Abstract
Long interspersed nuclear elements (LINE-1 or L1) comprise 17% of the human genome, although only 80–100 L1s are considered retrotransposition-competent (RC-L1). Despite their small number, RC-L1s are still potential hazards to genome integrity through insertional mutagenesis, unequal recombination and chromosome rearrangements. In this study, we provide several lines of evidence that the LINE-1 retrotransposon is susceptible to RNA interference (RNAi). First, double-stranded RNA (dsRNA) generated in vitro from an L1 template is converted into functional short interfering RNA (siRNA) by DICER, the RNase III enzyme that initiates RNAi in human cells. Second, pooled siRNA from in vitro cleavage of L1 dsRNA, as well as synthetic L1 siRNA, targeting the 5′-UTR leads to sequence-specific mRNA degradation of an L1 fusion transcript. Finally, both synthetic and pooled siRNA suppressed retrotransposition from a highly active RC-L1 clone in cell culture assay. Our report is the first to demonstrate that a human transposable element is subjected to RNAi.
INTRODUCTION
The majority of the human genome comprises DNA from repetitive sequences and mobile genetic elements. Retrotransposons, mobile genetic elements that move through an RNA intermediate in a process termed retrotransposition, are the most abundant mobile elements and comprise ∼40% of the human genomic DNA (1). Of these retrotransposons, the non-long terminal repeat (non-LTR) long interspersed nuclear elements (LINEs or L1) are referred to as autonomous retrotransposons because L1s encode their own proteins necessary for retrotransposition (2). Although over 99% of L1 sequences are inactive, either through deleterious mutations, 5′ end truncations, or through internal rearrangements, the human genome still contains ∼80–100 retrotransposition-competent LINE-1 elements (RC-L1s). The consensus RC-L1 is 6 kb in size and contains a 5′-untranslated region (5′-UTR) with an internal promoter, two non-overlapping open reading frames (ORF1 and ORF2) and a 3′-UTR that terminates with a poly(A) tail (2). ORF1 encodes a 40 kDa (p40) RNA binding protein that forms ribonucleoprotein particles (RNPs) with L1 RNA (3–5). ORF2 produces a 150 kDa protein with an N-terminal endonuclease and a C-terminal reverse transcriptase domain (6,7). A unique feature of L1s is that the two L1 ORF products act preferentially on the RNA that encodes them, a characteristic known as cis-preference, which might prevent rampant retrotransposition of other non-autonomous RNAs from disrupting the genome (8,9).
Despite the small number of RC-L1s, and the constraints placed upon their movement by cis-preference, a number of disease-causing mutations are attributed to insertional mutagenesis by L1 (2,10–12). In addition to insertional mutagenesis, unequal recombination between dispersed L1s and gene sequences is associated with several disease etiologies and has the potential to cause even more genomic damage due to the large number of L1 sequences (>500 000) that populate the human genome (13–15). Moreover, characterization of retrotransposition events using tagged RC-L1 clones in cultured cells indicate that ∼10% of L1 insertions are accompanied by large chromosomal rearrangements, suggesting that active L1s could also lead to genomic instability (16,17). In other eukaryotes, the RNA interference (RNAi) pathway suppresses the activity of transposable elements that predominate lower eukaryotic genomes. For example, RNAi is involved in silencing transposons in the germ line of Caenorhabditis elegans and silencing retroposons in Trypanosoma (18–22). Furthermore, disruption of the RNAi function in eight-cell stage mouse embryos results in increased accumulation of both IAP and MuERV-L transcripts (23). It is likely, therefore, that RNAi represents an evolutionary important mechanism whereby eukaryotic genomes are protected from the threat posed by mobile genetic elements.
RNAi is a conserved eukaryotic mechanism in which the double-stranded RNA (dsRNA) recognizes homologous mRNAs and causes sequence-specific degradation in a multi-step process. RNAi is initiated by the cleavage of endogenous long dsRNA or short hairpin RNA (shRNA) by the RNase III enzyme DICER into 21–23 nt small interfering RNA (siRNA) effector molecules (24–28). These siRNAs recognize their cognate mRNA and become associated with a large multi-protein complex referred to as the RNA-induced silencing complex (RISC) that destroys target mRNAs by endonucleolytic cleavage at regions homologous to the siRNA (29,30). Although only a few distinct components of the RNAi pathway are known, such as DICER, genetic and biochemical studies link members of the Argonaute family to the RISC complex (29,31,32). In addition, only a few endogenous siRNAs have been identified along with their targets. However, using both genetic and biochemical approaches several siRNAs derived from mobile genetic elements such as the Tc1 transposon in C.elegans and the Ingi retroposon in Trypanosoma are known to exist (18,22), although thus far there have not been any demonstrations of endogenous siRNAs targeting transposable elements in mammals.
In the present study, we show that dsRNA transcribed from a human L1 retrotransposon template is a substrate for in vitro cleavage by the RNase III enzyme DICER, and that the resulting L1 siRNA leads to RNAi-mediated degradation of a hybrid L1 fusion transcript. Moreover, both synthetic and in vitro processed L1 siRNA can inhibit L1 retrotransposon activity in an established cell culture retrotransposition assay. These results provide proof of principle that the human RNAi machinery can act on L1 transcripts and limit their movement.
MATERIALS AND METHODS
Cell culture
HeLa (CCL-2) and HCT116 (CCL-247) cells were obtained from ATCC. HeLa cells were grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 20 U/ml penicillin/streptomycin and 2 mM l-glutamine. HCT116 cells were grown in McCoy's 5A medium (Irvine Scientific, Irvine, CA) supplemented with 10% FBS, 20 U/ml penicillin/streptomycin and 1.5 mM l-glutamine. Both cell lines were cultured in a humidified 5% CO2 incubator at 37°C and split when they reached confluence.
Plasmid construction
Luciferase reporter vectors
Plasmid pGL3U1 was cloned by PCR amplification of the human U1 promoter and inserting it upstream of the firefly (FF) luciferase coding sequence in the pGL3 basic vector (Promega, Madison, WI). The wild-type 5′-UTR containing the L1 internal promoter was PCR amplified from either pCEP4/L1.3neo-ColE1 or pCEP4-L1RPneo using Pfu DNA polymerase (Stratagene, La Jolla, CA) and primers (HindIII-L1-F/HindIII-L1-R, Table 1). The resulting PCR products were digested with HindIII and subcloned into the pGL3 basic vector (Promega, Madison, WI), upstream of the luciferase cDNA to generate pHSS101/L1.3 and pHSS101/RP, respectively. The correct sequence of each luciferase construct was confirmed by DNA sequencing at the City of Hope DNA Sequencing Core.
