Abstract
Pathway- or disease-associated genes may participate in more than one transcriptional co-regulation network. Such gene groups can be readily obtained by literature analysis or by high-throughput techniques such as microarrays or protein-interaction mapping. We developed a strategy that defines regulatory networks by in silico promoter analysis, finding potentially co-regulated subgroups without a priori knowledge. Pairs of transcription factor binding sites conserved in orthologous genes (vertically) as well as in promoter sequences of co-regulated genes (horizontally) were used as seeds for the development of promoter models representing potential co-regulation. This approach was applied to a Maturity Onset Diabetes of the Young (MODY)-associated gene list, which yielded two models connecting functionally interacting genes within MODY-related insulin/glucose signaling pathways. Additional genes functionally connected to our initial gene list were identified by database searches with these promoter models. Thus, data-driven in silico promoter analysis allowed integrating molecular mechanisms with biological functions of the cell.
INTRODUCTION
The completion of several whole-genome sequencing projects has provided extensive lists of genes (DNA), RNAs and proteins of mammalian organisms (1–3). However, it quickly became evident that the complexity of higher organisms cannot be explained solely by the number of parts, but mainly arises from more sophisticated interactions and networks of the DNAs, RNAs and proteins (4). This triggered a new focus towards the analysis of gene groups, their products and their network interactions (e.g. signaling and metabolic networks), which is now defined as the ultimate goal of systems biology (5,6).
Part of that effort is the elucidation of transcriptional co-regulation networks, which can be seen as one of the most important levels at which network connections emerge (7,8). Considerable progress has been made in analysis of yeast regulatory networks from microarray experiments (9,10). However, those results cannot be generally transferred to the human system (11). Therefore, mammalian transcriptome analysis, which is a current focus of research (12,13), requires different strategies suitable for mammalian networks. A common theme to all analyses aiming at gene or gene product interactions is the definition of one or several interacting subsets associated by some evidence to a biological process, disease or condition. Such gene groups often are not well defined and contain several functionally distinct subgroups, which cannot be separated by conventional clustering methods (14). However, genes within such subgroups contributing to a particular biological pathway or process may be transcriptionally coupled to insure coordinated availability of the proteins. Transcription is primarily regulated by the binding of transcription factors to their specific binding sites in the promoter/enhancer of the genes (7). Therefore, one way to trace co-regulated transcription on the molecular level is by promoter analysis revealing shared organization of sets of transcription factor binding sites (referred to as frameworks hereafter). Such frameworks can be represented by computational models, which can be used to scan sequence databases for genes showing a similar promoter organization (15).
Unfortunately, promoter sequence conservation is not general (15) and even conserved sequence regions, called phylogenetic footprints (16) are not directly associated with functional conservation. Each mammalian promoter represents a mixture of conserved frameworks (associated with different signaling responses of the same promoter) necessary to ensure correct timing and spatial distribution of expression during development as well as correct function in the adult stage. Therefore, separation of individual functions by phylogenetic promoter analysis without further information about the biological context is usually not possible. On the other hand, horizontally co-regulated promoters (different genes within one mammalian species) often also share arbitrary frameworks that cannot be distinguished from those associated with the observed co-regulation.
We have designed a completely new strategy that combines phylogenetic analysis (inter-species analysis) with cross-gene analysis within one species (intra-species analysis) to identify single process-associated frameworks, overcoming the functional ambiguities of the individual approaches. We demonstrate on an example of a disease-related gene list that in silico promoter analysis contributes to bridging the gap between molecular mechanisms and biological functions of the cell.
METHODS
Terminology
Framework: Two or more transcription factor binding sites (TFBSs) arranged in a defined order, orientation and a defined distance range between adjacent TFBSs.
Model: Computational description of a framework for the purpose of computer-assisted detection of occurrences of frameworks in long DNA sequences.
Recall: Percentage of input gene promoters recognized by a model: 100% recall means all input gene promoters are found.
Selectivity: The ratio of recall versus the fraction (in %) of promoters from a large promoter database matched by the model (control).
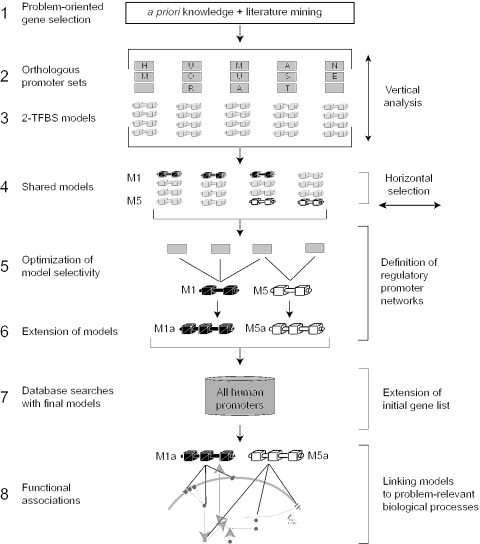
The step numbers below refer to the numbers in Figure 1.
Figure 1.
General strategy for problem-oriented promoter modeling. The bold numbers to the left of the short descriptions indicate the different steps of the strategy and correspond to the numbering used in Methods and Results. Step 2 indicates selection of orthologous promoters. Genes are symbolized by squares and the three species used are indicated (human, mouse, rat). Step 3 symbolizes the generation of models each containing two transcription factor binding sites (TFBSs) from orthologous promoter sets of individual genes obtained in Step 2. Horizontal optimization is done in Steps 4–6 across promoters from the initial problem-specific gene list (IPL). The links between promoter models and the functional association of genes in the cell is symbolized at the bottom (Step 8). For details of our application example, see Figure 4.
