Abstract
The Endosomal Sorting Complex Required for Transport (ESCRT)-III mediates membrane fission in fundamental cellular processes, including cytokinesis. ESCRT-III is thought to form persistent filaments that over time increase their curvature to constrict membranes. Unexpectedly, we found that ESCRT-III at the midbody of human cells rapidly turns over subunits with cytoplasmic pools while gradually forming larger assemblies. ESCRT-III turnover depended on the ATPase VPS4, which accumulated at the midbody simultaneously with ESCRT-III subunits, and was required for assembly of functional ESCRT-III structures. In vitro, the Vps2/Vps24 subunits of ESCRT-III formed side-by-side filaments with Snf7 and inhibited further polymerization, but the growth inhibition was alleviated by the addition of Vps4 and ATP. High-speed atomic force microscopy further revealed highly dynamic arrays of growing and shrinking ESCRT-III spirals in presence of Vps4. Continuous ESCRT-III remodeling by subunit turnover might facilitate shape adaptions to variable membrane geometries, with broad implications for diverse cellular processes.
Introduction
The Endosomal Sorting Complex Required for Transport-III (ESCRT-III) promotes membrane fission from the inner side of membrane necks in various cellular processes1, including the biogenesis of multivesicular bodies2, cytokinetic abscission3–7, nuclear envelope sealing8–11, plasma membrane repair12, HIV budding13,14, and exosome or microvesicle shedding15–17. ESCRT-III forms polymers that are thought to constrict membrane necks until they split18–25, but the mechanism underlying constriction is unknown.
ESCRT-III is evolutionary conserved from humans to archaea, and is composed of four structurally related core subunits with distinct functions18–27. Budding yeast Vps20 (human homolog is CHMP6) functions as a nucleation factor, Snf7 (human homolog CHMP4 has three isoforms, A-C) serves as a main polymer subunit, Vps24 (CHMP3 in humans) and Vps2 (CHMP2A and B isoforms in humans) inhibit Snf7 polymerization28–32 and recruit the ATPase Vps4, which is thought to predominantly disassemble ESCRT-III polymers30,31,33–38. How different ESCRT-III components coordinately assemble and remodel polymer structures has remained unclear.
Purified ESCRT-III subunits polymerize into filaments that form spirals on flat membranes or helices on membrane tubes36,39–45. ESCRT-III also forms filament spirals and helices in intact cells44,46,47, and it is required for the assembly of large filament helices that constrict the intercellular bridge during cytokinetic abscission4.
Prevailing models propose that ESCRT-III mediates membrane fission by sequential assembly of distinct subunits18–23,25, whereby late-binding Vps2/Vps24 (CHMP2/CHMP3) subunits might form a rigid dome-shaped scaffold to guide attached membranes towards the fission site36 or induce changes in the curvature of pre-assembled Snf7 filament spirals to promote membrane neck constriction41. These models rely on the sequential addition of distinct subunits and the persistence of ESCRT-III polymers, yet this has not been directly observed under physiological conditions. We hence set out to systematically quantify the assembly kinetics, dynamics, and structure of ESCRT polymers in live human cells and in an in vitro reconstitution system.
Results
ESCRT-III assemblies continuously turn over their subunits with the cytoplasm
To investigate the dynamics of ESCRT-III polymers at the abscission site, we generated stable HeLa cell lines expressing fluorescently tagged subunits. We found that CHMP4B tagged with GFP via a long flexible linker (Localization and Affinity Purification tag, LAP) and expressed close to endogenous levels4 did not perturb abscission (Fig. 1a). To probe the functionality of LAP-tagged CHMP4B, we depleted endogenous CHMP4B in wild-type HeLa cells or in HeLa cells stably expressing siRNA-resistant mouse CHMP4B-LAP (Supplementary Fig. 1a). Cytokinetic abscission was substantially perturbed upon depletion of endogenous CHMP4B in wild-type HeLa cells, but was not affected in mouse-CHMP4B-LAP-expressing HeLa cells (Fig. 1a), validating the functionality of CHMP4B-LAP.
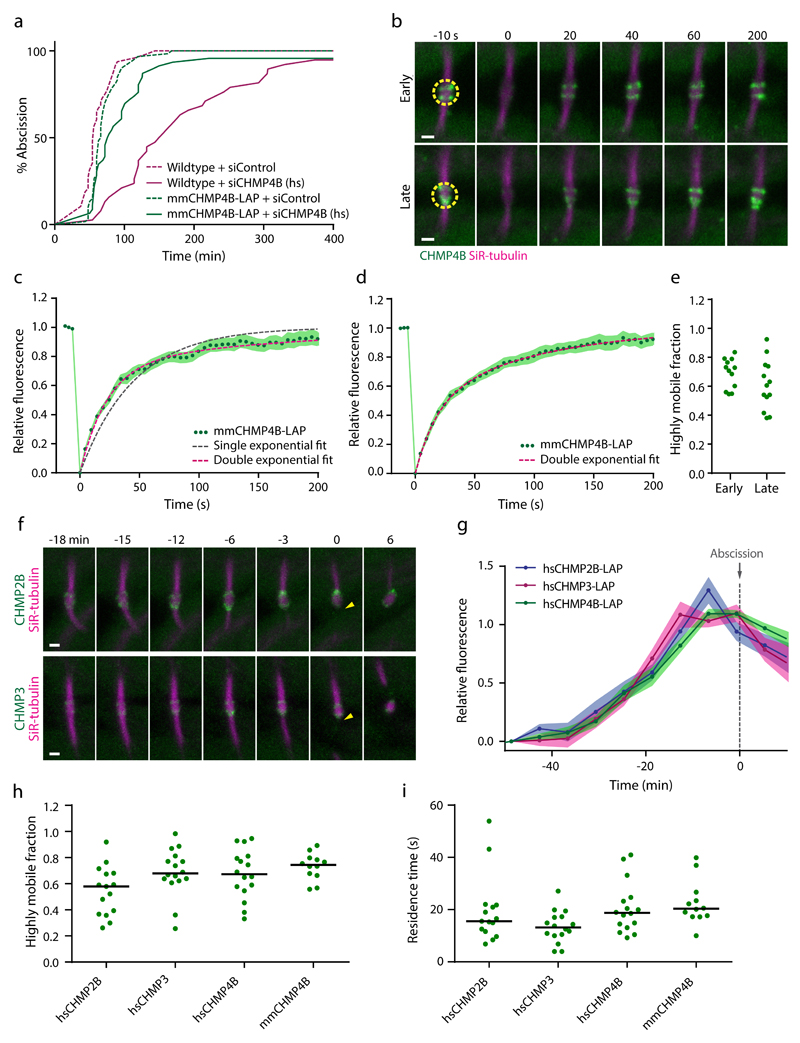
Figure 1. ESCRT-III assemblies at the midbody dynamically turn over subunits in early and late abscission stages.
(a) Validation of mmCHMP4B-LAP functionality by RNAi phenotype rescue. Cumulative histograms indicate duration from complete cleavage furrow ingression until abscission for wildtype HeLa cells and for HeLa cells expressing mmCHMP4B-LAP at 55-80 h after siRNA transfection (3 independent experiments with combined sample numbers of n = 48 cells for wildtype+siControl, n = 38 cells for wildtype+siCHMP4B, n = 60 cells for mmCHMP4B-LAP+siControl, and n = 46 cells for mmCHMP4B-LAP+siCHMP4B). siCHMP4B (hs) targets only endogenous human CHMP4B but not mmCHMP4B-LAP. (b) FRAP of mmCHMP4B-LAP at a HeLa cell midbody at early and late abscission stages, stained with SiR-tubulin. Dashed circles indicate photobleaching region; time 0 indicates first image after photobleaching. High-resolution example of experiment in c-e. (c-d) Fluorescence recovery curves for (c) early abscission (n = 18 cells from 4 independent experiments) or (d) late abscission stages (n = 17 cells from 4 independent experiments). Single exponential function f(t)=1-e^(-k*t), or double exponential function f(t)=A1*(1-e^(-k1*t))+(1-A1*)(1-e^(-k2*t)) were fitted to the data. Points and shaded areas indicate mean ± SEM of fluorescence; dashed lines indicate fits of exponential functions. (e) Quantification of highly mobile fractions by fitting double exponential functions to data from c, d. Dots represent individual cells. (f) 3D live-cell confocal microscopy of the intercellular bridge during telophase, in HeLa cells expressing hsCHMP2B-LAP or hsCHMP3-LAP, respectively. Arrowheads indicate abscission. High-resolution example of experiment in g. (g) Quantification of hsCHMP2B-LAP (n = 17 cells from 4 independent experiments), hsCHMP3-LAP (n = 13 cells from 3 independent experiments), and hsCHMP4B-LAP (n = 17 cells from 3 independent experiments) midbody accumulation. Points and shaded areas indicate mean ± SEM; normalized to intercellular bridge fluorescence after cleavage furrow ingression, and temporally aligned to abscission (time point 0). (h) Highly mobile fractions of LAP-tagged ESCRT-III subunits derived from double exponential fits to FRAP curves. Each dot represents a single FRAP experiment acquired in 3 independent experiments; bars indicate medians. (i) Residence times of highly mobile fractions for cells shown in h. Scale bars, 1 μm in b, f.
We next investigated the dynamics of midbody-localized ESCRT-III by fluorescence recovery after photobleaching (FRAP) experiments. Unexpectedly, we found that CHMP4B-LAP rapidly re-accumulated at the midbody following photobleaching (Fig. 1b, c and Supplementary Video 1). A single exponential function constrained to initial fluorescence values did not fit the FRAP kinetics (Fig. 1c), indicating the presence of two populations of CHMP4B-LAP with distinct residence times at the midbody. We determined the residence times for the two midbody-localized fractions by a double-exponential fit (Fig. 1b, h, i). A highly mobile fraction of 64 ± 6% (mean ± SEM, as in the rest of the paper, if not otherwise noted) of CHMP4B-LAP had a residence time of 19.5 ± 2.7 s, whereas a stably-bound fraction of 36 ± 2% had a residence time of 716.0 ± 91.3 s. Modeling the stably-bound fraction as completely immobile also yielded a good approximation (Supplementary Fig. 1b, c). Importantly, both methods of model fitting yielded consistent values for the fraction and residence time of the highly mobile pool (Supplementary Fig. 1d). Thus, the majority of CHMP4B-LAP molecules at the midbody continuously turns over with a cytoplasmic pool – at a rate up to two orders of magnitude faster than the macroscopic accumulation of ESCRT-III at the midbody4,5.
ESCRT-III initially localizes within two cortical regions adjacent to the midbody, which later constrict to split the plasma membrane4,5. Photobleaching of CHMP4B-LAP prior to or during constriction stages revealed similar recovery kinetics (Fig. 1b, d, e, and Supplementary Video 1), indicating that midbody-localized ESCRT-III dynamically turns over subunits with cytoplasmic pools during its macroscopic accumulation and constriction.
