Abstract
The TATA-binding protein (TBP) is a general factor that is involved in transcription by all three types of nuclear RNA polymerase. To delineate the roles played by the DNA-binding surface of TBP in these transcription reactions, we used a set of RNA aptamers directed against TBP and examined their ability to perturb transcription in vitro by the different RNA polymerases. Distinct responses to the TBP aptamers were observed for transcription by different types of polymerase at either the initiation, reinitiation or both stages of the transcription cycle. We further probed the TBP interactions in the TFIIIB•DNA complex to elucidate the mechanism for the different sensitivity of Pol III dependent transcription before and after preinitiation complex (PIC) formation. Lastly, the aptamers were employed to measure the time required for Pol III PIC formation in vitro. This approach can be generalized to define the involvement of a particular region on the surface of a protein at particular stages in a biological process.
INTRODUCTION
In the nucleus of eukaryotes, transcription is carried out by three different RNA polymerases, RNA polymerase I, II and III (Pol I, II and III) (1). Each of them is dedicated to the transcription of distinct sets of genes, but none is able to recognize its target promoters independently. Instead, general transcription factors (GTFs) recognize the promoter elements and recruit the correct RNA polymerase. The TATA-binding protein (TBP) is an essential GTF involved in transcription by all three eukaryotic RNA polymerases (2,3). TBP was first identified as a subunit of TFIID, a complex involved in transcription by Pol II (4–7), and recognizes an important eukaryotic core promoter motif, the TATA element (8). TBP is also present in two other complexes that are involved in Pol II dependent transcription, the B-TFIID complex and the TAC complex (9). Moreover, TBP is a component of complexes required for Pol I (SL1 complex) and Pol III (TFIIIB complex) dependent transcription (10–13).
Different RNA polymerases use diverse mechanisms to control and regulate transcription initiation and reinitiation. Pol I is reserved for the transcription of the large ribosomal RNA genes. To initiate Pol I dependent transcription, the polymerase interacts with promoters complexed with UBF and SL1. TBP and the Pol I-specific TBP-associated factors (TAFs) recruit Pol I directly to the promoter and remain bound to the DNA to support multiple rounds of transcription (14). A high density of Pol I molecules are loaded onto the rRNA gene when the rDNA template is being transcribed. Polymerases that read through from the upstream rDNA repeat can (in some cases) bypass the requirement for preinitiation complex (PIC) formation (15). These features contribute to the high transcription efficiency by Pol I.
Pol II is responsible for the transcription of the protein-coding genes and some small nuclear RNAs. Pol II depends on the basal transcription factors TFIID, IIB, IIF, IIE and IIH, and additional upstream activators for accurate and efficient initiation at different promoters. After Pol II leaves the promoter, TFIIB and TFIIF are released, whereas other factors such as activators, TBP, Mediator, TFIIH and TFIIE remain largely promoter-associated and form what is termed a reinitiation intermediate or scaffold, to facilitate subsequent rounds of transcription (16).
Pol III transcribes some structural and catalytic RNAs, including most small nuclear RNAs, tRNAs and 5S rRNA (17). It requires TFIIIB and IIIC for most of its promoters, and in addition, TFIIIA is essential for recognition of the 5S gene promoter (18). Transcription of Pol III genes also begins with the step-wise assembly of a PIC. When bound to the upstream region of 5S and tRNA genes, yeast TFIIIB correctly positions Pol III with respect to the promoter and supports multiple rounds of transcription (19). Reinitiation has a higher efficiency than does de novo initiation in Pol III dependent transcription (20). The stable association of TFIIIB with promoter, even after Pol III progresses into elongation, bypasses the need for PIC formation, thus accelerating the process of reinitiation.
TBP is a relatively small molecule with a divergent N-terminal domain and a highly conserved C-terminal domain. The C-terminal core domain of TBP is a pseudo-symmetric, saddle-shaped molecule with a concave surface that interacts primarily with the TATA element. This binding event induces a sharp bend in the DNA that is thought to be important for the juxtaposition of factors bound both upstream and downstream of the TATA element (21,22). Its convex side is recognized by many transcriptional activators and suppressors (23). The importance of TBP and, in particular, its DNA-binding surface seems to be distinct in different RNA polymerase systems. In an in vitro study, different types of transcription showed different sensitivity to TATA-containing DNA oligonucleotides (24), suggesting various roles played by the DNA-binding surface of TBP. Nevertheless, the functions of this surface area of TBP cannot be assessed accurately by using TATA-containing DNA, since RNA polymerase is known to associate with the ends of these DNA oligos non-specifically, thereby causing inhibition of transcription (24).