Literature analysis software (Step 1): Current data from literature on subject-related gene expression, gene function and gene–disease relationship were collected with the programs LitMiner (GSF, H. Maier, S. Döhr, K. Grote, S. O'Keeffe, T. Werner, M. Hrabé de Angelis and R. Schneider, in preparation), BiblioSphere™ (Genomatix), GeneCards™ (17), and OMIM (18).
The LitMiner is a web-based resource that was developed by the GSF group. It allows the generation of ranked lists of genes associated with diseases and tissues from abstracts of scientific publications, which are available from PubMed®.
Promoter extraction (Step 2): We extracted the promoter sequences from human, mouse and rat where available using the ‘Comparative Genomics’ task of the ElDorado™ database (Genomatix Suite–ElDorado™, release 3.0, Human Genome NCBI build 34, Mouse Genome MGSCv3, Rat Genome NCBI build 2). The promoter sequences used in this study are available as Supplementary Material.
Promoter selection and modeling (Steps 3–4): The DiAlign (19) task of GEMS Launcher was used for nucleotide sequence alignments to check overall promoter similarity for each orthologous promoter set. The GEMS Launcher task ‘FrameWorker’ using the available weight matrix library (GEMS Launcher Version 3.0, matrix library vertebrate section, Matrix Family Library 4.0 containing 535 matrices in 253 families, Genomatix software, Munich; http://www.genomatix.de) was applied.
Model optimization (Step 5): The FastM (20) task of GEMS Launcher was used to optimize models. ModelInspector (21) (a GEMS launcher task) was used to search databases with the optimized models. Selectivity was determined against the Eukaryotic Promoter Database (EPD, release 76, >4000 promoters) (22) and against the human promoter database (Genomatix Promoter Database, GPD, Genomatix software, Munich, release 3.0, >50 000 promoters).
Model extension (Step 6): The FastM task of GEMS Launcher was used to extend models by manually adding TFBSs (identified by MatInspector (23) analysis) to existing models.
Database search with final models (Step 7): ModelInspector database searches in the GPD were carried out with the final models.
Functional association (Step 8): Additional information about connections between the genes from the initial list and candidate genes found by the model search was taken from BiblioSphere™ analyses (basis for Figure 4).
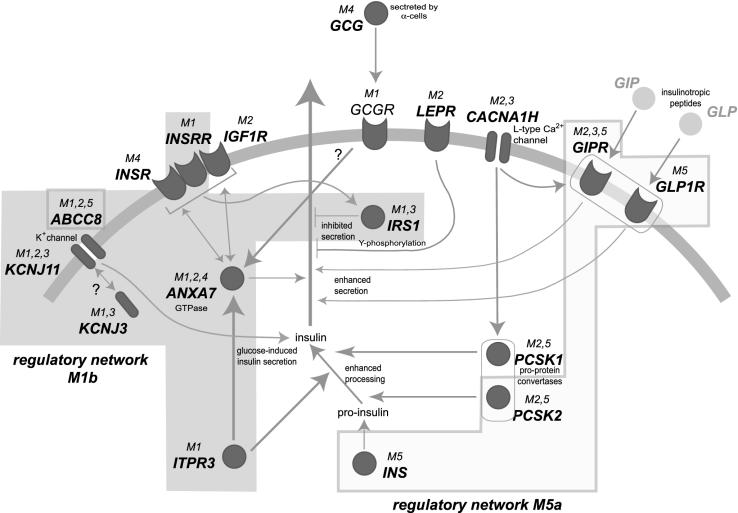
Figure 4.
Functional association between the biological networks and promoter model-derived regulatory networks. The gray arc symbolizes the cell membrane. Dark gray symbols indicate gene products. Membrane receptors are shown inserted into the membrane (with symbolized ligand docking site outside the membrane); ion channels are shown as bipartite structures crossing the membrane; gray circles indicate intracellular proteins. The functional connections between the genes from the IPL were derived by BiblioSphere™ analysis and are indicated by gray arrows; ‘?’ indicates putative connections. M1,2,3 above the gene symbols indicates that models 1, 2 and 3 all match within the promoter of the respective gene. Shaded areas underlying the graphics indicate potential regulatory networks, which are linked by shared promoter models (regulatory network M1b and regulatory network M5a).
Default parameters were applied for the initial analyses in all programs, if not indicated otherwise.
RESULTS
Rational of the strategy
Functional conservation of promoter organization is evident in two directions: vertically, in promoter sequences from orthologous genes (inter-species) and horizontally, in promoter sequences of co-regulated genes within one species (intra-species). Thus, selection of promoter substructures conserved vertically as well as horizontally should be best correlated with particular biological functions.
The only prerequisites for this strategy are a list of genes associated with the biological or medical question to be analyzed, and that the underlying biological processes are evolutionarily conserved. This allows generation of promoter models based on combined conservation (vertical and horizontal). Ensuring tight association of models with the biological problem requires further optimization. We propose to use selectivity for this purpose because biologically meaningful models are expected to show better association with the problem-correlated gene promoters. This resulted in the following strategy (Steps 1–8; Figure 1).