We next investigated the localization and dynamics of other ESCRT-III subunits. We tagged human CHMP2B, CHMP3 and CHMP4B with the same design used for tagging mouse CHMP4B. All three LAP-tagged ESCRT-III subunits localized to the midbody (Fig. 2f, g, and Supplementary Video 2) and did not perturb abscission (Supplementary Fig. 2a-d). CHMP2B, CHMP3, and CHMP4B subunits accumulated at the midbody with indistinguishable kinetics (Fig. 2f, g, and Supplementary Video 2). Hence, ESCRT-III assembles at the midbody with a fairly constant proportion of different core subunits during the progression of abscission.
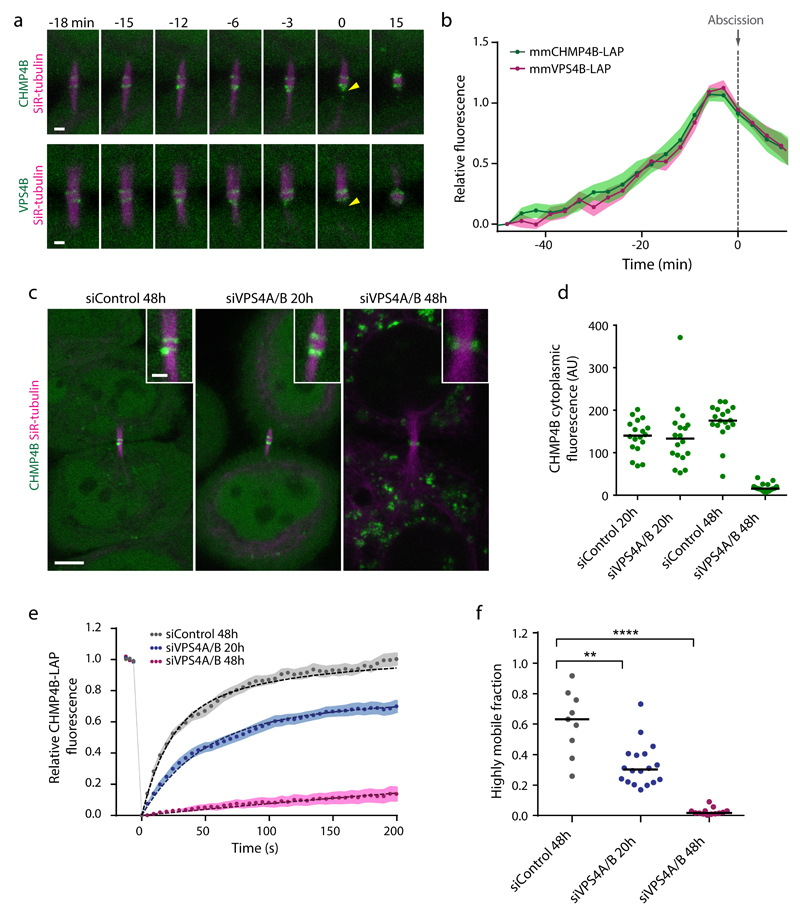
Figure 2. VPS4 is required for ESCRT-III accumulation and turnover.
(a) Confocal microscopy of the intercellular bridge in HeLa cells expressing mmCHMP4B-LAP or mmVPS4B-LAP, respectively. Arrowheads indicate abscission. High-resolution example of experiment in b. (b) Midbody accumulation of mmCHMP4B-LAP (n = 15 cells from 3 independent experiments) or mmVPS4B-LAP (n = 16 cells from 3 independent experiments) relative to abscission (time point 0). Points and shaded areas indicate mean and SEM. (c) Live-cell images of telophase cells expressing mmCHMP4B-LAP after transfection of a non-targeting control siRNA, or siRNAs targeting hsVPSP4A/B after 20 h or 48 h. Insets show enlarged midbody regions. The same contrast settings were used for all panels. High-resolution example of experiment in Supplementary Fig. 3c. (d) Quantification of cytoplasmic mmCHMP4B-LAP levels from data in e, f. Dots represent individual cells from 3 independent experiments; bars indicate medians. (e) FRAP curves and double exponential fits for mmCHMP4B-LAP at pre-constriction stages transfected with control siRNAs (n = 18 cells from 3 independent experiments) or siRNAs targeting VPS4A/B (n = 18 cells for siVPS4A/B 20h, and n = 17 cells for siVPS4A/B 48h from 3 independent experiments). Points and shaded areas indicate mean ± SEM. (f) Highly mobile fractions of mmCHMP4B-LAP determined by double exponential fits to FRAP curves shown in e (3 independent experiments with combined sample numbers of n = 9 cells for siControl 48h, n = 18 cells for siVPS4A/B 20h, and n = 13 cells for siVPS4A/B 48h). Statistical test using the two-sided Kolmogorov–Smirnov test yielded P = 6.562e-3 for siControl 48h relative to siVPS4A/B 20h, and P = 4.021e-6 for siControl 48h relative to siVPS4A/B 48h. Dots represent individual cells; bars indicate medians. Scale bars, 1 μm in a; 5 µm or 1 µm (inset) in c.
We next probed the dynamics of human CHMP2B, CHMP3, and CHMP4B at the midbody. FRAP experiments showed that all three subunits had highly mobile fractions with residence times similar to mouse CHMP4B (Fig. 1h, i, and Supplementary Fig. 2g-i). We noticed somewhat variable kinetics at late stages of FRAP recovery, which resulted in inaccurate fitting of single exponential functions (Supplementary Fig. 2e-g). Given the technical difficulty to accurately measure long residence times, the relevance of the observed variations remains unclear. Importantly, however, all FRAP experiments consistently show that highly mobile fractions of CHMP2B, CHMP3, and CHMP4B dynamically turn over with similar residence times. Overall, our experiments show that ESCRT-III forms highly dynamic assemblies at the midbody.
Dynamic subunit turnover in ESCRT-III assemblies depends on VPS4
We wondered if VPS4 could be responsible for ESCRT-III turnover, as it is the only known nucleotide hydrolase in the ESCRT-III pathway30,33. VPS4 was previously detected at the midbody only during late stages of abscission5, which would be inconsistent with its contribution to the high ESCRT-III turnover observed during early stages. However, previous measurements of VPS4 accumulation were based on overexpression from a viral promoter5, which could limit its detection at the midbody owing to high cytoplasmic background. We thus re-examined VPS4 accumulation in cells stably expressing LAP-tagged murine VPS4B from its endogenous promoter (Supplementary Fig. 3a). In these cells, VPS4B-LAP indeed accumulated at the midbody simultaneously with CHMP4B-LAP (Fig. 2a, b, and Supplementary Video 3). Thus, VPS4 is present at early stages and could contribute to ESCRT-III dynamics throughout the entire abscission process.
To investigate the role of VPS4 in ESCRT-III dynamics, we depleted both isoforms VPS4A and VPS4B in CHMP4B-LAP-expressing cells using RNAi. Depletion of endogenous VPS4A/B to undetectable levels at 48 h after siRNA transfection (Supplementary Fig. 3b) substantially reduced the amount of CHMP4B-LAP at the midbody (Fig. 2c, and Supplementary Fig. 3c). FRAP experiments revealed that under this condition, CHMP4B-LAP turnover at the midbody was almost completely suppressed (Fig. 2e, f). We considered that this phenotype may arise from the strong reduction of the cytoplasmic CHMP4B concentration, owing to the accumulation of ESCRT-III at endosomes upon complete VPS4 depletion47,48 (Fig. 3c, d). However, partial depletion of VPS4A/B at 20 h after siRNA transfection also reduced the fluorescence recovery after photobleaching (Fig. 2e, f) without altering the cytoplasmic CHMP4B-LAP concentration (Fig. 2c, d). The cytoplasmic levels of CHMP2B-LAP or CHMP3-LAP were also not affected upon partial VPS4A/B depletion (Supplementary Fig. 3d, e). Furthermore, microinjection of recombinant human CHMP4B protein into telophase cells resulted in rapid accumulation at the midbody in control cells, but a much slower rate in VPS4A/B-depleted cells (Supplementary Fig. 4). Together, these experiments show that VPS4 is required for dynamic turnover of ESCRT-III at the midbody.
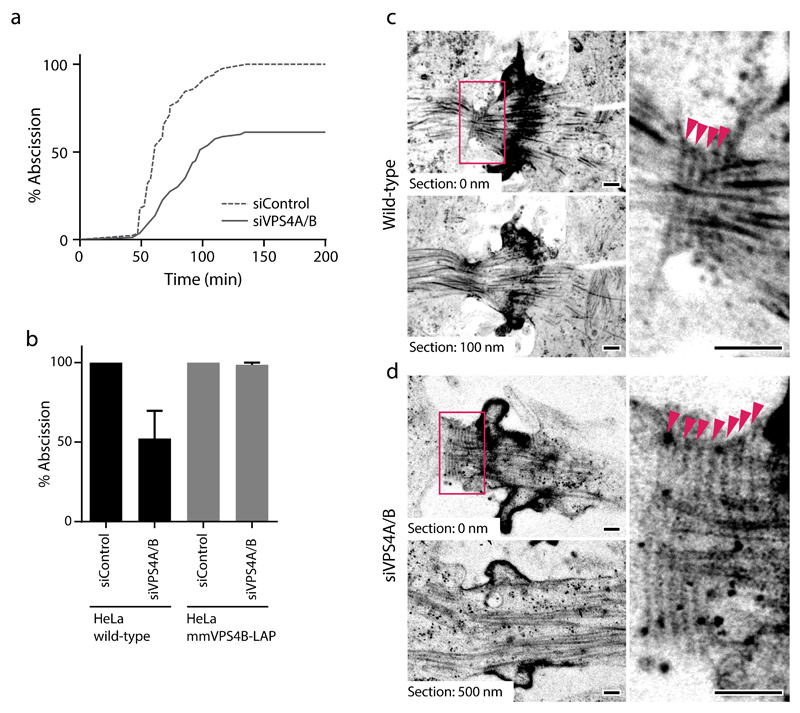
Figure 3. VPS4 is required for constriction of the intercellular bridge.
(a-b) Transfection of siRNAs targeting hsVPS4A/B causes abscission failure in wild-type HeLa cells, but not in HeLa cells stably expressing mmVPS4B-LAP. (a) Progression from cleavage furrow ingression (time point 0) until abscission in wildtype HeLa cells at 30-50 h after transfection of indicated siRNAs (n = 84 cells for wildtype+siControl, and n = 80 cells for wildtype+siVPS4A/B for 3 fields of view from 2 independent experiments). (b) Rescue of abscission failure in HeLa cells stably expressing mmVPS4B-LAP (data from a, n = 54 cells for mmVPS4B-LAP+siControl, and n = 45 cells for mmVPS4B-LAP+siVPS4A/B for 3 fields of view from 2 independent experiments). Bars and error bars indicate mean ± SEM. (c) Representative electron micrograph of an intercellular bridge of a control cell (n = 10 cells, out of which 3 cells had filaments without constriction, and 4 showed filaments with constriction). Arrowheads indicate 17 nm diameter filaments. (d) Intercellular bridge of a cell 26 h after transfection of VPS4A/B siRNA (n = 26 cells, out of which 4 cells showed filaments without constriction). Arrowheads indicate 17 nm diameter filaments. Scale bars, 200 nm in c, d.