Previously, we isolated and characterized a set of RNA aptamers that bind TBP tightly (25). These aptamers are well-characterized specific molecular probes: they all appear to bind to the concave side of TBP based on their ability to compete with TATA DNA for binding to TBP, yet their modes of interaction with TBP are distinct (25). Here, we describe the utility of these aptamers as novel reagents to probe transcription by the three eukaryotic RNA polymerases. The different RNA polymerases responded distinctively to these TBP aptamers. Pol I dependent transcription was completely resistant to all of the TBP aptamers tested. In contrast, Pol II dependent transcription was the most sensitive to TBP aptamers. In crude cell extracts, the aptamers inhibited Pol II dependent transcription even after PICs were formed. Although TBP aptamers inhibited Pol III dependent transcription when they were present during PIC formation, they failed to inhibit transcription after PIC formation. These results revealed that the DNA-binding surface of TBP is involved to different extents in the transcription by different RNA polymerases at both initiation and reinitiation stages. It also revealed a fundamental difference between the stability of the reinitiation intermediate in the Pol II system and its counterpart in the Pol III system. The results not only provide insights into the different involvement of TBP in transcription initiation by these RNA polymerases, but they also demonstrate the application of these aptamers for studies of complicated reaction mechanisms as in our analysis of TBP in Pol III transcription. Where aptamers are available, this approach can be generalized to define the role of a particular area on a protein molecule at particular stages of a biological process.
MATERIALS AND METHODS
RNA polymerase I transcription reactions
Preparation of whole-cell extract was described previously (25,26). Transcription reactions were carried out essentially according to (26), with minor modifications. The yeast 35S ribosomal gene promoter was used in 20 μl reaction mixtures each containing 100 μg of yeast whole-cell extract (containing about 20 nM of TBP). The buffer contained 20 mM HEPES–KOH pH 7.9, 50 mM potassium chloride, 10 mM magnesium chloride, 5 mM EGTA, 0.05 mM EDTA, 2.5 mM DTT, 10% glycerol, 100 μM each ribonucleoside triphosphate, 10 μg/ml α-amanitin and template DNA at 10 μg/ml (2 nM). The mixture without ribonucleoside triphosphates (NTPs) was incubated at room temperature for 30 min to allow PIC formation. Transcription was started by the addition of NTPs and allowed to proceed for 30 min at room temperature. To inhibit Pol II dependent transcription, 10 μg/ml α-amanitin was included in the reaction. Reactions were stopped by the addition of 180 μl 20 mM EDTA, 200 mM sodium chloride, 10 mM Tris–HCl, pH 7.6. After phenol/chloroform extraction, the products were precipitated together with 3 μg of glyco-blue. The transcripts were assayed by either S1 protection or primer extension assays.
S1 nuclease protection assays were performed essentially according to (26), with a 50 nt DNA oligonucleotide probe complementary to the template DNA from −15 to +35 (probe 35S). Correctly initiated transcripts yielded a 35 nt probe fragment. Read-through transcription originated upstream of the promoter yielded a 50 nt probe fragment. Probe 35S has the sequence of 5′-GGTCTTGACGAACTTGTCTTCAACTGCTTTCGCATGAAGTACCTCCCAAC-3′. The reaction was shown to be resistant to α-amanitin up to 100 μg/ml, thus confirming it as Pol I dependent transcription.
RNA polymerase II transcription reactions
Transcription was performed according to a previously described protocol (25) except that a plasmid bearing the yeast CYC1 promoter and a 390 nt G-less cassette was used as the template. Aptamers or DNA oligos containing a TATA box sequence were added to the mixture and incubated for 10 min before the addition of NTPs.
RNA polymerase III transcription reactions
Transcription reactions with the tRNA LEU3 template were carried out essentially as described previously (26). A typical 20-μl reaction mixture contained 20 μg of yeast whole-cell extract and 20 mM HEPES–KOH pH 7.9, 80 mM potassium chloride, 5 mM magnesium chloride, 1 mM EDTA, 1 mM DTT, 10% glycerol, 10 μg/ml α-amanitin and template DNA at 5 μg/ml. The mixture was incubated at room temperature for 30 min to let PIC form. Transcription was started by the addition of 250 μM ATP, CTP, GTP, 25 μM UTP and 10 μCi [α-32P]UTP, and allowed to proceed for 30 min at room temperature. Reactions were stopped by the addition of 180 μl stop buffer (10 mM Tris–HCl, pH 7.6, 20 mM EDTA, 0.2 M sodium chloride). After phenol/chloroform extraction, the products were precipitated together with 3 μg of glyco-blue. The template used in the experiment had wild-type tRNA LEU3 gene in the vector of pBlueScript SK+, and the tRNA gene was derived from pGE2.wt (3,27).
Conditions for transcription reactions with the 5S rRNA template were identical to those for tRNA LEU3 reactions, except that 100 μg extract and 100 ng template were used in the reaction. The template was pY5S that included the entire yeast 5S gene (3).
Conditions for transcription reactions with the U6 snRNA were also identical to those for tRNA LEU3 reactions, except that 100 μg extract and 200 ng template were used in the reaction. The template was pCH6 that contained the gene of SNR6 from −120 to +629 (28).