Table 1.
Primers and synthetic siRNAs used in this study
| L1 5′-UTR | HindIII-L1-F: AGACTGTAAAGCTTGGGGGAGGAGCCAAG |
| HindIII-L1-R: TGTTTTTTAAGCTTCTTTGTGGTTTTATC | |
| L1 5′-UTR dsRNA | HS25: GCGTAATACGACTCACTATAGGGAGAGGGGAGGAGCCAAGATGGC |
| HS26: GCGTAATACGACTCACTATAGGGAGAATCTTTGTGGTTTTATCTAC | |
| L1 ORF1 dsRNA | HS22: GCGTAATACGACTCACTATAGGGAGATGTCATTATGATGTTAGCTGG |
| HS19: GCGTAATACGACTCACTATAGGGAGACCATCTGTACATCACCATCATC | |
| L1 ORF2A dsRNA | HS17: GCGTAATACGACTCACTATAGGGAGAAGCTAACATCATAATGACAGG |
| HS20: GCGTAATACGACTCACTATAGGGAGAGTGGGTTTGTCATAGATAGC | |
| L1 ORF2B dsRNA | HS21: GCGTAATACGACTCACTATAGGGAGATATCTATGACAAACCCACAGC |
| HS18: GCGTAATACGACTCACTATAGGGAGAATACATGTGCCATGCTGG | |
| LacZ dsRNA | HS98: GCGTAATACGACTCACTATAGGGAGACTGGCGTAATAGGAAGAGG |
| HS99: GCGTAATACGACTCACTATAGGGAGACAGCAGCAGACCATTTTCAA | |
| GAPDH | HS66: CAGGTGGTCTCCTCTGACTT |
| HS67: TGCTGTAGCCAAATTCGTTGT | |
| Firefly luciferase | HS102: TATCCGCTGGAAGATGGAAC |
| HS103: CCAACACCGGCATAAAGAAT | |
| neoR | 437S: GAGCCCCTGATGCTCTTCGTCC |
| 1808AS: CATTGAACAAGATGGATTGCACGC | |
| L1 5′-UTR siRNA #749 | Sense: GGCACACUGACACCUCACAdTdT |
| Antisense: UGUGAGGUGUCAGUGUGCCdTdT | |
| Firefly luciferase siRNA | Sense: CUUACGCUGAGUACUUCGAdTdT |
| Antisense: UCGAAGUACUCAGCGUAAGdTdT | |
| HIV Rev siRNA | Sense: GCGGAGACAGCGACGAAGCGCdTdT |
| Antisense: GCUCUUCGUCGCUGUCUCCGCdTdT |
All primers and siRNA are listed 5′→3′.
Retrotransposon and expression vectors
Plasmids pCEP4-L1RPneo (wild type) and pCEP4-L1.3-JM111 (mutant) are LINE-1 constructs that contain the mneoI retrotransposition indicator cassette described previously and were generously provided by Drs Haig H. Kazazian and John V. Moran (33,34). Plasmid pEMD-GFP is a mammalian reporter vector expressing the green fluorescent protein (Packard Instrument Company, Meriden, CT). Plasmid pCIneo (Promega, Madison, WI) is a mammalian expression vector that contains an SV40-driven neomycin phosphotransferase gene allowing for stable selection of transfected cells.
siRNA
Generation of dsRNA
cDNA templates with opposing T7 phage RNA polymerase promoters were generated by PCR by incorporating the T7 sequence into both forward and reverse primers. A complete list of primers used to produce cDNA templates for in vitro transcription is given in Table 1. PCR products were amplified from the pCEP4-L1RPneo plasmid template using Pfu DNA polymerase and then gel extracted following agarose gel electrophoresis. Additional long dsRNA was generated from a β-galactosidase template (plasmid pCH110, Pharmacia, Piscataway, NJ) which served as a negative control in RNAi experiments. These cDNA templates were subjected to in vitro transcription using the Megascript T7 in vitro transcription kit (Ambion, Austin, TX) according to the manufacturer's directions. The resulting long dsRNA was heated to 75°C for 5 min in a heat block and allowed to cool slowly at room temperature to anneal the opposing RNA strands. Annealed dsRNA was purified using the RNAid kit (Bio101, Vista, CA) and examined for quality on a 2.5–3% agarose gel.
Diced siRNA production
About 60 μg of dsRNA was incubated with recombinant human DICER (Block-it Dicer kit, Invitrogen, Carlsbad, CA) as recommended by the manufacturer. The resulting siRNA was purified using siRNA purification columns (Block-it Dicer kit) and the quality and quantity of the siRNA was examined by spectrophotometry and agarose gel electrophoresis.
Synthetic siRNA
Synthetic siRNA was designed using a web-based algorithm found at http://proteas.uio.no/siRNAbeta.html (35). Briefly, the nucleotide sequences of the L1 5′-UTR and ORF1 regions from the RC-L1 clone L1RP were used to generate a list of siRNA candidates. Each candidate was then queried against the RefSeq database and the siRNAs that exhibited limited sequence homology to known mRNAs (<15/19 nt), especially at the 5′ end of the antisense strand, were chosen for synthesis. Individual siRNA strands were synthesized chemically (IDT, Coralville, IA) as 21mers with dTdT 3′ overhangs. Sense and antisense siRNA strands were annealed by combining equimolar amounts, heating to 75°C for 5 min and then cooling slowly to room temperature. In addition, synthetic siRNAs targeting the FF luciferase coding region and HIV Rev transcript were also produced (IDT, Coralville, IA). The sequence of L1 5′-UTR #749, FF luciferase and HIV Rev synthetic siRNAs are listed in Table 1.
Luciferase experiments
HCT116 (colon carcinoma) were seeded in 24-well culture dishes and transfected at 70–80% confluence with Lipofectamine 2000 according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA). Each well received 1 ng of the Renilla luciferase expression vector phRL (Promega, Madison, WI), 10 ng pEMD-GFP, 200 ng luciferase reporter vector (either pGL3U1 or pHSS101 derivatives) and 0–250 ng L1-derived siRNA. In addition, pBluescript II SK+ (Stratagene, La Jolla, CA) was added so that each well received a total of 800 ng of nucleic acid. All samples were transfected in duplicate or triplicate and the experiment was performed a minimum of three times. For control transfections, either synthetic siRNA targeting the FF luciferase transgene (positive control for the RNAi effect), synthetic siRNA targeting the HIV Rev transcript (negative control) or negative control diced siRNA targeting the β-galactosidase mRNA was included instead of siRNA produced from L1 dsRNA. The transfections were performed as follows: first, the serum-containing media was removed and the cell monolayer was incubated with 100 μl of the transfection mix and returned to the incubator. After 1 h, the transfection mix was removed and 500 μl of serum-containing media was added to the monolayer and the cells were returned to the 37°C incubator. Luciferase assays were performed 24 h after transfection using the Dual Luciferase Assay system according the manufacturer (Promega, Madison, WI).