VPS4 is required for constriction of the intercellular bridge
We next studied how VPS4 contributes to abscission. RNAi depletion of VPS4A/B delayed abscission and frequently caused cleavage furrow regression (Fig. 3a, b). These abscission failures did not occur in cells stably expressing murine VPS4B-LAP, which is resistant to siRNA targeting human VPS4A/B (Fig. 3b, and Supplementary Fig. 3a), validating that this phenotype is caused by on-target depletion. To gain further insight into the underlying defect, we investigated the ultrastructure of intercellular bridges in cryo-immobilized telophase cells. Most of the control cells contained constriction zones adjacent to the midbody with regularly spaced 17 nm diameter filaments and compressed bundles of microtubules (Fig. 3c; 4 out of 7 cells), as previously observed4. After partial VPS4A/B depletion at 26 h after siRNA transfection, only a small fraction of cells had 17 nm filaments (Fig. 3d; 4 out of 27 cells), and narrow constriction zones were never observed. Together, these data indicate that VPS4 is required for the formation and constriction of a functional ESCRT-III apparatus at intercellular bridges.
Vps2 and Vps24 inhibit Snf7 polymerization in vitro
The high ESCRT-III dynamics in cells prompted us to dissect the specific contribution of each subunit in vitro. As purified human ESCRT-III proteins are difficult to spontaneously polymerize on flat membranes under physiological concentrations49, we considered to use the evolutionarily conserved budding yeast proteins. We first tested whether budding yeast Snf7 (homolog of human CHMP4) can in principle recapitulate the cellular dynamics observed for human proteins and therefore expressed a LAP-tagged version in HeLa cells. Budding yeast Snf7-LAP specifically localized to the midbody during abscission and rapidly recovered after photobleaching similar to human and mouse CHMP4B-LAP (Supplementary Fig. 5a-d), validating the use of yeast proteins for in vitro analysis of ESCRT-III dynamics.
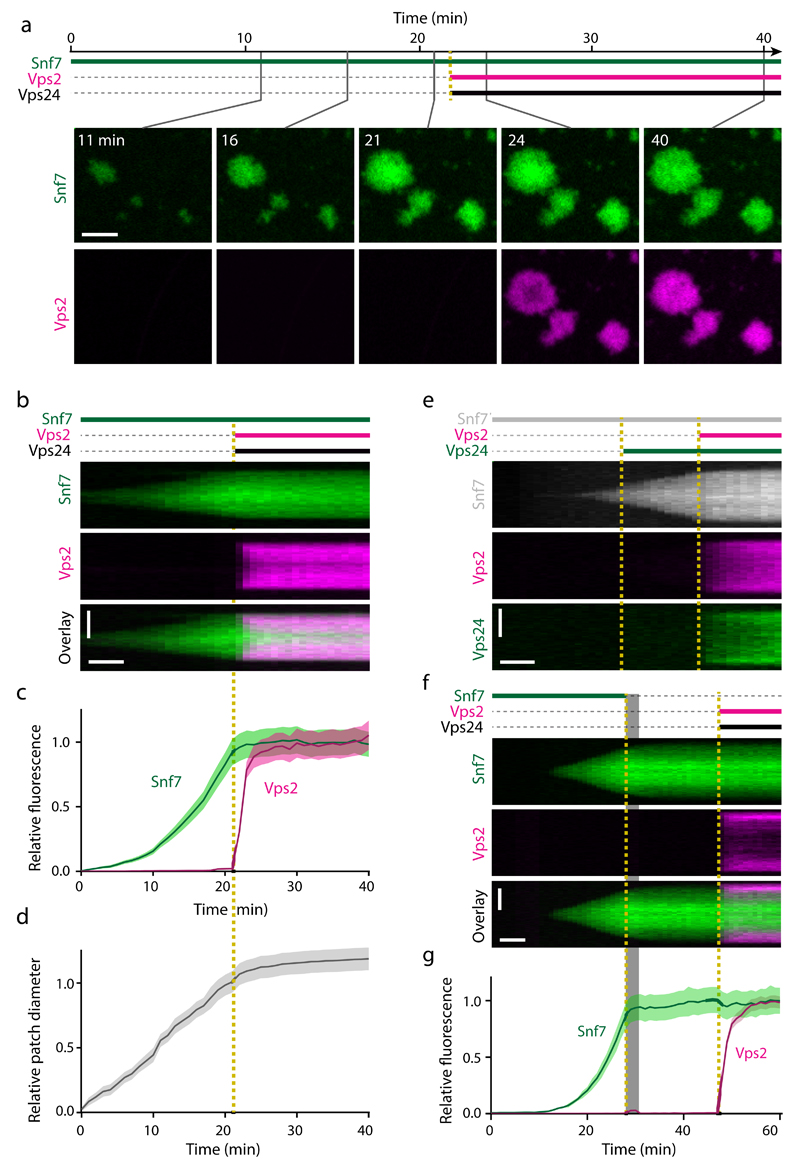
Previous work showed that Snf7 spontaneously polymerizes on membrane covered areas of coverslips to form large patches composed of densely packed filament spirals43. We studied patch growth kinetics only in central areas of membrane-covered regions, as patches stop growth at the edge of membrane-covered regions (Supplementary Fig. 5e-g). We first investigated how Vps24 (homolog of human CHMP3) and Vps2 (CHMP2) affected the kinetics of Snf7 polymerization. We therefore incubated supported lipid bilayers with fluorescently labeled Snf7 until patches formed and then simultaneously added Vps2 and Vps24 (Fig. 4a, b, and Supplementary Video 4; 22 min). Following rapid binding, Vps2 and Vps24 suppressed patch growth and strongly reduced further accumulation of Snf7 in patches (Fig. 4c, d, and Supplementary Fig. 6a-c). Sequential injection of Vps2 and Vps24 into the fluid chamber further showed that these subunits depend on each other in their Snf7 growth-inhibitory function (Fig. 4e, Supplementary Fig. 6d, e, and Supplementary Videos 5 and 6). Thus, prolonged phases of ESCRT-III assembly, as observed during cytokinetic abscission4,5, are not recapitulated by mixed solutions of Snf7, Vps2, and Vps24.
Figure 4. Vps2 and Vps24 cooperatively bind Snf7 patches and inhibit ESCRT-III polymerization.
(a) Time-lapse microscopy of ESCRT-III polymerization on supported lipid membranes in a microfluidic flow chamber. Recombinant Snf7-AlexaFluor-488 was injected at t = 0 min; Vps2-Atto-565 and Vps24 were added at t = 22 min. (b) Kymograph of a single ESCRT-III patch from a. (c-d) Quantification of (c) mean fluorescence and (d) patch diameters from 24 patches as in a-b (quantified from 4 fields of view within a representative experiment, and consistent results in 3 independent experiments using differently labeled proteins, e.g. Supplementary Fig. 6a-c). Curves and shaded areas represent mean ± SEM. (e) Kymograph of an experiment where Snf7-AlexaFluor-647N was added at t = 0 min, followed by sequential addition of Vps24-AlexaFluor-488 and Vps2-Atto-565 (representative image from 24 patches within the shown experiment, and 1 additional independent experiment). (f) Kymograph of an ESCRT-III patch, where Snf7-AlexaFluor-488 was added at t = 0 min, then washed out during 28-32 min (shaded area), followed by addition of Vps2-Atto-565 and Vps24 at t = 47 min. The transient increase of Vps2 signal during washout resulted from background ambient light. (g) Fluorescence quantification of 37 patches as in f (analyzed from 4 fields of view within the shown experiment, and 3 additional independent experiments). Curves and shaded areas represent mean ± SEM. Scale bars, 5 μm in a; 5 µm (vertical) and 5 min (horizontal) in b, e, f.
The inhibition of Snf7 patch growth by Vps2/Vps24 could be caused by lower rates of Snf7 subunit accumulation or by an increase of the Snf7 dissociation rate. To investigate this, we incubated supported lipid bilayers with fluorescently labeled Snf7 until patches formed and then washed out soluble Snf7. We subsequently added fluorescently labeled Vps2 and Vps24, which enriched at the edge of the patch, where newly growing Snf7 filament spirals localize43 (Fig. 4f, g, and Supplementary Video 7). Snf7 remained stably bound to patches throughout the entire imaging period, indicating that Snf7 polymers have extremely low intrinsic subunit dissociation rates irrespective of their association with Vps2 and Vps24. The inhibition of Snf7 patch growth imposed by Vps2 and Vps24 is thus independent of Vps4 and caused by a reduced rate of Snf7 subunit incorporation.
Vps2 and Vps24 form filament bundles with Snf7
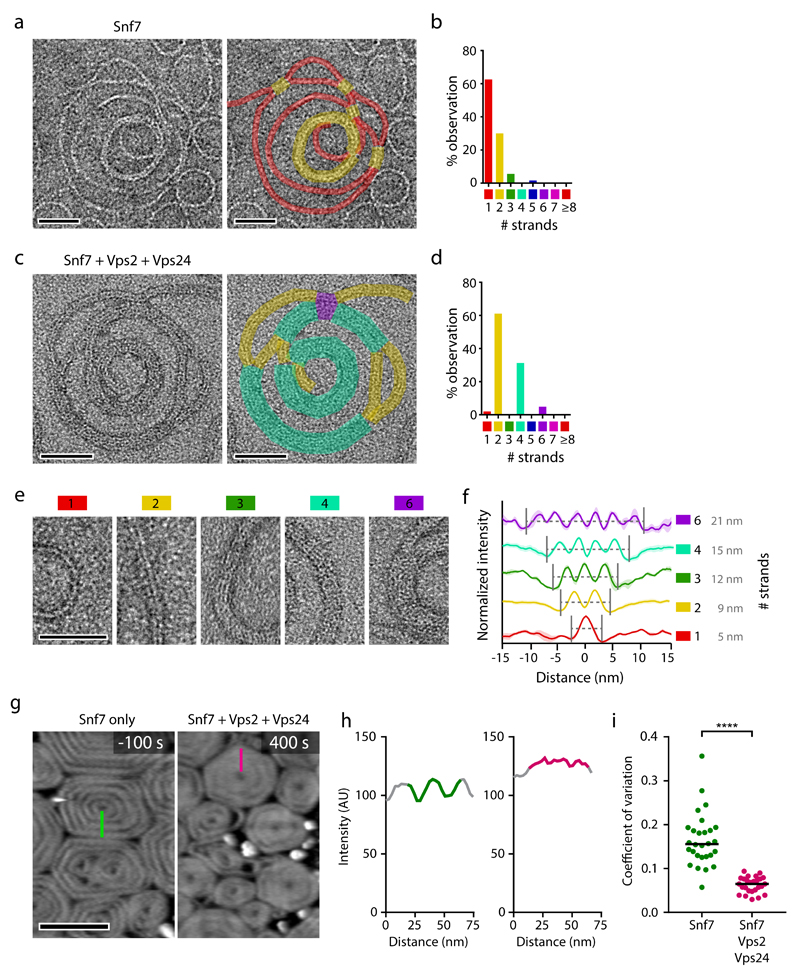
We next investigated if growth inhibition by Vps2 and Vps24 could arise from an ultrastructural change in ESCRT-III polymers. Transmission electron microscopy showed that Snf7 alone polymerized on liposomes to form one-start spirals containing a single 4.5 nm wide filament, which occasionally paired between neighboring turns (Fig. 5a, b), as previously observed43. When Vps2 and Vps24 were added after Snf7 polymerization, filaments appeared double-stranded and neighboring spiral turns occasionally bundled to form quadruple strands with an approximate width of 15 nm (Fig. 5c, d) – close to the width of ESCRT-III-dependent filaments observed at the abscission site in vertebrate cells4 (Fig. 5c-f; compare Fig. 3c). Given the one-start single-stranded geometry of Snf7 spirals prior to addition of Vps2/Vps24, the paired filaments likely represent lateral copolymers of Vps2/Vps24 along Snf7.