The lengths and patterns of the transcripts from the three promoters were confirmed to be consistent with those reported in the literature (3), and the transcription was Pol III-specific (insensitive to low concentration of α-amanitin). Different strengths of transcription were observed for different promoters. The amount of template and cell extracts used in the experiment was determined empirically to give reasonable signals.
Electrophoretic mobility shift assay (EMSA)
For TFIIIB•DNA complex, the TATA-DNA used in the binding assays was produced by annealing deoxyoligonucleotides bearing a 30 bp DNA segment derived from the yeast SNR6 promoter with the following sequence: 5′-TTTTCGGCTACTATAAATAAATGTTTTTTT-3′. The TFIIIB•DNA complex was resolved on a 6% polyacrylamide gel run in 2× TE (20 mM Tris–HCl pH 8, 2 mM EDTA) buffer. In the competition experiment, DNA probe was incubated with different competitors before the addition of protein mixture containing 10 nM of TBP, Brf1 and Bdp1. In the disruption experiment, the TFIIIB•DNA complexes were formed at room temperature for 30 min before the addition of different competitors, the mixtures were loaded onto a native gel after another 30 min incubation.
RESULTS
Pol I dependent transcription is resistant to TBP aptamers
The core Pol I promoter is located immediately upstream of the initiation site, and is both necessary and sufficient for initiation of basal transcription in most species (14). Pol I promoters are less conserved than their counterparts in the Pol II and Pol III systems. There is very little sequence similarity between rRNA promoters from different species. The ribosomal initiator is an AT-rich sequence surrounding the initiation site, but it is not a binding site for TBP. Nonetheless, TBP is a subunit of the SL1 complex, which is required for Pol I dependent transcription (10), and in yeast, TBP itself has been shown to be required for transcription by Pol I (2,3). To assay the effects of TBP aptamers on Pol I dependent transcription, a template containing the promoter of yeast 35S rRNA gene was used in an in vitro transcription reaction and the transcripts were quantified either by S1 protection or primer extension assays (26). Control experiments showed that the transcription is specific for Pol I and was both template and extract dependent (see Materials and Methods).
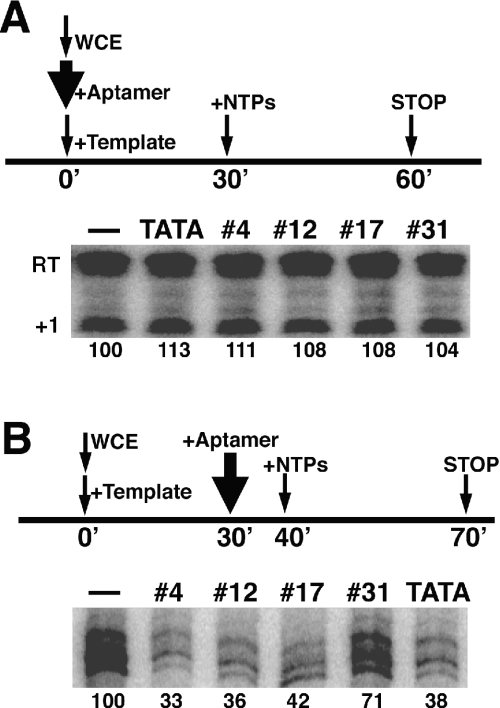
We attempted to perturb Pol I dependent transcription using three aptamers, named #4, #12 and #17, which bind tightly to TBP, and a control RNA called #31, which does not bind TBP (25). The effects of these aptamers were compared to those of a DNA fragment containing the TATA element. As shown in Figure 1A, no inhibition of transcription was observed when 160 nM RNA aptamers were added to cell extract simultaneously with the DNA template (RNA aptamers were incubated with the DNA template before mixing with the whole-cell extract). At this concentration, the RNA aptamers completely abolished transcription by Pol II [Figure 1B and (25)]. An even higher concentration of RNA aptamers (1.6 μM) also had no detectable effect on Pol I dependent transcription (data not shown). This result is consistent with a previous report that Pol I dependent transcription is very resistant to DNA oligos containing a TATA element (24) in a purified system. Based on this information and the fact that the TBP aptamers bind tightly to TBP and block DNA binding (25), our results indicate that the DNA-binding surface of TBP is not critical to rDNA template recognition, nor is it likely to function in a critical protein interaction associated with Pol I.
Figure 1.
Pol I and Pol II dependent in vitro transcription. The procedure is schematically represented at the top of each panel. The thin arrows indicate when the different components of the assay were added, and the thick arrow indicates when aptamers were added. The aptamers used are listed on the top of the corresponding lanes. The numbers under the lane indicate the relative intensity of the transcript signal, with the reaction without aptamer set at 100. TATA-DNA and the RNA #31 were used as controls. (A) Transcription from the 35S rRNA promoter. Transcripts were quantified by the S1 nuclease protection assay. The sign ‘+1’ indicates the transcripts starting from the correct site, while ‘RT’ indicates the read-through products of transcription from upstream (both products are Pol I dependent). (B) Transcription from the CYC1 promoter.