L1 retrotransposition experiments
Transient retrotransposition experiments using pCEP4-L1RPneo were performed essentially as described previously (36,37). HCT116 or HeLa cells were seeded in 6-well plates and transfected with Lipofectamine 2000 when they reached 70–80% confluence. Each well received 1 μg of RC-L1 construct, 100 ng of plasmid pEMD-GFP to normalize transfection efficiency, 0–250 ng siRNA and pBluescript II SK+ up to 2 μg of total nucleic acid. Transfections were performed in the absence of serum similar to the luciferase assay transfections. At 24 h post-transfection, each well was trypsinized, counted with a hemocytometer and serial dilutions were plated onto 100 mm dishes. The remaining cells were subjected to flow cytometry to normalize for transfection efficiency between samples. At 36 h post-transfection, the serially plated cells were selected using G418 at a concentration of 400 μg/ml (HCT116) and 600 μg/ml G418 (HeLa).
RT–PCR and genomic DNA PCR
Semi-quantitative RT–PCR
RNA was harvested from HCT116 cells 24 h post-transfection and treated two times with RNAse-free DNase I to remove any contaminating genomic and plasmid DNA. About 3 μg of RNA was used for cDNA preparation with M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA) and random hexamers to prime first-strand synthesis. Parallel cDNA reactions were carried out for each sample, except M-MLV reverse transcriptase was omitted from the reaction. Separate PCR reactions for GAPDH and FF luciferase transcripts were carried out in a total volume of 50 μl containing 1× Taq buffer, 1.5 mM MgCl2, 0.2 mM of each dNTP, 0.2 μM of each primer, 2.5 U Taq DNA polymerase (Brinkmann Instruments, Inc., Westbury NY) and 10 μl (∼100 ng) of the appropriate cDNA. The PCR was carried out at 95°C for 5 min (1 cycle) followed by 94°C for 30 s, 55°C for 30 s and 72°C for 30 s. Primers used to amplify GAPDH and FF luciferase transcripts are listed in Table 1. PCR cycles for GAPDH and FF luciferase were 22 and 24, respectively, as determined by cycle study analysis to determine the linear range of amplification. PCR products were separated on a 1.5% agarose gel and visualized by ethidium bromide staining. The amplified products were quantified using an Alpha-innotech imager and software.
Genomic DNA PCR
Genomic DNA was harvested from HCT116 G418R clones using the QiaAmp DNA Blood Kit (Qiagen, Valencia, CA). Each PCR reaction was performed in a 50 μl vol containing 1× Taq buffer, 1.5 mM MgCl2, 10 U of Taq polymerase, 0.2 mM dNTPs, ∼100 ng of genomic DNA template and 200 ng of primers 437S and 1808AS (Table 1), which are specific for the neoR gene. PCR was performed using a cycling program of 95°C, 5 min (1 cycle); 94°C, 1 min; 64°C, 30 s; 72°C, 1 min (30 cycles); a final extension of 72°C for 10 min. One-tenth of the PCR reaction was loaded onto a 1.5% agarose gel and visualized by ethidium bromide staining.
RESULTS
Recombinant human DICER can convert L1 dsRNA into siRNA
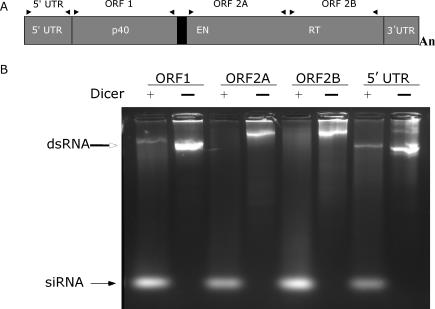
To investigate whether the human RNAi machinery has the potential to limit the activity of the human LINE-1 (L1) non-LTR retrotransposon, we first determined if long dsRNA generated from the L1s 5′-UTR, ORF1 and ORF2 regions, could be converted into siRNA by recombinant human DICER (Figure 1A). As shown in Figure 1B, L1 dsRNA was converted to siRNA of the appropriate size in an efficient and processive manner. Almost all of the long L1 dsRNA was converted into siRNA, yielding a substantial pool of siRNA for gene silencing experiments. However, before the gene silencing experiments could begin, the pool of siRNA needed to be purified from any contaminating long dsRNA because of the likelihood that the transfection of contaminating dsRNA would lead to non-specific reduction of mRNA and translation inhibition through interferon induction. We experimented with a number of ways to separate the siRNA from residual long dsRNA, including purification from both PAGE and agarose gels following electrophoresis. However, we found that using size-exclusion spin columns was the most efficient method and produced the highest quantity of siRNA, free of contaminating long dsRNA, as determined by agarose gel electrophoresis.
Figure 1.
L1 dsRNA is a substrate for recombinant human DICER. (A) Schematic representation of a retrotransposition-competent LINE-1 (RC-L1) illustrating the 5′-UTR with internal promoter, ORF1 product p40, inter-ORF region (bold black line), ORF2 domains EN (endonuclease) and RT (reverse transcriptase), and 3′-UTR. Arrowheads above the diagram indicate the positions of the primers (see Table 1) used to generate cDNA templates for in vitro transcription (B) About 1 μg of each L1 dsRNA was incubated in the presence (+) or absence (−) of recombinant human DICER for 18 h and resolved on a 2.5% agarose gel. Open arrowhead, long dsRNA; closed arrowhead, L1 siRNA, measuring ∼21–23 nt.
