Abstract
Prokaryotic chromosomes code for toxin–antitoxin (TA) loci, often in multiple copies. In E.coli, experimental evidence indicates that TA loci are stress-response elements that help cells survive unfavorable growth conditions. The first gene in a TA operon codes for an antitoxin that combines with and neutralizes a regulatory ‘toxin’, encoded by the second gene. RelE and MazF toxins are regulators of translation that cleave mRNA and function, in interplay with tmRNA, in quality control of gene expression. Here, we present the results from an exhaustive search for TA loci in 126 completely sequenced prokaryotic genomes (16 archaea and 110 bacteria). We identified 671 TA loci belonging to the seven known TA gene families. Surprisingly, obligate intracellular organisms were devoid of TA loci, whereas free-living slowly growing prokaryotes had particularly many (38 in Mycobacterium tuberculosis and 43 in Nitrosomonas europaea). In many cases, TA loci were clustered and closely linked to mobile genetic elements. In the most extreme of these cases, all 13 TA loci of Vibrio cholerae were bona fide integron elements located in the V.cholerae mega-integron. These observations strongly suggest that TA loci are mobile cassettes that move frequently within and between chromosomes and also lend support to the hypothesis that TA loci function as stress-response elements beneficial to free-living prokaryotes.
INTRODUCTION
All cells possess quality control mechanisms that ensure degradation of defective protein and mRNA. Eukaryotic cells have a surveillance system called nonsense-mediated mRNA decay (NMD) that removes aberrant mRNAs containing a premature termination codon in their protein coding regions. The NMD pathway is triggered during the first round of translation of the aberrant mRNA thus ensuring that synthesis of truncated, potentially harmful proteins is prevented (1,2). A related mechanism is operational in bacteria. Here, quality control of gene expression is accomplished by an interplay between tmRNA (3,4) and ‘toxins’ that cleave mRNA at the ribosomal A-site (5–8). tmRNA is both a tRNA and an mRNA that recognizes ribosomes that are locked by translation of broken (or non-stop) mRNAs. Ribosomes trapped on non-stop mRNAs cannot terminate translation by the regular termination pathway. Such ribosomes are rescued by tmRNA in a reaction called trans-translation that simultaneously mediates ribosome recycling and tagging of incomplete proteins for degradation by cellular proteases. Recently, two families of ribonucleases, RelE and MazF, which block translation by cleavage of mRNAs, were identified (5,6,8,9). Such mRNAs lack their natural stop-codons and tmRNA is needed to release ribosomes locked at their termini. Consistently, tmRNA counteracted the toxic effect of RelE and MazF overexpression and cells devoid of tmRNA became hypersensitive to the toxins (5,6). Together these results suggest that the RelE and MazF toxins function in quality control of gene expression.
The relBE locus of E.coli encodes RelE toxin and RelB antitoxin. RelB counteracts RelE activity by direct protein–protein interaction (10). RelB also represses relBE transcription and RelE acts as a co-repressor of relBE transcription (10). The mazEF locus has a very similar genetic organization (11). Nutritional stresses, such as amino acid and glucose starvation, activates RelE and MazF to inhibit translation by mRNA cleavage (5,12). Activation of RelE and MazF depends on Lon protease (5,12). During nutritional stress, Lon degrades RelB and most likely also MazE (5,12). In wild-type cells, the simultaneous degradation of RelB and MazF antitoxins has two effects that act in concert: it increases RelE and MazE activities and it increases the transcription rates of the relBE and mazEF operons (5,12). In turn, theincreased transcription rates sustain toxin synthesis during the stress period. Ectopic expression of RelE or MazF inhibited translation and conferred rapid loss of colony formation (5,12). However, cell viability could be fully regained by later induction of relB transcription, thus indicating that even efficient overproduction of RelE or MazF did not confer cell death (12,13).
Toxin–antitoxin loci were discovered due to their ability to stabilize plasmids by post-segregational killing (PSK) (14,15). Plasmid stabilization is a consequence of the differential stabilities of the toxins and antitoxins: since the antitoxins are metabolically unstable, cells that lose a TA locus experience activation of the toxin that, in turn, prevents further cell growth of the plasmid-free cells. In a growing bacterial population, this results in phenotypic stabilization of plasmids that carry a TA locus (16). During the years, seven plasmid-encoded TA families have been described (15,17). The TA loci belonging to these seven families are listed in the order of discovery: ccd of F (14), parD/pem of R1/R100 (18), vapBC of a Salmonella dublin virulence plasmid (19), phd/doc of P1 (20), parDE of RK2 (21), higBA of Rts1 (22) and relBE of P307 (23). All TA loci belonging to these seven families have the same modular genetic set-up and overall similar regulatory and phenotypic properties, except for higBA that has a reversed gene order (higB toxin gene is located upstream of higA that encodes the antitoxin) (17). The elucidation of the cellular targets of the toxins has been of particular interest: CcdB of F and ParE of RK2 inhibit DNA replication by inhibiting DNA gyrase (24,25) and PemK of R1/R100 and RelE of P307 inhibit translation by mRNA cleavage (23,26). Indirect evidence suggests that Doc inhibits translation (27), whereas the targets of VapC and HigB are not yet known.
Toxin–antitoxin loci are also present on bacterial chromosomes, often in multiple copies. Thus, the chromosome of E.coli K-12 encodes three relBE homologous loci (relBE, dinJ yafQ and yoefM yoeB) (17,28) and two mazEF homologous loci (initially called chpA and chpB for chromosomal homologs of plasmid-encoded genes) (29). Recent work has shown that chromosomal TA loci are surprisingly abundant in both bacteria and archaea, and exhibit very complex phylogenetic patterns (17,30–32). While we performed this work, it was described that the RelE, ParE and HigB toxins constitute a large superfamily of toxins (28). It was also proposed that the VapC PIN-domain proteins are ribonucleases that may constitute an evolutionary link between NMD in eukaryotes and quality control of gene expression in prokaryotes (28). More recently, the structure of the first VapC toxin was solved (33).
Here, we present an exhaustive search for TA loci in 126 totally sequenced prokaryotic genomes. We identify 671 complete TA loci belonging to the seven known TA gene families. Strikingly, we find that TA loci are surprisingly abundant in free-living prokaryotes, but are virtually absent from restricted and obligate host-associated organisms. The marine bacterium Vibrio cholerae has 13 TA loci, all located within the mega-integron on chromosome II. All 13 TA loci have closely linked attC sites, strongly suggesting that they are bona fide integron elements that are transposed via the integron-encoded integrase. The overall phylogenetic pattern supports that TA loci are stress-response elements that function in quality control of gene expression particularly beneficial to slowly growing free-living prokaryotes. The extensive database mining presented here will be highly useful in the further characterization of prokaryotic TA loci.
