Abstract
The phosphorylation of threonine residues in proteins regulates diverse processes in eukaryotic cells, and thousands of threonine phosphorylations have been identified. An understanding of how threonine phosphorylation regulates biological function will be accelerated by general methods to bio-synthesize defined phospho-proteins. Here we address limitations in current methods for discovering aminoacyl-tRNA synthetase/tRNA pairs for incorporating non-natural amino acids into proteins, by combining parallel positive selections with deep sequencing and statistical analysis, to create a rapid approach for directly discovering aminoacyl-tRNA synthetase/tRNA pairs that selectively incorporate non-natural substrates. Our approach is scalable and enables the direct discovery of aminoacyl-tRNA synthetase/tRNA pairs with mutually orthogonal substrate specificity. We biosynthesize phosphothreonine in cells, and use our new selection approach to discover a phosphothreonyl-tRNA synthetase/tRNACUA pair. By combining these advances we create an entirely biosynthetic route to incorporating phosphothreonine in proteins and biosynthesize several phosphoproteins; enabling phosphoprotein structure determination and synthetic protein kinase activation.
Introduction
The phosphorylation of threonine residues in proteins is a key post-translational modification that regulates diverse biological processes including energy metabolism1, cell cycle2, apoptosis3 and signal transduction pathways4–6. Analytical methods have identified thousands of threonine phosphorylations, but in many cases their function remains unknown7,8. General routes to synthesize proteins that incorporate phosphothreonine at specified sites would accelerate and deepen our understanding of the functional consequences of threonine phosphorylation.
Genetic code expansion, using orthogonal aminoacyl-tRNA synthetase/tRNA pairs9,10, forms the basis of powerful approaches for synthesizing proteins bearing defined post-translational modifications11 and complements other approaches, including protein semi-synthesis 12. Two orthogonal synthetase/tRNA pairs – the Methanocaldococcus jannaschii tyrosyl-tRNA synthetase/tRNACUA and the pyrrolysyl-tRNA synthetase/tRNACUA pairs from methanosarcina species (M. barkeri and M. mazei) – have been extensively developed for non-natural amino acid incorporation. Evolved variants of these pairs enable the synthesis of proteins bearing several key post translational modifications, including lysine acetylation, methylation and ubiquitination11.
Phosphoseryl-tRNA synthetase (SepRS)/tRNACUA pairs13,14, derived from M. janaschii and Methanococcus maripaludis SepRS/tRNACysGCA 15,16, have been developed for the incorporation of O-phospho-L-serine (pSer, 1, for structures of all amino acids used in this study, see Fig. S1), an amino acid present in the cell as a biosynthetic pre-cursor to serine. We discovered a mutant SepRS/tRNA pair (referred to herein as the SepRSv1.0/tRNAv1.0CUA pair) that incorporates pSer, and its phosphonate analog, with an efficiency and fidelity comparable to that of other established orthogonal aminoacyl-tRNA synthetase/tRNA pairs that are widely used for genetic code expansion 13,17. However, this pair has not been evolved to incorporate non-natural substrates.
Evolving the SepRSv1.0/tRNAv1.0CUA pair for the site-specific incorporation of O-phospho-L-threonine (pThr, 2) into recombinant proteins is challenging: the active site of the SepRS/tRNA complex selects pSer over pThr by a factor of 104 18, and pThr only differs from pSer by the addition of a methyl group. Moreover, pSer is present in E.coli at a concentration that allows its incorporation into proteins with the SepRSv1.0/tRNAv1.0CUA pair (Fig. S2), while the pThr concentration in Escherichia. coli is low. These features suggest that it would be extremely challenging to implement existing aminoacyl-tRNA synthetase selection methods for the discovery of a pThr synthetase from SepRSv1.0 10,19.
Here we generate millimolar concentrations of pThr in E. coli and improve the orthogonality of the SepRSv1.0/tRNAv1.0CUA pair 4000-fold. We develop and validate a rapid, parallelized and scalable approach for discovering aminoacyl-tRNA synthetase/tRNA pairs that direct the incorporation of non-natural amino acids into proteins. Our new strategy for synthetase discovery enables the identification of a phosphothreonyl-tRNA synthetase/tRNACUA pair. We demonstrate the biosynthetic incorporation of pThr into recombinant proteins, and exemplify the utility of our approach by solving the structure of a phosphorylated protein and synthetically activating a protein kinase via encoded phosphorylation.
Results
Biosynthesis of pThr in E. coli
The in vivo, co-translational incorporation of pThr into proteins requires the amino acid to be present in the cell. We did not detect pThr in E. coli (Fig. S3), suggesting a low intracellular concentration (<40μM) of pThr. Adding 1mM pThr to the cell’s media did not lead to a substantial increase of pThr in cells (Fig. S3). We conclude that either pThr does not enter the cell, or enters the cell and is rapidly metabolized.
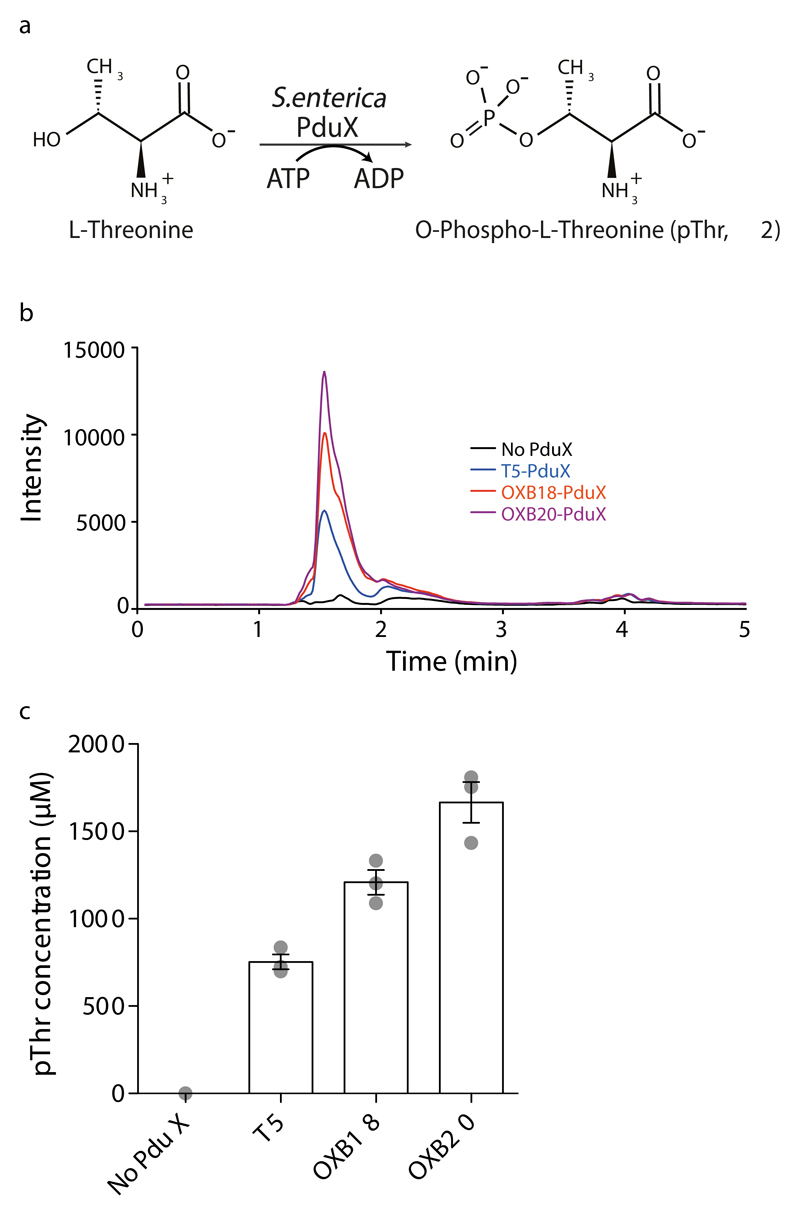
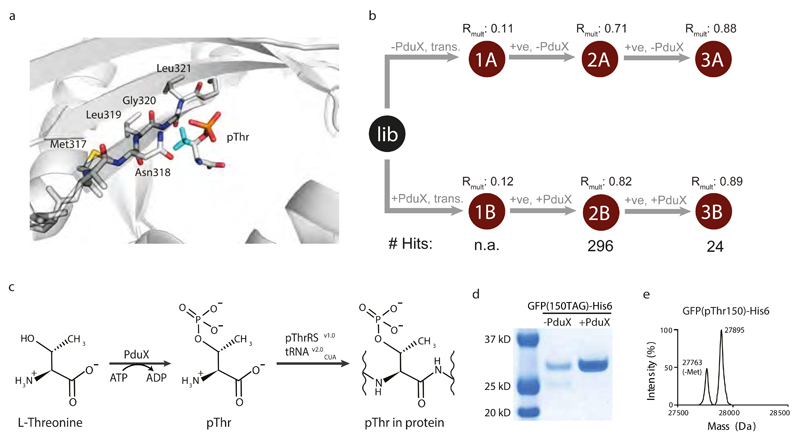
To increase the intracellular concentration of pThr, we aimed to biosynthesize this amino acid in E.coli. In Salmonella enterica a kinase, PduX, converts L-threonine to pThr using ATP (Fig.1a) as part of the de novo biosynthesis of vitamin B12 and the assimilation of corbyric acid 20,21. We found that expressing the pduX gene in E. coli led to high levels (up to 1.7mM) of pThr in cells, with pThr levels tracking known promoter strength (Fig.1b, c).
Figure 1. Expression of S.enterica PduX in E. coli enables the intracellular biosynthesis of phosphothreonine at millimolar concentrations.
a, S.enterica PduX uses ATP to phosphorylate threonine to produce pThr. b, Expression of PduX in E.coli leads to an intracellular pool of pThr. Cells were transformed with PduX on the indicated promoter and lysates analysed. c, PduX overexpressed in E.coli produces a high level of pThr. The intracellular concentration of pThr in each sample was generated from the data in panel b and a standard curve (Fig.S3). The bar shows the mean from three independent cultures, the individual data points are shown as grey dots, and the error bars represent the standard deviation.