Strategy
Problem-oriented gene selection: The first step is the identification of an Initial Problem-specific List (IPL) of genes correlated with a disease, a signaling pathway, a metabolic pathway or any other gene group linked by a biological function.
Orthologous promoters: Orthologous promoter sets from three mammalian species (human, mouse and rat where available) are collected for every gene in the IPL.
2-TFBS-models: Orthologous promoter sets are analyzed for frameworks consisting of two elements, resulting in initial models (each model representing one vertically conserved framework).
Shared models: Networks have to contain at least three members. Therefore, models from the orthologous promoter sets of the genes in the IPL are selected for further analysis if they match at least two additional promoters of the IPL.
Optimization of model selectivity: The models are refined solely based on promoters present within the IPL using the following restrictions: each TFBS is oriented strand-specific, and distance range variability between the two TFBSs is minimized. Selectivity (defined in methods) versus a genome-wide promoter database is used as the sole optimization criterion.
Extension of models: In this step, models resulting from Step 5 are extended by at least one additional TFBS (missed by standard parameters) resulting in models of more than two elements. Optimization of models proceeds as in Step 5. At this point, orthologous conservation of the extended models in the additionally identified genes is no longer required.
Database search with final models: Next, the complete match list for the models defined in Step 6 is determined from a database of all available human promoters. This provides the basis for extension of the initial gene list. Hitherto unrelated genes can be linked to the original problem on the basis of their promoter organization, and subsequent verification of the connection from an independent source justifies extending the initial list.
Functional associations: The regulatory networks of IPL-genes defined by matches to shared promoter models are then superimposed onto the literature-derived biological process network of all IPL genes to assess concurrence between these independently derived networks.
Application example
We have applied this strategy to identify genes and their transcriptional networks important in the context of maturity onset diabetes of the young (MODY). We were able to identify at least two potential co-regulation networks clearly associated with different biological networks directly connected to insulin/glucose signaling. We also extended the original gene list by several new candidate genes for these networks.
Problem-oriented gene selection by automated literature analysis (Step 1)
Mechanisms of glucose homeostasis are disturbed in the MODY-syndromes (diabetes mellitus type II) that were used as model system. We initiated the analysis with an exhaustive automatic literature search using all available PubMed® abstracts. LitMiner was used to extract disease-associated genes. The following queries were used independently: <MODY>, <Non-insulin-dependent diabetes mellitus>, <islets of Langerhans>, <beta cell>, <glucose homeostasis insulin signaling>. The result of each query was a separate list of genes. All of these were merged to compile the list shown in Table 1.
Table 1.
Problem-oriented gene selection: MODY
| Gene | LocusID | Description | Ortholog | Functional data (Literature) |
|---|---|---|---|---|
| ABCC8* | 6833 | ATP-binding cassette, subfamily C (CFTR/MRP), member 8 | hmr | Insulin release |
| ANXA7 | 310 | Annexin VII: calcium-channel, voltage-gated | hmr | Membrane fusion |
| CACNA1A | 773 | Calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | hr | Hormone release |
| CACNA1D | 776 | Calcium channel, voltage-dependent, L type, alpha 1D subunit | h | Calcium signaling |
| CACNA1H | 8912 | Calcium channel, voltage-dependent, L type, alpha 1H subunit | h | Calcium signaling |
| GCG* | 2641 | Glucagon | hm | Glucose metabolism |
| GCGR | 2642 | Glucagon receptor | hm | Carbohydrate metabolism |
| GCK* | 2645 | Glucokinase | hmr | Glucose metabolism |
| GCKR | 2646 | Glucokinase regulatory peptide | hm | Glucose metabolism |
| GIPR* | 2696 | Gastric inhibitory polypeptide receptor | hmr | Stimulates insulin release |
| GLP1R* | 2740 | Glucagon-like peptide 1 receptor | hmr | Stimulates insulin release |
| IGF1* | 3479 | Insulin-like growth factor 1 | hmr | Glucose metabolism |
| IGF1R* | 3480 | Insulin-like growth factor 1 receptor | hmr | Carbohydrate metabolism |
| INS | 3630 | Insulin | hmr | Glucose metabolism |
| INSR* | 3643 | Insulin receptor | hmr | Carbohydrate metabolism |
| INSrR | 3645 | Insulin related receptor | hmr | Carbohydrate metabolism |
| IRS1* | 3667 | Insulin receptor substrate 1 | hmr | Inhibition of insulin signaling |
| ITPR3 | 3710 | Inositol 1,4,5-triphosphate receptor 3 | hm | Calcium channel, signaling |
| KCNJ3* | 3760 | Potassium inwardly rectifying channel, subfamily J, member 3 | hmr | Insulin release (assumed) |
| KCNJ5 | 3762 | Potassium inwardly rectifying channel, subfamily J, member 5 | hr | Insulin release (assumed) |
| KCNJ6* | 3763 | Potassium inwardly rectifying channel, subfamily J, member 6 | hm | Insulin release |
| KCNJ11* | 3767 | Potassium inwardly rectifying channel, subfamily J, member 11 | hmr | Insulin release |
| LEPR | 3953 | Leptin receptor | h | Adipose-tissue regulation |
| NPY1R | 4886 | Neuropeptide Y/peptide YY receptor Y1 | h | Gastrointestinal signaling |
| PCSK1* | 5122 | EC 3.4.21.93, proprotein convertase 1 | hmr | Insulin processing |
| PCSK2* | 5126 | EC 3.4.21.94, proprotein convertase 2 | hmr | Insulin processing |
| SLC2A2* | 6514 | Solute carrier family 2 | hmr | Carbohydrate metabolism |
The initial problem-specific list (IPL) of 27 genes; all gene names are according to HUGO officially preferred symbols (46). Availability of orthologous gene promoters is indicated by single-letter abbreviations in column 4. h = human, m = mouse, r = rat. The 15 final orthologous promoter sets used for promoter modeling are indicated by asterisks (*).