METHODS
As of August 1, 2003, the fully sequenced genomes of 126 prokaryotic organisms (110 bacteria and 16 archaea) were downloaded from the NCBI website and defined the DNA and protein sequence spaces used throughout this work (Table S1). For simplicity, we did not include plasmid-encoded genes even though some plasmid sequences were present in some of the organisms sequenced.
Searches with toxin protein query sequences
Using standard BLASTP, we searched the protein sequence spaces exhaustively for toxins and antitoxins belonging to the seven known TA families. The Gene Identifiers (GIs) were collected and added to the list of query toxins and antitoxins and the BLASTP procedure continued until it converged. In many cases, we could identify closely linked, annotated toxin or antitoxin partners. In cases which there was no annotated partner was apparent, we looked for unannotated open reading frames in the DNA. The cut-off E-value used in this analysis was 10−4.
In those genomes in which we did not find annotated toxins or antitoxins belonging to a given family by the BLAST procedure, we used the TBLASTN algorithm to exhaustively search for the presence of toxin and antitoxin genes. Thus, the genomes in which we found no TA loci (Table 3) were queried with all known toxin sequences listed in Table S2. The cut-off E-value used in this analysis was also 10−4.
Table 3.
List of organisms with more than 8 TA loci
| Organism | No. of TA locia | Organism lifestyle |
|---|---|---|
| Archaea | ||
| Sulfolobus_tokodaii | 32 | Chemolithotrophic and hyperthermophilic |
| Archaeoglobus_fulgidus | 28 | Chemolithotrophic and hyperthermophilic |
| Sulfolobus_solfataricus | 23 | Chemolithotrophic and hyperthermophilic |
| Pyrococcus_abyssi | 17 | Chemoorganotrophic and hyperthermophilic |
| Pyrococcus_furiosus | 17 | Chemoorganotrophic and hyperthermophilic |
| Pyrococcus_horikoshii | 14 | Chemoorganotrophic and hyperthermophilic |
| Methanosarcina_acetivorans | 12 | Chemolithotrophic |
| Pyrobaculum_aerophilum | 11 | Chemolithotrophic and hyperthermophilic |
| Aeropyrum_pernix | 9 | Hyperthermophilic |
| Methanococcus_jannaschii | 9 | Methylotrophic and hyperthermophilic |
| Methanosarcina_mazei | 9 | Methylotrophic and hyperthermophilic? |
| Gram-positive bacteria | ||
| Mycobacterium_tuberculosis_H37Rv | 38 | Human pathogen, intra- and extracellular |
| Mycobacterium_tuberculosis_CDC1551 | 36 | Human pathogen, intra- and extracellular |
| Nostoc_sp_PCC_7120 | 29 | Marine and phototrophic |
| Synechocystis_PCC6803 | 14 | Marine and phototrophic |
| Proteobacteria | ||
| Nitrosomonas_europaea | 45 | Chemolithotrophic and mesophilic |
| Xylella_fastidiosa | 17 | Plant pathogen |
| Agrobacterium_tumefaciens_str_C58_Cereon | 14 | Plant pathogen |
| Agrobacterium_tumefaciens_C58_Washington | 14 | Plant pathogen |
| Vibrio_cholerae | 13 | Water-borne human pathogen |
| Sinorhizobium_meliloti_1021 | 12 | Plant pathogen |
| Caulobacter_crescentus | 11 | Waterborne |
| Mesorhizobium_loti | 10 | Plant pathogen |
| Pseudomonas_syringae | 9 | Plant pathogen |
| Pseudomonas_putida_KT2440 | 8 | Soil bacterium |
| Escherichia_coli_O157 | 8 | Human pathogen, mesophilic |
| Escherichia_coli_O157 EDL933 | 8 | Human pathogen, mesophilic |
| Salmonella_typhimurium_LT2 | 8 | Human pathogen, mesophilic |
| Xylella_fastidiosa_Temecula1 | 8 | Plant pathogen |
aNumbers include solitary toxin genes.
Exhaustive genome search using TBLASTN
We developed a method to exhaustively search all 126 selected chromosomes for the presence of unannotated toxins. In principle, the output from a TBLASTN query would immediately identify novel unannotated toxins in any given genome. However, to do the analysis batchwise, we searched with all known toxin GIs as compiled in Table S2 (∼600 toxin GIs). This procedure in many cases yielded vast and intractable amounts of data. To avoid this problem, we first annotated all known toxin genes in the 126 genomes using Vector NTI (version 7.0), deleted them from the 126 chromosomes and then searched the ‘crippled’ chromosomes using TBLASTN. By this exhaustive method, we identified an additional 37 TA loci and a number of solitary toxin genes, corresponding to 7% of the total number of loci identified.
RESULTS
Phyletic and phylogenetic distribution of the seven known TA families
Using BLASTP and TBLASTN, we exhaustively searched the completely sequenced genomes of 126 prokaryotic organisms for their content of members belonging to the seven known TA gene families as described above. We identified 671 complete TA loci and 37 toxin genes without a closely linked antitoxin gene (here called solitary toxin genes). The distribution of TA loci in the 126 organisms is given in Table S1. The detailed information (GIs, DNA coordinates, gene sizes and gene distances) of all TA loci and solitary toxin genes analyzed is compiled in Table S2. Of the 1379 toxin and antitoxin genes listed in Table S2, 66 and 183, respectively, were not annotated. The sequences of these toxins and antitoxins are listed in Table S3. In 25 cases, the annotations of toxin and antitoxin genes were corrected. These corrections, which were based primarily on multiple sequence alignments of toxins and antitoxins, are also listed in Table S2.