Removing misaminoacylation of tRNAv1.0CUA
tRNAv1.0CUA, like its parent tRNA, is a measureable substrate for endogenous synthetases in E. coli in the absence of SepRSv1.0 (Fig.S4, for all full gels of this study see Fig.S5), and we demonstrated that glycine and valine are the amino acids that are mis-incorporated by tRNAv1.0CUA in the absence of SepRS (Fig.S4, for all raw MS data of this study, see Fig.S6). To improve the orthogonality of the pair, and the dynamic range of subsequent selections for synthetases, that incorporate pThr we aimed to remove the misaminoacylation of tRNAv1.0CUA by endogenous synthetases in E. coli.
To discover variants of tRNAv1.0CUA that are not aminoacylated by endogenous synthetases we created a library, tRNAv1.0CUA (N10 IE); this library targets ten nucleotides (N10) in tRNAv1.0CUA, for mutagenesis to all four bases. Nucleotide positions within this library (2:71, 3:70, 4:69) form identity elements (IEs), by which the E. coli aminoacyl-tRNA synthetases for glycine and valine recognize their cognate tRNAs 22, but are not identity elements for SepRS 23 (Fig.2a). The library was created with 3x109 clones, oversampling the theoretical diversity of 106 by 3,000-fold.
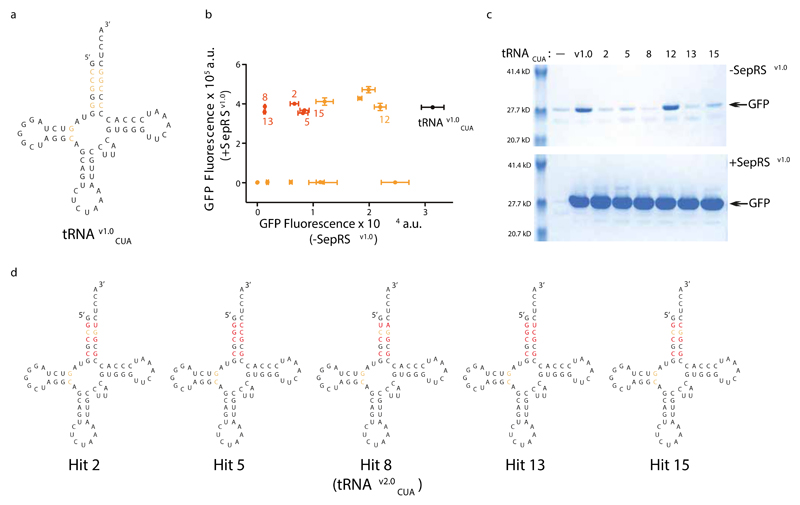
Figure 2. tRNAv1.0CUA evolution removes aminoacylation of tRNAv1.0CUA by natural aminoacyl-tRNA synthetases in E.coli.
a, Bases on tRNAv1.0CUA selected for randomization to remove mis-aminoacylation. Sequence of tRNAv1.0CUA is shown in black, and bases that were randomized in the N10 library are shown in orange. b, Certain evolved tRNAv1.0CUA variants show drastically decreased SepRSv1.0 independent amber suppression but maintain full activity with SepRSv1.0. The indicated hits were used to express GFP(150TAG)His6 in the presence or absence of SepRSv1.0. Hits that show substantially reduced SepRSv1.0-independent GFP(150TAG)His6 read-through are shown in red, the remaining hits are shown in orange. The data show the mean of three replicates from independent cultures, and the error bars represent the standard deviation. c, Characterization of the hits shown in b by expression and purification of recombinant GFP from GFP(150TAG)His6. e, Sequences of evolved tRNAv1.0CUA variants. Bases targeted in the N10 library that remain unchanged in the selected clones are shown in orange, while those that were mutated are shown in red. Hit 8 tRNA is renamed as tRNAv2.0CUA and was used in further experiments.
We selected active tRNAs from the tRNAv1.0CUA (N10 IE) library in the presence of SepRSv1.0/EF-Sep (a variant of EF-Tu) 14 on the basis of their ability to direct read through of an amber codon in chloramphenicol acetyl transferase (cat), and confer chloramphenicol resistance (Fig.S4 and Fig.S7). We screened surviving tRNAv1.0CUA (N10 IE) library members for minimal read through of an amber stop codon at position 150 in GFP in the absence of SepRSv1.0 (Fig.S4 and Fig.S7).
We identified tRNAs, clones 8 and 13, from this two step process that are as active as tRNAv1.0CUA with SepRSv1.0, but show a 4000-fold decrease in read-through of the amber stop codon in the absence of SepRSv1.0 (Fig.2b,c). Our data demonstrate that amber stop codon read-through resulting from misaminoacylation of the tRNA by endogenous aminoacyl-tRNA synthetases has been minimized. When GFP (150TAG)-His6 was expressed and purified using the evolved pair pSer was incorporated, as expected (Fig.S8). In all the selected tRNAs (Fig.2d) the C2:G71 base pair - a canonical identity element for Glycyl-tRNA synthetase 22 - is mutated. We conclude that the selection identifies nucleotides that are important for the activity of SepRS and/or discrimination against endogenous synthetases. We used clone 8, which we renamed tRNAv2.0CUA, in further experiments.
Rapid identification of amino acid selective synthetases via parallel positive selection
One challenge for evolving the SepRSv1.0/tRNAv2.0CUA pair for the site-specific incorporation of pThr comes from the serial rounds of positive and negative selection currently used to discover synthetases with altered specificity 10,19.
Despite the clear utility of current approaches, they do have certain limitations: i) it is not possible to follow the enrichment of individual clones through the selection to identify when substantial enrichment has taken place or which clones are substantially enriched, ii) the activity and specificity of synthetase clones isolated by the selection is restricted by the dynamic range of negative selections; indeed, the most active clones may often be deleted from the gene pool by negative selections, even though the activity of the synthetase they encode in the presence of an added unnatural amino acid may be sufficient to outcompete endogenous amino acid incorporation 24; iii) it is not possible to directly identify mutually orthogonal synthetases that have specificity with respect to many other unnatural substrates, and iv) the individual clones isolated at the end of the selection are picked randomly without regard to their relative enrichment or sequence. Our ability to find hits upon screening selection outputs is a function of the number of clones we examine and the frequency of the hit in the library following selection; this is in turn a function of the library size, the power of the selection steps to enrich the desired hits, the unknown frequency of hits in the library, and stochastic events.
The serial positive and negative selection approach is challenging to implement for the discovery of pThr specific synthetases from a SepRS library because i) positive selections would enrich both SepRS variants that still incorporate pSer and those that might incorporate pThr, with the former SepRS variants likely to be substantially more abundant and ii) synthetases that direct the clean incorporation of phosphothreonine when endogenous pSer is present may incorporate pSer (that differs from pThr by only a single methyl group) in the absence of pThr. This would lead to substantial read through of the amber codon in the negative selection step and deletion of desirable clones from the selection.
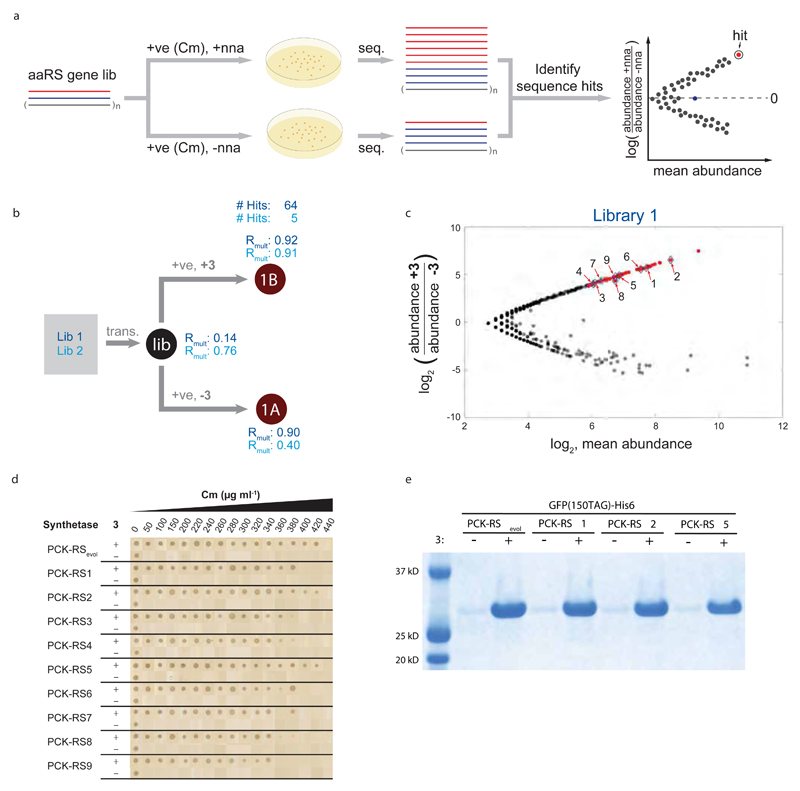
To address the limitations of current selection approaches we proposed selecting synthetases using parallel positive selections in the presence and absence of non-natural amino acid, coupled to deep sequencing and statistical analyses that compare sequencing read counts for each synthetase clone in the presence and absence of non-natural amino acid (Fig.3a). To demonstrate the power of this approach we selected aminoacyl synthetase/tRNACUA pairs that incorporate Nε-((1-(6-nitrobenzo[d][1,3]dioxol-5-yl)ethoxy)carbonyl)-L-lysine (3, an amino acid that has been extensively used to optically control diverse protein function25–28 or Nε-(carbobenzoyloxy)-L-lysine (CbzK, 4), using parallel positive selections on PylS libraries in which four (A276, Y271, L274, C313) or five (M241, A276, Y271, L274, C313) residues in the active site were mutated to all possible combinations (Fig.S9, Fig.3b, and Fig.S10). We identified multiple statistically significant hits from each selection (Fig.3c and Fig.S10-13, see Table S1 for sequences of all the aminoacyl-tRNA synthetase mutants in this study).