The diced L1 siRNA is functional at targeting a 5′-UTR-driven hybrid reporter transcript
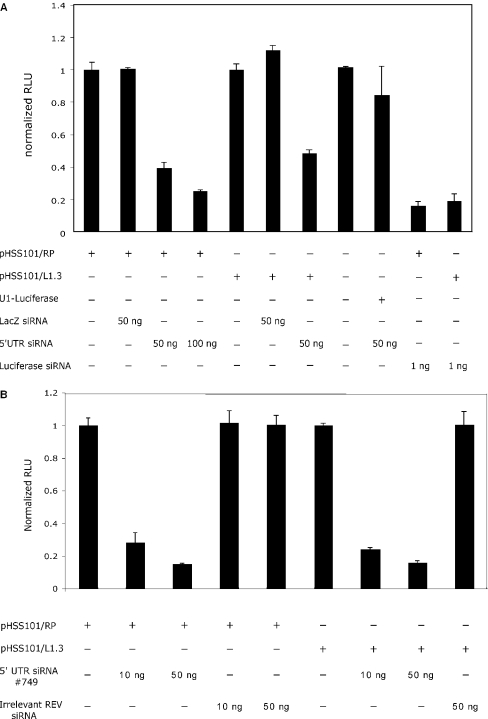
Next, we assessed whether L1 siRNA was capable of RNAi-mediated gene silencing of homologous L1 sequences. For this purpose, we cloned the 5′-UTR from the RC-L1 clones L1RP and L1.3 upstream of the FF luciferase reporter gene, to generate pHSS101/RP and pHSSS101/L1.3, respectively. The L1 5′-UTR is ∼910 nt and contains an internal promoter with transcription beginning at or near the first nucleotide of the element (38,39). Thus, the 5′-UTR-driven luciferase construct produces a hybrid mRNA with L1 sequence at the 5′ end of the transcript followed by the luciferase coding region and a poly (A) tail. Diced 5′-UTR L1RP siRNA, along with the respective pHSS101 construct, was co-transfected into HCT116 and the FF luciferase activity was determined after 24 h. A limiting amount of Renilla luciferase was included in each transfection to normalize for transfection efficiency between wells. No differences in Renilla luciferase activity were observed between different siRNA treatments, suggesting that the diced 5′-UTR siRNA did not produce any measurable off-target effects (data not shown).
In the presence of 50 ng diced 5′-UTR L1RP siRNA, the FF luciferase activity was reduced by 62% (P < 0.001), indicating that diced L1 siRNA could efficiently inhibit expression of the hybrid 5′-UTR-luciferase transcript. L1 siRNA inhibited the expression of the hybrid transcript in a dose-dependent manner and reached a maximum of 75% inhibition with 100 ng diced 5′-UTR siRNA (P = 0.01) (Figure 2). In addition to targeting the L1RP 5′-UTR from which the dsRNA template for in vitro dicing was generated, we investigated whether L1RP 5′-UTR siRNA could inhibit the expression of the hybrid transcript driven by the 5′-UTR from another RC-L1 clone, L1.3. Because the L1RP 5′-UTR and L1.3 5′-UTR exhibit greater than 98% sequence identity, we suspected that L1RP-derived 5′-UTR siRNA could reduce luciferase expression of the L1.3-driven hybrid transcript (40). As expected, L1RP-derived 5′-UTR siRNA inhibited the FF luciferase activity from a construct driven by the L1.3 5′-UTR by more than 51% (P = 0.002) (Figure 2). This amount of inhibition of the L1.3-driven transcript is significantly less (P = 0.001) than the 62% inhibition measured for the L1RP-driven transcript. To rule out the possibility that the lower inhibition of the L1.3-driven transcript is due to increased promoter activity of L1.3 compared with L1RP, we calculated the promoter strength of each 5′-UTR-driven vector in the absence of siRNA. Following normalization to Renilla luciferase, transcription from the L1RP 5′-UTR internal promoter was 1.4-fold higher (P = 0.009, data not shown) than the expression of FF luciferase from the L1.3 5′-UTR. Thus, the increased 5′-UTR siRNA-mediated inhibition of the L1RP-driven transcript may reflect the perfect complementarity of the L1RP-derived siRNA, whereas the L1.3-driven hybrid mRNA is predicted to contain small sequence differences at the 5′ end.
Figure 2.
L1 siRNA specifically inhibits the expression of a hybrid luciferase transcript driven by the L1 5′-UTR internal promoter. HCT116 cells were co-transfected with the indicated firefly (FF) luciferase expression vector and increasing amounts of L1 siRNA or control siRNA. Relative FF luciferase activity in the absence of siRNA was set at 1.0. The amount of each specific siRNA is indicated below the graphs. (A) Luciferase assays performed with in vitro ‘diced’ siRNA. Diced LacZ siRNA and synthetic FF luciferase siRNA served as negative and positive controls, respectively. (B) Luciferase assays performed with synthetic siRNA targeting nucleotides 749–769 of the L1 5′-UTR. Irrelevant synthetic siRNA targeting the HIV Rev transcript was included as the negative control.
Because the siRNA used in these experiments represents a pool derived from in vitro dicing of long 5′-UTR dsRNA, we wished to exclude the possibility that the observed inhibition in luciferase activity was due to the 5′-UTR siRNA acting against the luciferase region of the hybrid transcript. Thus, we co-transfected the L1RP 5′-UTR siRNA with a separate FF luciferase expression construct driven by the human U1 promoter. This U1-driven luciferase transcript contains no significant homology to the L1RP siRNA. As shown in Figure 2A, no significant decrease (P = 0.404) in luciferase activity was observed for the U1-driven luciferase construct in the presence of 100 ng L1RP 5′-UTR siRNA. However, synthetic luciferase siRNA significantly reduced expression from the U1-driven construct by >74%, indicating that this transcript is also susceptible to RNAi-mediated inhibition (data not shown). As an additional control for non-specific inhibition by diced siRNA, we ‘diced’ long dsRNA derived from the β-galactosidase coding region and co-transfected this β-gal siRNA with both pHSS101/L1.3 and pHSS101/L1RP. The diced β-gal siRNA showed no reduction of FF luciferase activity for both the L1RP- (P = 0.903) and L1.3-driven (P = 0.103) hybrid transcripts (Figure 2A). Thus, the decrease in luciferase activity mediated by L1 siRNA was specific to the hybrid transcript, and not to the luciferase sequence itself.