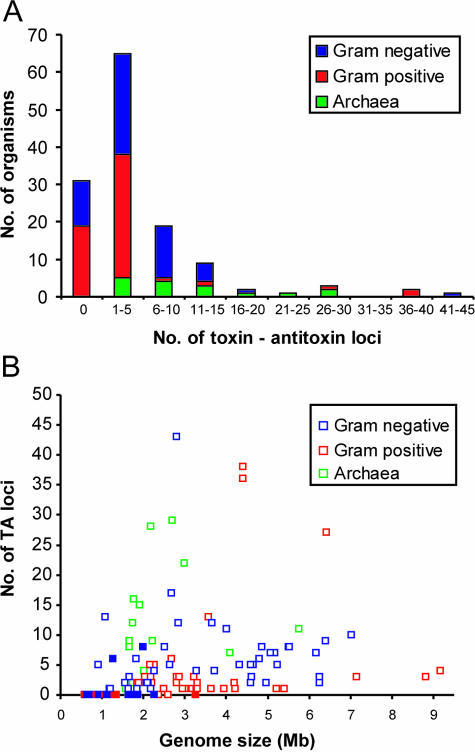
Table 1 summarizes the phyletic distribution of the seven TA families. The two largest gene families, vapBC and relBE, were abundantly represented in bacteria and archaea, constituting 42% and 23% percent of the 671 loci, respectively. Four TA gene families (mazEF, parDE, higBA and ccd) were confined to the bacterial domain whereas 3 out of 25 phd/doc loci were archaeal. Table S1 gives an overview of the distribution of a number of individual TA family members in the 126 prokaryotic organisms. As seen, vapBC loci were present in all 16 archaeal genomes, often in surprisingly high numbers. Similarly, relBE loci were present in most Archaea. The parDE, higBA, mazEF and phd/doc loci were present both in Gram-positive and Gram-negative bacteria, whereas ccd loci were confined to Gram-negative bacteria. Figure 1A graphically illustrates the distribution of the numbers of TA loci in all 126 genomes. As seen, 31 had none, whereas a large fraction (64/126) had between 1 and 5 TA loci. A significant number of genomes had more than 5 TA loci. Figure 1B shows the number of TA loci as a function of genome size. As seen, there was no simple correlation between the two parameters: some relatively small organisms had particularly many TAs whereas some of the very large genomes had few. Interestingly, the relatively small genomes of a number of archaea had remarkably many TAs (green squares in Figure 1B), whereas the genomes of all obligate intracellular organisms had none or few (filled squares in Figure 1B).
Table 1.
Phyletic distribution of TA loci and solitary toxin genes in 126 organisms
| Gene family | relBE | parDE | higBA | vapBC | mazEF | phd/doc | ccdAB | Total |
|---|---|---|---|---|---|---|---|---|
| Total in bacteria | 129 | 59 | 74 | 139 | 67 | 22 | 5 | 495 |
| Total in archaea | 27 | 0 | 0 | 146 | 0 | 3 | 0 | 176 |
| Total TAs in 126 organisms | 156 | 59 | 74 | 285 | 67 | 25 | 5 | 671 |
| Solitary toxins | 13 | 0 | 2 | 13 | 7 | 2 | 0 | 37 |
Figure 1.
Distribution of TA loci in 126 organisms. (A) Number of organisms as a function of their number of TA loci. (B) Number of TA loci in individual genomes as a function of genome size. Filled symbols indicate obligate intracellular organisms.
Obligate host-associated organisms do not retain TA loci
We anticipated that the comprehensive survey would reveal a distinct phylogenetic pattern of TA loci. Table 2 lists the names and the habitats of the 31 organisms that have no TA loci. As seen, the vast majority of these organisms is obligate host-associated parasites or live in close association with other organisms, either parasitically or symbiotically. Thus, bacteria living in constant environments do not retain TA loci. A notable exception that, at a first glance violated this rule, was the finding that the obligate intracellular pathogens Rickettsia conorii and Coxiella burnetii do contain TA loci (Tables S1 and S2). Intracellular pathogens, such as Rickettsia, evolved from extracellular organisms (34,35) and the genomes of R.conorii and C.burnetii are still undergoing reductive evolution (36,37). However, R.prowazekii, which has an even more reduced genome than the two former Rickettsiales, has no TA loci (Table 2), thus supporting the conclusion that obligate host-associated organisms do not retain TA loci.
Table 2.
Organisms with no identifiable TA
| Organism | Size of chromosome (Mb) | Organism lifestyle | Toxin pseudogenes detectedb |
|---|---|---|---|
| Archaea | |||
| All archaea analyzed have TA loci | |||
| Gram-positive bacteria | |||
| Mycobacterium_leprae | 3.3 | Obligate host-associated | + |
| Tropheryma_whipplei_Twist | 0.9 | Obligate host-associated | − |
| Tropheryma_whipplei_TW08_27 | 0.9 | Obligate host-associated | − |
| Chlamydia_muridarum | 1.1 | Obligate host-associated | − |
| Chlamydia_trachomatis | 1.0 | Obligate host-associated | − |
| Chlamydophila_caviae_GPIC | 1.2 | Obligate host-associated | − |
| Chlamydophila_pneumoniae_CWL029 | 1.2 | Obligate host-associated | − |
| Chlamydophila_pneumoniae_TW_183 | 1.2 | Obligate host-associated | − |
| Chlamydophila_pneumoniae_AR39 | 1.2 | Obligate host-associated | − |
| Chlamydophila_pneumoniae_J138 | 1.2 | Obligate host-associated | − |
| Prochlorococcus_marinus | 1.8 | Obligate host-associated | − |
| Mycoplasma_gallisepticum | 1.0 | Obligate host-associated | − |
| Mycoplasma_genitalium | 0.6 | Obligate host-associated | − |
| Mycoplasma_penetrans | 1.4 | Obligate host-associated | − |
| Mycoplasma_pneumoniae | 0.8 | Obligate host-associated | − |
| Mycoplasma_pulmonis | 1.0 | Obligate host-associated | + |
| Ureaplasma_urealyticum | 0.8 | Obligate host-associated | − |
| Lactococcus_lactis | 2.4 | Fastidious lifestyle | − |
| Thermosynechococcus_elongatus | 2.6 | Thermophilic and phototropic | − |
| Gram-negativea | |||
| Rickettsia_prowazekii | 1.1 | Obligate host-associated | − |
| Campylobacter_jejuni | 1.6 | Habitat is the lower bowel | − |
| Helicobacter_hepaticus | 1.8 | Habitat is the lower bowel | + |
| Buchnera_sp_APS | 0.6 | Obligate host-associated | − |
| Buchnera_aphidicola | 0.6 | Obligate host-associated | − |
| Buchnera_aphidicola_Sg | 0.6 | Obligate host-associated | − |
| Wigglesworthia_brevipalpis | 0.7 | Obligate host-associated | − |
| Borrelia_burgdorferi | 0.9 | Obligate host-associated | − |
| Treponema_pallidum | 1.1 | Obligate host-associated | + |
| Thermotoga_maritima | 1.9 | Thermophilic | − |
| Haemophilus_ducreyi_35000HP | 1.7 | Obligate human pathogen, extracellular | − |
| Pasteurella_multocida | 2.3 | Pathogen—grows best on blood–agar | + |
aThe deep branching Aquifex_aeolicus and Thermotoga_maritima were here classified as Gram-negative bacteria for simplicity.
bSearch for pseudogenes was done with TBLASTN using individual chromosome sequences as search spaces and all toxin GIs (Table S2) as query sequences.