Figure 3. Parallel positive selections, with and without non-natural amino acid, coupled to deep sequencing and analysis rapidly identifies efficient and specific aminoacyl-tRNA synthetases for genetic code expansion.
a, Parallel positive selection, deep sequencing and analysis strategy for identifying evolved orthogonal aminoacyl-tRNA synthetases for genetic code expansion. A library of aminoacyl-tRNA synthetase genes (aaRS gene lib.) of distinct sequences is subjected to parallel positive selections on chloramphenicol (Cm), with and without an non-natural amino acid (n.n.a), in cells containing the cognate tRNACUA and a chloramphenicol acetyl transferase (cat) gene bearing an amber codon at a permissive position. For illustration purposes, two sequences in the library (colour coded red and blue) are followed through the process, the other (n) sequences in the library are coloured grey. b, Implementing the strategy described in a for the identification of PylS variants that direct the incorporation of 3. PylS libraries that randomize four (Lib1) or five (Lib2) positions in the active site of PylS were transformed (trans.) and then subjected to the indicated positive (+ve) selections in presence or absence of 3. Each selection was performed in three biological replicates. The resulting synthetase gene pools 1A and 1B were deep sequenced. Correlations of sequence abundances in the three independent transformations and selections are represented by Rmult. Aminoacyl-tRNA synthetase sequences that are selectively enriched in presence (1B) versus absence (1A) of 3 were identified by DESeq. The number of sequences identified as hits is provided. c, DESeq analysis of the selection shown in b, significant hits hits (FDR<0.01) are shown in red. The positions of sequences characterized further are indicated by their identifying number. d, Synthetases identified in panel c have comparable activity and specificity to the best previously identified synthetase for 3. Data show Cm resistance resulting from read through of an amber codon in cat. e, The activity and specificity of PylS/tRNACUA variants selected for 3 is comparable to that of the best previously evolved system in recombinant protein production.
We compared the efficiency and specificity of nine synthetases that were significantly enriched from both Lib1 (and present in selected Lib2 sequences) in the presence of 3. The efficiency and specificity of the synthetases we identified in a single round were comparable to that of PCKRSevol, the best synthetase we previously discovered for this amino acid by three rounds of classical positive and negative selection followed by screening of 96 clones25 (Fig. 3d,e, and Fig.S11). These experiments demonstrate the power of our approach for rapidly identifying synthetases for efficient and specific non-natural amino acid incorporation.
Direct selection of synthetases with mutually orthogonal substrate specificity
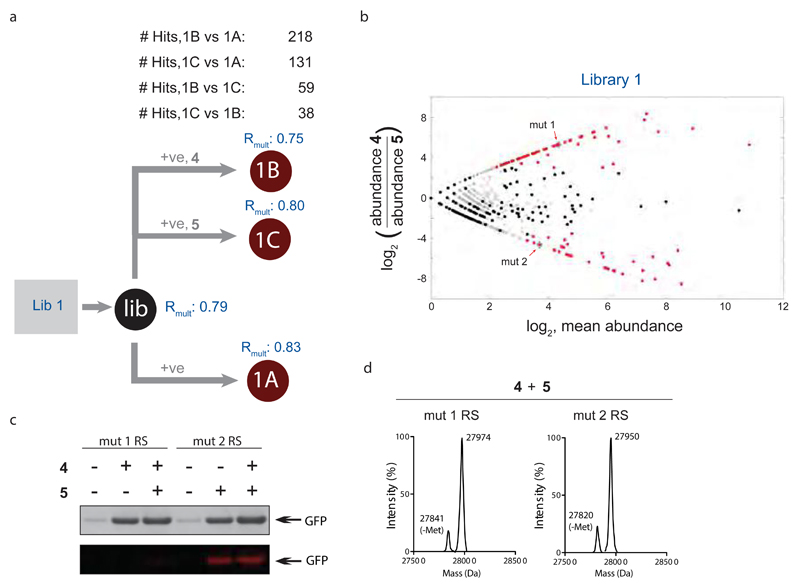
To demonstrate that the parallel positive selection approach can be extended to directly identify aminoacyl-tRNA synthetases that discriminate between non-natural amino acids we performed parallel positive selections on Lib1 in the absence of non-natural amino acid, in the presence of non-natural amino acid 4, or in the presence of Nε-(((2-methylcycloprop-2-en-1-yl) methoxy)carbonyl)-L-lysine (5) (Fig.4a). We compared the abundance of each sequence following positive selection in the presence of 4 or 5 and following positive selection in the absence of any non-natural amino acid. (Fig.4b) and, identified sequences that are only significantly enriched in the presence of one non-natural amino acid or the other.
Figure 4. Scalable parallel positive selections, deep sequencing and analysis directly identifies aminoacyl-tRNA synthetases with mutually orthogonal non-natural substrate specificity.
a, Lib 1 DNA was transformed (trans.) into cells, yielding (Lib). Parallel positive selections were performed in presence of 4, presence of 5 and absence of both 4 and 5. Each step was performed in independent duplicates. The resulting library pools (1B, 1C, and 1A, respectively) were deep sequenced, correlations for the independent duplicates (Rmult) were calculated and the number of hit mutants was determined using DESeq. b, Identifying PyRS variants that are mutually orthogonal in their substrate specificity and do not recognize natural amino acids by deep sequencing and analysis of the parallel positive selections in a. Significant hits (FDR<0.01) are indicated in red. c, Characterizing the mutual orthogonality of evolved PylS/tRNACUA pairs identified in b. Expression of GFP(150TAG)His6 with amino acids 4, 5, neither or both. The purified protein was subjected to SDS-PAGE, and visualized by InstantBlue staining (top). Each protein sample was incubated with 6 and visualised by fluorescence scanning. Only the sample containing 5 is fluorescently labeled, consistent with the established inverse electron demand Diels Alder reaction of 5 and a tetrazine-fluorophore conjugate (6). d, ESI-MS analysis of GFP from GFP(150TAG)His6, expressed in presence of 4 and 5 with the indicated synthetase and its cognate tRNA. The detected mass with mut 1 RS corresponds to GFP-3 (expected mass: 27,977 Da, measured: 27,975 Da), the detected mass with mut 2 RS corresponds to GFP-4 (expected: 27,953 Da, measured: 27,952 Da).
We characterized the amino acids incorporated by hits predicted to be selective for 4 (mutRS1, which contains the mutations Y271M, L274G and C313T), and 5 (mutRS2, which contains an A267S mutation) (Fig.4c, d). These experiments demonstrated that the mut1RS/tRNAPylCUA pair and the mut2RS/tRNAPylCUA pair direct the selective incorporation of 4 and 5 respectively. As predicted, when cells containing the mut1RS/tRNAPylCUA pair were provided with both 4 and 5, they selectively incorporated 4 in response to the amber codon in GFP(150TAG)-His6, as judged by mass spectrometry of the purified protein (Fig.4d). In contrast, cells containing the mut2RS/tRNAPylCUA pair provided with both 4 and 5, selectively incorporated 5 in GFP(150TAG)-His6, as judged by mass spectrometry.
These experiments demonstrate that our approach provides a route to the direct identification of synthetases with defined selectivity not only with respect to the twenty natural amino acids, but also with respect to defined non-natural amino acids. It is not possible to systematically select such mutually orthogonal synthetases using existing methods. Since positive selections may be performed with many different substrates in parallel, the approach provides a scalable route to the discovery of mutually orthogonal systems. The ability to rapidly discover synthetases with mutually orthogonal specificity may enable cell and tissue specific incorporation of distinct unnatural amino acids into proteins for multi-colour imaging or multiplexed cell-specific proteomics 29,30.
Selecting a pThrRS/tRNAv2.0CUA pair
Next, we aimed to use our selection approach to select a variant of the SepRSv1.0/tRNAv2.0CUA pair for the site-specific incorporation of pThr. We used the structure of Archaeoglobus fulgidus SepRS/phosphoseryl-tRNACys to model pThr in the active site of SepRS (Fig.5a) and identify residues to target for mutation to accommodate the methyl group of pThr. We designed a library, SepRSv1.0 (317-321 lib) in which M317, N318, L319, G320, and L321 (M. maripaludis residue numbering is used throughout), that lie along a beta strand, are mutated to all twenty amino acids 31. We created this library with 109 independent clones, exceeding the theoretical diversity of the library by 15-fold.
Figure 5. Discovery and characterization of a pThrRS/tRNAv2.0CUA pair for the biosynthesis and genetic encoding of pThr in recombinant proteins.
a, Active site of SepRS with pThr modeled. Binding pocket of A. fulgidus SepRS–tRNACys–O-phosphoserine ternary complex (PDB ID: 2DU3) is shown. The methyl group (Cyan) of pThr was built onto pSer in the complex using Pymol. AfSepRS is shown in gray cartoon. The side chains of the randomized residues on M. maripaludis SepRS, Met317, Asn318, Leu319, Gly320, and Leu321 (corresponding to Met324, Asn325, Leu326, Gly327, and Leu328 in A. fulgidus SepRS), in the library are shown. b, The SepRSv1.0 (324-328) library was subjected to two rounds of positive selection in the presence and absence of PduX. Each selection was performed in independent triplicates. Correlations of sequence abundances in the three independent transformations and selections are represented by Rmult. For each selection step the number of significantly (FDR < 0.01), differentially enriched mutants in presence versus absence of 2 was identified by DESeq. The number of sequences identified as hits is provided. c, Coupling the biosynthesis and genetic encoding of phosphothreonine into proteins. d, PduX dependent production of GFP incorporating 2 at position 150. Purified proteins from an equal number of cells were separated by SDS-PAGE and visualized by InstantBlue staining. e, Purified proteins were also analyzed by electrospray ionization mass spectrometry. The observed mass of 27894.0Da compares well to the predicted mass of 27894.3Da for pThr incorporation.
To identify SepRSv1.0 variants that incorporate pThr we transformed the SepRSv1.0 (317-321 lib)/tRNAv2.0CUA library into cells with or without pduX, in parallel transformations, and performed two rounds of positive selection on each transformation (Fig.5b, and Fig.S14). This experiment differs from our previous parallel positive selections with PylS libraries because it uses parallel transformations in two different genetic backgrounds (+pduX and –pduX) to generate the parallel positive selection conditions. This distinction means that there may be a difference in the abundance of any clone in the +pduX cells and -pduX cells before positive selection; which may increase the frequency of false positives or negatives in statistical analyses that assume a clone is present at the same abundance before selection in the presence or absence of the non-natural amino acid. While this variation could in principle be addressed by further statistical analysis, in this case only sixteen clones are selectively enriched in the +pduX sample with respect to the -pduX sample after both rounds of positive selection and we therefore decided to remove false positives by testing the sixteen clones for pThr incorporation. One pair, which we named pThrRS/tRNAv2.0CUA, led to the quantitative site-specific incorporation of pThr into recombinant proteins (see below). pThrRS contains the mutations G320A and L321Y, with respect to SepRSv1.0.
