun. 2004;317:321–329. doi: 10.1016/j.bbrc.2004.03.061. [DOI] [PubMed] [Google Scholar]
- 41.Cherepanov A.V., de Vries S. Kinetic mechanism of the Mg2+-dependent nucleotidyl transfer catalyzed by T4 DNA and RNA ligases. J. Biol. Chem. 2002;277:1695–1704. doi: 10.1074/jbc.M109616200. [DOI] [PubMed] [Google Scholar]
- 42.Horton N.C., Perona J.J. DNA cleavage by EcoRV endonuclease: two metal ions in three metal ion binding sites. Biochemistry. 2004;43:6841–6857. doi: 10.1021/bi0499056. [DOI] [PubMed] [Google Scholar]
- 43.DasGupta S., Fernandez L., Kameyama L., Inada T., Nakamura Y., Pappas A., Court D.L. Genetic uncoupling of the dsRNA-binding and RNA cleavage activities of the Escherichia coli endoribonuclease RNase III—the effect of dsRNA binding on gene expression. Mol. Microbiol. 1998;28:629–640. doi: 10.1046/j.1365-2958.1998.00828.x. [DOI] [PubMed] [Google Scholar]
- 44.Conrad C., Schmitt J.G., Evguenieva-Hackenberg E., Klug G. One functional subunit is sufficient for catalytic activity and substrate specificity of Escherichia coli endoribonuclease III artificial heterodimers. FEBS Lett. 2002;518:93–96. doi: 10.1016/s0014-5793(02)02653-4. [DOI] [PubMed] [Google Scholar]
- 45.Calin-Jageman I., Amarasinghe A.K., Nicholson A.W. Ethidium-dependent uncoupling of substrate binding and cleavage by Escherichia coli ribonuclease III. Nucleic Acids Res. 2001;29:1915–1925. doi: 10.1093/nar/29.9.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beese L.J., Steitz T.A. Structural basis for the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I; a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng L., Li M., Shan J., Krishnamoorthi R., Shen B. Distinct roles of two Mg2+ binding sites in regulation of murine flap endonuclease-1 activities. Biochemistry. 2002;41:10323–10331. doi: 10.1021/bi025841s. [DOI] [PubMed] [Google Scholar]
- 48.Cooperman B.S. The role of divalent metal ions in phosphoryl and nucleotidyl transfer. In: Sigel H., editor. Metal Ions in Biological Systems. Vol. 5. New York: Marcel Dekker; 1976. pp. 79–125. [Google Scholar]
- 49.Knowles J.R. Enzyme-catalyzed phosphoryl transfer reactions. Annu. Rev. Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 50.Gerlt J.A. Mechanistic principles of enzyme-catalyzed cleavage of phosphodiester bonds. In: Linn S.M., Lloyd R.S., Roberts R.J., editors. Nucleases 2nd edn. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1993. pp. 1–34. [Google Scholar]
- 51.Zhang H., Kolb F.A., Jaskiewicz L., Westhof E., Filipowicz W. Single processing center models for human dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 52.Aphasizhev R., Aphasizheva I., Nelson R.E., Gao G., Simpson A.M., Kang X., Falick A.M., Sbicego S., Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang H.W., Cowan J.A. Metallobiochemistry of the magnesium ion. Characterization of the essential metal-binding site in Escherichia coli ribonuclease H. Eur. J. Biochem. 1994;219:253–260. doi: 10.1111/j.1432-1033.1994.tb19936.x. [DOI] [PubMed] [Google Scholar]
- 54.Uchiyama Y., Iwai S., Ueno Y., Ikehara M., Ohtsuka E. Role of the Mg2+ ion in the Escherichia coli ribonuclease HI reaction. J. Biochem. (Tokyo) 1994;116:1322–1329. doi: 10.1093/oxfordjournals.jbchem.a124682. [DOI] [PubMed] [Google Scholar]
- 55.Tsunaka Y., Haruki M., Morikawa M., Oobatake M., Kanaya S. Dispensability of glutamic acid 48 and aspartic acid 134 for Mn2+-dependent activity of Escherichia coli ribonuclease HI. Biochemistry. 2003;42:3366–3374. doi: 10.1021/bi0205606. [DOI] [PubMed] [Google Scholar]
- 56.Sun W., Li G., Nicholson A.W. Mutational analysis of the nuclease domain of Escherichia coli ribonuclease III. Identification of conserved acidic residues that are important for catalytic function in vitro. Biochemistry. 2004;43:13054–13062. doi: 10.1021/bi049258i. [DOI] [PubMed] [Google Scholar]