Further support for this contention came from the pattern of TA loci in Mycobacteria. Mycobacterium tuberculosis (Mtb) H37Rv and CDC1551 have 38 and 36 TA loci, respectively (Table 3, Table S1). In stark contrast, M.leprae did not have a single intact TA locus (Table 2). Using exhaustive TBLASTN searches with all toxin sequences (Table S2), we identified five toxin pseudogenes in M.leprae (data not shown). In all these cases, closely related, intact toxin genes were present in Mtb. M.leprae evolved from Mtb by massive reductive evolution and its extant genome contains a large number of pseudogenes, indicating that its genome is still undergoing decay (38). Our findings here raise the obvious question as to why Mtb has many and M.leprae no functional TA loci. M.leprae is an obligate intracellular pathogen, whereas Mtb has an extra- and an intracellular growth phase. Thus, the striking phylogenetic pattern in Mycobacteria supports the conclusion that obligate host-associated organisms do not retain TA loci while they are beneficial to free-living organisms. A similar pattern was seen in spirochetes: the obligate parasitic spirochetes Treponema pallidum and Borrelia burgdorferi have no TA loci (Table 2) whereas the free-living spirochete Leptospira interrogans has five (Table S2).
Toxin–antitoxin loci are ubiquitously present in free-living prokaryotes, often in high numbers
Only very few free-living organisms do not have TA loci (Table 2), encompassing the fastidious Lactococcus lactis, that thrives in rich medium only (milk), two pathogens, one phototroph and the deeply branching thermophile Maritima thermotoga. Thus, almost all free-living bacteria that live in changing environments have TA loci.
Many organisms have multiple or even many TA loci (Table 3). Strikingly, Nitrosomonas europaea has 43 intact TA loci and 2 solitary toxin genes, and most archaeal chromosomes have vapBC and relBE loci in high numbers. Is there a common trait for the organisms that have many TAs? Apparently, they live in very different environments: N.europaea is an obligate chemolithotrophic soil organism whereas many of the archaeal organisms are chemolithoautotrophs or extremophiles, and Mtb is a human pathogen. However, many of these organisms are characterized by very low growth rates. Our observations thus raise the possibility that, in free-living bacteria, the number of TA loci may be correlated with the cell growth rate, i.e. TA loci may be beneficial for organisms characterized by slow growth. Further support for this hypothesis comes from the observation that the chromosome of M.smegmatis, a fast-growing close relative of Mtb, encodes two TA loci only (mazEF and phd/doc; data not shown).
Phylogenetic and functional relationships of the seven TA families
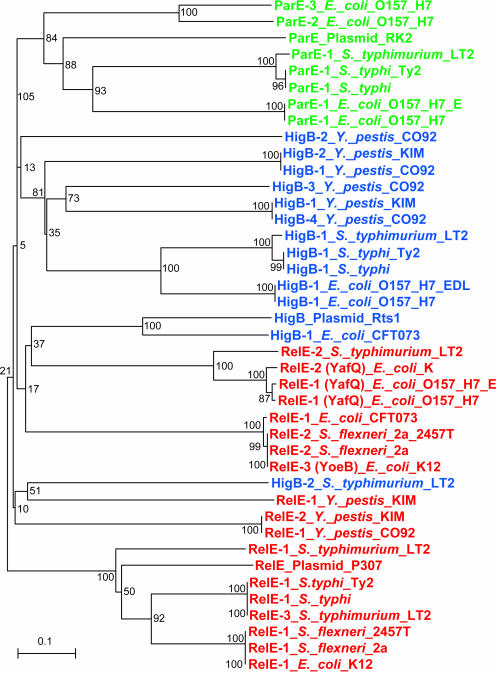
While this work was ongoing, it was reported that RelE, ParE and HigB toxins are phylogenetically related (28). During our BLAST searches, we also noted that members of the RelE, ParE and HigB families exhibit weak but significant sequence similarities supporting that they constitute a superfamily of toxins with a common ancestral origin. This conclusion is perhaps surprising, given that the RelE and ParE toxins are known to have different cellular targets (8,39). A phylogenetic tree of the RelE/ParE/HigB superfamily of toxins from enteric bacteria is shown in Figure 2. The boot-strap values support that RelE and ParE are distinct families but do not, however, allow for discrimination between the RelE and HigB families.
Figure 2.
Phylogenetic tree (Chladogram) of chromosomal RelE, HigB and ParE toxins from enteric bacteria. The RelE, HigB and ParE sequences were retrieved from Table S2 (as GIs) and Table S3 (raw sequences). For comparison, one plasmid-encoded toxin homolog from each group was included in the analysis (RelE of P307, HigB of Rts1 and ParE of RK2). The tree was calculated using Clustal W version 1.83. The lengths of the horizontal lines indicate relative evolutionary distances. A scale bar is also shown.
In some cases, the sequence similarities between RelE, ParE and HigB toxins made it difficult to assign a given toxin sequence to a distinct toxin family. In case of HigB family of toxins, we used the reversed gene order of higBA loci to discriminate HigBs from the other two families. This criterion could not be applied to discriminate between RelE and ParE toxins. In these cases, discrimination was based solely on BLAST E-values. Therefore, we do not exclude that a few ParEs and/or RelEs homologs later may be reassigned to the other family.
Toxin–antitoxin gene statistics
The large number of TA loci identified here yielded the possibility to statistically analyze their numerical properties. Figure S1 shows histograms of gene lengths and of distances between the toxin and antitoxin genes of the five most abundant TA gene families. The length of the toxin genes belonging to the relBE, parDE and higBA families are comparable (average 92, 99 and 101 codons per gene, respectively). In contrast, mazF and vapC toxin genes were larger (average 114 and 130 codons). For all families, the antitoxin genes were smaller than the toxin genes (Figure S1).
The genes of TA loci are located in operons in which the antitoxin gene is upstream of the toxin gene except in the case of higBA loci that invariably have a reversed gene order (higB toxin gene upstream of higA). The distances between the toxin and antitoxin genes vary from locus to locus (Figure S1). In the regular TA loci, a 4 bp gene overlap, in which the upstream antitoxin TGA stop-codon overlaps with the ATG start-codon of the toxin gene in a ATGA sequence, is the most common situation. An overlap of 1 bp (−1) is also frequent. Gene overlap is common in prokaryotes and often reflects translational coupling. Translational coupling between toxin and antitoxin genes occurs in the parD/pem locus of plasmid R1/R100 (40) and may be common to the typical TA loci. The higBA loci exhibited a quite different distance pattern with respect to gene differences, which may be a consequence of the reversed gene order.