Expression of recombinant phospho-proteins for structural and biochemical studies
To create a system for producing recombinant proteins that site-specifically incorporate pThr we cloned the genes encoding pThrRS/tRNAv2.0CUA, EF-Sep and PduX into a high copy number pUC-based plasmid. In this system, PduX catalyzes the biosynthesis of pThr from L-threonine, and pThrRS catalyzes the aminoacylation of tRNAv2.0CUA with pThr for co-translational incorporation into proteins in response to an amber stop codon in a gene of interest (Fig.5c).
To test for pThr incorporation, we co-transformed this pUC-based plasmid with a pNHD GFP (150TAG)-His6 plasmid, for production of sfGFP, into BL21 (DE3) ΔserC E. coli (deletion of serC minimizes intracellular pSer). Using this system we expressed 5 mg of GFP per L of culture (Fig.5d); thus the evolved pThrRS directs the incorporation of pThr with an efficiency comparable to that with which SepRS incorporates its natural substrate, a conclusion supported by several additional experiments (Fig.S15). GFP expression was strongly PduX dependent, and weakly EF-Sep dependent (Fig.S16), consistent with previous observations 13. pThr was incorporated with high fidelity in response to the amber stop codon (Fig.5e, for all ESI-MS/MS analysis, see Fig.S17). Additional experiments demonstrated the genetically encoded incorporation of two phosphothreonines in a single polypeptide (Fig.S17 and Fig.S18).
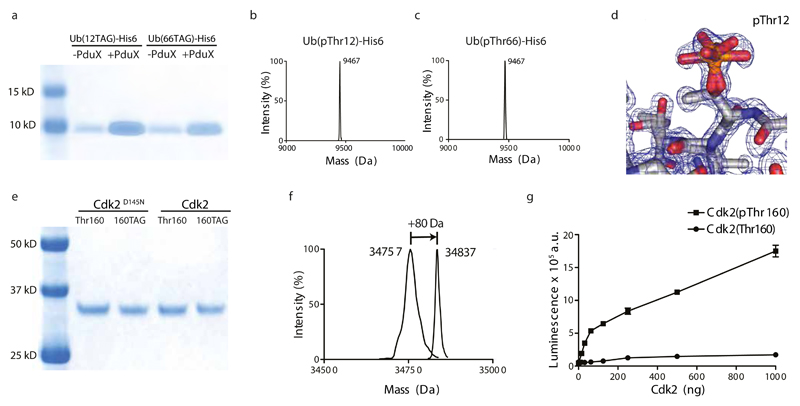
To investigate the generality of our approach, and to install pThr at native sites of phosphorylation identified in eukaryotic cells, we expressed ubiquitin (Ub) bearing pThr at position 12 or 66 32. The kinases that install these phosphorylations are unknown, and these phosphoproteins have not previously been synthesized.
Production of Ub bearing pThr at position 12 was strongly PduX dependent, consistent with the pThrRS aminoacylating tRNAv2.0CUA with pThr. However, the resulting protein contained a mixture of threonine and phosphothreonine, and similar results were obtained when we directed the incorporation of pThr at position 66 of Ub, or into other proteins (Fig.S19). We hypothesized that an E. coli phosphatase dephosphorylates phosphothreonine incorporated into proteins (or aminoacylated onto tRNAv2.0CUA) and that deletion of the relevant phosphatase would enable the production of proteins that retain encoded phosphorylation.
To identify the phosphatase responsible for pThr dephosphorylation we deleted individual candidate genes in E. coli and screened these deletion strains for the production of ubiquitin that is homogeneously phosphorylated at position 12 (Fig. S20). Deleting a single putative phosphatase, ycdX, enabled the production of homogenously phosphorylated ubiquitin, and additional experiments demonstrated the generality of this approach for homogeneous pThr incorporation at different sites and in different proteins (Fig.S19, Fig.6 and Fig. S17).
Figure 6. Biosynthesis of proteins with genetically encoded pThr enables structural and biochemical characterization of biologically relevant phospho-proteins (Ub (pThr12), Ub (pThr66), Cdk2(pThr160).
a, PduX dependent production of Ub incorporating pThr at position 12 or at position 66. Proteins were purified, from equal numbers of cells in +PduX and -PduX samples, were separated by SDS-PAGE and visualized by InstantBlue staining. b,c, Purified phospho-ubiquitins were also analyzed by electrospray ionization mass spectrometry. The observed masses were 9467 Da and compare well to the predicted mass of 9467.7Da for pThr incorporation. d, The 1.07Å crystal structure of Ub (pThr12). The density in the region of pThr12 with two conformations of the phosphate is shown. e, Expression of Cdk2 with or without the D145N mutation that removes residual catalytic activity. Each protein is expressed from a gene with a threonine codon or the amber codon, which is decoded by the phosphothreonine incorporation system, in the activation loop of the kinase (codon 160). Protein loading from the amber mutants and non-amber controls was normalized. f, ESI-MS of Cdk2(D145N) in which threonine or phosphothreonine is genetically encoded at position 160. Genetically encoded phosphorylation leads to the expected 80 Da shift in protein mass. g, Synthetically activating Cdk2 by genetically encoding phosphothreonine at position 160. Cdk2 activity was measured by following the release of ADP upon Histone H1 phosphorylation in three independent kinase reactions. The error bars represent the standard deviation.
To demonstrate the utility of our approach for producing homogeneously phosphorylated proteins for structural biology we grew crystals of ubiquitin incorporating phosphothreonine at position 12 and solved the structure of this phosphoprotein at 1.07Å (Fig.6d, Fig.S21). The structure shows clear density for two rotamers of phosphothreonine, and demonstrates that the amino acid we have biosynthesized and incorporated into proteins has the correct stereochemistry at both the alpha and beta carbon of phosphothreonine. The r.m.s.d. from unmodified Ub is small, 0.595Å, as expected (Fig. S21, and Table S2).
To demonstrate that our system enables the efficient incorporation of pThr in the activation loop of a protein kinase, we expressed cyclin-dependent kinase 2 (Cdk2), a key kinase that controls the eukaryotic cell cycle and is activated by phosphorylation on Thr160 (Fig.6e). We expressed and purified the catalytically inactive Cdk2(D145N, 160TAG) mutant (we used this mutant to avoid autophosphorylation and aid characterization of pThr incorporation by mass spectrometry) in cells expressing the pThr incorporation machinery and demonstrated the genetically encoded incorporation of pThr at position 160 by ESI-MS and MS/MS (Fig.6f and Fig.S17). We then expressed and purified Cdk2 (pThr160) from cells containing Cdk2(160TAG) and the pThr incorporation machinery and confirmed pThr at position 160 by MS/MS. Cdk2 (pThr160) was much more active than wild-type Cdk2 in phosphorylating Histone H1 (Fig.6g), demonstrating synthetic kinase activation through encoding pThr in the activation loop.
Discussion
We have generated a rapid, scalable approach for discovering orthogonal aminoacyl-tRNA synthetases that incorporate non-natural amino acids into proteins. Our approach addresses many of the limitations of previous strategies, and allows the direct identification of synthetases with mutually orthogonal substrate specificity. We anticipate that our approach will be extended to the selection of other translational components with new specificities. More generally, we provide a robust foundation for coupling directed evolution, deep sequencing and statistical analysis to the direct and scalable identification of mutants with desired specificities.
We have evolved the SepRSv1.0/tRNAv2.0CUA pair for a first non-natural substrate, and it will be interesting to investigate the scope of non-natural substrates that may be incorporated by future directed evolution of this pair. The pThr/tRNAv2.0CUA pair enables the biosynthesis of proteins containing pThr and we explicitly demonstrate utility for structural biology and synthetic kinase activation. We anticipate that this approach will facilitate an understanding of how threonine phosphorylation regulates diverse biological processes.
Online Methods
Plasmid generation
The DNA fragment encoding Salmonella enterica PduX was cloned into a pCDF-based vector. A T5 promoter was inserted before PduX by inverse PCR to give pCDF_T5-PduX; OXB18 and OXB20 promoter sequences were cloned from pSF-OXB vectors (Oxford Genetics) and were inserted upstream of PduX by Gibson assembly to give pCDF_OXB18-PduX and pCDF_OXB20-PduX. For tRNA selection, the engineered Methanocaldococcus jannaschii (Mj) tRNA(B4)CUA 13(hereafter referred to as tRNAv1.0CUA) was inserted into a pKW vector 33 by Gibson cloning and was named as pKW_SeptRNAv1.0CUA. DNA encoding the engineered Methanococcus maripaludis SepRS(2) 13 (hereafter referred to as SepRSv1.0), EF-Sep, and CAT112TAG were assembled into pCDF vector, which was named as pCDF_SepRSv1.0_EF-Sep_CAT112TAG. We refer to sfGFP as GFP throughout; the DNA fragment encoding sfGFP150TAG was cloned into pNHD vector 17 with a C-terminal 6xHis tag to give pNHD_sfGFP150TAG-His6. The 158th codon of sfGFP ORF was further mutated to TAG to give pNHD_sfGFP150TAG/158TAG-His6. For SepRS library selection, SeptRNAv2.0CUA, evolved from SeptRNAv1.0CUA in this study, as well as SepRSv1.0, was assembled into pKW vector to give pKW_SepRSv1.0_SeptRNAv2.0CUA. EF-Sep, and CAT112TAG were assembled into pCDF_OXB18-PduX to give pCDF_OXB18-PduX_EF-Sep_CAT112TAG. For efficient protein expression, pThrRS, tRNAv2.0CUA, OXB20-PduX, and EF-Sep were assembled into a pUC vector. Another pUC based vector comprised of pThrRS, tRNAv2.0CUA, and EF-Sep was made as control. DNA fragments encoding human Ubiquitin and Cdk2 were cloned into a pNHD vector as described 17. The 12th codon of Ubiquitin ORF was mutated to TAG to give pNHD_Ub12TAG; the 66th codon of Ubiquitin ORF was mutated to TAG to give pNHD_Ub66TAG; The 160th codon of Cdk2 ORF was mutated to TAG to give pNHD_Cdk2160TAG. The 145th codon of Cdk2 ORF was further mutated give pNHD_Cdk2D145N and pNHD_Cdk2D145N/160TAG. A CDF based vector overexpressing serB, pCDF_SerB_EF-Sep, was described earlier 13. pRSF vectors containing GST(TAG)CaM or riboQ1 rRNA with O-GST(TAG)CaM were described earlier 33,34.