Figure 5. Vps2 and Vps24 polymerize side-by-side with Snf7 to form filament bundles.
(a) Transmission electron microscopy of Snf7 spirals polymerized on liposomes. Colored overlays indicate the number of parallel filament strands. (b) Distribution of filament bundle lengths quantified in 11 spirals from 3 independent experiments as in a. (c) Snf7 was polymerized on liposomes, followed by Vps2 and Vps24 addition. Colored overlays indicate the number of parallel filament strands. (d) Quantification of 17 spirals from 2 independent experiments as in c. (e) Examples of filament morphologies with different strand numbers, corresponding to colored overlays used in a-d. (f) Averaged line profiles across ESCRT-III filament bundles from a-d (n = 3 filaments for 1 strand, n = 8 filament bundles for 2 strands, n = 3 filament bundles for 3 strands, n = 8 filament bundles for 4 strands; and n = 3 filament bundles for 6 strands). Curves and shaded areas indicate mean ± SEM. (g-i) HS-AFM imaging of ESCRT-III polymers on supported lipid membranes. Snf7 was polymerized on lipid membranes, followed by addition of Vps2 and Vps24 at t = 0. (g) Spiral morphology before and after addition of Vps2 and Vps24. Green and magenta lines indicate line profiles used to (h) measure height variability. (i) Height variability was measured as coefficient of variation along radial line profiles within spirals before and after addition of Vps2 and Vps24, respectively, as shown in g, h (n = 28 spirals for Snf7, and n= 26 spirals for Snf7+Vps2+Vps24 from 2 independent experiments). Statistical test using the two-sided Kolmogorov–Smirnov test yielded P = 2.875e-14 for Snf7 relative to Snf7+Vps2+Vps24. Dots represent individual line profile measurements; bars indicate medians. Scale bars, 50 nm in a, c, e; 200 nm in g.
To further characterize the morphological changes of Snf7 filaments upon addition of Vps2/24, we visualized ESCRT-III assemblies by high-speed atomic force microscopy (HS-AFM) (Fig. 5g). Snf7 alone formed spirals with pronounced filaments, but subsequent addition of Vps2/Vps24 induced a compact disc-like morphology (Fig. 5g-i). This is consistent with a filament thickening and bundling limiting access of the AFM tip in between neighboring spiral turns. Together, these data suggest that Vps2/Vps24 might reduce the rate of Snf7 polymerization through the formation of bundled filaments.
Vps4 induces subunit turnover and net growth of ESCRT-III assemblies in vitro
In cells, Vps2 and Vps24 are both present in the cytoplasm, raising the question of how ESCRT-III polymerization is sustained over prolonged periods. Our in vivo observations imply the possibility that Vps4 might leverage Vps2/Vps24-mediated growth inhibition by ESCRT-III turnover. To explore whether Vps4 promotes ESCRT-III turnover in vitro, we separately measured ESCRT-III subunit association and dissociation kinetics. We first determined the rate by which Vps4 disassembles Snf7 patches in the absence of Vps2 and Vps24. We polymerized Snf7 on supported lipid membranes, washed out soluble Snf7, and then added Vps4 and ATP. This did not cause detectable disassembly of Snf7 patches even at very high Vps4 concentrations (Fig. 6a, b), indicating that Vps4-mediated Snf7 depolymerization strictly depends on Vps2/Vps24, as shown before37. We thus quantified the rate of Vps4-mediated ESCRT-III patch disassembly in presence of Vps2 and Vps24. We first polymerized Snf7 patches on supported lipid membranes, then removed the soluble pool of Snf7, and subsequently added a mix of fluorescently labeled Vps2 and unlabeled Vps24, Vps4, and ATP. Vps2 first bound to Snf7 patches and subsequently partially disassembled together with Snf7 (Fig. 6c, d, Supplementary Fig. 7a, b, and Supplementary Video 8). Thus, Vps4-mediated Snf7 depolymerization is fast enough to account for dynamic turnover of its homolog CHMP4B at the midbody in human cells.
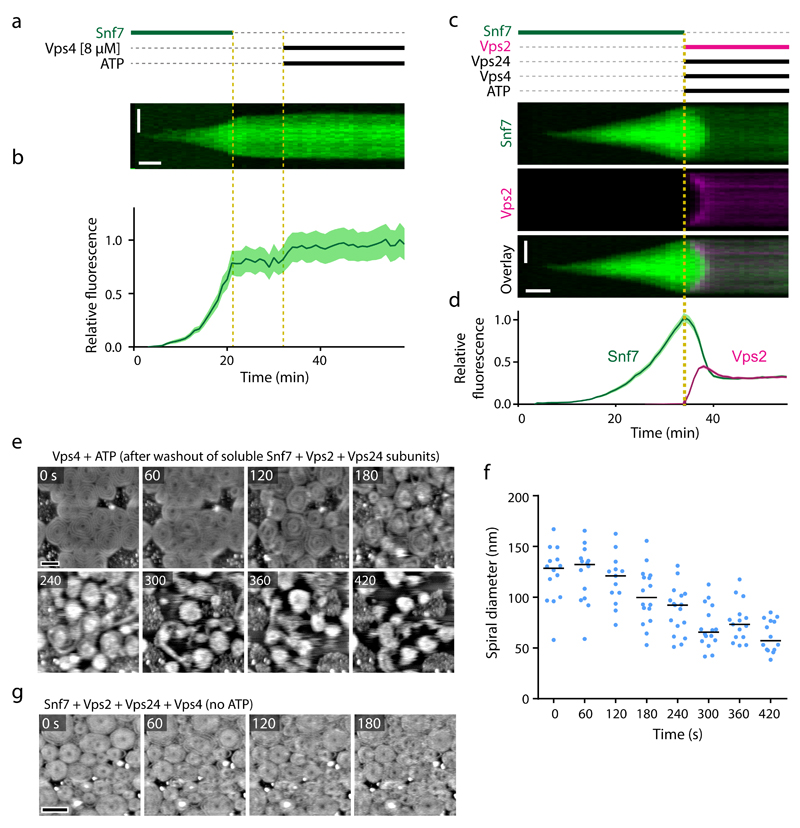
Figure 6. Kinetics of ESCRT-III patch disassembly by Vps4.
(a) Kymograph of a representative patch from time-lapse microscopy of Snf7 patches on supported lipid membranes in a flow chamber. A solution of Snf7-AlexaFluor-488 was injected into the flow chamber and incubated until patches polymerized on the membrane. Snf7 was washed out at t = 22 min, followed by addition of ATP and 8 µM Vps4 at t = 33 min. (b) Quantification of mean fluorescence of 13 patches as in a (4 different fields of view within the shown experiment, and 1 additional independent experiment). Curves and shaded areas indicate mean ± SEM. (c) A solution of Snf7-AlexaFluor-488 was injected into the flow chamber and incubated until patches polymerized. At t = 34 min, Snf7 was removed from the solution and Vps2-Atto-565, Vps24, Vps4, and ATP were injected. (d) Quantification of mean fluorescence of 33 patches as in c (4 fields of view within the shown experiment, and 1 additional independent experiment). Curves and shaded areas indicate mean ± SEM. (e-g) Vps4-induced dynamic turnover and lateral mobility of ESCRT-III filament spirals depends on ATP. (e) HS-AFM imaging of ESCRT-III spirals. Assemblies were generated by polymerization of Snf7 on supported lipid membranes, followed by addition of Vps2 and Vps24. After washout of all soluble components, Vps4 was injected. Then, ATP and Mg2+ were added, and imaging was started 22 s later (t = 0). Images represent averages of 2 consecutive time frames to improve signal-to-noise ratio. (f) Quantification of spiral diameters from e (119 spirals from a representative experiment, out of 6 independent experiments). Dots represent single spirals; bars indicate medians. (g) Snf7 was polymerized on supported lipid membranes, followed by addition of Vps2 and Vps24, and subsequent addition of Vps4, as in Fig. 8a but without ATP. Imaging was started 30 s after addition of Vps4 (t = 0). Images represent averages of 3 consecutive time frames. Corresponding spiral diameter quantification is shown in Fig. 8c. Scale bars, 5 µm (vertical) and 5 min (horizontal) in a, c; 100 nm in e; 200 nm in g.
To characterize the Snf7-disassembly process at the molecular level, we visualized morphological changes of individual ESCRT-III spirals by HS-AFM. We polymerized Snf7 patches on supported lipid bilayers, then added Vps2 and Vps24, and subsequently washed out the soluble components. We then added Vps4 and ATP and acquired HS-AFM movies, which showed that ESCRT-III spirals reduced their diameter (Fig. 6e, f, and Supplementary Video 9). When omitting ATP from the reaction, Vps4 did not disassemble ESCRT-III spirals (Fig. 6g, and Supplementary Video 10), confirming that ESCRT-III disassembly is an energy-consuming process. Given that the ESCRT-III spirals did not depolymerize below a certain diameter, these data suggest that Vps4 mediates Snf7 filament disassembly predominantly from the outer regions of spirals, whereby inner spiral segments are refractory to disassembly.