Toxin–antitoxin gene localization patterns in individual chromosomes
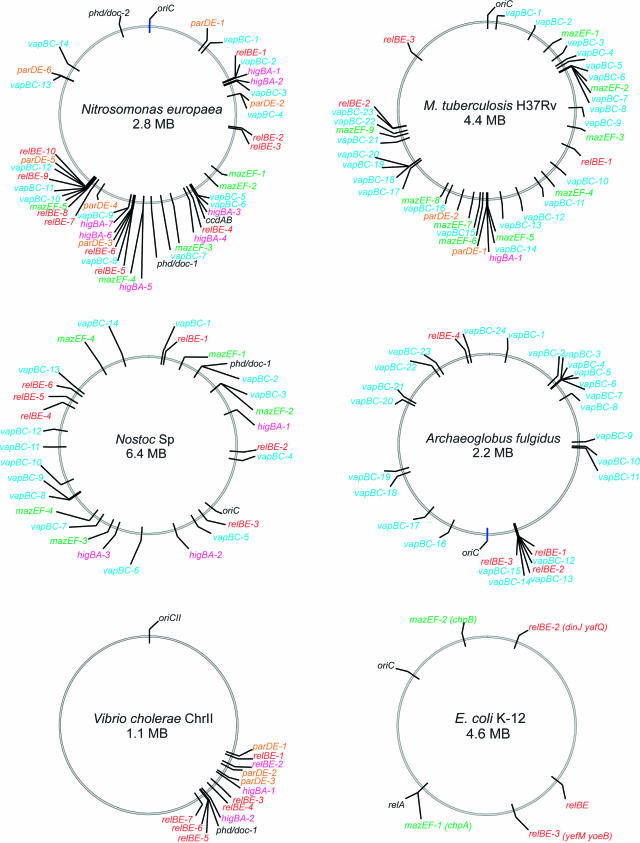
Next, we investigated the numbers and relative positions of TA loci in individual chromosomes. In N.europaea, Mtb, Nostoc, Archaeoglobus fulgidus, V.cholerae, TA loci were located nonrandomly and clustered in particular chromosomal regions (Figure 3). Nonrandom positioning was also evident for many other chromosomes (data not shown). The nonrandom positioning could reflect local gene duplication events. However, since TA loci belonging to different (unrelated) families cluster together, it is possible that TA locus clustering reflects a common mechanism for DNA movement (see below). The model organism E.coli K-12 has five TA loci (three relBE and two mazEF) scattered throughout its genome (Figure 3).
Figure 3.
Chromosomal maps of TA loci in individual chromosomes. Information was derived from Table S2. The maps were created using Vector NTI version 7 (Informax). oriC denotes the origin of replication. All TA loci in the two strains of Mtb were identical except that mazEF-1 and mazEF-7 of Mtb H37Rv were not present in Mtb CDC1551. Note that solitary toxin genes are also shown here as TA loci.
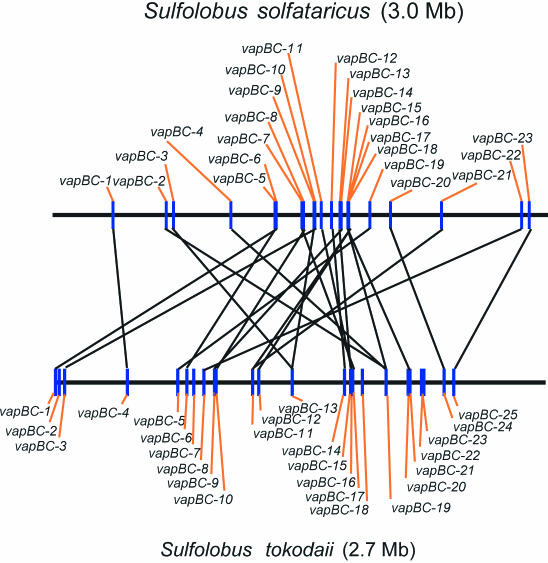
The two sequenced genomes of Sulfolobus spp. encode 23 and 25 vapBC loci (Table 3) and thus yielded the possibility to compare TA gene localization across the genomes of two different species of the same genus. Figure 4 shows a linear representation of the positions of vapBC loci in the two chromosomes. Using BLASTP, each vapC gene of S.solfataricus was paired with its most similar vapC gene in S.tokodaii. As seen, most (21/25) of the vapC genes in S.tokodaii have a potential ortholog in S.solfataricus. However, gene synteny was not conserved, supporting the previous notion that the chromosomes of the two Sulfolobales are scrambled (41). Four of the 25 vapBC loci apparently do not have an ortholog in the other species, thus indicating rapid evolutionary change or lateral gene transfer.
Figure 4.
Comparison of the genomic locations of the 23 and 25 vapC loci of S.solfataricus and S.tokodaii. Using BLASTP, each vapC gene of S.solfataricus was paired with its most similar vapC ortholog in S.tokodaii.
Toxin–antitoxin loci in V.cholerae are all located within the mega-integron, flanked by attC sites
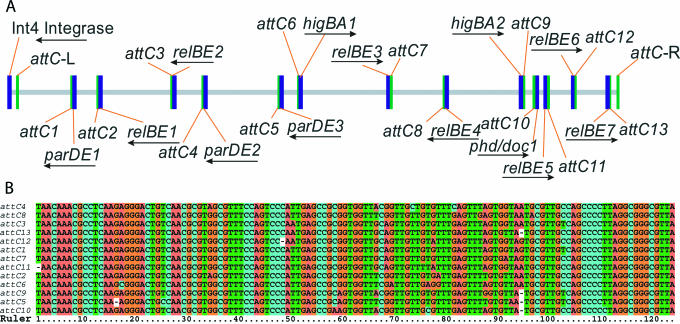
V.cholerae has 13 TA loci all of which are located in the mega-integron of Chromosome II (Fig. 3). The V.cholerae mega-integron contains 7 relBE, 3 parDE, 2 higBA and 1 phd/doc locus (Figure 5). A number of these loci were also identified by others (42). All 13 loci have closely linked attC sites (43) that are target sites for cassette integration by the integron integrase. The relative position of the 13 TA loci in the mega-integron is shown in Figure 5A and the corresponding attC sites are aligned in Figure 5B. The association between TA loci and attC sites argues strongly that the TA loci of V.cholerae are mobile cassettes that frequently move from one location to another. Frequent TA gene duplication and movement is also reflected in the fact that there are two identical TA pairs in the mega-integron (relBE-2 and relBE-7, and parDE-1 and parDE-3, respectively).