Strain generation
The serC gene from DH10B cells (NEB) was disrupted using the GeneBridges Counter-Selection Kit to give DH10B ΔserC. DH10B ΔserB cells, and BL21 (DE3) ΔserB cells were described 13. Genes in BL21 (DE3) were deleted using lambda red recombination (GeneBridges E. coli Gene Deletion Kit). The serC gene was disrupted in BL21 (DE3) (Invitrogen) to give BL21 (DE3) ΔserC by replacing the ORF of serC with a DNA fragment containing a chloramphenicol resistance gene (CmR) and sacB gene 35. The selection cassette was removed by recombination and selection on LB agar plates supplemented with sucrose to a concentration of 7.5% (w/v). Then, each of five phosphatase genes aphA, phoA, ycdX, pgpA, and pgpC was individually knocked out by replacing the ORF with CmR-SacB selection cassette to give BL21 (DE3) ΔserC ΔaphA, BL21 (DE3) ΔserC ΔphoA, BL21 (DE3) ΔserC ΔycdX, BL21 (DE3) ΔserC ΔpgpA, and BL21 (DE3) ΔserC ΔpgpC.
PylS Library creation
PylS library 2 (M. barkeri PylS residues M241, A267, Y271, L274, and C313 randomised to NNK) corresponded to the PCK-RS “up” library reported previously 25. To derive PylS library 1 (M. barkeri PylS residues A267, Y271, L274, and C313 randomised to NNK), position M241 was fixed by EI-PCR using PrimeStar Max DNA polymerase (Clontech) with the following primer pair: 5’-GCGCAGGTCTCAGAACGTNDTGGCATTAACAACGACACCGAACTGAGCAAA C-3' (F) and 5’-gcgcaGAGTAGGTCTCAGTTCCACATATTCCGCCGGAATCAGAATC-3' (R).
The PCR products were gel extracted, digested by BsaI, re-circularized by T4 DNA ligase, and electroporated into DH10B competent cells to create a library with a diversity of ≈109 colonly forming units (c.f.u), surpassing the required 107 c.f.u. The DNA pool was then extracted by DNA midiprep and frozen.
Parallel positive selections on PylS libraries
PylS library 1 or 2 was transformed into DH10B cells with pREP_PylT_CAT112TAG to a diversity of 5x109 c.f.u. The transformations were done in triplicates and incubated at 37°C. After overnight incubation, plasmid DNA was extracted, representing the unselected library sample and was stored at -20°C until further use. From each overnight culture, 1ml of cells were diluted into 10ml fresh LB medium supplemented with 1mM unnatural amino acid (3, 4, or 5) and 1ml of cells were diluted into 10mL fresh LB medium not supplemented with unnatural amino acid. The cultures were grown until OD600=0.6. From each culture, 2ml of cells were spread onto LB plates supplemented with 100μg/ml chloramphenicol, in the presence or absence of 1mM unnatural amino acid. The plates were incubated at 37°C for 40hrs. After incubation, colonies on each plate were washed off and collected in PBS buffer and plasmid DNA was extracted.
For sequential rounds of positive selection in Fig.S10, the extracted plasmid DNA from the first round of selection was electroporated into a fresh batch of DH10B cells with pREP_PylT_CAT112TAG and the positive selection protocol was restarted. Each positive selection replicate was transformed separately.
SeptRNAv1.0CUA (N10 IE) library generation
To create the SeptRNAv1.0CUA (N10 IE) library, 10 bases including C2:G71, C3:G70, G4:C69, G6:C67, and G10:C25 were randomized by enzymatic inverse PCR (EI-PCR) using PrimeStar Max DNA polymerase (Clontech). The PCR product was gel extracted, digested with BsaI, re-circularized using T4 DNA ligase, and electroporated into DH10B competent cells to create a library with a diversity of ≈3x109 c.f.u, surpassing the required theoretical diversity of 106 c.f.u. The DNA pool was then extracted by DNA midiprep and frozen.
tRNAv1.0CUA (N10 IE) library selection
The tRNAv1.0CUA (N10 IE) library was co-transformed into DH10B cells with pCDF_SepRSv1.0_EFSep_CAT112TAG to a diversity of 5x109 c.f.u. Transformed cells were grown overnight in LB at 37°C. Then, 10ml of the overnight culture was diluted into 500 ml fresh LB medium supplemented with 1mM IPTG and incubated at 37°C until OD600=0.6. 2ml of this culture was spread onto LB plates with 1mM IPTG and 100μg/ml chloramphenicol. The plate was incubated at 37°C for 40hrs.
After incubation, colonies on the plate were washed off and collected in PBS buffer and the plasmids were extracted by DNA midiprep. To remove the pCDF vector, the extracted DNA was digested with NcoI and KpnI restriction endonucleases and re-purified using PCR purification kit (Qiagen). The remaining SeptRNAv1.0CUA library vector was further co-transformed into DH10B cells with pCDF_EFSep and pNHD_sfGFP150TAG, and the transformed cells were spread onto LB plates and incubated at 37°C for 16hrs.1,890 colonies were picked from the plates using a Qpix 420 automatic colony picking system and inoculated into 80μl fresh LB supplemented with 1mM IPTG and 0.2% L-arabinose in five 384-well plates. The plates were incubated at 37°C, and the OD600 and GFP signals (λex = 485 nm, λem = 520 nm) of each well were recorded after 24hrs by Tecan plate reader (Thermofisher).
Cells from 15 wells with the lowest GFP/OD600 ratios were diluted and grown in fresh LB medium, and the vector containing SeptRNAv1.0CUA mutants was extracted by miniprep. The 15 SeptRNAv1.0CUA mutants were co-transformed into DH10B cells with pNHD_sfGFP150TAG and with pCDF_SepRSv1.0_EFSep or pCDF_EFSep. Transformed cells were grown at 37°C overnight and 4μl of each cell culture was diluted into 200μl fresh LB supplemented with 1mM IPTG and 0.2% L-arabinose in 96-well plates in triplicates. The OD600 and GFP signals (λex = 485 nm, λem = 520 nm) of each well were recorded after 24hrs by Tecan plate reader (Thermofisher).
SepRS library generation
Guided by the structure of the Archaeoglobus fulgidus SepRS–tRNACys–O-phosphoserine ternary complex (PDB ID: 2DU3), residues M317, N318, L319, G320, and L321 on Methanococcus maripaludis SepRS were chosen for randomization by EI-PCR using primers mixed based on “small-intelligent” principles 31. The PCR products were gel extracted, digested by BsaI, re-circularized by T4 DNA ligase, and electroporated into DH10B competent cells to create a library with a diversity of ≈109 c.f.u, surpassing the required 1.3x107 c.f.u. The DNA pool was then extracted by DNA midiprep and frozen.
SepRSv1.0 (317-321) library parallel positive selection
For the first round selection, SepRSv1.0 (317-321) library was co-transformed into DH10B ΔserC cells with pCDF_OXB18-PduX_EFSep_CAT112TAG or pCDF_EFSep_CAT112TAG to a diversity of 109 c.f.u. The transformations were done in triplicates and incubated at 37°C. After overnight incubation, 10ml of cells from each culture were diluted into 500ml fresh LB medium supplemented with 1mM IPTG and grown until OD600=0.6. 2ml of cells from each culture were spread onto LB plates supplemented with 1mM IPTG and 100μg/ml chloramphenicol. The plates were incubated at 37°C for 40hrs. After incubation, colonies on each plate were washed off and collected in PBS buffer and the plasmids were extracted by DNA midiprep, yielding six plasmid pools with three replicates for +PduX and three for -PduX.
For the second round selection, a plasmid pool from the first round selection in the presence of PduX was transformed again into DH10B ΔserC cells with pCDF_OXB18-PduX_EFSep_CAT112TAG; and a plasmid pool from the first round selection in the absence of PduX was transformed again into DH10B ΔserC cells with pCDF_EFSep_CAT112TAG. Transformation diversities were all above 109 c.f.u. The rest of the selection process was identical to the first round selection. Finally, the plasmids were extracted from cells on each plate by DNA midiprep, yielding six plasmid pools with three replicates for +PduX and three for –PduX. Plasmid pools from two rounds of parallel positive selections were then prepared for deep sequencing.
Deep sequencing sample prep and sequencing
Illumina adapter sequences were attached by PCR of purified, plasmid-borne DNA selection samples using primers that contained Illumina adapter sequences on their 5’ overhangs. From 5’ to 3’ both the forward and the reverse primer consisted of the following: 1) Illumina adapter (forward ‘P5’ primer sequence: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT, reverse ‘P7’ primer sequence: CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT), 2) four degenerate nucleotides (NNNN), 3) hexanucleotide barcode sequence to allow for sample multiplexing (Table S3), and 4) primer binding sites specific to the sequences flanking the library sites (for PylS sequencing: F: CGGCGGAATATGTGGAAC, R: CAGCCGCTGCCCATT; for SepRS sequencing: F: CATCCGAAACTTAAGGAATGGCTGGA, R: GCTAATCATCGCTAGCCGCTCA). Barcode sequences were designed using an approach reported previously 36. For PCR, samples of 20µL volume were prepared using the KAPA HiFi HotStart ReadyMix (Kapa Biosystems) according to the manufacturer’s instructions. Thermal cycling was conducted in a TAdvanced (Biometra) cycler as follows: 1) 95 °C for 3 min, 2) 98 °C for 20 s, 3) 60 °C for 15 s, 4) 72 °C for 1 min, 5) repeat steps 2 to 4 for 18 cycles, 6) 72 °C for 10 min. PCR products were purified by gel extraction using the E-Gel SizeSelect Agarose Gel 2% kit according to the manufacturer’s instructions. For quantification of DNA concentration, quantitative PCR (qPCR) was used. PCR products were diluted 1:106 and qPCR was performed in triplicates using the Illumina Complete Kit Universal (Kapa Biosystems). DNA concentration was determined based on CT values by linear fitting of the sample data with the log-transformed values obtained for the kit’s concentration standards. DNA samples were diluted to 2 nM. Per sequencing lane, up to 40 samples were multiplexed. The samples were denatured to single stranded DNA and prepared for deep sequencing according to the instructions given in the MiSeq Reagent Kit v2 (300-cycles) by Illumina. To reduce the sequence homogeneity, the sequencing-ready sample was spiked with Illumina PhiX Control v3 to a final ratio of 10%. Deep sequencing was performed on a MiSeq Sequencer (Illumina), which yielded 16 Million reads per lane. Subtracting the fraction of reads aligning to the PhiX genome and assuming 40x multiplexing, up to 360,000 reads per sample were obtained.