Orthologous promoter sets (Step 2)
Promoters were identified and extracted for all the genes in our list (Table 1). For the majority of genes, promoter sequences were available from all species (human, mouse and rat) that were chosen for the interspecies comparison. For seven genes, promoters were only available in two species (human and mouse or human and rat) and for four genes, promoters were available only for human (CACNA1D, CACNA1H, LEPR, NPY1R).
We obtained 23 sets of orthologous gene promoters from a total of 62 promoter sequences, some of which consisted only of two sequences (see Table 1, column 4). Promoter sequences were extracted from ElDorado™.
Functionally conserved frameworks cannot be distinguished from trivial occurrences caused by sequence identity in case of high overall sequence similarity (every sequence-associated feature is necessarily ‘conserved’ when the sequence is identical). Therefore, we first checked the degree of sequence similarity for each orthologous promoter set by sequence alignment. Overall sequence similarity ranged from 36% to 77% for human versus mouse/rat and from 62% to 95% for mouse versus rat. Twenty-one sets with an overall sequence similarity up to 60% (empirical limit) were accepted for further analysis. Models of 2-TFBS-frameworks represent the smallest functional transcriptional units as known from composite elements (24) and transcriptional modules (25). Therefore, 2-TFBS-frameworks were generated within these orthologous promoter sets (interspecies comparison). Each promoter set was subjected to three separate FrameWorker runs using distances of 5–150 bp between elements. These models were required to be present in all orthologous promoters of each set. The remaining 15 suitable promoter sets fulfilling both criteria (up to 60% sequence similarity and matching all available orthologous promoters, marked by * in Table 1) yielded 89 different models.
Shared models (Step 4) and optimization of model selectivity (Step 5)
Five of the 89 models recognized at least two additional gene promoters in the IPL and were selected for further optimization (M1–M5 depicted on top in Figure 2). The different parameters for matrix similarity, matrix orientation (strand specificity), model similarity and distance variation between weight matrices could be adjusted manually for three models to maximize selectivity against the EPD (Figure 3). We found that all five 2-TFBS-models contain at least one TFBS associated with endocrine tissues, and four of the eight transcription factors associated with weight matrices in our models are described as being expressed in endocrine tissues (V$FKHD, V$HOXF, V$MAZF, V$NEUR, BiblioSphere™ analysis).
Figure 2.
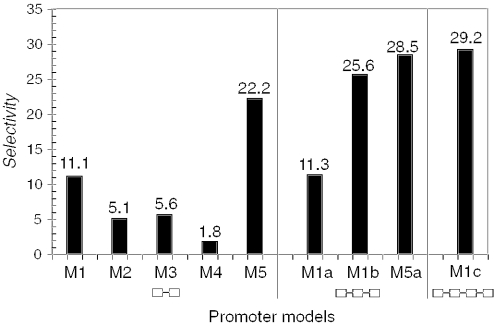
Model descriptions. The selected five 2-TFBSs-models (TFBSs symbolized by gray boxes) generated from promoter analysis are shown on the top (M1–M5). Naming of TFBSs is according to vertebrate matrix families in MatInspector (Genomatix). The threshold used (opt = optimized; −0.02 = optimized − 0.02) is indicated above the boxes. ‘+’ and ‘−’ signs inside the boxes indicate strand orientation of the respective TFBS. Numbers centered below the boxes denote distances between TFBSs. Extended models (M1a, M1b, M1c, M5a) are shown below models M1–M5 (newly added TFBSs are indicated by open boxes).
Figure 3.
Optimization of model selectivity. The histogram shows the increase in selectivity (as defined in Methods) determined for the gene list against the Genomatix Human Promoter Database (see also Table 2). The joined boxes below the histogram indicate the different model structures with 2-, 3- or 4-TFBSs.
Extension of models (Step 6)
Models containing 3-TFBSs were generally found to be more selective than 2-TFBS models (26,27). Therefore, we inspected the orthologous promoter sets for the genes KCNJ11, ABCC8, GIPR, GCG and GLP1R (models M1–M5, Table 2) by MatInspector™ for additional less well-conserved TFBSs in all three organisms, and within a distance range limit of 100 bp from one of the two initial TFBSs. Again as in Step 3, this range was manually adjusted for individual models. This process resulted in extension of model M1 and model M5 by a third TFBS leading to models M1a, M1b (one additional EBOX binding site each) and M5a (additional SP1 binding site). We noticed that model M1a and M1b extended the same model in two directions and then merged them into model M1c (schematic drawing in Figure 2), which now consists of four TFBSs.
Table 2.