To conclusively demonstrate that the L1 5′-UTR is susceptible to RNAi, we designed three synthetic siRNAs against different regions of the 5′-UTR (5′-UTR #319, #454, #749, respectively). We carefully selected siRNA sequences that showed no significant homology to endogenous mRNAs by querying the RefSeq database. In preliminary co-transfection experiments with pHSS101/RP, 5′-UTR #749 siRNA was determined to be the most effective at silencing the hybrid transcript and chosen for additional experiments (data not shown). In transient co-transfection experiments with both pHSS101/RP and pHSS101/L1.3, as little as 10 ng of 5′-UTR #749 could decrease the luciferase activity by >75%, reaching a maximum suppression of 85% at 50 ng of the synthetic 5′-UTR #749 (Figure 2B). We did not observe increased silencing of luciferase activity at siRNA levels >50 ng, suggesting that the RNAi pathway is saturated at higher amounts of 5′-UTR #749. Irrelevant siRNA targeting the HIV Rev mRNA produced no inhibition of both 5′-UTR-driven transcripts (41).
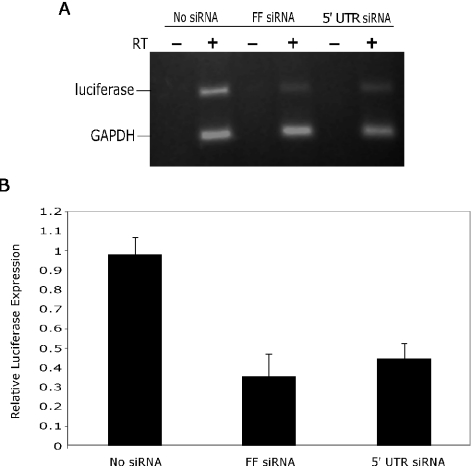
The diced siRNA reduces luciferase activity by decreasing mRNA levels of the 5′-UTR-driven transcript
To confirm that the observed decrease in FF luciferase activity was due to 5′-UTR siRNA targeting the hybrid transcript for RNAi-mediated degradation, we measured the expression of the remaining hybrid mRNA levels by semi-quantitative RT–PCR. As shown in Figure 3, the 5′-UTR siRNA reduced the hybrid transcript levels by 56% compared with that in the absence of siRNA. In addition, synthetic FF luciferase siRNA reduced the hybrid transcript levels by 64%. Thus, the pool of 5′-UTR siRNA is acting on the hybrid transcript by targeting it for degradation, presumably through an RNAi-mediated mechanism, and the reduced luciferase activity observed in the presence of 5′-UTR siRNA is not due to translational inhibition. While we measured a 56% reduction in hybrid mRNA levels in response to 5′-UTR siRNA, we observed a greater reduction in luciferase activity from the same sample using the commercial dual luciferase assay. This discrepancy most likely arises from the short half-life of luciferase mRNAs or the decreased sensitivity of semi-quantitative RT–PCR compared with the highly sensitive luciferase assay.
Figure 3.
L1 siRNA inhibits luciferase expression through RNAi-mediated degradation of the hybrid transcript. (A) RNA harvested from HCT116 cells co-transfected with pHSS101/RP and the indicated siRNA was subjected to semi-quantitative RT–PCR analysis with primers set to amplify the FF luciferase transgene and a separate set of primers to amplify the GAPDH gene as an internal loading control. FF luciferase expression was normalized to GAPDH levels. Relative luciferase expression in the absence of siRNA was set at 1.0. (B) Graphical representation of the semi-quantitative RT–PCR data. Columns represent mean FF luciferase expression (±standard deviations) of three independent PCR experiments.
L1 siRNA can reduce the activity of an RC-L1 in a transient retrotransposition assay
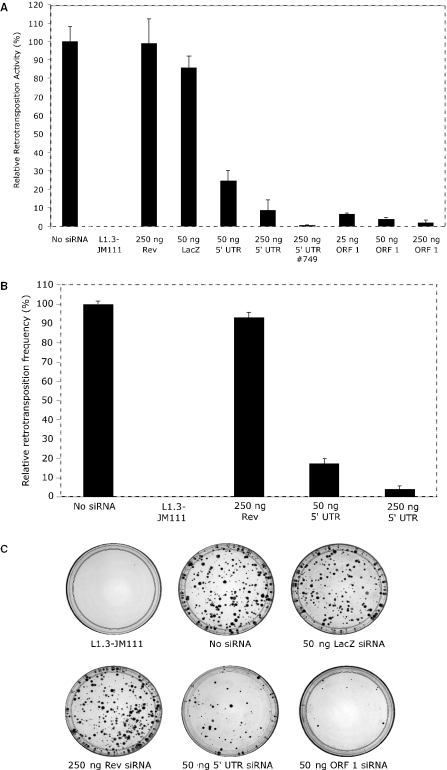
Next, we investigated whether L1 retrotransposition activity is reduced in the presence of siRNA homologous to different regions of an RC-L1 transcript by employing an established transient retrotransposition assay. In this assay, a tagged RC-L1 expression construct containing an indicator cassette (mneoI) is transiently co-transfected with a limiting amount of a GFP expression vector to monitor transfection efficiency. The mneoI retrotransposition indicator cassette consists of the neomycin phosphotransferase (neoR) selectable marker, a heterologous promoter and polyadenylation signal and is cloned in the 3′-UTR in the antisense orientation with respect to the L1 transcript. In addition, the neoR gene is interrupted by intron 2 of the γ-globin gene, which is in the sense orientation. Thus, expression of the mneoI cassette will occur only if RC-L1 is expressed from its 5′-UTR, and the intron is correctly spliced, reverse transcribed and integrated into chromosomal DNA. Expression of the neoR gene from the heterologous promoter will then give rise to G418 resistance (42). We chose the RC-L1 clone L1RP because it is one of the most active RC-L1 clones examined to date and would not only provide a high baseline level of retrotransposition from which to observe a decrease due to siRNA targeting the L1 transcript, but would also afford a good measurement of the potential for RNAi to limit L1 activity (34).
Before proceeding with the retrotransposition assays, we performed preliminary experiments to ensure that the diced L1 siRNA did not produce any off target effects which would pose problems during G418 selection, such as targeting the neoR mRNA. Therefore, we co-transfected HCT116 cells with the plasmid pCIneo, a mammalian expression vector that contains an SV40-driven neoR selectable marker, and diced siRNA from the L1s 5′-UTR, ORF1, or irrelevant LacZ siRNA. Serial dilutions of cells were plated and G418 selection was carried out 36 h after transfection for 14 days. No difference in stable G418 selection was observed for any of the diced siRNAs relative to pCIneo transfected without siRNA, demonstrating that the diced L1 and irrelevant LacZ siRNA do not produce non-specific inhibition (data not shown).