PylS phenotyping by bacterial culture fluorescence assay
E. coli strain DH10B bearing plasmid pBAD-pylT-sfGFPTAG150 was electroporated with pBk-PylS plasmid variant bearing a PylS mutant. Single colonies were derived and grown to density overnight in LB medium. Each overnight culture was used to inoculate LB medium in a multi-well plate supplemented with 0.2% L-arabinose + 1 mM non-natural amino acid and LB medium supplemented with 0.2% L-arabinose, no non-natural amino. The multi-well plate was incubated at 37 °C, 400 rpm for 16 h. Readings of green fluorescence (λex = 485 nm, λem = 520 nm) and OD600 were taken for each well using a M200 Pro Plate Reader (Tecan).
Protein labeling with tetrazine-conjugated dye
Protein labeling was performed as previously described 37.
E.coli cells extraction for intracellular pThr concentration measurement
A cell extraction method was developed38. In 50-ml Falcon tubes, 15-ml cultures of DH10B ΔserC cells were incubated in LB at 37°C for 16hrs. To measure the import of pThr, 1mM pThr (Sigma-Aldrich) was supplemented into the LB medium. After incubation, the OD600 was measured from a 10-fold dilution, and the cells were harvested by centrifugation at 4,000g for 15 min. The resulting cell pellets were resuspended in 1ml of ice-cold LB media. The suspension was transferred to a 2-ml eppendorf tube. The cells were pelleted by centrifugation at 6,000g for 5min. The supernatant was removed by carefully pipetting off of the top. The pellet was washed three more times by the same suspension-centrifugation method. The final pellet was resuspended in 400µl of 40:60 methanol:water. To the suspension was added 300mg of 0.1mm cell disruption beads. The samples were vortexed for 12min to lyse the cells. The resulting lysate was centrifuged at 21,000g for 30min at 4°C. From the supernatant, 300µl were pipetted into a fresh 1.5-ml conical eppendorf tube. The samples were centrifuged again at 21,000g for 2-3h at 4°C. From the top of the supernatant, 100µl were removed for LC-MS analysis.
To measure the intracellular concentration of pThr with PduX overexpressed, DH10B ΔserC cells transformed with pCDF_T5-PduX, pCDF_OXB18-PduX, or pCDF_OXB20-PduX were incubated in LB at 37°C for 16 hrs. DH10B ΔserC cells without transformation were incubated under the same conditions as control. The cytosol extraction was as described above for other samples.
LC-MS method and data analysis (for amino acids)
The clarified lysates were pipetted into 250-µl glass inserts (Agilent). An Agilent 1260 Infinity equipped with an Agilent 6130 Quadrupole LC-MS unit was used for analysis of all samples. From each sample, 5µl was injected onto a Zorbax SB C18 column, 4.6 x 150 mm equipped with a guard column (Agilent). The sample was eluted from the column using a mobile phase gradient from 0.5% to 95% acetonitrile containing 0.02% formic acid. The mass spectrometer was set to selected ion monitoring (SIM) mode. The ions monitored were 200 M/z in the positive mode and 198 M/z in the negative mode. Standards of 5, 10, 20, 50, and 70µM of phosphothreonine in water were prepared and analyzed on the same day. A lysate sample obtained from E. coli grown in the absence of the NNA was spiked with phosphothreonine to a final concentration of 10µM. The peak areas corresponding to phosphothreonine were compared between the phosphothreonine-spiked sample and the 10µM phosphothreonine standard. There was negligible difference between these peak areas, indicating that minimal ion suppression occurs during ionization of the clarified lysates.
A linear fit of the standard samples was used to determine the concentration of phosphothreonine in the lysate. Using the following equation, the intracellular concentration was determined from the lysate concentration and the OD600 measurements for each sample.
Approximate values were used for the cell concentration as a function of OD600 (8x108 cells per OD600) and for the E. coli cell volume (0.6fL).
Protein expression and purification
For confirmation of PylS mutants with mutual orthogonality, E. coli strain DH10B bearing plasmid pBAD_pylT_sfGFPTAG150 was electroporated with plasmid pBk-PylS bearing a PylS mutant. A single colony was derived and used to inoculate 5 mL of LB supplemented with 50 µg mL-1 ampicillin and 7.5 µg mL-1 tetracycline. The bacterial culture was incubated at 37 °C for 16 h. OD600 was monitored and the culture was used to inoculate two fresh cultures at OD600 = 0.1. The two inoculated cultures each contained 20 mL LB supplemented with 50 µg mL-1 ampicillin and 7.5 µg mL-1 tetracycline. The cultures were incubated at 37 °C until OD600 reached 0.3 and one culture was supplemented with the relevant non-natural amino acid to a concentration of 1 mM. The cultures were incubated at 37 °C for 20 min and 200µg mL-1 L-arabinose was added. Culture incubation continued at 37 °C for 6 h. OD600 and green fluorescence (λex = 485 nm, λem = 520 nm) readings were taken using a M200 Pro Plate Reader (Tecan). Cultures were centrifuged at 4 °C, 4000 rpm for 20 min. Supernatant was discarded, the pellet was resuspended in 800 µL PBS and transferred into microcentrifuge tubes. The suspensions were centrifuged at 4 °C, 10,000 rpm for 10 min. Supernatant was discarded, the pellet was resuspended in 800 µL PBS and the suspensions were centrifuged at 4 °C, 10,000 rpm for 10 min. Supernatant was discarded and 800µL lysis buffer (1 mg mL-1 lysozyme [Sigma Aldrich], 0.1 mg mL-1 DNAse I [Sigma-Aldrich], 20 mM imidazole, cOmplete Protease Inhibitor Cocktail [Roche]) was added to each pellet. Lysis occurred by incubation at 25°C, 1000 rpm for 1 h. Lysates were centrifuged in a table-top centrifuge at 4°C, 10000 rpm for 30 min. During centrifugation 100 µL Ni-NTA Magnetic Agarose Beads (Qiagen) were washed three times in 800 µL wash buffer (50 mM Tris, 300 mM NaCl, and 20 mM imidazole) by binding the beads to a magnetic rack and discarding the unbound liquid. Lysate supernatants were transferred each to 50 µL pre-washed Ni-NTA Magnetic Agarose Beads and incubated at 4°C, 30 rpm for 16 h. Lysates were transferred to a magnetic rack and the unbound fraction was discarded. Beads were washed three times with wash buffer. For elution, the beads were incubated with 50 µL elution buffer (wash buffer supplemented with 300 mM imidazole) and separated from the eluted fraction on the magnetic rack. Elution was repeated a second time to yield a total of 100 µL per sample.
For SeptRNAv1.0CUA library hits validation, pNHD_sfGFP150TAG was co-transformed into DH10B cells with SeptRNAv1.0CUA mutant and pCDF_SepRSv1.0_EFSep or pCDF_EFSep. For pThrRS hits validation, pNHD_sfGFP150TAG was co-transformed into DH10B ΔserC cells with pCDF_OXB18-PduX_EFSep and SepRS(317-321) mutant. The transformed cells were spread onto LB plates, and a single colony was picked and inoculated into LB for overnight incubation. 0.5ml overnight culture was diluted into 25ml fresh LB supplemented with 1mM IPTG and grown until OD600=0.6, and L-arabinose was added to a final concentration of 0.2%. Cells were incubated at 37°C for 16hrs and pelleted.
For overexpression of sfGFP incorporating 2, BL21 (DE3) ΔserC cells were transformed with pNHD_sfGFP150TAG and pUC_pThrRS_SeptRNAv2.0CUA_OXB20-PduX_EFSep or pUC_pThrRS_SeptRNAv2.0CUA_EFSep. We picked a single colony and inoculated LB for overnight incubation. 5ml overnight cultures were diluted into 250ml fresh LB and grown until OD600=0.6, and 1mM IPTG together with 1% L-arabinose was added. Cells were incubated at 37°C for 3hrs and pelleted.
To lyse cells, 5ml Bugbuster Protein Extraction Reagent (Novagen) was used to resuspend the cell pellet and incubated on ice for 30min. Cell debris was then pelleted by centrifuging at 26,000g for 30min at 4°C. NaCl was added to supernatant to 500mM and imidazole was added to 20mM. Then, the supernatant was incubated with 0.3ml Ni-NTA resin (Qiagen) for 1hr at 4°C with end-to-end rotation. After binding, the resin was transferred to 10ml Poly Prep Column (Bio-Rad) and allowed to settle. The resin was then washed with 3x10ml washing buffer (50mM HEPEs pH=7.5, 500mM NaCl, 20mM imidazole, and 1.4mM β-mercaptoethanol). The bound protein was then eluted with 1ml elute buffer (50mM HEPEs pH=7.5, 50mM NaCl, 300mM imidazole, and 2mM DTT). The eluted protein was dialyzed against buffer (20mM HEPEs pH=7.5, 25mM NaCl, 2mM DTT) overnight and was further purified by anion exchange chromatography using a HiTrap Q HP column (AKTA explorer). Pure fractions were confirmed by SDS-PAGE and were pooled and concentrated. The entire purification process was carried out at 4°C. An aliquot of the protein was boiled in LDS sample buffer and analyzed by SDS-PAGE (4-12% gel). The protein gel was visualized by InstantBlue staining.
For overexpression of Ubiquitin incorporating 2, BL21 (DE3) ΔserC cells were co-transformed with pNHD_Ub12TAG or pNHD_Ub66TAG and pUC_pThrRS_SeptRNAv2.0CUA_OXB20-PduX_EFSep or pUC_pThrRS_SeptRNAv2.0CUA_EFSep. We picked a single colony and inoculated LB for overnight incubation. 20ml overnight culture was diluted into 1L fresh LB and grown until OD600=0.6, and 1mM IPTG was added. Cells were further incubated at 37°C for 6hrs and pelleted before purification as described 13.
For overexpression of Cdk2 and Cdk2D145N, BL21(DE3) cells were transformed with pNHD_Cdk2-His6 or pNHD_Cdk2D145N-His6. We picked a single colony and inoculated LB for overnight incubation. 20ml overnight culture was diluted into 1L fresh LB and grown until OD600=0.6, and 0.2 mM IPTG was added. Cells were further incubated at 37°C for 4hrs and pelleted. For overexpression of Cdk2160TAG and Cdk2D145N/160TAG, pNHD_Cdk2160TAG-His6 or pNHD_Cdk2D145N/160TAG-His6 was co-transformed with pUC_pThrRS_SeptRNAv2.0CUA_OXB20-PduX_EFSep into BL21(DE3) ΔserC ΔycdX cells. We picked a single colony and inoculated LB for overnight incubation. 20ml overnight culture was diluted into 1 L fresh LB and grown until OD600=0.6, and 1 mM IPTG was added. Cells were further incubated at 37°C for 6hrs and pelleted.