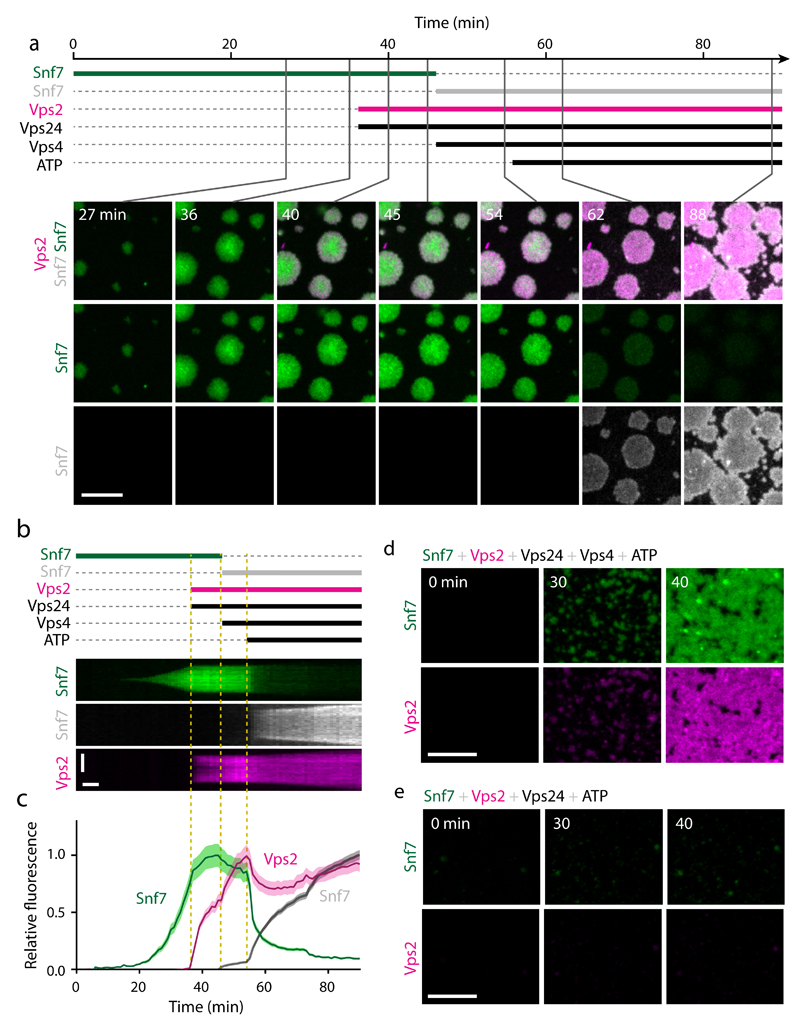
We next tested whether Vps4 can mediate Snf7 turnover in vitro. For this purpose, we used Snf7 subunits labeled with two distinct fluorophores. We first incubated supported lipid bilayers with AlexaFluor-488-labeled Snf7 until patches formed and then added Vps24 and Vps2 to inhibit further patch growth (Fig. 7a-c, see 0 - 45 min, and Supplementary Video 11). We then exchanged the soluble pool of Snf7-AlexaFluor-488 with Snf7-Atto-647N while maintaining Vps2 and Vps24 in the reaction. At this point, we also added Vps4. Snf7 polymer patches maintained constant size and did not incorporate Atto-647N-Snf7, presumably because ATP was not yet present (Fig. 7a-c, see 45 - 54 min). Indeed, subsequent ATP addition caused dissociation of AlexaFluor-488-Snf7 from patches, but also concomitant binding of Atto-647N-Snf7 (Fig. 7a-c, see 54 - 88 min). Thus, an ATP-dependent activity of Vps4 promotes not only disassembly of ESCRT-III but also the formation of new ESCRT-III polymers on membranes.
Figure 7. Vps4 induces subunit turnover and net growth of ESCRT-III assemblies.
(a) Time-lapse microscopy of ESCRT-III polymerization on supported lipid membranes. Snf7-AlexaFluor-488 was injected at t = 0 min. Vps2-Atto-565 and Vps24 were added at t = 36 min while maintaining Snf7 in the solution. At t = 45 min, Snf7-AlexaFluor-488 was removed and a mix containing Snf7-Atto-647N, Vps2-Atto-565, Vps24 and Vps4 was added, followed by addition of ATP at t = 54 min. (b) Kymograph of a single patch from a. (c) Mean fluorescence quantification of 35 patches as in b (4 fields of view within the shown experiment, and consistent results in 2 additional independent experiments using differently labeled proteins). Curves and shaded areas represent mean ± SEM. (d-e) Time-lapse microscopy of in vitro polymerization as in a, but for a mixed solution containing Snf7-AlexaFluor-488, Vps2-Atto-565, Vps24, and ATP (d) in the presence of Vps4, or (e) without Vps4. Representative images of 2 independent experiments per condition are shown. Scale bars, 10 μm in a, d, e; 5 µm (vertical) and 5 min (horizontal) in b.
Interestingly, Vps4 and ATP also restored the macroscopic growth of Snf7 patches despite the continued presence of Vps2 and Vps24 (Fig. 7a, b, see 58 - 88 min, and Supplementary Video 11). We thus tested if a solution containing Vps4, ATP, and all three ESCRT-III subunits supports nucleation and growth of ESCRT-III polymers as observed in vivo. Strikingly, this combination resulted in efficient nucleation and growth of ESCRT-III patches, in contrast to a mix that lacked Vps4 (Fig. 7d, e). Furthermore, the net ESCRT-III assembly rate increased with higher concentrations of Vps4 (Supplementary Fig. 7c). Thus, Vps4 promotes net growth of ESCRT-III assemblies in presence of growth-inhibiting Vps2 and Vps24 subunits.
Vps4 induces dynamic growth and shrinkage of ESCRT-III filament spirals
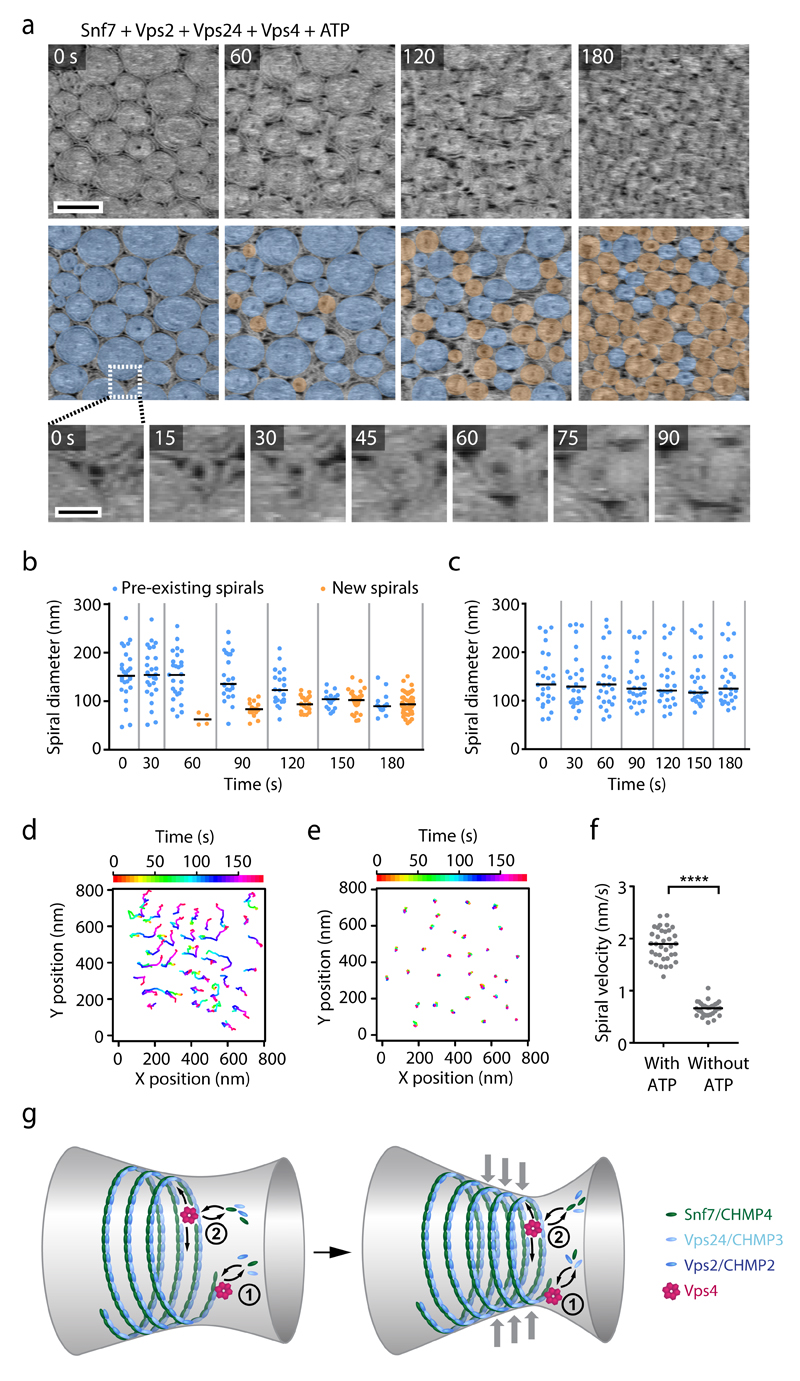
To study Vps4-mediated polymer remodeling at the molecular level, we visualized ESCRT-III spirals by HS-AFM. We assembled Snf7 spirals on supported lipid bilayers and then added Vps2, Vps24, Vps4, and ATP. This induced a drastic reorganization of ESCRT-III polymers: preexisting spirals reduced their size, while many new spirals nucleated and grew in between the original spirals (Fig. 8a, b, and Supplementary Video 12). The innermost parts of ESCRT-III spirals appeared refractory to disassembly and thus might represent the stably-bound fraction of ESCRT-III observed by fluorescence microscopy. In contrast, pre-existing spirals remained unchanged in the absence of ATP (Fig. 8c, and Fig. 6g). Thus, Vps4 induces a dynamic steady-state with both growing and shrinking ESCRT-III spirals when Snf7, Vps2, Vps24 are present in solution.
Figure 8. Vps4 induces dynamic reorganization of ESCRT-III assemblies.
HS-AFM imaging of ESCRT-III polymers on supported lipid membranes. (a) Snf7 was polymerized on supported lipid membranes, followed by addition of Vps2, Vps24, and Vps4. Then, Mg2+ and ATP were added and imaging was started 5.5 min later (t = 0). Overlays highlight pre-formed spirals (blue) or newly formed spirals (orange). Bottom panels show a close-up of the nucleation of a new spiral. Images represent averages of 3 consecutive time frames to improve the signal-to-noise ratio. Scale bars, 200 nm (top panel) or 5 nm (bottom panel). (b) Quantification of spiral diameters from a (274 spirals from a representative experiment, from a total of 3 independent experiments). Dots represent single spirals; bars indicate medians. (c) Quantification of spiral diameters from Fig. 6g (175 spirals from a representative experiment, from a total of 6 independent experiments). Dots represent single spirals; bars indicate medians. (d) Tracking of spiral centers from a. (e) Tracking as in d, but for an experiment without ATP and Mg2+ as shown in Fig. 6g. (f) Quantification of mean velocity of spiral centers from d, e (n = 34 spirals for with ATP, and n = 31 spirals for without ATP, for a representative experiment per condition, from a total of 3 or 6 independent experiments, respectively). Statistical test using the two-sided Kolmogorov–Smirnov test yielded P = 4.441e-16 for spiral velocities in the presence of ATP relative to velocities in the absence of ATP. Dots represent single tracks; bars indicate medians. (g) Model of dynamic ESCRT-III assembly and constriction. Vps4 mediates continuous subunit turnover in ESCRT-III assemblies during growth and constriction. (1) At the tip, dynamic turnover of growth-inhibitory Vps2 and Vps24 subunits could sustain extension of inward-curving filaments. (2) At the core of filament bundles, Vps4-mediated subunit turnover could facilitate sliding of neighboring helical turns to promote constriction.