Pol II dependent transcription from the CYC1 promoter is sensitive to TBP aptamers
It has been proposed based on in vitro analysis that different Pol II promoters might use different sets of basal transcription factors (29). Previously, we showed that TBP aptamers #4, #12 and #17 were able to inhibit Pol II dependent transcription efficiently from an Adenovirus major late (AdML) promoter (25). To test the generality of the effect of TBP aptamers on the Pol II dependent transcription, here we employed a native yeast promoter, CYC1, which has distinct properties and can support high levels of transcription in vitro (30,31). In vivo, TBP has been shown to associate with the CYC1 promoter before gene activation (32–34). Unlike the AdML promoter, which directs transcription from a single start site, the CYC1 promoter directs transcription from multiple start sites both in vitro and in vivo (35,36).
As shown in Figure 1B, when the DNA template containing a CYC1 promoter was used in an in vitro transcription system, all three TBP aptamers (present at 160 nM), inhibited transcription efficiently even when they were added after the PIC had formed, while the negative control, RNA #31, had little effect. The same concentration of yeast tRNA also had no effect on transcription. Apparently, the different features of the CYC1 and AdML promoters did not cause detectable difference in their sensitivity to TBP aptamers. These results, and the previous observation that these RNA aptamers inhibited transcription from the AdML promoter before and after PIC was formed (25), indicate that it is possible to use these TBP aptamers as inhibitors for Pol II dependent transcription from TATA-containing promoters. As observed previously with the AdML promoter (25), here the TBP aptamers (and the TATA-DNA) act even after PIC formation and appear to gain access to the DNA-binding surface of TBP during rounds of reinitiation (Figure 1B). Therefore, while TBP has been shown to remain promoter-associated and part of a reinitiation scaffold (16), the interaction of TBP with DNA is sufficiently dynamic that these inhibitors are effective in blocking most Pol II transcription from both CYC1 and AdML promoters.
Pol III dependent transcription shows different sensitivity to TBP aptamers before and after PIC formation
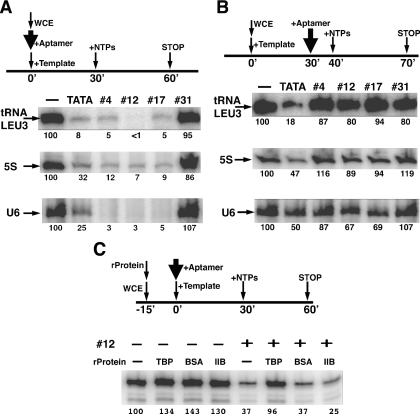
To test the effects of TBP aptamers on Pol III dependent transcription, we used three different templates, representing each category of genes transcribed by yeast Pol III. The promoter of the 5S rRNA gene is a type I promoter that requires the internal DNA sequence elements, the conserved A and C blocks, and the transcription factors TFIIIA, IIIB and IIIC for efficient transcription (14). Promoters of tRNA genes are type II promoters that require two highly conserved sequence blocks, A and B, within the transcribed region and transcription factors TFIIIB and TFIIIC (14). Yeast U6 snRNA genes have functional A and B blocks that are located in unusual positions and a functional TATA box upstream of the gene (28).
When RNA aptamers were present at 160 nM during PIC formation, transcription of all three Pol III genes was inhibited efficiently by all three TBP aptamers (Figure 2A). However, when we added the aptamers after incubating the template with the extract to allow the PIC to form, Pol III dependent transcription became resistant to the RNA aptamers (Figure 2B). In some cases, TATA containing DNA oligos had greater inhibitory effect than TBP RNA aptamers, even though the RNA aptamers bind more tightly to TBP and are more effective at disrupting TBP/DNA complexes (25). This may be due to the effect of double-stranded DNA on RNA polymerases—even double-stranded DNA without a TATA element were found to inhibit in vitro transcription by Pol II and Pol III (24). TBP aptamers are more specific inhibitors of TBP than TATA element-containing DNA oligos, as the RNA aptamers appear to act only on TBP but not on the RNA polymerase (25). A higher concentration of RNA aptamers (1.6 μM) also had no detectable inhibitory effect after PIC was formed (data not shown).
Figure 2.
Pol III dependent transcription from different promoters. Sensitivity of transcription to the aptamers before (A) and after (B) PIC formation. The procedure is schematically represented at the top. The aptamers used are listed on the top of the corresponding lanes. The genes assayed are denoted to the left of the gel. (C) Rescuing aptamer inhibition of Pol III dependent transcription by excess TBP. In each reaction, 2 nM tRNA LEU3 template was incubated with 20 μg yeast whole-cell extract (WCE) for 30 min to allow PIC formation. The transcription reaction was started by the addition of NTPs and allowed to proceed for 30 min. Where indicated, 30 ng of different recombinant proteins, TBP, BSA or TFIIB (about 50 nM for TBP), were mixed with WCE before mixing with the DNA template; aptamer #12 at final concentration of 20 nM was mixed with the DNA template before incubating with WCE.