To purify recombinant Cdk2-His6 proteins, a cell pellet from 1 L culture was resuspended in 20 ml lysis buffer (50 mM Tris-HCl pH=8.0, 300 mM NaCl, 20 mM imidazole, 50 μg/ml DNase1, 0.1% (v/v) 2-mercaptoethanol, supplemented with cOmplete Protease Inhibitor Cocktail Tablet (Roche)). Cells were lyzed by sonication (40% energy input, 4 min with 5s pause after every 5s sonication). The lysate was clarified by centrifugation at 39,000 × g for 30 min and filtration through a 0.4 μm polyethersulfone (PES) membrane. Ub was purified using nickel affinity chromatography (HisTrap HP column, GE Healthcare) with a linear gradient of imidazole (30–500 mM). Fractions containing Cdk2-His6 protein were pooled and dialyzed against buffer A (20 mM Tris-HCl pH=8.0, 50 mM NaCl, 1 mM DTT) overnight at 4°C. Then, the protein was passed through HiTrap Q column, and the flow through was directly loaded onto HiTrap S column (GE Healthcare). The protein was then eluted with a linear gradient of 50-1000 mM NaCl. Pure fractions were analyzed by SDS-PAGE, pooled, concentrated, snap frozen in liquid nitrogen, and stored at -80°C.
GST(TAG)CaM read-through assay
For expression of GST(TAG)CaM incorporating pSer, DH10B ΔserB cells were transformed with a pKW vector containing SepRS/tRNACUA and EF-Sep and a pRSF vector containing riboQ1 rRNA and GST(TAG)CaM downstream of an orthogonal ribosomal binding site (o-GST(TAG)CaM); or a pRSF vector containing GST(TAG)CaM downstream of an WT ribosomal binding site (GST(TAG)CaM) as described earlier 13. For expression of GST(TAG)CaM incorporating pThr, DH10B ΔserC cells were transformed with a pUC vector containing pThrRS/tRNACUA, PduX, and EF-Sep together with the relevant pRSF vector.
For each cell, we picked a single colony and inoculated LB for overnight incubation. 0.5 ml overnight culture was diluted into 25 ml fresh LB and grown until OD600=0.3, and 1 mM IPTG was added. Cells were further incubated at 37°C for 4hrs and pelleted. Cells were lyzed by resuspending in 1 ml Bugbuster solution supplemented with 1 mM DTT and 50 μg/ml DNase 1 and incubating at RT for 30 min. After pelleting at 17,000 g for 20 min, the supernatant was transferred to incubate with 100 μl pre-equilibrated glutathione resin. After incubation for 30 min at RT, resin was washed five times with 1x PBS supplemented with 1 mM DTT. After the last wash, proteins were eluted by incubating in 200 μl 1x PBS with 15 mM reduced glutathione. All eluted samples were then normalized by OD280 readings, boiled in LDS sample buffer, analyzed by SDS-PAGE, and visualized by InstantBlue staining.
Crystallographic Analysis of Ub (pThr12)
Purified Ub (pThr12) in 20 mM Tris (pH=7.4) was crystallized at 11.5 mg/ml in a sitting-drop setup using the vapor diffusion method. Crystals grew in 20% (w/v) PEG 1K, 20% (v/v) ethanol, and 100 mM phosphate-citrate (pH=4.2). Data were collected at the ID-23.1 beamline at the European Synchrotron Radiation Facility (ESRF). The structure was determined by molecular replacement in Phaser 39, using a search model of Ub (PDB: 1UBQ 40) that lacked the last five flexible C-terminal residues. Model building was carried out in Coot 41. Refinement was performed using Phenix 42,43 and REFMAC5 44. The final statistics are shown in Table S2.
Cdk2 kinase assay
Cdk2 kinase assay reactions were setup according to manufacturer protocol of ADP-Glo kinase assay (Promega). Basically, each 25 μl reaction contains 1x kinase buffer (40 mM Tris-HCl pH=7.4, 20 mM MgCl2, 0.1 mg/ml BSA, and 1 mM DTT), 1 μg Histone H1 (Sigma), 100 μM ultra pure ATP (Promega). Each reaction was then added with 1000, 500, 250, 125, 62.5, 31.25, 15.625, or 0 ng Cdk2WT-His6 or Cdk2160TAG-His6. Biological triplicates were performed at each concentration. Reactions were then incubated at 30°C for 40 min, cooled down at RT for 10 min, and mixed with 25 μl ADP-Glo reagent. After 40 min incubation at RT, 50 μl Kinase Detection Reagent was further added to each reaction and incubated at RT for 40 min. The luminescence for each reaction was then measured by Pherastar Plate Reader.
Electrospray ionization mass spectrometry (For proteins)
Mass spectra for protein samples were acquired on an Agilent 1200 LC-MS system that employs a 6130 Quadrupole spectrometer. The solvent system used for liquid chromatography (LC) was 0.2 % formic acid in H2O as buffer A, and 0.2 % formic acid in acetonitrile (MeCN) as buffer B. Samples were injected into Phenomenex Jupiter C4 column (150 × 2 mm, 5 μm) and subsequently into the mass spectrometer using a fully automated system. Spectra were acquired in the positive mode and analyzed using the MS Chemstation software (Agilent Technologies). The deconvolution program provided in the software was used to obtain the mass spectra. The minimum ion numbers were set at 4 for Ubiquitin proteins and 8 for other proteins. Theoretical average molecular weight of proteins with unnatural amino acids was calculated by first computing the theoretical molecular weight of wild-type protein using an online tool (http://www.peptidesynthetics.co.uk/tools/), and then manually correcting for the theoretical molecular weight of non-natural amino acids.
Electrospray ionization tandem mass spectrometry
Polyacrylamide gel slices (1-2 mm) containing the purified proteins or proteins in solution were prepared for mass spectrometric analysis by manual in situ enzymatic digestion. Briefly, the excised protein gel pieces were placed in a well of a 96-well microtitre plate and destained with 50% v/v acetonitrile and 50 mM ammonium bicarbonate, reduced with 10 mM DTT, and alkylated with 55 mM iodoacetamide. After alkylation, proteins were digested with 6 ng/μL Trypsin (Promega, UK) overnight at 37 °C. The resulting peptides were extracted in 2% v/v formic acid, 2% v/v acetonitrile. The digest was analysed by nano-scale capillary LC-MS/MS using a Ultimate U3000 HPLC (ThermoScientific Dionex, San Jose, USA) to deliver a flow of approximately 300 nL/min. A C18 Acclaim PepMap100 5 μm, 100 μm × 20 mm nanoViper (ThermoScientific Dionex, San Jose, USA), trapped the peptides prior to separation on a C18 Acclaim PepMap100 3 μm, 75 μm × 150 mm nanoViper (ThermoScientific Dionex, San Jose, USA). Peptides were eluted with a gradient of acetonitrile. The analytical column outlet was directly interfaced via a modified nano-flow electrospray ionisation source, with a hybrid dual pressure linear ion trap mass spectrometer (Orbitrap Velos, ThermoScientific, San Jose, USA). Data dependent analysis was carried out, using a resolution of 30,000 for the full MS spectrum, followed by ten MS/MS spectra in the linear ion trap. MS spectra were collected over a m/z range of 300–2000. MS/MS scans were collected using a threshold energy of 35 for collision induced dissociation. LC-MS/MS data were then searched against an in house protein sequence database, containing Swiss-Prot and the protein constructs specific to the experiment, using the Mascot search engine programme (Matrix Science, UK). Database search parameters were set with a precursor tolerance of 5 ppm and a fragment ion mass tolerance of 0.8 Da. Two missed enzyme cleavages were allowed and variable modifications for oxidized methionine, carbamidomethyl cysteine, pyroglutamic acid and phosphorylated serine, threonine, tyrosine, were included. MS/MS data were validated using the Scaffold programme (Proteome Software Inc., USA). All data were additionally interrogated manually.
Processing of raw deep sequencing data
Processing of raw deep sequencing reads was performed using bespoke scripts written in the programming language Python. Each paired end read was first trimmed to 141 nucleotides. Paired end read 1 was concatenated with the reverse-complement of paired end read 2. The assembly was matched using regular expressions against a pattern specifying the expected sequence string with degeneracy specified for the sequence barcode and for each library site. For matching sequences, the degenerate sites were extracted and collected into a comma-separated file together with the detected barcode ID.
An m x n counts table N of m unique mutants in n conditions was constructed. Each entry kij corresponded to mutant i in condition j. To identify the m mutants and their corresponding read counts, all unique rows in the comma-separated file were collected and counted.
Pearson correlation
The Pearson correlation coefficient denoted by R indicates the linear dependence of two vectors {x1…xn} and {y1…yn}. It was calculated as follows: xi
In the formula, x̄ and ȳ stand for the mean values of vectors x and y, respectively. The coefficient R takes values in the range between -1 (for negatively correlated datasets) and +1 (for positively correlated datasets).
Correlation of ≥ 2 replicates: Rmult
A coefficient denoted Rmult was calculated on the basis of variation explained by the principal components (PCs) of a m x n read counts table N of m mutants measured in n replicates. The coefficient Rmult represents the ratio of the variation explained by PC1 to the sum of variation explained by all principal components PC1 … PCn, scaled to a range of Rmult = {0 … 1}. Principal Component Analysis (PCA) was performed using the Matlab R2014a software (Mathworks) function princomp. The output variable latent contained vector with Eigenvalues {λ1 … λn} of the covariance matrix of N. The Eigenvalues corresponded to the share of variation explained by each PC. The ratio of the variation explained by λ1 against the sum of all Eigenvalues was calculated and gave ν1 :
Because the principal components share the amount of variability in a dataset and are arranged by decreasing variability, the contribution of the first component λ1 minimally corresponds to 1 / n, observed for datasets where replicates n are uncorrelated and each component explains an equal amount of variation. Values for ν1, therefore, lie in the range of {1/n … 1}. To normalise for the varying number of replicates n, such that values in the range of {0 … 1} are attained, ν1 was transformed to yield Rmult:
The coefficient Rmult can be used to assess variation in datasets consisting of any number of replicates. Given a dataset of identical replicates, then Rmult = 1. If however the replicates differ, some of the variation will be attributable to the measurement noise. The sum of {λ2 … λn} will be greater than 0, and Rmult will take values such that 0 < Rmult < 1.