Furthermore, while ESCRT-III spirals formed an immobile array in the absence of ATP, the addition of ATP induced substantial lateral mobility of individual ESCRT-III spirals relative to each other (Fig. 8d-f, and Fig. 6g). Thus, in the presence of Vps4 and ATP, ESCRT-III forms highly dynamic polymer structures on membranes.
Discussion
Our study shows that membrane-bound ESCRT-III polymers rapidly turn over subunits with cytoplasmic pools while they assemble into larger structures. The dynamic subunit turnover is driven by Vps4 and is necessary to sustain efficient net growth of ESCRT-III assemblies in presence of inhibitory Vps2 and Vps24 subunits.
Most previous models for ESCRT-III-mediated membrane fission imply sequential recruitment of different ESCRT-III subunits and Vps4 to establish distinct phases of polymer growth, reorganization/maturation, and disassembly5,14,18–21,23–25,28,30,31. However, our study shows that different ESCRT-III subunits and VPS4 accumulate at the midbody with indistinguishable kinetics. This does not rule out sequential subunit binding at the single-filament level, but indicates that the distinct stages of ESCRT-III accumulation, constriction, and disassembly during abscission cannot be explained by a sequence of subunit binding.
Our findings call for re-interpretation of the terminal phenotype resulting from Vps4 depletion –the accumulation of ESCRT-III polymers in endosomes (termed class E compartment in yeast) that has led to the model of Vps4 serving predominantly as a disassembly factor 26,28,33,50. We show that Vps4 is important for efficient ESCRT-III assembly, yet we also note that residual slow ESCRT-III polymerization in the absence of Vps4 is sufficient to ultimately capture all cytoplasmic subunits in class E compartments – owing to the extremely low intrinsic subunit dissociation rates.
At the molecular level, Vps4 might promote the net growth of ESCRT-III assemblies by constant turnover of Vps2 and Vps24 in side-by-side co-polymers along Snf7 filaments – to create growth-competent free Snf7 filament tips (Fig. 8g). Furthermore, Vps4 might induce subunit turnover at the core of ESCRT-III filament bundles, in analogy to interaction of the Vps4 homolog Spastin with the lattice of microtubules51 (Fig. 8g). In contrast to previous models5,52, however, Vps4 does not cut an ESCRT-III helix during a single definite time point to induce tension release, but rather continuously remodels filaments. Within bundled filaments, such turnover does not necessarily lead to a complete breakage of larger structures.
The innermost parts of ESCRT-III spirals appear refractory to disassembly even at high concentrations of Vps4, suggesting that ESCRT-III interaction with Vps4 might depend on mechanical stress owing to low filament curvature or on filament polarity. Potential stochastic fluctuations in subunit turnover rates could then lead to dynamic growth and shrinkage of ESCRT-III spirals.
Vps4-induced subunit turnover in ESCRT-III assemblies might directly contribute to membrane constriction. Indeed, macroscopic shape changes of many other cellular polymer structures critically depend on dynamic subunit turnover within the constituent filaments, as for example mitotic spindles53 or actomyosin rings54. In ESCRT-III assemblies, Vps4-induced subunit turnover might facilitate sliding of adjacent helix turns, thereby promoting constriction of mechanically pre-stressed, low-curved filaments into more relaxed high-curvature states43. The underlying bending forces could be generated by binding of Vps2 and Vps24 to Snf741, or by shortening-induced increase of filament rigidity. By revealing dynamic subunit turnover in ESCRT-III assemblies, our study provides a framework for understanding how this highly conserved membrane fission machinery adapts to diverse membrane geometries.
Methods
Cloning and generation of stable cell lines
A Gateway recombination cassette was inserted into pcDNA5/FRT/TO (Invitrogen) to generate a Tet-inducible pTO_mCherry_N-DEST. Next, mCherry was exchanged with a LAP tag using KpnI and XbaI (ThermoFisher Scientific), resulting in pTO_LAP_N-DEST, a Gateway destination vector for C-terminal LAP tags that allows FRT recombination and Tet-inducible expression. Expression vectors were generated by recombining pTO_LAP_N-DEST with Gateway entry clones containing cDNA of human ESCRT-III subunits in pDONR223 (obtained from hORFeome Database) or S. cerevisiae SNF7 (amplified from genomic DNA and cloned into pDONR223) using Gateway LR Clonase II (Invitrogen), to generate pTO_CHMP2B_LAP, pTO_CHMP3_LAP, pTO_CHMP4B_LAP and pTO_SNF7_LAP.
Cell lines for inducible expression of ESCRT-III subunits were generated from HeLa Kyoto FRT_H2B-mRED55. To remove H2B-mRED, cells were grown without selection for H2B-mRED in medium containing 0.5 µg/mL puromycin (Merck), 400 µg/mL zeocin (Invitrogen), but no G418. Cells were then FACS-sorted for the absence of red fluorescence, and single clones were isolated. Inducible ESCRT-III constructs were then integrated into the FRT site by co-transfection with pOG44 (Invitrogen) at a ratio of 1:10, and selected in medium containing 300 μg/mL hygromycin B (Roche) and 0.5 μg/mL puromycin (Merck).
To generate a cell line expressing mmVPS4B-LAP, the bacterial artificial chromosome (BAC) RP24-139J22 was used to GFP-tag mouse VPS4B using BAC-TransGeneOmics56,57. Homologous recombination yielded BAC DNA containing VPS4B-LAP, which was isolated and transfected into HeLa cells using Effectene (Qiagen), and selected in medium containing 400 µg/mL G418 (Invitrogen). All HeLa BAC-GFP cell lines were cultured in medium supplemented with 300 μg/mL G418 (Invitrogen).
No cell line used in this study is listed in the database of commonly misidentified cell lines that is maintained by ICLAC and NCBI Biosample. The HeLa Wildtype cell line (‘Kyoto’ strain) was obtained from S. Narumiya (Kyoto University, Japan) and validated by a Multiplex human Cell line Authentication test (MCA), 21.04.16. All cell lines have been regularly tested for mycoplasma, and a contamination has never been detected.
Cell culture, live-cell microscopy and image analysis
HeLa cells were cultured at 37°C and 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM, Invitrogen), supplemented with 10% (v/v) Fetal Calf Serum (FCS, Invitrogen) and 1% (v/v) penicillin-streptomycin (Sigma-Aldrich). For live-cell imaging, cells were plated in LabTek II chambered coverslips (ThermoFisher Scientific) or glass bottom dishes (µ-Dish35 mm, high Glass Bottom, Ibidi) in cell culture medium without riboflavin and phenol red to minimize background fluorescence58. Microtubules were stained with 100 nM SiR-tubulin59 and 10 µM verapamil (Sigma-Aldrich; to inhibit efflux of SiR-tubulin from cells), at least 1 h before imaging. Protein expression in cell lines bearing hsCHMP2B-LAP, hsCHMP3-LAP or hsCHMP4B-LAP was induced with 0.2 ng/mL doxycycline (Sigma-Aldrich), titrated to match levels of mmCHMP4B-LAP from the BAC construct. For expression of yeast Snf7-LAP, HeLa Kyoto FRT were transfected with pTO_SNF7_LAP using X-tremeGENE 9 (Roche) 24 h prior to imaging, and induced with 0.05 ng/mL doxycycline (Sigma-Aldrich).
Cells were imaged on a Zeiss LSM780 confocal microscope controlled by ZEN 2011 and an autofocus macro (AutofocusScreen) kindly provided by Jan Ellenberg (EMBL). Imaging was performed in a humidified atmosphere of 5% CO2 and 37°C in an incubation chamber (EMBL).
For analysis of accumulation kinetics, cells were imaged with a 63x 1.4 NA oil DIC Plan-Apochromat objective (Zeiss). ESCRT-III levels at the midbody were quantified in single optical sections using Fiji60. For normalization, the fluorescence signal prior to ESCRT-III accumulation was set to zero. Due to the variable duration of abscission, we chose the earliest time point for which we had data in all cells, which was at 48 min before abscission. Peak signals were normalized to the average midbody fluorescence from 12 min before to 6 min after abscission. Accumulation curves in VPS4A/B depletion experiments were normalized to 18-30 min after complete cleavage furrow ingression.
For quantification of abscission timing, cells were imaged with a 40x 1.4 NA oil DIC Plan-Apochromat objective (Zeiss). Abscission timing was measured as the duration from complete cleavage furrow ingression until the first disassembly of the midbody microtubules. For VPS4B rescue experiments, only cells expressing mmVPS4B-LAP were analyzed.
FRAP experiments
Midbodies were imaged using a 63x 1.4 NA oil DIC Plan-Apochromat objective (Zeiss). Photobleaching was performed in circular regions with 2 iterations of 488 nm at 42-fold higher laser intensity compared to that used for imaging. Fluorescence was measured in midbody regions followed by background subtraction. For normalization, acquisition photobleaching and ESCRT-III net accumulation during the recording interval was measured separately for early and late-stage midbodies. FRAP curves were then normalized to the mean of the pre-bleach fluorescence and to the first frame after photobleaching. For all different constructs, cells with expression levels similar to that of mmCHMP4B-LAP cells were used.
To determine FRAP recovery kinetics, we fitted single exponential functions f(t) = 1 − e−kt, single exponential functions with a variable immobile fraction f(t) = A(1 − e−kt), or double exponential functions f(t) = A1(1 − e−k1t) + (1 − A1)(1 − e−k2t), using the Levenberg-Marquardt nonlinear least-squares algorithm (minpack.lm package for R, version 3.0.2). Convergence to minimal residuals was reached in ~76% of the curves. Residence times were calculated from the dissociation rates as 1/k1 and 1/k2. Because the slow dissociation rate k2 was frequently fitted as 0 due to a completely stable fraction, we derived the slow residence times from the Taylor expansion to calculate the reciprocal mean from and standard deviation from , where µ is the mean and σ is the standard deviation. Since the long residence time could not be measured accurately due to the relatively short duration of measurements, we constrained k2 to the mean ± SD of mmCHMP4B-LAP in all experiments. For comparison of highly mobile fractions, we additionally constrained k1 to the mean k1 of mmCHMP4B-LAP, which corresponds to a residence time of 19.5 s.
siRNA treatment
Cells were transfected using Lipofectamine RNAiMAX (Invitrogen) with non-targeting siRNA siControl (ambion, sense strand: UUCUCCGAACGUGUCACGUtt), or siRNAs targeting CHMP4B (Microsynth, sense strand: AGAAAGAAGAGGAGGACGAtt)4,61, VPS4A (Microsynth, sense strand: CCGAGAAGCUGAAGGAUUAtt)61, or VPS4B (Microsynth, sense strand: CCAAAGAAGCACUGAAAGAtt)61, at a final concentration of 80 nM. Samples were analyzed at the indicated times after transfection.
Generation and microinjection of CHMP4B
To generate a CHMP4B bacterial expression plasmid containing an N-terminal 6xHis tag followed by a maltose binding protein (MBP) and a tobacco etch virus (TEV) protease cleavage site, human CHMP4B was cloned into pMBP-HIS2-Snf7 (Addgene #21492), by replacing Snf7 with PCR-amplified CHMP4B using BamHI and NotI.