To confirm that the inhibition of transcription by RNA aptamers was solely through the inhibition of TBP function, we tested the ability of TBP and additional different recombinant proteins to rescue the aptamer-inhibited transcription. As expected, recombinant TBP rescued the tRNA LEU3 transcription that was inhibited by aptamers (#12 is shown here, similar data was obtained for #4 and #17—not shown), but none of the other proteins tested were able to do so (Figure 2C). Like the BSA and TFIIB controls shown, Brf1, TFIIA and VP16 were also tested and had no ability to overcome the aptamer inhibition (data not shown). Inhibition of 5S rRNA and U6 snRNA by aptamer #12 can also be rescued by addition of recombinant TBP (data not shown). These experiments suggest that TBP was indeed the target and the only transcription-dependent target that RNA aptamers bind in this complex system.
TFIIIB•promoter interaction probed by RNA aptamers
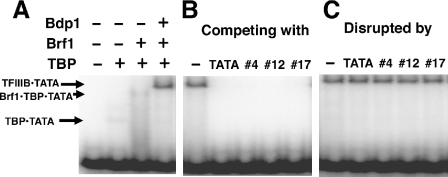
To probe the mechanism responsible for the difference in sensitivity of Pol III dependent transcription to the aptamers before and after the PIC formation, we examined directly the effects of RNA aptamers on the TFIIIB•promoter DNA binding. A short DNA fragment bearing the TATA element of the U6 promoter was used in EMSA experiments, and this fragment has an affinity for TBP similar to that of the AdML promoter-containing fragments. Two of our aptamers, #4 and #12, not only compete with TATA for forming TBP complex, but they also actively disrupt preformed TBP-TATA complexes formed with either AdML (25) or U6 promoters (data not shown). In the Pol II system, these aptamers (especially #4) also disrupted higher order complexes that contain TFIIA, or TFIIB, or both (25). In the TFIIIB•promoter system, the aptamers behaved differently.
We examined the effects of the three different aptamers on the formation and disruption of TFIIIB complexes. Functional TFIIIB was reconstituted by mixing recombinant TBP, Brf1 and Bdp1 (37). Without Bdp1, the resulting Brf1•TBP•U6 TATA–DNA complexes could be formed but they were not stable under the gel conditions established for the assay. Addition of Bdp1 to make full TFIIIB complexes produced a stable complex (Figure 3A). When RNA aptamers were incubated with the DNA probe before the addition of protein, RNA aptamers prevented the formation of TBP-containing complexes efficiently (Figure 3B), as was seen for the higher order Pol II GTF complexes (25). This effect can also be observed at a lower concentration of aptamers (40 nM, data not shown). In contrast, the preformed TFIIIB•U6 TATA–DNA complex was very resistant to all three RNA aptamers even when the aptamers were present at 320 nM (Figure 3C). This indicates that the aptamer-binding surface of TBP in the TFIIIB•DNA complex is not accessible to any of the RNA aptamers, or this surface of TBP is no longer critical for the stability of the protein•DNA complex once TFIIIB is associated with the promoter. This can explain why TBP aptamers were able to inhibit Pol III dependent transcription only prior to PIC formation; but they were not able to inhibit the transcription after PIC formation.
Figure 3.
TFIIIB•DNA interactions perturbed by aptamers. In the binding reaction, a DNA probe containing the TATA box and its flanking region of the yeast SNR6 gene was radiolabeled by kinase and used to form the TFIIIB•DNA complex (Bdp1•Brf1•TBP•TATA complex). (A) Assembly of the TFIIIB•DNA complex. The components of the various complexes are given to the left of the gel. (B) Prevention of the formation of the TFIIIB•DNA complex by aptamers present during assembly. In the binding reaction, 320 nM of the aptamers or TATA containing DNA oligos were included. (C) Resistance of the preformed TFIIIB•DNA complex to aptamers. The TFIIIB•DNA complex was formed as in (A), then a fraction of the mixture was taken out and mixed with 320 nM of different aptamers or TATA-DNA and incubated for 30 min at room temperature before being loaded onto a native gel.
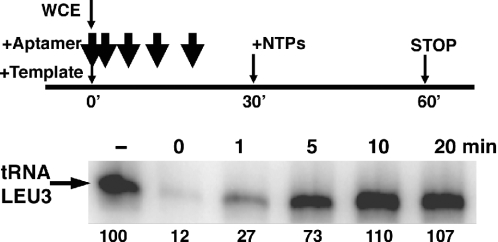
The kinetics of PIC formation in Pol III dependent transcription
The distinct responses of Pol III dependent transcription initiation and reinitiation to TBP aptamers allowed us to determine the duration of time required for Pol III transcription to become resistant to TBP aptamers, which should be indicative of the time required for PIC formation. Cell extracts were incubated with DNA template for 30 min before the reaction was started by the addition of NTPs. In each reaction, RNA aptamer #12 was added to the system at a time point before transcription was started. As shown in Figure 4, during the first minute of incubation of DNA template with cell extracts, the transcription is very sensitive to the aptamer. As time elapsed, the reaction becomes progressively resistant to the TBP aptamer. After a 10 min incubation, no difference can be discerned between reactions with or without aptamer added. This time course suggests that at least 5 min is required for forming an aptamer-resistant complex for Pol III dependent transcription. In a purified transcription system, adding aptamer at different times could be used further to reveal the dynamics of the interactions between DNA template and TFIIIC, TFIIIB and Pol III.