Model evaluation
| Model | Origin | Model matches in IPL(27 genes) | Recall in IPL | Hits in EPD | Hits in GPD | Selectivity | |||
|---|---|---|---|---|---|---|---|---|---|
| % | N | % | N | % | EPD | GPD | |||
| M1 | KCNJ11 | KCNJ11, ABCC8, ANXA7, GCGR, INSRR, IRS1, ITPR3, KCNJ3 | 30.0 | 96 | 3.2 | 1335 | 2.7 | 9.4 | 11.1 |
| M2 | ABCC8 | ABCC8, ANXA7, CACNA1H, GIPR, IGF1R, KCNJ11, LEPR, PCSK1, PCSK2 | 33.0 | 253 | 8.4 | 3283 | 6.5 | 3.9 | 5.1 |
| M3 | GIPR | GIPR, KCNJ3, CACNA1H, IRS1, KCNJ11 | 18.5 | 95 | 3.2 | 1650 | 3.3 | 5.8 | 5.6 |
| M4 | GCG | GCG, ANXA7, INSR | 11.1 | 145 | 4.8 | 3093 | 6.2 | 2.3 | 1.8 |
| M5 | GLP1R | GLP1R, ABCC8, GIPR, INS, PCSK1, PCSK2 | 22.2 | 35 | 1.2 | 484 | 1.0 | 18.5 | 22.2 |
| M1a | KCNJ11 | KCNJ11, ABCC8, ITPR3 | 11.1 | 34 | 1.1 | 490 | 1.0 | 9.8 | 11.3 |
| M1b | KCNJ11 | KCNJ11, ABCC8, ANXA7, INSRR, IRS1, ITPR3, KCNJ3 | 25.9 | 36 | 1.2 | 505 | 1.0 | 21.6 | 25.6 |
| M5a | GLP1R | GLP1R, GIPR, INS, PCSK2 | 14.8 | 20 | 0.7 | 260 | 0.5 | 22.1 | 28.5 |
| M1c | KCNJ11 | KCNJ11, ABCC8, ITPR3 | 11.1 | 15 | 0.5 | 191 | 0.4 | 22.2 | 29.2 |
Selected models and their matches found in the list (IPL) of 27 genes and in two different databases (EPD and GPD). All gene names are according to HUGO officially preferred symbols (46). Origin of the model (column 2) denotes the respective set of orthologous gene promoters used for modeling. Promoters of four genes (ABCC8, ANXA7, GIPR, KCNJ11) match to three different models indicating highly interconnected networks. Models with three TFBSs show higher selectivity than models with two TFBSs (columns 5, 6 and 7, absolute match numbers, percentage recognized of all sequences in database and selectivity).
The model selectivity was assessed against the GPD Database. The most selective model (model M5) matched in 484 (1.0%, Table 2) gene promoters and the least specific model (model M2) matched in 3283 (6.5%, Table 2) gene promoters. Model M2 exhibited the best recall (33%, Table 2). The recall of the 3-TFBS-models was lower as compared to models with 2-TFBSs, but showed increased selectivity (Figure 3). The increase in selectivity of the 3- and 4-TFBS-models based on the GPD (>50 000 promoters) is clearly evident (Figure 3), which was essentially paralleled in an analysis based on EPD (>4000 promoters, data not shown).
Database search with final models (Step 7)
The GPD was searched with all models M1–M5 as well as models M1a, M1b, M1c and M5a (Table 2). A clear reduction in the number of matches in the database (3- to 6-fold) can be seen between the 2-TFBS-models and the extended models, which is reflected in a corresponding increase in selectivity. Inspection of the matches of the extended models also allowed extension of the IPL. We found additional genes already known to be involved in insulin/glucose signaling that were not contained in the IPL, as they did not match our LitMiner queries (PRKAA1, ADRB3, PPARGC1B, CLIC3, RyR2, VIPR).
Functional association (Step 8)
Biological links between the genes of the IPL were identified from BiblioSphere™, which is a gene-centered approach combining literature with sequence analysis (used to compile the scheme shown in Figure 4). This biological network revolving around insulin/glucose signaling is overlaid with gray areas indicating the groups of IPL genes identified by the two models M1b and M5a, which are extensively linked in the biological networks (summarized in Figure 4). Briefly, the ATP-sensitive K+ channels composed of KCNJ11 and ABCC8 (28) (probably extended by KCNJ3 through models 1 and 3) are involved in glucose-induced insulin secretion (29), and seem to be co-regulated as indicated by their shared promoter framework. INSRR is known to form heterodimers with INSR and IGF1R (30) and is involved in tyrosine-phosphorylation of the IRS1 product (31), which in turn inhibits insulin secretion (32). The CACNA1H gene encodes the L-type voltage-dependent calcium channel VDCC, which is linked to other genes: It may be involved in the actions of two insulin pro-protein convertases PCSK1, PCSK2 (33). VDCC might also influence the GIPR and GLP1R receptor genes both of which enhance insulin secretion (34).
DISCUSSION
We show that promoter modeling can link disease-associated genes to potential regulatory networks. The most important result obtained in this study is achieving this by using a generally applicable strategy based on optimization of selectivity of promoter models that also identifies regulatory subgroups when necessary. We were able to identify putative regulatory networks within the initial gene list, adding another level of evidence derived from promoter analysis to links known from the literature. We also identified novel members of the putative regulatory networks, which were clearly associated with the biological processes analyzed. Thus, a link between known biological networks and regulatory networks described by molecular promoter organization became evident. Although such links have been established in previous studies (27,35), these depended on particular expert knowledge and/or problem-specific conditions preventing generalization of the approach.