Figure 5.
All 13 TA loci in the mega-integron of V.cholerae Chromosome II have juxtaposed attC sites. (A) The locations of the 13 TA loci in the V.cholera mega-integron are shown relative to the closest attC site. Arrows indicate direction of transcription of the TA operons. The boundaries of the integron were determined here by the Int4 integrase gene and the right-most attC site (attC-R). attC-L is the leftmost attC site of a total of ∼150 attC sites of the integron. (B) Nucleotide sequences of the attC sites [123 bp elements with inverted repeats at their ends as defined in (56)] located near TA loci in the V.cholerae mega-integron. The coordinates of the attC sites in the V.cholerae genome and their distances to the TA structural genes (start codons of antitoxin genes or stop codons of toxin genes): attC1: 322548..322671, +14 bp; attC2: 327878..328000, +17 bp; attC3: 343101..343224C, +18 bp; attC4: 349509..349631, −3 bp; attC5: 365215..365336, +15 bp; attC6: 369203..369325, +35 bp; attC7: 388220..388342, −80 bp; attC8: 399166..399288, +7 bp; attC9: 415580..415701, −2 bp; attC10: 417953..418074, +18 bp; attC11: 420535..420656, +6 bp; attC12: 426191..426312, +6 bp; attC13: 432894..433016, +18 bp). In all cases but two (higBA-1 and phd/doc-1), the attC sites were located downstream and close to the toxin structural genes.
N.europaea and X.fastidiosa, which have 43 and 17 TA loci, respectively, both encode integrases of the integron type (44). Therefore, our observations raise the possibility that TA loci in other cases as well may be mobile elements that undergo rapid evolution and horizontal transfer.
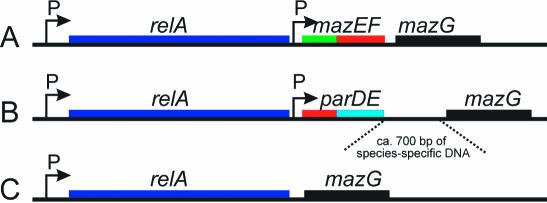
The region downstream of relA of enteric bacteria contains a hotspot for TA integration
The large number of TA loci identified here allowed us to search for different TA loci located at similar gene neighborhoods. In E.coli K-12, the mazEF locus is located 80 bp downstream of relA (29,45). The relA gene is located in overall similar genetic contexts in all enteric genomes sequenced so far (data not shown). We discovered that Salmonella typhimurium LT2, which does not contain a mazEF locus (Table S1), however does contain a parDE locus just downstream of relA (Figure 6). A more systematic survey of enteric bacteria revealed that E.coli K-12, E.coli O157, E.coli O157 EDL933 and Shigella flexneri 2a all have a mazEF locus downstream of relA (Figure 6). In contrast, Salmonella typhimurium LT2, S.typhi and S.typhi 2A all have parDE, whereas E.coli CFT073 and S.flexneri 2a and S.flexneri 2a 2457T have no TA locus at this position. Thus, in enteric bacteria, the region downstream of relA contains a hotspot for TA insertion.
Figure 6.
Comparison of the genetic composition of the relA region of enteric bacteria. Three different genetic contexts of the relA in enterics: (A) E.coli K-12, E.coli O157, E.coli O157 EDL933 and Shigella flexneri 2a; (B) Salmonella typhimurium LT2, S.typhi and S.typhi 2A; (C) E.coli CFT073 and S.flexneri 2a 2457T.
DISCUSSION
We show here that members of the seven known TA families are surprisingly abundant in free-living prokaryotes and virtually absent from obligate host-associated organisms. The few obligate intracellular organisms that have TA loci still undergo reductive evolution. This striking pattern raises important questions: what are the functions of all these genes and why do some organisms have so many whereas others have none? In the following text, we discuss the phylogenetic pattern of TA loci in the context of the three current models that have been proposed to explain the function of TA loci.
Programmed cell death hypothesis
We have previously suggested that plasmid-encoded TA loci mediate programmed cell death (15). Moreover, the chromosome-encoded mazEF locus of E.coli is thoroughly described as a system that confers programmed cell death during amino acid starvation and other stressful conditions (11,46). However, in our hands, the cell death hypothesis did not survive closer scrutiny. Most importantly, we obtained evidence that cells inhibited by ectopic overexpression of either RelE or MazF could be resuscitated if transcription of their cognate antitoxins was induced at a later time (13). In fact, even though protein synthesis was strongly inhibited, the cells stayed fully viable during 5 h of ectopic overexpression of the toxins. Furthermore, in wild-type cells, amino acid starvation induced strong transcription of the relBE and mazEF operons, and concomitant toxin activation reduced the global rate-of-translation by mRNA cleavage (12). However, the viability of cells of three standard E.coli strains did not decrease during 5 h of amino acid starvation (5,6). Thus, we did not find experimental support for the connection between programmed cell death and TA loci.
Plasmids encode another type of TA loci that mediate post-segregational killing (47) and this may have misled us and others to suggest that chromosome-encoded TA loci mediate programmed cell death, a phenotype akin to apoptosis in higher organisms. The phylogenetic pattern seen here neither supports nor rejects the cell death hypothesis, but it is counter-intuitive that single-celled organisms should encode a plethora of suicide genes.
Selfish gene hypothesis/gene stabilization
As discussed above, TA loci can stabilize plasmids by reducing the growth or, in some cases, even kill plasmid-free cells. This is a kind of selfish gene behavior and confers a selective advantage to cells that retain the loci. Clearly, the function of chromosome-encoded TA loci is not to stabilize plasmids. However, it is perhaps more than a formal possibility that TA loci might function to stabilize the vertical transmission of neighboring gene regions similar to what has been proposed for restriction–modification systems (RM) (48–50). In V.cholerae at least, stabilization would be restricted to genes in the mega-integron. If true, mega-integrons with many TA loci should be genetically stable—a hypothesis that now can be challenged experimentally.