Figure 4.
Kinetics of PIC formation in Pol III dependent transcription. Transcription reactions were performed as in Figure 3, except that at different time points; 160 nM of aptamer #12 was added to the template and whole-cell extract mixture. Transcription of tRNA LEU3 gradually becomes resistant to the aptamer after the PIC is allowed to form.
DISCUSSION
Binding of RNA aptamers to a protein occludes a particular area on the surface of that protein, and thereby interferes with its normal interactions. Our RNA aptamers to TBP appear to bind to the concave, DNA-binding surface of TBP (25). Here, we test the ability of these aptamers to interfere with transcription of six different promoters. The results are summarized in Table 1. It is clear that different types of transcription responded distinctly to the TBP aptamers, whereas various promoters used by the same type of polymerase, but having distinct features, showed the same type of response. Consequently, like α-amanitin, these aptamers can be utilized to distinguish transcription by different polymerases.
Table 1.
Responses of transcription to TBP aptamers
| RNA polymerase | Promoter | Stage(s) of transcription | Sensitivity to aptamers |
|---|---|---|---|
| Pol I | 35S rRNA | Initiation and reinitiation | No |
| Pol II | CYC1 | Initiation and reinitiation | Yes |
| AdML | Initiation and reinitiation | Yes | |
| Pol III | tRNA LEU3 | Initiation | Yes |
| Reinitiation | No | ||
| 5S | Initiation | Yes | |
| Reinitiation | No | ||
| U6 | Initiation | Yes | |
| Reinitiation | No |
The different sensitivities of Pol I, II and III to RNA aptamers provide insights to the role of the concave surface of TBP in the processes of both PIC formation and the reinitiation of transcription. The resistance of Pol I transcription to aptamers supports the hypothesis that the DNA-binding surface of TBP is not involved in either DNA binding, complex assembly, or reinitiation. Alternatively, and less likely, it is also possible that the DNA-binding surface is buried and inaccessible to RNA aptamers in the SL1 complex, but if so, it must remain so both before and after SL1 binding to DNA. The sensitivity of Pol III dependent initiation and the resistance of Pol III reinitiation to aptamers are mirrored at the level of TFIIIB•DNA complex formation. These complementary experiments suggest that the concave surface of TBP is required to set up the Pol III PIC, but is either unnecessary or inaccessible to aptamers after the PIC formation. The resilience of a Pol III reinitation platform demonstrated here is reminiscent of work by Geiduschek and colleagues, who showed that 5S and tRNA gene promoters that contained a heparin-resistant TFIIIB complex is sufficient to allow reinitiation (19). In contrast, both Pol II PIC formation and reinitiation are sensitive to aptamers. The dynamic association of transcription factors with rounds of Pol II transcription, which is revealed by this susceptibility to aptamer inhibition, may allow the reinitiation process to serve as a checkpoint for transcription regulation.
While TBP is a subunit of the general transcription factor TFIIIB that is essential for Pol III dependent transcription, most Pol III promoter regions contain no TATA box or TATA-like DNA element. The binding of aptamers to TBP may block Brf1 from associating with TBP, thus prevent the assembly of TFIIIB, as Brf1 has been shown to contact both stirrups of TBP (38,39) and induce TBP dimer dissociation (40). Taking into account the fact that TBP uses overlapping surfaces for dimerization and DNA binding (41,42), it is possible that the binding of Brf1 to TBP needs the DNA-binding surface of TBP. After the slow step of PIC formation, which is susceptible to aptamer inhibition, and the first round of transcription, TFIIIB remains associated with the promoter and functions during the following reinitiation events, in which Pol III is the only factor that is recycled (15,20). This mechanism of reinitiation bypasses steps required for the initial transcription cycle, and potentially facilitates highly efficient RNA production. TFIIIB may also become more stable when it is associated with DNA, thus resisting challenge by the aptamers.
The RNA aptamers used here could also be used to study gene activation or repression in the context of a chromatin-based transcription system. Furthermore, we have demonstrated that RNA aptamers can be produced in living organisms as protein antagonists (43); however, studying transcription mechanisms using aptamers in vivo requires further refinement of the techniques for delivering active aptamers to cells with sufficient temporal resolution.