As shown in Figure 4, literature analysis identified a group of genes, which are tightly linked in larger functional networks. Furthermore, for nine genes (ABCC8, KCNJ11, PCSK1, PCSK2, INS, INSR, GCG, IGF1R, LEPR) the BiblioSphere™ literature co-citation analysis revealed a connection to one of the transcription factors that are part of the 2-TFBS-models. However, we used the literature analysis solely to compile the IPL, and then relied entirely on sequence analysis to find and improve subgroups of potentially co-regulated genes as exemplified by shared promoter frameworks. This allowed us to use a systematic approach, purging the huge list of possible frameworks to only five. The final extended models M1a,b,c and M5a preferentially link the promoters of genes that are also functionally connected, such as binding to each other (e.g. receptor complexes) or acting in a common pathway (e.g. insulin processing, Figure 4). This further supports the idea that promoter organizational models can indeed provide the link between the genomic sequence and their biological function.
We found at least six new candidate genes for the insulin/glucose signaling network by searching the human promoter database with models M5a and M1c that were not in the IPL, but clearly associated with insulin/glucose signaling (PRKAA1, ADRB3, PPARGC1B, CLIC3, RyR2, VIPR). They were not included into the IPL either because the literature was not yet available at the time of IPL compilation or they ranked too low in the initial list (e.g. no explicit link to beta cells). The PPARGC1B gene (coding for PGC-1beta) for example is clearly affected in diabetes (36,37). However, this gene is not solely associated with beta-cells and, for example, may be involved in diabetes-related events in the liver (38), further extending the range of the regulatory network. Promoter analysis added another line of evidence for the relevance of these newly identified genes, which allows better experimental setup for further evaluation of these signaling networks. This should help to gain a better understanding of complex biological processes.
Our strategy described here has several advantages over problem-specific approaches. Compilation of a complete gene list from literature would require a priori knowledge of the solution in order to define the correct queries. In our approach, the initial problem-oriented list of genes does not need to be complete, and it can be compiled semiautomatically. When starting with a single gene or even just a disease name, it is possible to collect a list of genes definitely related to the topic of interest. This was shown using the literature tools described here for mammalian systems. There is also no need to exactly know how the selected genes are linked. Our strategy successfully analyzed mixed data sets not restricted to a single transcriptional mechanism, and identified subsets connected by shared promoter frameworks (see Figure 4). Mixed data sets usually present an obstacle to pattern analysis and only recently the problem has been approached successfully in mammalian systems (39). However, this and other approaches (40,41) focused on individual elements rather than complete promoter organization, which is the focus of this study.
Throughout the analysis, selectivity was evaluated against databases, which were orders of magnitude larger than our training set. Selectivity was chosen, as sensitivity and specificity require knowledge about the true positive and false negative, both not available for whole-genome promoter databases. Evaluation of results against the background of all promoters in the human genome is desirable as it excludes any artificial bias on control sampling, supporting biological relevance of our findings. Selectivity proved to be a suitable optimization criterion as demonstrated in Figure 4. The importance of combinations of TFBSs for biological function was also well established before (20), and the particular organization of frameworks has been used successfully to describe individual functions already (42). Phylogenetic conservation of TFBSs was used for promoter analysis as well (43,44). However, the combination of vertical (inter-species) and horizontal (intra-species) framework conservation has so far not been exploited to the extent implemented here.
The key to success was the extension from single gene analysis (orthologous sets) towards non-orthologous gene groups providing the basis to separate different gene groups matching to distinct models. This required to limit the first step (orthologous promoter analysis) to frameworks of two elements, which are usually neither selective nor necessarily linked to a particular function. Larger models of four or more TFBSs in orthologous promoter sets begin to show over-fitting (we generally found them recognizing only the training set), a feature not desirable in this context. Selectivity and functional association were brought to these models by the interactive optimization process. Gain in selectivity almost always causes a loss of recall. Models containing three TFBSs turned out to represent a good balance between selectivity and recall in our example, which is required for a successful search for potential new candidates in a regulatory network.
This strategy currently requires interactive decisions (such as which models to extend and how). However, such decisions are reached in a data-driven approach and the selectivity analysis provides an objective measure of improvement. Thus, model finding and optimization are principally suitable for automation, which could be achieved by systematic parameter range variation. Detailed expert knowledge of the problem is only required for the functional assessment in Step 8, but will also facilitate compilation of the IPL.
The systematic extraction of promoter structures (frameworks) from a group of genes related to a wide variety of questions or fields of interest and linking these frameworks to biological functions becomes possible by our strategy. However, as the input gene list may be incomplete, so may the result. This strategy will probably not identify all the models or all the functions hidden in the input genes.