Testing for differential enrichment between selection conditions
The DESeq algorithm 45 was implemented in the Matlab R2014a (Mathworks) software using instructions provided online (http://uk.mathworks.com/help/bioinfo/examples/identifying-differentially-expressed-genes-from-rna-seq-data.html). The resulting p-values were corrected for multiple testing using the procedure by Benjamini and Hochberg, yielding the False Discovery Rate (FDR) 46.
Receiver operating characteristic (ROC)
A contingency table or classification table reported the outcomes of two categorical response variables X and Y. As an example, the responses in X could report the measured, true phenotypes of a aminoacyl-tRNA synthetase mutant (given by activity and substrate-selectivity) and the responses in Y the deep sequencing -based outcomes of statistical testing, thresholded for significance. An example of a contingency table is provided (Table S4). Responses are then classified into true positives, false positives, true negatives, and false negatives depending on the congruency of variables X and Y.
Specificity and sensitivity were calculated as follows.
Sensitivity is the ratio of true positives over all positives (including those that were detected as false negatives) and specificity is the ratio of true negatives over all negatives (including those that were detected as false positives).
To determine the ROC plot (Fig. S13), FDR thresholds as determined by the DESeq method were used to determine response Y and thresholded ratios of fluorescence in presence versus absence of 3 represented response X. Sensitivity and specificity were calculated for a range of FDR thresholds as determined using the DESeq method: FDR = 10-n, with{0 ≤ n ≤ 100}. The ROC was evaluated for three different fluorescence threshold ratios: 2x, 10x, 20x.
A step by step protocol for parallel positive selection can be found in Supplementary Note 1 and in Ref. 47.
Data availability
The datasets generated during the current study are available from the corresponding author on reasonable request. The sequences of all selected synthetases are available in Supplementary Table 1. The sequences of all plasmids are provided in Supplementary Data 1.
Supplementary Material
Acknowledgements
This work was supported by the Medical Research Council, UK (MC_ U105181009 and MC_UP_A024_1008, BBSRC (BB/M000842/1, for automation) and an ERC Advanced Grant (SGCR, 669351), all to JWC. MSZ was supported by an EMBO Fellowship (ALTF 297-2015), S.F.B. was supported by a Boehringer Fonds PhD fellowship, A.D.L. was supported by an NSF fellowship (1523390). We are grateful to D. Cervettini, M. Mahesh (MRC-LMB), Y.-H. Tsai for contributions, and to T. Elliott (MRC-LMB) for compound 4. We are grateful to the MRC-LMB mass spectrometry facility for ESI-MS/MS. We are grateful to M. Yu, P. Emsley, G. Murshudov, O. Perisic, and R. Williams for help in crystallization and structural data analysis.
Footnotes
Accession codes
The structure of Ub (pThr12) has the PDB ID XXXX.
Author contributions
J.W.C. defined the direction of research. D.T.R suggested using PduX, M.S.Z. demonstrated pThr biosynthesis in E. coli, and optimized the tRNA with the help of W.H.S.. A.D.L. developed the assay for determining intracellular amino acid concentration. S.F.B. developed the parallel selection and sequencing approach. M.S.Z. and S.F.B. applied the approach to pThrRS discovery. M.S.Z. performed protein expression and biochemical experiments. N.H.D. did protein expression purification and structural studies with assistance from M.S.Z.. M.S.Z., S.F.B. and J.W.C. wrote the paper with input from all authors.
Competing Financial Interest
The authors declare no competing financial interest.
References
- 1.Oakhill JS, Scott JW, Kemp BE. AMPK functions as an adenylate charge-regulated protein kinase. Trends Endocrinol Metab. 2012;23:125–132. doi: 10.1016/j.tem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Kelly AE, et al. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho DH, et al. Leucine-Rich Repeat Kinase 2 (LRRK2) phosphorylates p53 and induces p21(WAF1/CIP1) expression. Mol Brain. 2015;8:54. doi: 10.1186/s13041-015-0145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, et al. EYA1's conformation-specificity in dephosphorylating phosphothreonine in Myc and its activity on Myc stabilization in breast cancer. Molecular and Cellular Biology. 2016 doi: 10.1128/MCB.00499-16. MCB.00499-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinardy JL, et al. Phosphorylation of Threonine 794 on Tie1 by Rac1/PAK1 Reveals a Novel Angiogenesis Regulatory Pathway. PLoS ONE. 2015;10:e0139614. doi: 10.1371/journal.pone.0139614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahajan A, et al. Structure and function of the phosphothreonine-specific FHA domain. Sci Signal. 2008;1:re12–re12. doi: 10.1126/scisignal.151re12. [DOI] [PubMed] [Google Scholar]
- 7.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y-W, Lai H-Y, Tang H, Chen W, Lin H. Prediction of phosphothreonine sites in human proteins by fusing different features. Sci Rep. 2016;6:34817. doi: 10.1038/srep34817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 10.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu Rev Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 11.Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 12.Masania J, Li J, Smerdon SJ, Macmillan D. Access to phosphoproteins and glycoproteins through semi-synthesis, Native Chemical Ligation and N→S acyl transfer. Org Biomol Chem. 2010;8:5113–5119. doi: 10.1039/c0ob00363h. [DOI] [PubMed] [Google Scholar]
- 13.Rogerson DT, et al. Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog. Nat Chem Biol. 2015;11:496–503. doi: 10.1038/nchembio.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park H-S, et al. Expanding the genetic code of Escherichia coli with phosphoserine. Science. 2011;333:1151–1154. doi: 10.1126/science.1207203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sauerwald A, et al. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 16.Fukunaga R, Yokoyama S. Structural insights into the first step of RNA-dependent cysteine biosynthesis in archaea. Nat Struct Mol Biol. 2007;14:272–279. doi: 10.1038/nsmb1219. [DOI] [PubMed] [Google Scholar]
- 17.Huguenin-Dezot N, et al. Synthesis of Isomeric Phosphoubiquitin Chains Reveals that Phosphorylation Controls Deubiquitinase Activity and Specificity. Cell Rep. 2016;16:1180–1193. doi: 10.1016/j.celrep.2016.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauenstein SI, Hou Y-M, Perona JJ. The homotetrameric phosphoseryl-tRNA synthetase from Methanosarcina mazei exhibits half-of-the-sites activity. J Biol Chem. 2008;283:21997–22006. doi: 10.1074/jbc.M801838200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie J, Schultz PG. An expanding genetic code. Methods. 2005;36:227–238. doi: 10.1016/j.ymeth.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 20.Fan C, Fromm HJ, Bobik TA. Kinetic and functional analysis of L-threonine kinase, the PduX enzyme of Salmonella enterica. J Biol Chem. 2009;284:20240–20248. doi: 10.1074/jbc.M109.027425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan C, Bobik TA. The PduX enzyme of Salmonella enterica is an L-threonine kinase used for coenzyme B12 synthesis. J Biol Chem. 2008;283:11322–11329. doi: 10.1074/jbc.M800287200. [DOI] [PubMed] [Google Scholar]
- 22.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hohn MJ, Park H-S, O'Donoghue P, Schnitzbauer M, Söll D. Emergence of the universal genetic code imprinted in an RNA record. Proc Natl Acad Sci USA. 2006;103:18095–18100. doi: 10.1073/pnas.0608762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooley RB, et al. Structural basis of improved second-generation 3-nitro-tyrosine tRNA synthetases. Biochemistry. 2014;53:1916–1924. doi: 10.1021/bi5001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gautier A, et al. Genetically encoded photocontrol of protein localization in mammalian cells. J Am Chem Soc. 2010;132:4086–4088. doi: 10.1021/ja910688s. [DOI] [PubMed] [Google Scholar]
- 26.Gautier A, Deiters A, Chin JW. Light-activated kinases enable temporal dissection of signaling networks in living cells. J Am Chem Soc. 2011;133:2124–2127. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A. Optical Control of CRISPR/Cas9 Gene Editing. J Am Chem Soc. 2015;137:5642–5645. doi: 10.1021/ja512664v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker OS, et al. Photoactivation of Mutant Isocitrate Dehydrogenase 2 Reveals Rapid Cancer-Associated Metabolic and Epigenetic Changes. J Am Chem Soc. 2016;138:718–721. doi: 10.1021/jacs.5b07627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elliott TS, et al. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat Biotechnol. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikić I, et al. Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew Chem Int Ed Engl. 2014;53:2245–2249. doi: 10.1002/anie.201309847. [DOI] [PubMed] [Google Scholar]
- 31.Tang L, et al. Construction of ‘small-intelligent’ focused mutagenesis libraries using well-designed combinatorial degenerate primers. BioTechniques. 2012;52:149–158. doi: 10.2144/000113820. [DOI] [PubMed] [Google Scholar]
- 32.Herhaus L, Dikic I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015;16:1071–1083. doi: 10.15252/embr.201540891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang K, et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat Chem. 2014;6:393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 35.Wang K, et al. Defining synonymous codon compression schemes by genome recoding. Nature. 2016;539:59–64. doi: 10.1038/nature20124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faircloth BC, Glenn TC. Not all sequence tags are created equal: designing and validating sequence identification tags robust to indels. PLoS ONE. 2012;7:e42543. doi: 10.1371/journal.pone.0042543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sachdeva A, Wang K, Elliott T, Chin JW. Concerted, rapid, quantitative, and site-specific dual labeling of proteins. J Am Chem Soc. 2014;136:7785–7788. doi: 10.1021/ja4129789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinfeld JB, Aerni HR, Rogulina S, Liu Y, Rinehart J. Expanded cellular amino acid pools containing phosphoserine, phosphothreonine, and phosphotyrosine. ACS Chem Biol. 2014;9:1104–1112. doi: 10.1021/cb5000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society Series B. 1995 doi: 10.2307/2346101. [DOI] [Google Scholar]
- 47.Zhang MS, et al. Parallel positive selections for the discovery of selective aminoacyl-tRNA synthetase. Protocol Exchange. 2017 doi: XXXX. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study are available from the corresponding author on reasonable request. The sequences of all selected synthetases are available in Supplementary Table 1. The sequences of all plasmids are provided in Supplementary Data 1.