Next, we transfected HCT116 and HeLa cells with the tagged RC-L1RP construct alone or with serial dilutions of siRNA targeting the 5′-UTR. In the presence of as little as 50 ng diced 5′-UTR siRNA, the retrotransposition efficiency in HCT116 cells was significantly reduced by 75% relative to the no siRNA control (P < 0.001). The decrease in retrotransposition was dose-dependent and reached a maximum reduction (91%, P = 0.004) with 250 ng of diced 5′-UTR siRNA (Figure 4A). Although we measured a small 15% decrease in the retrotransposition activity with 50 ng of diced LacZ siRNA (P = 0.02), this difference may be the result of short regions of homology between the diced LacZ and both the 5′-UTR and ORF2 regions of L1RP. Retrotransposition activity was almost completely abolished with 250 ng synthetic 5′-UTR #749 siRNA (>99%, P < 0.001). This large decrease was specific to the 5′-UTR, as we did not detect any decrease in the retrotransposition activity in HCT116 cells in the presence of 250 ng synthetic irrelevant siRNA targeting the HIV Rev transcript (P = 0.92). We observed a more pronounced reduction in the retrotransposition frequency in HeLa cells transfected with both 50 ng (84%, P = 0.02) and 250 ng (94%, P = 0.009) of the diced 5′-UTR siRNA relative to the no siRNA control (Figure 4B). Again, irrelevant HIV Rev siRNA produced no significant reduction in L1 activity (P = 0.16).
Figure 4.
L1 siRNA can limit the activity of the RC-L1 clone, L1RP, in a transient retrotransposition assay. HCT116 or HeLa cells were transfected with pCEP4-L1RPneo alone or co-transfected with different siRNA. The retrotransposition activity was determined using a transient retrotransposition assay following G418 selection. The relative retrotransposition frequency of pCEP4-L1RPneo in the absence of siRNA was set at 100%. The negative control pCEP4-L1.3neo-JM111, an RC-L1 clone with two missense mutations in ORF1 rendering it inactive, showed no retrotransposition activity. (A) Results of the retrotransposition assay in HCT116 cells. Control siRNA used for transfection included 250 ng synthetic HIV Rev and 50 ng diced LacZ siRNA. Diced 5′-UTR siRNA (50 and 250 ng), diced ORF1 siRNA (25, 50 and 250 ng) and 250 ng 5′-UTR #749 synthetic siRNA were assayed separately. (B) Results of the retrotransposition assay in HeLa cells. Synthetic HIV Rev siRNA (250 ng) was included as a control. Diced 5′-UTR siRNA (50 and 250 ng) was also assayed. (C). The results of the retrotransposition assay achieved in HCT116 cells. The wild-type pCEP4-L1RPneo (or the negative control pCEP4-L1.3neo-JM111) was transfected alone or with the indicated amount of siRNA. G418 colonies were fixed and stained 14 days post-selection with 4% Giemsa for visualization.
To further study the potential of RNAi to limit the activity of the L1 retrotransposon, we also measured retrotransposition activity in HCT116 cells exposed to diced ORF1 siRNA. ORF1 siRNA also reduced retrotransposition activity in a dose-dependent manner relative to the no siRNA control (Figure 4A). The observed reduction in response to 50 ng diced ORF1 siRNA was significantly greater than the reduction in the retrotransposition activity measured with the same concentration of diced 5′-UTR siRNA (75% versus 96%, P < 0.02). We were unable to design synthetic ORF1 siRNA using the web-based algorithm that lacked significant homology to known endogenous mRNAs after querying the RefSeq database. To confirm that the G418 resistance in HCT116 clones was derived from a retrotransposition event, proper splicing of the γ-globin intron disrupting the neoR gene was confirmed by PCR analysis of genomic DNA from several stable clones (data not shown).
DISCUSSION
We have several lines of evidence providing proof of principle that the RNAi machinery can act on L1s and limit their activity in a transient retrotransposition assay. First, long dsRNA transcribed in vitro from an RC-L1 clone is efficiently processed into functional siRNA by the RNase III enzyme DICER. Second, both synthetic and in vitro ‘diced’ L1 siRNA inhibit the expression of an L1 hybrid transcript by RNAi-mediated degradation. Third, siRNA targeting different regions of the L1 can limit the activity of an RC-L1 in a cultured cell retrotransposition assay.
We initially decided to target the 5′-UTR of the L1 retrotransposon because this region is presumably part of most, if not all, remaining RC-L1s that populate the human genome. In addition, the L1 5′-UTR contains both a sense promoter, as well as an antisense promoter (ASP), and thus might play a larger role in the transcriptional regulation (43) through the production of L1 dsRNA. Furthermore, because the 5′-UTR contains the L1s internal promoter with transcription beginning at or near the first nucleotide of the L1 at an unconventional start site (5′-GGGGG-3′), we reasoned that it would be easy to test the efficacy of siRNAs targeting the 5′-UTR by creating a fusion construct with a measurable reporter gene such as FF luciferase (38,39). Indeed, we found that the 5′-UTR-driven reporter construct achieved high levels of luciferase expression in both HCT116 and HeLa cells, consistent with the data from other studies examining the promoter activity of the L1 5′-UTR, suggesting that the factors necessary for efficient transcription are present in many somatic cells types (38,39,44,45). Even with the robust expression of the 5′-UTR driven constructs, we observed a sharp decrease in the luciferase activity from the hybrid transcript in the presence of either in vitro ‘diced’ or synthetic siRNA targeting the 5′-UTR. The decrease in the luciferase activity was not due to non-specific targeting of the luciferase coding region, as a control construct expressing FF luciferase from the human U1 snRNA promoter showed no decrease in luciferase activity in the presence of 5′-UTR siRNA.
While the in vitro ‘diced’ siRNA represents a pool of many 21–23 nt siRNAs spanning the 910 nt 5′-UTR region, and thus it is not possible to know which fraction of the pool can actually function to target the homologous regions of the transcript for RNAi-mediated degradation, we have demonstrated that the RNAi effect occurs only with transcripts that include the 5′-UTR and not the luciferase coding region itself. In addition, in vitro ‘diced’ irrelevant siRNA from the β-galactosidase coding region did not reduce the expression of the hybrid transcript, providing further evidence that the significant reduction in FF luciferase activity observed with the pool of 5′-UTR assay is due to sequence-specific targeting of the hybrid transcript. Furthermore, the reduced luciferase mRNA levels detected by semi-quantitative RT–PCR provide additional evidence that the decreased luciferase activity is due to reduction in the hybrid transcript, and not due to altered translation of the hybrid reporter gene. Thus, our data is in agreement with two other published reports that in vitro cleavage of long dsRNA by recombinant DICER is a viable method to produce sequence-specific siRNAs for gene silencing experiments (46,47). Interestingly, the use of a single siRNA (5′-UTR #749) provided a more potent silencing effect than a similar concentration of diced 5′-UTR siRNA. Furthermore, the silencing effect of the synthetic 5′-UTR siRNA was very pronounced even at low concentrations, but we did not observe the same silencing effect with diced siRNA at these lower concentrations. One speculation for the increased potency of the synthetic siRNA is that the functional 21mers in the diced siRNA pool compete with the many non-functional 21mers for target binding and active RISC formation. Indeed, we have observed a similar phenomenon when targeting endogenous genes; namely that two weakly functional siRNAs do not perform together as well as a single efficient siRNA at lower concentrations (H. S. Soifer, unpublished data).