Nevertheless, even being aware that the result will only be a partial analysis of the problem, this strategy can be used for most problems involving evolutionarily conserved mechanisms of gene regulation. Elucidation of regulatory mechanisms (45) through functional models as demonstrated here, significantly contributes to the functional annotation of mammalian genomes.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Kornelie Frech, Kerstin Cartharius and Matthias Scherf for critical reading of the manuscript and for many discussions. This work was supported by DHGP grant 01KW9909 (S.D. and R.S.) and in part by grants WE2370/1-2 and SCH746/1-3 (DFG) and 031U112B/031U212B (BFAM). Funding to pay the Open Access publication charges for this article was provided by GSF.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 4.Levine M., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 5.Oksman-Caldentey K.M., Inze D., Oresic M. Connecting genes to metabolites by a systems biology approach. Proc. Natl Acad. Sci. USA. 2004;101:9949–9950. doi: 10.1073/pnas.0403636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–1664. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 7.Werner T., Fessele S., Maier H., Nelson P.J. Computer modeling of promoter organization as a tool to study transcriptional coregulation. FASEB J. 2003;17:1228–1237. doi: 10.1096/fj.02-0955rev. [DOI] [PubMed] [Google Scholar]
- 8.Dean A. Chromatin remodelling and the interaction between enhancers and promoters in the beta-globin locus. Brief Funct. Genomic Proteomic. 2004;2:344–354. doi: 10.1093/bfgp/2.4.344. [DOI] [PubMed] [Google Scholar]
- 9.Bader G.D., Heilbut A., Andrews B., Tyers M., Hughes T., Boone C. Functional genomics and proteomics: charting a multidimensional map of the yeast cell. Trends Cell Biol. 2003;13:344–356. doi: 10.1016/s0962-8924(03)00127-2. [DOI] [PubMed] [Google Scholar]
- 10.Pilpel Y., Sudarsanam P., Church G.M. Identifying regulatory networks by combinatorial analysis of promoter elements. Nature Genet. 2001;29:153–159. doi: 10.1038/ng724. [DOI] [PubMed] [Google Scholar]
- 11.Figeys D. Combining different ‘omics’ technologies to map and validate protein–protein interactions in humans. Brief Funct. Genomic Proteomic. 2004;2:357–365. doi: 10.1093/bfgp/2.4.357. [DOI] [PubMed] [Google Scholar]
- 12.Sugars K.L., Rubinsztein D.C. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003;19:233–238. doi: 10.1016/S0168-9525(03)00074-X. [DOI] [PubMed] [Google Scholar]
- 13.Brazhnik P., de la Fuente A., Mendes P. Gene networks: how to put the function in genomics. Trends Biotechnol. 2002;20:467–472. doi: 10.1016/s0167-7799(02)02053-x. [DOI] [PubMed] [Google Scholar]
- 14.Altman R.B., Raychaudhuri S. Whole-genome expression analysis: challenges beyond clustering. Curr. Opin. Struct. Biol. 2001;11:340–347. doi: 10.1016/s0959-440x(00)00212-8. [DOI] [PubMed] [Google Scholar]
- 15.Werner T. Identification and functional modelling of DNA sequence elements of transcription. Brief Bioinform. 2000;1:372–380. doi: 10.1093/bib/1.4.372. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z., Gerstein M. Of mice and men: phylogenetic footprinting aids the discovery of regulatory elements. J. Biol. 2003;2:11. doi: 10.1186/1475-4924-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Safran M., Chalifa-Caspi V., Shmueli O., Olender T., Lapidot M., Rosen N., Shmoish M., Peter Y., Glusman G., Feldmesser E., et al. Human Gene-Centric Databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res. 2003;31:142–146. doi: 10.1093/nar/gkg050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamosh A., Scott A.F., Amberger J., Bocchini C., Valle D., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2002;30:52–55. doi: 10.1093/nar/30.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgenstern B., Frech K., Dress A., Werner T. DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics. 1998;14:290–294. doi: 10.1093/bioinformatics/14.3.290. [DOI] [PubMed] [Google Scholar]
- 20.Klingenhoff A., Frech K., Quandt K., Werner T. Functional promoter modules can be detected by formal models independent of overall nucleotide sequence similarity. Bioinformatics. 1999;15:180–186. doi: 10.1093/bioinformatics/15.3.180. [DOI] [PubMed] [Google Scholar]
- 21.Frech K., Danescu-Mayer J., Werner T. A novel method to develop highly specific models for regulatory units detects a new LTR in GenBank which contains a functional promoter. J. Mol. Biol. 1997;270:674–687. doi: 10.1006/jmbi.1997.1140. [DOI] [PubMed] [Google Scholar]
- 22.Schmid C.D., Praz V., Delorenzi M., Perier R., Bucher P. The Eukaryotic Promoter Database EPD: the impact of in silico primer extension. Nucleic Acids Res. 2004;32:D82–D85. doi: 10.1093/nar/gkh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quandt K., Frech K., Karas H., Wingender E., Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kel A., Kel-Margoulis O., Babenko V., Wingender E. Recognition of NFATp/AP-1 composite elements within genes induced upon the activation of immune cells. J. Mol. Biol. 1999;288:353–376. doi: 10.1006/jmbi.1999.2684. [DOI] [PubMed] [Google Scholar]
- 25.Werner T. Promoters can contribute to the elucidation of protein function. Trends Biotechnol. 2003;21:9–13. doi: 10.1016/s0167-7799(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 26.Klingenhoff A., Frech K., Werner T. Regulatory modules shared within gene classes as well as across gene classes can be detected by the same in silico approach. In Silico Biol. 2002;2:S17–S26. [PubMed] [Google Scholar]