Stress-response/quality control hypothesis
The relBE and mazEF loci of E.coli inhibit translation during nutritional stresses such as amino acid and glucose starvation (5,12). They do so by cleavage of mRNA and may thus be regarded as stress-response elements that function in parallel with ppGpp during the stringent response (5,6,8). In keeping with the observed mRNA cleavage, tmRNA counteracted RelE and MazF cell toxicity (5,6). The trans-translation reaction is strictly dependent on a truncated mRNA positioned at the ribosomal A-site, thus establishing a firm link between tmRNA and mRNA cleaving enzymes such as RelE and MazF (51). During limiting growth conditions, it obviously becomes highly important to rescue ribosomes stalled on broken mRNAs and to optimize quality control of gene expression by securing rapid degradation of newly synthesized, truncated proteins. The trans-translation reaction mediated by tmRNA also confers rapid degradation of the damaged mRNAs (52).
The fact that almost all obligate host-associated and fastidious organisms lack TA loci whereas free-living organisms have plentiful, lends further support to the stress-response/quality control hypothesis. Obligate intracellular organisms thrive in constant environments and are thus expected to encounter minimal metabolic stress. In keeping with this notion, many obligate intracellular organisms have also lost the relA/spoT gene that encodes ppGpp synthetase (data not shown). However, it is also known that obligate host-associated organisms have exceptionally stable genomes due to loss of transposable elements, plasmids and enzymes involved in DNA rearrangements (35,53). In support of the gene stabilization hypothesis, it could be argued that organisms with highly stable genomes would not need TA loci to accomplish further gene stabilization. That TA loci function as stress-response elements does not, to our view, exclude that they in some cases also may function to stabilize genes, which certainly is the case for plasmid-borne TA loci. In fact, a gene stabilization effect may accelerate the horizontal spread of the genes. This is analogous to RM systems, which can stabilize DNA segments and plasmids, but whose main function is to reduce invasion of foreign DNA (50).
The multiplicity of TA loci deserves a comment: 30 organisms have 8 or more TA loci (Table 3). We can only speculate why some organisms have so many. One attractive possibility is that the number of TA loci is correlated with cell growth rate such that free-living, slowly growing organisms characterized by low translation rates benefit from having many TA loci. Most of the organisms that have many TA loci grow in nutrient-limited environments or are chemolithoautotrophs (Table 3). These organisms grow very slowly and, intuitively, optimization of quality control of gene expression seems highly important for such organisms. We find that the phylogenetic pattern of TA loci described here is most easily reconciled with our previous hypothesis that TA loci are stress-response elements and/or quality control elements that increase the fitness of free-living prokaryotes (17). This interpretation is supported by recent excellent reviews by Thomas Nyström (54,55).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Siv G. Andersson for valuable comments to the manuscript. This work was supported by the Danish Biotechnology Instrument Center (DABIC). Funding to pay the Open Access publication charges for this article was provided by The Danish Natural Research Council.
REFERENCES
- 1.Le Hir H., Nott A., Moore M.J. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 2003;28:215–220. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 2.Singh G., Lykke-Andersen J. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem. Sci. 2003;28:464–466. doi: 10.1016/S0968-0004(03)00176-2. [DOI] [PubMed] [Google Scholar]
- 3.Karzai A.W., Roche E.D., Sauer R.T. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nature Struct. Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 4.Keiler K.C., Waller P.R., Sauer R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 5.Christensen S.K., Pedersen K., Hansen F.G., Gerdes K. Toxin–antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol Biol. 2003;332:809–819. doi: 10.1016/s0022-2836(03)00922-7. [DOI] [PubMed] [Google Scholar]
- 6.Christensen S.K., Gerdes K. RelE toxins from bacteria and archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol. 2003;48:1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x. [DOI] [PubMed] [Google Scholar]
- 7.Hayes C.S., Sauer R.T. Toxin–antitoxin pairs in bacteria: killers or stress regulators? Cell. 2003;112:2–4. doi: 10.1016/s0092-8674(02)01282-5. [DOI] [PubMed] [Google Scholar]
- 8.Pedersen K., Zavialov A.V., Pavlov M.Y., Elf J., Gerdes K., Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Zhang J., Hoeflich K.P., Ikura M., Qing G., Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 10.Gotfredsen M., Gerdes K. The Escherichia coli relBE genes belong to a new toxin–antitoxin gene family. Mol. Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 11.Aizenman E., Engelberg-Kulka H., Glaser G. An Escherichia coli chromosomal ‘addiction module’ regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl Acad. Sci. USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen S.K., Mikkelsen M., Pedersen K., Gerdes K. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl Acad. Sci. USA. 2001;98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen K., Christensen S.K., Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 14.Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl Acad. Sci. USA. 1983;80:4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen R.B., Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 16.Jensen R.B., Grohmann E., Schwab H., Diaz-Orejas R., Gerdes K. Comparison of ccd of F, parDE of RP4, and parD of R1 using a novel conditional replication control system of plasmid R1. Mol. Microbiol. 1995;17:211–220. doi: 10.1111/j.1365-2958.1995.mmi_17020211.x. [DOI] [PubMed] [Google Scholar]
- 17.Gerdes K. Toxin–antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bravo A., de Torrontegui G., Diaz R. Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol. Gen. Genet. 1987;210:101–110. doi: 10.1007/BF00337764. [DOI] [PubMed] [Google Scholar]
- 19.Pullinger G.D., Lax A.J. A Salmonella dublin virulence plasmid locus that affects bacterial growth under nutrient-limited conditions. Mol. Microbiol. 1992;6:1631–1643. doi: 10.1111/j.1365-2958.1992.tb00888.x. [DOI] [PubMed] [Google Scholar]
- 20.Lehnherr H., Maguin E., Jafri S., Yarmolinsky M.B. Plasmid addiction genes of bacteriophage P1: doc, which causes cell death on curing of prophage, and phd, which prevents host death when prophage is retained. J. Mol. Biol. 1993;233:414–428. doi: 10.1006/jmbi.1993.1521. [DOI] [PubMed] [Google Scholar]