The binding sites of all three aptamers used in this study have been proposed to map to the concave side of TBP (25). If this submolecular specificity can be attained for different patches on the surface of the same protein, it would be possible to mechanistically dissect the roles played by these different surfaces. We have developed general methods to isolate aptamers to different sites on a single target molecule (44) and have recently isolated aptamers that bind to the convex side of TBP at a site overlapping with that recognized by TFIIA (to be documented elsewhere). As demonstrated here with three distinct functional assays, these reagents should allow the function of multiple sites on a protein molecule to be assessed in well-defined stages of a biological process.
Acknowledgments
We are very grateful to Drs E.P. Geiduscheck and G. Kassavetis for kindly providing recombinant TBP, Brf1 and Bdp1; to Drs J. Fu and M.H. Suh for purified TFIIB; to A. Sevilimedu for help with purification of TFIIA; to Dr K. Adelman for help with in vitro transcription assays; and to Drs S. Hahn, A. Berk, M.C. Schultz, D. Brow, Z.S. Juo and J.A. Jaehning for kindly providing strains and plasmids. This work was supported by an NIH grant GM40918 to JTL. Funding to pay the Open Access publication charges for this article was provided by NIH.
REFERENCES
- 1.Sentenac A. Eukaryotic RNA polymerases. CRC Crit. Rev. Biochem. 1985;18:31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- 2.Cormack B.P., Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 3.Schultz M.C., Reeder R.H., Hahn S. Variants of the TATA-binding protein can distinguish subsets of RNA polymerase I, II, and III promoters. Cell. 1992;69:697–702. doi: 10.1016/0092-8674(92)90233-3. [DOI] [PubMed] [Google Scholar]
- 4.Cavallini B., Huet J., Plassat J.L., Sentenac A., Egly J.M., Chambon P. A yeast activity can substitute for the HeLa cell TATA box factor. Nature. 1988;334:77–80. doi: 10.1038/334077a0. [DOI] [PubMed] [Google Scholar]
- 5.Hahn S., Buratowski S., Sharp P.A., Guarente L. Isolation of the gene encoding the yeast TATA binding protein TFIID: a gene identical to the SPT15 suppressor of Ty element insertions. Cell. 1989;58:1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- 6.Horikoshi M., Wang C.K., Fujii H., Cromlish J.A., Weil P.A., Roeder R.G. Cloning and structure of a yeast gene encoding a general transcription initiation factor TFIID that binds to the TATA box. Nature. 1989;341:299–303. doi: 10.1038/341299a0. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt M.C., Kao C.C., Pei R., Berk A.J. Yeast TATA-box transcription factor gene. Proc. Natl Acad. Sci. USA. 1989;86:7785–7789. doi: 10.1073/pnas.86.20.7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butler J.E., Kadonaga J.T. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- 9.Mitsiou D.J., Stunnenberg H.G. TAC, a TBP-sans-TAFs complex containing the unprocessed TFIIAalphabeta precursor and the TFIIAgamma subunit. Mol. Cell. 2000;6:527–537. doi: 10.1016/s1097-2765(00)00052-6. [DOI] [PubMed] [Google Scholar]
- 10.Comai L., Tanese N., Tjian R. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell. 1992;68:965–976. doi: 10.1016/0092-8674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- 11.Kassavetis G.A., Joazeiro C.A., Pisano M., Geiduschek E.P., Colbert T., Hahn S., Blanco J.A. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 12.Taggart A.K., Fisher T.S., Pugh B.F. The TATA-binding protein and associated factors are components of pol III transcription factor TFIIIB. Cell. 1992;71:1015–1028. doi: 10.1016/0092-8674(92)90396-t. [DOI] [PubMed] [Google Scholar]
- 13.White R.J., Jackson S.P. Mechanism of TATA-binding protein recruitment to a TATA-less class III promoter. Cell. 1992;71:1041–1053. doi: 10.1016/0092-8674(92)90398-v. [DOI] [PubMed] [Google Scholar]
- 14.Paule M.R., White R.J. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dieci G., Sentenac A. Detours and shortcuts to transcription reinitiation. Trends Biochem. Sci. 2003;28:202–209. doi: 10.1016/S0968-0004(03)00054-9. [DOI] [PubMed] [Google Scholar]
- 16.Yudkovsky N., Ranish J.A., Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez N. TBP, a universal eukaryotic transcription factor. Genes Dev. 1993;7:1291–1308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 18.Schramm L., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 19.Kassavetis G.A., Braun B.R., Nguyen L.H., Geiduschek E.P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 20.Dieci G., Sentenac A. Facilitated recycling pathway for RNA polymerase III. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 21.Nikolov D.B., Hu S.H., Lin J., Gasch A., Hoffmann A., Horikoshi M., Chua N.H., Roeder R.G., Burley S.K. Crystal structure of TFIID TATA-box binding protein [see comments] Nature. 1992;360:40–46. doi: 10.1038/360040a0. [DOI] [PubMed] [Google Scholar]