Although we demonstrated that siRNA homologous to the 5′-UTR could target a hybrid reporter transcript for RNAi-mediated degradation, it was not readily apparent that the full length L1 transcript would be susceptible to RNAi. This concern grew from the fact that the L1 RNA exists in the cytoplasm as RNPs with multimerized p40 protein encoded by its own ORF1 (3,4). Although the function of the human p40 has not been assigned, one possibility is that the RNPs protect the L1 from degradation by cytoplasmic RNAses. Despite the co-localization of L1 RNA to RNPs, however, siRNA targeting the 5′-UTR produced >80% reduction in L1 retrotransposition activity in HCT116 cells at concentrations of ∼10 nM. We observed a greater reduction of retrotransposition activity in HeLa cells with the same concentrations of diced 5′-UTR siRNA relative to the no siRNA control. The increased response of HeLa cells to 5′-UTR siRNA might result from the hyper-triploid karyotype of HeLa cells, thus providing more RNAi components compared with HCT116 cells, which possess a near diploid karyotype (48,49). Since we have little understanding for the role of human p40 in the retrotransposition process, we can only speculate that the siRNA is working at a stage before the RNP assembly with L1 RNA. If the siRNA is capable of targeting the L1 RNA after p40 multimerization and RNP assembly, this would require association of the large RNP with RISC producing a very large complex approaching 800 kDa. While p40 does contain a leucine zipper that could mediate interactions with other proteins, there is no evidence to suggest that the human L1 p40 interacts with the RISC machinery.
In addition to targeting the 5′-UTR, we also produced in vitro ‘diced’ siRNA from the ORF1 region of the RC-L1 clone L1RP. The level of inhibition achieved with 10 nM of diced ORF1 siRNA was significantly greater than the inhibition observed with a similar concentration of 5′-UTR siRNA. A number of possibilities exist to explain the more pronounced decrease in L1 retrotransposition in response to ORF1 siRNA. The most obvious explanation is that the ORF1 siRNA pool contains more functional siRNA molecules than the pool of 5′-UTR siRNA. While there is some indication as to the thermodynamic characteristics that distinguish a functional siRNA duplex from a non-functional siRNA, such as 5′-end duplex stability, GC content, and internal Tm, we are still unable to predict which set of 21mers from a given long dsRNA will be effective siRNAs (50,51). Additionally, we cannot identify the population of ‘diced’ siRNA based on the sequence of the starting long dsRNA. Although biochemical evidence suggests that DICER recognizes the stem loop structure of both shRNA and micro RNA precursors and measures from the end of the stem to cleave a 21–23 nt duplex, the manner in which DICER cleaves long dsRNA ex vivo has not been established (52) (H. S. Soifer, unpublished data). Another possibility is that the stable secondary structure of the 5′-UTR resulting from higher GC content (58% 5′-UTR versus 42% ORF1) poses a barrier to RNAi. Indeed, strong localized secondary structure surrounding the target site of the mRNA is known to negatively affect RNAi (53).
In summary, we have presented evidence that the L1 retrotransposon is susceptible to RNA interference. While our study does not provide direct proof that RNAi functions to control the activity of the remaining RC-L1s, we have provided proof of principle that the RNAi machinery can act on an active RC-L1 and limit its retrotransposition activity using a cell culture retrotransposition assay. Direct evidence that RNAi limits the activity of L1s awaits the cloning of endogenous LINE-1 siRNAs from human cells. Efforts to clone the small RNA population from cultured human cells failed to detect L1 siRNA, suggesting that if endogenous L1 siRNAs are produced, they may be present only during specific developmental stages (54). One requirement for the production of L1 siRNA would be transcription of antisense L1 RNA that could hybridize with L1 sense RNA to form either dsRNA or siRNA. In fact, there is experimental evidence demonstrating that large quantities of both sense and antisense L1 RNA of variable size are present in the total RNA of a human teratocarcinoma cell line (55). As the human genome is populated by more than 500 000 L1 sequences, it is possible that a subset is transcribed by a nearby promoter into ds- or siRNA. This arrangement, however, would still require the existence of two opposing promoters to transcribe both sense and antisense L1 sequence. Alternatively, the production of sense/antisense L1 dsRNA might take advantage of a unique feature of the L1 5′-UTR, namely the existence of an internal promoter that transcribes L1 sense RNA and an antisense promoter within nucleotides +400 to +600 of the 5′-UTR that transcribes L1 sequence in the opposite direction. In cell lines where the 5′-UTR sense promoter shows transcriptional activity, the L1 ASP is also transcriptionally active, albeit at lower levels (43). A recent report demonstrates that transcription from the L1 ASP is dependent on the transcription factor RUNX3 (44). In addition to characterizing endogenous L1-derived siRNAs that may arise from the L1s 5′-UTR, the establishment of conditional RNAi knock-out mice and derived fibroblast lines will permit more extensive analysis of the retrotransposon activity than can be achieved with the current DICER and AGO2 null mouse models (31,56).