- 27.Gailus-Durner V., Scherf M., Werner T. Experimental data of a single promoter can be used for in silico detection of genes with related regulation in the absence of sequence similarity. Mamm. Genome. 2001;12:67–72. doi: 10.1007/s003350010219. [DOI] [PubMed] [Google Scholar]
- 28.Yamada M., Kurachi Y. The nucleotide-binding domains of sulfonylurea receptor 2A and 2B play different functional roles in nicorandil-induced activation of ATP-sensitive K+ channels. Mol. Pharmacol. 2004;65:1198–1207. doi: 10.1124/mol.65.5.1198. [DOI] [PubMed] [Google Scholar]
- 29.Ball A.J., McCluskey J.T., Flatt P.R., McClenaghan N.H. Chronic exposure to tolbutamide and glibenclamide impairs insulin secretion but not transcription of K(ATP) channel components. Pharmacol. Res. 2004;50:41–46. doi: 10.1016/j.phrs.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Kitamura T., Kido Y., Nef S., Merenmies J., Parada L.F., Accili D. Preserved pancreatic beta-cell development and function in mice lacking the insulin receptor-related receptor. Mol. Cell. Biol. 2001;21:5624–5630. doi: 10.1128/MCB.21.16.5624-5630.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirayama I., Tamemoto H., Yokota H., Kubo S.K., Wang J., Kuwano H., Nagamachi Y., Takeuchi T., Izumi T. Insulin receptor-related receptor is expressed in pancreatic beta-cells and stimulates tyrosine phosphorylation of insulin receptor substrate-1 and -2. Diabetes. 1999;48:1237–1244. doi: 10.2337/diabetes.48.6.1237. [DOI] [PubMed] [Google Scholar]
- 32.Greene M.W., Sakaue H., Wang L., Alessi D.R., Roth R.A. Modulation of insulin-stimulated degradation of human insulin receptor substrate-1 by Serine 312 phosphorylation. J. Biol. Chem. 2003;278:8199–8211. doi: 10.1074/jbc.M209153200. [DOI] [PubMed] [Google Scholar]
- 33.Janssen S.W., Hoenderop J.G., Hermus A.R., Sweep F.C., Martens G.J., Bindels R.J. Expression of the novel epithelial Ca2+ channel ECaC1 in rat pancreatic islets. J. Histochem. Cytochem. 2002;50:789–798. doi: 10.1177/002215540205000605. [DOI] [PubMed] [Google Scholar]
- 34.Preitner F., Ibberson M., Franklin I., Binnert C., Pende M., Gjinovci A., Hansotia T., Drucker D.J., Wollheim C., Burcelin R., et al. Gluco-incretins control insulin secretion at multiple levels as revealed in mice lacking GLP-1 and GIP receptors. J. Clin. Invest. 2004;113:635–645. doi: 10.1172/JCI20518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kel A.E., Kel-Margoulis O.V., Farnham P.J., Bartley S.M., Wingender E., Zhang M.Q. Computer-assisted identification of cell cycle-related genes: new targets for E2F transcription factors. J. Mol. Biol. 2001;309:99–120. doi: 10.1006/jmbi.2001.4650. [DOI] [PubMed] [Google Scholar]
- 36.Patti M.E., Butte A.J., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R., et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc. Natl Acad. Sci. USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ek J., Andersen G., Urhammer S.A., Gaede P.H., Drivsholm T., Borch-Johnsen K., Hansen T., Pedersen O. Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to Type II diabetes mellitus. Diabetologia. 2001;44:2220–2226. doi: 10.1007/s001250100032. [DOI] [PubMed] [Google Scholar]
- 38.Lin J., Tarr P.T., Yang R., Rhee J., Puigserver P., Newgard C.B., Spiegelman B.M. PGC-1beta in the regulation of hepatic glucose and energy metabolism. J. Biol. Chem. 2003;278:30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 39.Elkon R., Linhart C., Sharan R., Shamir R., Shiloh Y. Genome-wide in silico identification of transcriptional regulators controlling the cell cycle in human cells. Genome Res. 2003;13:773–780. doi: 10.1101/gr.947203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolfertstetter F., Frech K., Herrmann G., Werner T. Identification of functional elements in unaligned nucleic acid sequences by a novel tuple search algorithm. Comput. Appl. Biosci. 1996;12:71–80. doi: 10.1093/bioinformatics/12.1.71. [DOI] [PubMed] [Google Scholar]
- 41.Wolff H., Brack-Werner R., Neumann M., Werner T., Schneider R. Integrated functional and bioinformatics approach for the identification and experimental verification of RNA signals: application to HIV-1 INS. Nucleic Acids Res. 2003;31:2839–2851. doi: 10.1093/nar/gkg390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fessele S., Maier H., Zischek C., Nelson P.J., Werner T. Regulatory context is a crucial part of gene function. Trends Genet. 2002;18:60–63. doi: 10.1016/s0168-9525(02)02591-x. [DOI] [PubMed] [Google Scholar]
- 43.Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E. Human-mouse genome comparisons to locate regulatory sites. Nature Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 44.Levy S., Hannenhalli S., Workman C. Enrichment of regulatory signals in conserved non-coding genomic sequence. Bioinformatics. 2001;17:871–877. doi: 10.1093/bioinformatics/17.10.871. [DOI] [PubMed] [Google Scholar]
- 45.Santagati F., Gerber J.K., Blusch J.H., Kokubu C., Peters H., Adamski J., Werner T., Balling R., Imai K. Comparative analysis of the genomic organization of Pax9 and its conserved physical association with Nkx2-9 in the human, mouse, and pufferfish genomes. Mamm. Genome. 2001;12:232–237. doi: 10.1007/s003350010267. [DOI] [PubMed] [Google Scholar]
- 46.Wain H.M., Lush M.J., Ducluzeau F., Khodiyar V.K., Povey S. Genew: the Human Gene Nomenclature Database, 2004 updates. Nucleic Acids Res. 2004;32:D255–D257. doi: 10.1093/nar/gkh072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.