For production of recombinant protein, the plasmid was transformed into E. coli Rosetta cells and expressed for 3 h at 30°C after induction with 0.5 mM IPTG. Bacterial were lysed by sonication at 4°C in lysis buffer containing 20 mM Hepes pH8, 100 mM NaCl, 1% Triton X-100, and cOmplete Protease Inhibitor Cocktail (Roche). CHMP4B was purified using affinity chromatography (MBPTrap HP 5 mL column, GE Healthcare), washed with 20 mM Hepes pH8, 250 mM NaCl, 0.1% Triton X-100, and eluted in 20 mM Hepes pH8, 10 mM maltose, followed by removal of maltose using Zeba Spin Desalting Columns (7K MWCO 5 mL, ThermoFisher Scientific). After cleavage with TEV protease, the resulting His-MBP fragment was removed by incubation with Ni-NTA Agarose beads (ThermoFisher Scientific) for 1 h, followed by incubation with Dextrin Sepharose beads (GE Healthcare) for 1 h to remove remaining protein that was not cleaved by TEV protease.
Purified CHMP4B was labeled with Atto-565-NHS ester (Sigma-Aldrich), by incubation with two molar excess of reactive dye in the presence of 100 mM NaHCO3 pH9. Excess free dye was removed by dialysis against 20 mM Hepes pH8 at 4°C using a MWCO membrane of 12-14 kDa, and the sample was further purified using Zeba spin desalting columns (ThermoFisher Scientific). The resulting labeling density was ~80%. Protein aggregates were removed by ultracentrifugation at 100,000 g for 10 min at 4°C, and the protein stored at -80°C after flash freezing.
For microinjection into cells, CHMP4B was ultracentrifuged for 30 min at 80,000 g at 4°C and loaded into Femtotips (Eppendorf). Microinjection was performed using InjectMan 4 micromanipulator (Eppendorf) and the FemtoJet 4i (Eppendorf) microinjection system, with an injection pressure of 100 hPa, an injection time of 0.2-0.5 s, and a maintenance pressure of 20 hPa. The microinjection system was mounted on the Zeiss LSM780 system described above, and cells were imaged with a 40x 1.4 NA oil DIC Plan-Apochromat objective (Zeiss) immediately prior and after injection. Fluorescence intensity of injected CHMP4B-Atto-565 was measured in midbody regions containing a CHMP4B-LAP signal. The mean intensity of the surrounding region was subtracted for background correction, and the first time point after injection was normalized to 0.
Western blot analysis
Cells were seeded and transfected as described above in LabTek II chambered coverslips (ThermoScientific). Protein extracts were separated on 4-12% NuPAGE Bis-Tris gradient gels (Life Technologies) and transferred to Nitrocellulose (Protran, GE Healthcare) or PVDF membranes (Amersham Hybond, GE Healthcare). Western blotting was performed by standard methods using antibodies against CHMP4B (1:1000, Abcam, ab105767), actin (1:30000, Merck Millipore, MAB1501), VPS4B (1:500, Abcam, ab137027), GFP (1:5000, Abcam, ab290) and GAPDH (1:2500, Abcam, ab9485).
Electron microscopy of high-pressure frozen cells
HeLa cells were seeded in T75 flasks and transfected with siRNAs using Lipofectamine RNAiMAX (Invitrogen). At 23 h post-transfection, a primary shake-off was performed to remove dead cells and debris. Mitotic cells were harvested ~2 h later with a second shake-off, and seeded on 3 mm sapphire discs (M. Wohlwend GmbH) coated with 0.01% Poly-L-lysine (Sigma) in PBS. 97 min later, sapphire discs were transferred to a 3 mm aluminum specimen carrier (type B, flat side up) and covered with a second carrier (type A), which had a 50 µm cavity filled with 10% (w/v) BSA (AppliChem) in cell culture medium, followed by high-pressure freezing in an HPF Compact 01 (M. Wohlwend GmbH). Freeze substitution was performed using a Leica EM AFS-2, in a medium of anhydrous acetone (Merck) containing 1% osmium tetroxide (Electron Microscopy Sciences) and 0.1% uranyl acetate (Merck) according to the following protocol: 21 h at -90°C, warm up at a rate of 5°C per hour to -30°C, 6 h at -30°C, warm up at a rate of 5°C per hour to 0°C, 2 h at 0°C. Specimen were washed 3 times in anhydrous acetone (Merck) at 4°C and embedded in an Epon/Araldite resin (Electron Microscopy Sciences). Serial thin sections of a nominal thickness of 100 nm were collected on Formvar-coated (Electron Microscopy Science) copper slot grids (PLANO). Sections were post-stained with 2% aqueous uranyl acetate (Polyscience) and Reynold’s lead citrate (Electron Microscopy Science), and imaged either on a Morgagni 268D transmission electron microscope (FEI) at 80 kV with a Morada CCD camera (Olympus-SIS), or on an EM 906 transmission electron microscope (Zeiss) at 80 kV with a slow-scan CCD camera (Albert-Tröndle Systems).
ESCRT-III protein purification
ESCRT-III yeast proteins were expressed from plasmids encoding Snf7 (Addgene #21492), Vps2 (Addgene #21494), Vps24 (gift from James Hurley lab), Vps4 (Addgene #21495) and purified as previously described 31. Snf7 with a cysteine residue at the N-terminus of the protein was purified as in 62. Snf7 was labeled either with maleimide-Atto-647N (Atto-Tec AD 647N-4), or with TFP-AlexaFluor-488 (ThermoFisher Scientific, A-30005). Vps2 was labeled with Atto-565 (Atto-Tec AD 565-3), and Vps24 with Atto-488 (Atto-Tec AD 488-3). The proteins were used at the following concentrations: [Snf7] = 300 nM, [Vps2] = 100 nM, [Vps24] = 100 nM, [Vps4] = 2 µM, except in Fig 6a, b, Supplementary Fig. 5e-g and Supplementary Fig. 7, where concentrations are indicated in the figures. When added to the reaction, ATP concentration was 2 mM. Fluorescent proteins were mixed with unlabeled proteins at the following ratios: Snf7-Atto-647N, Vps2-Atto-565, Vps24-Atto-488: 1/3 labeled with 2/3 unlabeled; Snf7-AlexaFluor-488: 2/3 labeled with 1/3 unlabeled.
Preparation of giant unilamellar vesicles (GUVs)
GUVs were prepared by electroformation using 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) mixtures (Avanti Polar Lipids). 20 µl of 1 mg/mL lipid mix (DOPC:DOPS, 6:4, mol:mol) were deposited on two indium tin oxide-coated glass slides (70-100 Ω resistivity, Sigma-Aldrich) and placed in a vacuum drying oven for 1 h for complete solvent evaporation. An opened tor rubber joint of ~1 mm thickness was used as a non-leaky spacer between the two ITO slides, and the chamber was formed by compressing the two slides with spring metal tweezers. The chamber was filled with 400 µl of 500 mM sucrose solution and exposed to 1 V AC-current (10 Hz sinusoidal) at room temperature for 1 h. The resulting suspension was collected in a tube and used within the next days for experiments.
Supported lipid bilayer assay
The assay is described in detail in 43. Briefly, to prepare supported membranes, a cleaned coverslip was assembled to a flow chamber (Sticky-Slide VI 0.4, Ibidi), with one entry connected to a syringe pump (Aladdin, World Precision Instruments), and the other left open for sequential introduction of solutions. The flow chamber is initially filled with 200 µL of buffer and 5 µl of GUV suspension. After ~5 min, bursting of multiple vesicles was observed on the surface of the coverslip. Then 100 µl of casein (Sigma-Aldrich) solution at 1 mg/mL was incubated for 30 min in the flow chamber to block the bare glass around membrane patches. The chamber was then washed with 200 µl of buffer before starting the experiment. The buffer used for all in vitro experiments was composed of 20 mM Tris HCl (pH 6.8), 200 mM NaCl and 1 mM MgCl2. An anti-bleaching mix composed of 10 mM glucose, 12 KU/mL catalase, 49 U/mL Glucose Oxidase and 0.75 mg/mL Trolox (Sigma-Aldrich) was used. When the incubation duration was longer than 20 min, the solution was renewed to avoid loss of the antibleaching mix efficiency (Fig. 7, and Supplementary Fig. 7).
In vitro imaging and data analysis
Imaging was performed using a spinning disc confocal inverted microscope assembled by 3i (Intelligent Imaging Innovation) including a Nikon base (Eclipse C1, Nikon), a 100x 1.49 NA oil immersion objective with a CSU-W1 confocal head and a photometrics evolve camera. Nikon perfect focus system was used in combination with a multi-positioning system (ASI) to acquire multiple fields of view over time.
Imaging was performed only in membrane-covered areas of the coverslip, which were distinguishable from the non-membrane-covered glass surface based on an unspecific background fluorescence of the labeled ESCRT-III subunits adsorbing onto glass. For all experiments, a 2 µm thick volume stack surrounding the coverslip surface was acquired with one minute time lapse.
The raw data was processed in 3 steps. First, the 3D stacks were projected in 2D by a Fiji plugin63, which determines the right focal surface plane along a z-stack. Second, the field illumination inhomogeneity was corrected by dividing all the images by an image of a homogeneously labeled surface. Third, the x-y stage drift throughout the image acquisition was corrected using the plugin Turboreg. For each experiment, patches that did not fuse completely with neighboring patches were manually selected for quantification. For each patch, a slice was manually selected to extract a kymograph. The integrated fluorescence intensity of these kymographs was measured and maximum values were normalized to 1 for all experiments, except for Fig. 7c, where curves were adjusted to match the end point of the experiment. To calculate kinetics of patch disassembly, Snf7 mean patch intensity curves were fitted to exponential decay function .
Electron microscopy of ESCRT-III filaments
For electron microscopy, multilamellar vesicles made of DOPC:DOPS, 6:4 (mol:mol), were prepared at 5 mg/mL. For Snf7 only experiments, vesicles at 0.5 mg/mL were incubated for 1 h with Snf7 at 750 nM. For Snf7+Vps2+Vps24 experiments, Snf7 was first incubated as before, then Vps2 and Vps24 were added, each at 500 nM final concentration for 1 h. All samples were then centrifuged for 4 min at 4000 g, washed, adsorbed onto Formvar-coated EM grids, then negatively stained for 30 s with 2% uranyl acetate. Images were acquired using a Tecnai G2 Sphera (FEI) electron microscope.
Sample preparation for HS-AFM
1 µL of adsorption buffer (10 mM Hepes, 220 mM NaCl, 25 mM MgCl2, pH 7.4) was added to freshly cleaved mica, followed by addition of 1 µL of the GUVs suspension. GUVs were incubated for 10 min, gently rinsed with imaging buffer (10 mM Tris-HCl, 150 mM KCl, pH 7.4), before immersion into the HS-AFM fluid chamber (volume: ~120 µL). After SLB assessment and image acquisition area positioning, Snf7 was added to the fluid chamber to a final concentration of 1 µM. After ~20 min, Snf7 filaments started to form on the SLB and assembled into spiral structures in the following minutes. Vps2 and Vps24 were then added to the AFM fluid chamber to a final concentration of 100 nM. Next, Vps4 was injected to a final concentration of 1-2 µM. Finally, ATP and MgCl2 were added to the fluid chamber to a final concentration of 200 µM and 1 mM, respectively.