Acknowledgments
We would like to thank Dr John V. Moran (University of Michigan) and Dr Haig H. Kazazian (University of Pennsylvania) for generously providing the pCEP4-based retrotransposon plasmids used in this study. We also thank members of the Rossi Lab for helpful comments. This work was supported by a post-doctoral fellowship awarded to H.S.S. by the Arnold and Mabel Beckman Foundation and a grant from NLB HL074704 to J.J.R. Funding to pay the Open Access publication charges for this article was provided by grant NLB HL074704 to J.J.R.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Ostertag E.M., Kazazian H.H., Jr Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 3.Hohjoh H., Singer M.F. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hohjoh H., Singer M.F. Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. J. Mol. Biol. 1997;271:7–12. doi: 10.1006/jmbi.1997.1159. [DOI] [PubMed] [Google Scholar]
- 5.Hohjoh H., Singer M.F. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Q., Moran J.V., Kazazian H.H., Jr, Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 7.Mathias S.L., Scott A.F., Kazazian H.H., Jr, Boeke J.D., Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 8.Esnault C., Maestre J., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nature Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 9.Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kazazian H.H., Jr, Wong C., Youssoufian H., Scott A.F., Phillips D.G., Antonarakis S.E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 11.Schwahn U., Lenzner S., Dong J., Feil S., Hinzmann B., van Duijnhoven G., Kirschner R., Hemberger M., Bergen A.A., Rosenberg T., et al. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nature Genet. 1998;19:327–332. doi: 10.1038/1214. [DOI] [PubMed] [Google Scholar]
- 12.Brouha B., Meischl C., Ostertag E., de Boer M., Zhang Y., Neijens H., Roos D., Kazazian H.H., Jr Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am. J. Hum. Genet. 2002;71:327–336. doi: 10.1086/341722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burwinkel B., Kilimann M.W. Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J. Mol. Biol. 1998;277:513–517. doi: 10.1006/jmbi.1998.1641. [DOI] [PubMed] [Google Scholar]
- 14.Kumatori A., Faizunnessa N.N., Suzuki S., Moriuchi T., Kurozumi H., Nakamura M. Nonhomologous recombination between the cytochrome b558 heavy chain gene (CYBB) and LINE-1 causes an X-linked chronic granulomatous disease. Genomics. 1998;53:123–128. doi: 10.1006/geno.1998.5510. [DOI] [PubMed] [Google Scholar]
- 15.Suminaga R., Takeshima Y., Yasuda K., Shiga N., Nakamura H., Matsuo M. Non-homologous recombination between Alu and LINE-1 repeats caused a 430-kb deletion in the dystrophin gene: a novel source of genomic instability. J. Hum. Genet. 2000;45:331–336. doi: 10.1007/s100380070003. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert N., Lutz-Prigge S., Moran J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 17.Symer D.E., Connelly C., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 18.Sijen T., Plasterk R.H. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 19.Tabara H., Sarkissian M., Kelly W.G., Fleenor J., Grishok A., Timmons L., Fire A., Mello C.C. The rde-1 gene, RNA interference, and transposon silencing in C.elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 20.Ketting R.F., Haverkamp T.H., van Luenen H.G., Plasterk R.H. Mut-7 of C.elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 21.Djikeng A., Shi H., Tschudi C., Ullu E. RNA interference in Trypanosoma brucei: cloning of small interfering RNAs provides evidence for retroposon-derived 24–26-nucleotide RNAs. RNA. 2001;7:1522–1530. [PMC free article] [PubMed] [Google Scholar]
- 22.Shi H., Djikeng A., Tschudi C., Ullu E. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol. Cell. Biol. 2004;24:420–427. doi: 10.1128/MCB.24.1.420-427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svoboda P., Stein P., Anger M., Bernstein E., Hannon G.J., Schultz R.M. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev. Biol. 2004;269:276–285. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- 24.Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 25.Provost P., Dishart D., Doucet J., Frendewey D., Samuelsson B., Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 28.Paddison P.J., Caudy A.A., Bernstein E., Hannon G.J., Conklin D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 30.Martinez J., Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 32.Doi N., Zenno S., Ueda R., Ohki-Hamazaki H., Ui-Tei K., Saigo K. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 2003;13:41–46. doi: 10.1016/s0960-9822(02)01394-5. [DOI] [PubMed] [Google Scholar]
- 33.Sassaman D.M., Dombroski B.A., Moran J.V., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr Many human L1 elements are capable of retrotransposition. Nature Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 34.Kimberland M.L., Divoky V., Prchal J., Schwahn U., Berger W., Kazazian H.H., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 35.Amarzguioui M., Prydz H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 36.Han J.S., Boeke J.D. A highly active synthetic mammalian retrotransposon. Nature. 2004;429:314–318. doi: 10.1038/nature02535. [DOI] [PubMed] [Google Scholar]
- 37.Wei W., Morrish T.A., Alisch R.S., Moran J.V. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal. Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 38.Swergold G.D. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Athanikar J.N., Badge R.M., Moran J.V. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32:3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostertag E.M., Prak E.T., DeBerardinis R.J., Moran J.V., Kazazian H.H., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee N.S., Dohjima T., Bauer G., Li H., Li M.J., Ehsani A., Salvaterra P., Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 42.Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 43.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang N., Zhang L., Zhang Y., Kazazian H.H., Jr An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hata K., Sakaki Y. Identification of critical CpG sites for repression of L1 transcription by DNA methylation. Gene. 1997;189:227–234. doi: 10.1016/s0378-1119(96)00856-6. [DOI] [PubMed] [Google Scholar]
- 46.Kawasaki H., Suyama E., Iyo M., Taira K. siRNAs generated by recombinant human Dicer induce specific and significant but target site-independent gene silencing in human cells. Nucleic Acids Res. 2003;31:981–987. doi: 10.1093/nar/gkg184. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Myers J.W., Jones J.T., Meyer T., Ferrell J.E., Jr Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat. Biotechnol. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- 48.Macville M., Schrock E., Padilla-Nash H., Keck C., Ghadimi B.M., Zimonjic D., Popescu N., Ried T. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 1999;59:141–150. [PubMed] [Google Scholar]
- 49.Masramon L., Ribas M., Cifuentes P., Arribas R., Garcia F., Egozcue J., Peinado M.A., Miro R. Cytogenetic characterization of two colon cell lines by using conventional G-banding, comparative genomic hybridization, and whole chromosome painting. Cancer Genet. Cytogenet. 2000;121:17–21. doi: 10.1016/s0165-4608(00)00219-3. [DOI] [PubMed] [Google Scholar]
- 50.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 51.Schwarz D.S., Tomari Y., Zamore P.D. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr. Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 53.Yoshinari K., Miyagishi M., Taira K. Effects on RNAi of the tight structure, sequence and position of the targeted region. Nucleic Acids Res. 2004;32:691–699. doi: 10.1093/nar/gkh221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 55.Skowronski J., Singer M.F. Expression of a cytoplasmic LINE-1 transcript is regulated in a human teratocarcinoma cell line. Proc. Natl Acad. Sci. USA. 1985;82:6050–6054. doi: 10.1073/pnas.82.18.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. Dicer is essential for mouse development. Nature Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]