- 22.Chasman D.I., Flaherty K.M., Sharp P.A., Kornberg R.D. Crystal structure of yeast TATA-binding protein and model for interaction with DNA. Proc. Natl Acad. Sci. USA. 1993;90:8174–8178. doi: 10.1073/pnas.90.17.8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burley S.K., Roeder R.G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu. Rev. Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 24.Radebaugh C.A., Matthews J.L., Geiss G.K., Liu F., Wong J.M., Bateman E., Camier S., Sentenac A., Paule M.R. TATA box-binding protein (TBP) is a constituent of the polymerase I-specific transcription initiation factor TIF-IB (SL1) bound to the rRNA promoter and shows differential sensitivity to TBP-directed reagents in polymerase I, II, and III transcription factors. Mol. Cell. Biol. 1994;14:597–605. doi: 10.1128/mcb.14.1.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan X., Shi H., Adelman K., Lis J.T. Probing TBP interactions in transcription initiation and reinitiation with RNA aptamers that act in distinct modes. Proc. Natl Acad. Sci. USA. 2004;101:6934–6939. doi: 10.1073/pnas.0401523101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz M.C., Choe S.Y., Reeder R.H. Specific initiation by RNA polymerase I in a whole-cell extract from yeast. Proc. Natl Acad. Sci. USA. 1991;88:1004–1008. doi: 10.1073/pnas.88.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker R.E., Camier S., Sentenac A., Hall B.D. Gene size differentially affects the binding of yeast transcription factor tau to two intragenic regions. Proc. Natl Acad. Sci. USA. 1987;84:8768–8772. doi: 10.1073/pnas.84.24.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brow D.A., Guthrie C. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev. 1990;4:1345–1356. doi: 10.1101/gad.4.8.1345. [DOI] [PubMed] [Google Scholar]
- 29.Parvin J.D., Shykind B.M., Meyers R.E., Kim J., Sharp P.A. Multiple sets of basal factors initiate transcription by RNA polymerase II. J. Biol. Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 30.Lue N.F., Kornberg R.D. Accurate initiation at RNA polymerase II promoters in extracts from Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1987;84:8839–8843. doi: 10.1073/pnas.84.24.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woontner M., Jaehning J.A. Accurate initiation by RNA polymerase II in a whole cell extract from Saccharomyces cerevisiae. J. Biol. Chem. 1990;265:8979–8982. [PubMed] [Google Scholar]
- 32.Chen J., Ding M., Pederson D.S. Binding of TFIID to the CYC1 TATA boxes in yeast occurs independently of upstream activating sequences. Proc. Natl Acad. Sci. USA. 1994;91:11909–11913. doi: 10.1073/pnas.91.25.11909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuras L., Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 34.Li X.Y., Virbasius A., Zhu X., Green M.R. Enhancement of TBP binding by activators and general transcription factors. Nature. 1999;399:605–609. doi: 10.1038/21232. [DOI] [PubMed] [Google Scholar]
- 35.Hahn S., Hoar E.T., Guarente L. Each of three ‘TATA elements’ specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1985;82:8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W.Z., Sherman F. Two types of TATA elements for the CYC1 gene of the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:666–676. doi: 10.1128/mcb.11.2.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kassavetis G.A., Kumar A., Letts G.A., Geiduschek E.P. A post-recruitment function for the RNA polymerase III transcription-initiation factor IIIB. Proc. Natl Acad. Sci. USA. 1998;95:9196–9201. doi: 10.1073/pnas.95.16.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kassavetis G.A., Han S., Naji S., Geiduschek E.P. The role of transcription initiation factor IIIB subunits in promoter opening probed by photochemical cross-linking. J. Biol. Chem. 2003;278:17912–17917. doi: 10.1074/jbc.M300743200. [DOI] [PubMed] [Google Scholar]
- 39.Juo Z.S., Kassavetis G.A., Wang J., Geiduschek E.P., Sigler P.B. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature. 2003;422:534–539. doi: 10.1038/nature01534. [DOI] [PubMed] [Google Scholar]
- 40.Alexander D.E., Kaczorowski D.J., Jackson-Fisher A.J., Lowery D.M., Zanton S.J., Pugh B.F. Inhibition of TBP dimerization by RNA polymerase III transcription initiation factor Brf1. J. Biol. Chem. 2004;9:9. doi: 10.1074/jbc.M405782200. [DOI] [PubMed] [Google Scholar]
- 41.Kim Y., Geiger J.H., Hahn S., Sigler P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 42.Kim J.L., Nikolov D.B., Burley S.K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 43.Shi H., Hoffman B.E., Lis J.T. RNA aptamers as effective protein antagonists in a multicellular organism. Proc. Natl Acad. Sci. USA. 1999;96:10033–10038. doi: 10.1073/pnas.96.18.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi H., Fan X., Ni Z., Lis J.T. Evolutionary dynamics and population control during in vitro selection and amplification with multiple targets. RNA. 2002;8:1461–1470. doi: 10.1017/s1355838202029941. [DOI] [PMC free article] [PubMed] [Google Scholar]