HS-AFM and image analysis
Imaging was performed at room temperature on an HS-AFM 1.0 (RIBM) using ~8 µm long cantilevers (NanoWorld) with resonance frequency of ~0.7 MHz in liquid and a spring constant of ~0.15 Nm-1. Typically, pixel samplings ranged between 300 x 300 pixels and 200 x 200 pixels and frame acquisition rates ranged between 1 s-1 and 2 s-1.
For HS-AFM movie analysis, we corrected stage drift using the StackReg plugin in Fiji, and performed a moving average of 2 to 4 consecutive time frames. ESCRT-III spiral diameters and line profiles were manually measured using Fiji. Coefficients of variation were determined for 45 nm-long sections of the line profiles from the ratio of SD to mean. TrackMate was used for manual and semi-automated tracking of spiral centers with a ~5 s interval. Tracks shorter than 55 s were omitted from the analysis.
Statistics and reproducibility
To account for non-normal distributions and unequal variances, statistical analysis was performed using the two-sided Kolmogorov–Smirnov test using R (version 3.0.2) or GraphPad Prism 7.
Numbers of repeated and experiments are indicated in the respective figure legends, and always refer to independent biological experiments, with all samples showing the same behavior. All experiments were performed 3 or more times independently under identical or similar conditions, except experiments shown in panels Fig. 3a, b, Fig. 4e, Fig. 5d, g-i, Fig. 6a-d, Fig. 7d, e, Supplementary Fig. 1a, Supplementary Fig. 3a, b, d, and Supplementary Fig. 6d, e, that were performed twice, and experiments in panels Fig. 3c, d, and Supplementary Fig. 7a, b, that were performed once.
Supplementary Material
Acknowledgments
D.W.G. has received financial support from the European Community’s Seventh Framework Programme FP7/2007-2013 under grant agreements no 241548 (MitoSys) and no 258068 (Systems Microscopy), from an ERC Starting Grant (agreement no 281198), from the Wiener Wissenschafts-, Forschungs- und Technologiefonds (WWTF; project nr. LS14-009), and from the Austrian Science Fund (FWF; project nr. SFB F34-06). B.E.M. has received a PhD fellowship from the Boehringer Ingelheim Fonds. A.R. acknowledges funding from: Human Frontier Science Program (HFSP), Young Investigator Grant #RGY0076-2008: the European Research Council (ERC), starting (consolidator) grant #311536-MEMFIS: the Swiss National Fund for Research, grants #131003A_130520 and #131003A_149975. NC acknowledges the European Commission for the Marie-Curie post-doctoral fellowship CYTOCUT #300532-2011. J.M.F. acknowledges funding by an EMBO long-term fellowship (ALTF 1065-2015). T.M.R. has received funding from the Deutsche Forschungsgemeinschaft (DFG) grant MU1423/4-1. S.S. acknowledges funding by an ANR grant ANR-Nano (ANR-12-BS10-009-01) and a European Research Council (ERC) Starting Grant (#310080, MEM-STRUCT-AFM). The authors thank D. Teis, M. Alonso Y Adell, C. Campsteijn, and J. Gruenberg for comments on the manuscript, the IMBA/IMP/GMI BioOptics core facility for technical support, the EM Facility of the Vienna Biocenter Core Facilities (VBCF), who performed parts of the sample preparation and electron microscopy, F. Humbert for protein purification, C. Sommer and R. Höfler for statistical advice, C. Blaukopf for technical support, W. Reiter for providing S. cerevisiae genomic DNA, and Life Science Editors for editing assistance.
Footnotes
Author Contributions
B.E.M. designed, conducted, and analyzed all cell biological experiments, and analyzed part of the HS-AFM data. N.C. designed, conducted, and analyzed in vitro reconstitution experiments based on fluorescence microscopy. L.R-M. designed, conducted, and analyzed HS-AFM experiments. J.M-v-F. and N.C. designed, conducted, and analyzed electron microscopy of in vitro-assembled ESCRT-III polymers. J.K. and T.M-R designed, conducted, and analyzed electron microscopy experiments of intercellular bridges. J.L. established the CHMP4B purification and produced labelled CHMP4B. I.P. generated HeLa cells stably expressing mmVPS4B-LAP. B.E.M., N.C., D.W.G., A.R. and S.S. conceived the project, analyzed data, and wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Data availability
All data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–2407. doi: 10.15252/embj.201592484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanson PI, Cashikar A. Multivesicular body morphogenesis. Annu Rev Cell Dev Biol. 2012;28:337–362. doi: 10.1146/annurev-cellbio-092910-154152. [DOI] [PubMed] [Google Scholar]
- 3.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 4.Guizetti J, et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 5.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci U S A. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lafaurie-Janvore J, et al. ESCRT-III assembly and cytokinetic abscission are induced by tension release in the intercellular bridge. Science. 2013;339:1625–1629. doi: 10.1126/science.1233866. [DOI] [PubMed] [Google Scholar]
- 7.Mierzwa B, Gerlich DW. Cytokinetic abscission: molecular mechanisms and temporal control. Dev Cell. 2014;31:525–538. doi: 10.1016/j.devcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Vietri M, et al. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 2015;522:231–235. doi: 10.1038/nature14408. [DOI] [PubMed] [Google Scholar]
- 9.Olmos Y, Hodgson L, Mantell J, Verkade P, Carlton JG. ESCRT-III controls nuclear envelope reformation. Nature. 2015;522:236–239. doi: 10.1038/nature14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raab M, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 11.Denais CM, et al. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jimenez AJ, et al. ESCRT machinery is required for plasma membrane repair. Science. 2014;343:1247136. doi: 10.1126/science.1247136. [DOI] [PubMed] [Google Scholar]
- 13.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 14.Bleck M, et al. Temporal and spatial organization of ESCRT protein recruitment during HIV-1 budding. Proc Natl Acad Sci U S A. 2014;111:12211–12216. doi: 10.1073/pnas.1321655111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baietti MF, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 16.Choudhuri K, et al. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature. 2014;507:118–123. doi: 10.1038/nature12951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matusek T, et al. The ESCRT machinery regulates the secretion and long-range activity of Hedgehog. Nature. 2014;516:99–103. doi: 10.1038/nature13847. [DOI] [PubMed] [Google Scholar]
- 18.Henne WM, Stenmark H, Emr SD. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCullough J, Colf LA, Sundquist WI. Membrane fission reactions of the mammalian ESCRT pathway. Annu Rev Biochem. 2013;82:663–692. doi: 10.1146/annurev-biochem-072909-101058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peel S, Macheboeuf P, Martinelli N, Weissenhorn W. Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem Sci. 2011;36:199–210. doi: 10.1016/j.tibs.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 21.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guizetti J, Gerlich DW. ESCRT-III polymers in membrane neck constriction. Trends Cell Biol. 2012;22:133–140. doi: 10.1016/j.tcb.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt O, Teis D. The ESCRT machinery. Curr Biol. 2012;22:R116–120. doi: 10.1016/j.cub.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoneberg J, Lee IH, Iwasa JH, Hurley JH. Reverse-topology membrane scission by the ESCRT proteins. Nat Rev Mol Cell Biol. 2017;18:5–17. doi: 10.1038/nrm.2016.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem Sci. 2017;42:42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 27.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teis D, Saksena S, Emr SD. Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell. 2008;15:578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Teis D, Saksena S, Judson BL, Emr SD. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 2010;29:871–883. doi: 10.1038/emboj.2009.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlson LA, Hurley JH. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc Natl Acad Sci U S A. 2012;109:16928–16933. doi: 10.1073/pnas.1211759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obita T, et al. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 35.Stuchell-Brereton MD, et al. ESCRT-III recognition by VPS4 ATPases. Nature. 2007;449:740–744. doi: 10.1038/nature06172. [DOI] [PubMed] [Google Scholar]
- 36.Lata S, et al. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adell MA, et al. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J Cell Biol. 2014;205:33–49. doi: 10.1083/jcb.201310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang B, Stjepanovic G, Shen Q, Martin A, Hurley JH. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat Struct Mol Biol. 2015;22:492–498. doi: 10.1038/nsmb.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghazi-Tabatabai S, et al. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Pires R, et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henne WM, Buchkovich NJ, Zhao Y, Emr SD. The Endosomal Sorting Complex ESCRT-II Mediates the Assembly and Architecture of ESCRT-III Helices. Cell. 2012;151:356–371. doi: 10.1016/j.cell.2012.08.039. [DOI] [PubMed] [Google Scholar]
- 42.Shen QT, et al. Structural analysis and modeling reveals new mechanisms governing ESCRT-III spiral filament assembly. J Cell Biol. 2014;206:763–777. doi: 10.1083/jcb.201403108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiaruttini N, et al. Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell. 2015;163:866–879. doi: 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCullough J, et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 2015;350:1548–1551. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMillan BJ, et al. Electrostatic Interactions between Elongated Monomers Drive Filamentation of Drosophila Shrub, a Metazoan ESCRT-III Protein. Cell Rep. 2016;16:1211–1217. doi: 10.1016/j.celrep.2016.06.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180:389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cashikar AG, et al. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. Elife. 2014;3 doi: 10.7554/eLife.02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Babst M, Sato TK, Banta LM, Emr SD. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 1997;16:1820–1831. doi: 10.1093/emboj/16.8.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee IH, Kai H, Carlson LA, Groves JT, Hurley JH. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc Natl Acad Sci U S A. 2015;112:15892–15897. doi: 10.1073/pnas.1518765113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cashikar AG, et al. Structure of cellular ESCRT-III spirals and their relationship to HIV budding. Elife. 2014:e02184. doi: 10.7554/eLife.02184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roll-Mecak A, Vale RD. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 2008;451:363–367. doi: 10.1038/nature06482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elia N, Fabrikant G, Kozlov MM, Lippincott-Schwartz J. Computational model of cytokinetic abscission driven by ESCRT-III polymerization and remodeling. Biophys J. 2012;102:2309–2320. doi: 10.1016/j.bpj.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kline-Smith SL, Walczak CE. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 54.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 55.Zemp I, et al. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J Cell Biol. 2009;185:1167–1180. doi: 10.1083/jcb.200904048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poser I, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hein MY, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 58.Schmitz MH, Gerlich DW. Automated live microscopy to study mitotic gene function in fluorescent reporter cell lines. Methods Mol Biol. 2009;545:113–134. doi: 10.1007/978-1-60327-993-2_7. [DOI] [PubMed] [Google Scholar]
- 59.Lukinavicius G, et al. Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods. 2014;11:731–733. doi: 10.1038/nmeth.2972. [DOI] [PubMed] [Google Scholar]
- 60.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morita E, et al. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci U S A. 2010;107:12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aguet F, Van De Ville D, Unser M. Model-based 2.5-d deconvolution for extended depth of field in brightfield microscopy. IEEE Trans Image Process. 2008;17:1144–1153. doi: 10.1109/TIP.2008.924393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.