- 21.Roberts R.C., Helinski D.R. Definition of a minimal plasmid stabilization system from the broad-host-range plasmid RK2. J. Bacteriol. 1992;174:8119–8132. doi: 10.1128/jb.174.24.8119-8132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian Q.B., Ohnishi M., Tabuchi A., Terawaki Y. A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 1996;220:280–284. doi: 10.1006/bbrc.1996.0396. [DOI] [PubMed] [Google Scholar]
- 23.Gronlund H., Gerdes K. Toxin–antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J. Mol. Biol. 1999;285:1401–1415. doi: 10.1006/jmbi.1998.2416. [DOI] [PubMed] [Google Scholar]
- 24.Bernard P., Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 25.Miki T., Park J.A., Nagao K., Murayama N., Horiuchi T. Control of segregation of chromosomal DNA by sex factor F in Escherichia coli. Mutants of DNA gyrase subunit A suppress letD (ccdB) product growth inhibition. J. Mol. Biol. 1992;225:39–52. doi: 10.1016/0022-2836(92)91024-j. [DOI] [PubMed] [Google Scholar]
- 26.Lemonnier M., Ziegelin G., Reick T., Munoz G.A., Diaz-Orejas R., Lanka E. Bacteriophage P1 Ban protein is a hexameric DNA helicase that interacts with and substitutes for Escherichia coli DnaB. Nucleic Acids Res. 2003;31:3918–3928. doi: 10.1093/nar/gkg463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazan R., Sat B., Reches M., Engelberg-Kulka H. Postsegregational killing mediated by the P1 phage ‘addiction module’ phd-doc requires the Escherichia coli programmed cell death system mazEF. J. Bacteriol. 2001;183:2046–2050. doi: 10.1128/JB.183.6.2046-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anantharaman V., Aravind L. New connections in the prokaryotic toxin–antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masuda Y., Miyakawa K., Nishimura Y., Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherny I., Gazit E. The YefM antitoxin defines a family of natively unfolded proteins: implications as a novel antibacterial target. J. Biol. Chem. 2004;279:8252–8261. doi: 10.1074/jbc.M308263200. [DOI] [PubMed] [Google Scholar]
- 31.Grady R., Hayes F. Axe-Txe, a broad-spectrum proteic toxin–antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 2003;47:1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x. [DOI] [PubMed] [Google Scholar]
- 32.Mittenhuber G. Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J. Mol. Microbiol. Biotechnol. 1999;1:295–302. [PubMed] [Google Scholar]
- 33.Arcus V.L., Backbro K., Roos A., Daniel E.L., Baker E.N. Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J. Biol. Chem. 2004;279:16471–16478. doi: 10.1074/jbc.M313833200. [DOI] [PubMed] [Google Scholar]
- 34.Andersson S.G., Zomorodipour A., Andersson J.O., Sicheritz-Ponten T., Alsmark U.C., Podowski R.M., Naslund A.K., Eriksson A.S., Winkler H.H., Kurland C.G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 35.Tamas I., Klasson L., Canback B., Naslund A.K., Eriksson A.S., Wernegreen J.J., Sandstrom J.P., Moran N.A., Andersson S.G. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 36.Ogata H., Audic S., Renesto-Audiffren P., Fournier P.E., Barbe V., Samson D., Roux V., Cossart P., Weissenbach J., Claverie J.M., et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- 37.Seshadri R., Paulsen I.T., Eisen J.A., Read T.D., Nelson K.E., Nelson W.C., Ward N.L., Tettelin H., Davidsen T.M., Beanan M.J., et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl Acad. Sci. USA. 2003;100:5455–5460. doi: 10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cole S.T., Eiglmeier K., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y., Pogliano J., Helinski D.R., Konieczny I. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 2002;44:971–979. doi: 10.1046/j.1365-2958.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz-Echevarria M.J., de la C.G., Diaz-Orejas R. Translational coupling and limited degradation of a polycistronic messenger modulate differential gene expression in the parD stability system of plasmid R1. Mol. Gen. Genet. 1995;248:599–609. doi: 10.1007/BF02423456. [DOI] [PubMed] [Google Scholar]
- 41.Brugger K., Torarinsson E., Redder P., Chen L., Garrett R.A. Shuffling of Sulfolobus genomes by autonomous and non-autonomous mobile elements. Biochem. Soc. Trans. 2004;32:179–183. doi: 10.1042/bst0320179. [DOI] [PubMed] [Google Scholar]
- 42.Rowe-Magnus D.A., Guerout A.M., Biskri L., Bouige P., Mazel D. Comparative analysis of superintegrons: engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 2003;13:428–442. doi: 10.1101/gr.617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall R.M., Brookes D.E., Stokes H.W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol. Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 44.Vaisvila R., Morgan R.D., Posfai J., Raleigh E.A. Discovery and distribution of super-integrons among pseudomonads. Mol. Microbiol. 2001;42:587–601. doi: 10.1046/j.1365-2958.2001.02604.x. [DOI] [PubMed] [Google Scholar]
- 45.Aizenman E., Engelberg-Kulka H., Glaser G. An Escherichia coli chromosomal ‘addiction module’ regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl Acad. Sci. USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sat B., Reches M., Engelberg-Kulka H. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 2003;185:1803–1807. doi: 10.1128/JB.185.6.1803-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerdes K., Rasmussen P.B., Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl Acad. Sci. USA. 1986;83:3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulakauskas S., Lubys A., Ehrlich S.D. DNA restriction–modification systems mediate plasmid maintenance. J. Bacteriol. 1995;177:3451–3454. doi: 10.1128/jb.177.12.3451-3454.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kusano K., Naito T., Handa N., Kobayashi I. Restriction–modification systems as genomic parasites in competition for specific sequences. Proc. Natl Acad. Sci. USA. 1995;92:11095–11099. doi: 10.1073/pnas.92.24.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Naito T., Kusano K., Kobayashi I. Selfish behavior of restriction–modification systems. Science. 1995;267:897–899. doi: 10.1126/science.7846533. [DOI] [PubMed] [Google Scholar]
- 51.Ivanova N., Pavlov M.Y., Felden B., Ehrenberg M. Ribosome rescue by tmRNA requires truncated mRNAs. J. Mol. Biol. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto Y., Sunohara T., Jojima K., Inada T., Aiba H. SsrA-mediated trans-translation plays a role in mRNA quality control by facilitating degradation of truncated mRNAs. RNA. 2003;9:408–418. doi: 10.1261/rna.2174803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suyama M., Bork P. Evolution of prokaryotic gene order: genome rearrangements in closely related species. Trends Genet. 2001;17:10–13. doi: 10.1016/s0168-9525(00)02159-4. [DOI] [PubMed] [Google Scholar]
- 54.Nyström T. Conditional senescence in bacteria: death of the immortals. Mol. Microbiol. 2003;48:17–23. doi: 10.1046/j.1365-2958.2003.03385.x. [DOI] [PubMed] [Google Scholar]
- 55.Nyström T. Annu. Rev. Microbiol. Vol. 58. 2004. Stationary-Phase Physiology; pp. 161–181. [DOI] [PubMed] [Google Scholar]
- 56.Mazel D., Dychinco B., Webb V.A., Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.