Abstract
We present a profile–profile multiple alignment strategy that uses database searching to collect homologues for each sequence in a given set, in order to enrich their available evolutionary information for the alignment. For each of the alignment sequences, the putative homologous sequences that score above a pre-defined threshold are incorporated into a position-specific pre-alignment profile. The enriched position-specific profile is used for standard progressive alignment, thereby more accurately describing the characteristic features of the given sequence set. We show that owing to the incorporation of the pre-alignment information into a standard progressive multiple alignment routine, the alignment quality between distant sequences increases significantly and outperforms state-of-the-art methods, such as T-COFFEE and MUSCLE. We also show that although entirely sequence-based, our novel strategy is better at aligning distant sequences when compared with a recent contact-based alignment method. Therefore, our pre-alignment profile strategy should be advantageous for applications that rely on high alignment accuracy such as local structure prediction, comparative modelling and threading.
INTRODUCTION
Protein sequences mutate to varying degrees of divergence through evolution. In order to identify homologous proteins and reveal important similarities, sequence alignment methods are commonly used [for recent review see (1)]. These methods rely mainly on approximated evolutionary models that aim at reflecting as accurately as possible the evolutionary paths that connect two or more protein sequences. Most state-of-the-art alignment methods align sequence pairs by dynamic programming (2) and for three or more sequences they apply the progressive strategy (3), where sequences (or profiles) are hierarchically aligned in pairs according to a pre-generated tree (dendrogram), based on sequence similarity. However, when aligning the sequences or profiles to estimate their sequence similarity, pre-determined substitution scores are commonly employed [e.g. the scores from the BLOSUM (4) and PAM (5) series and more recently the JTT (6), GONNET (7), VT (8) and VTML (9) series] that have been derived using a specific set of ‘true’ alignments. Such a generalization presents a problem because these substitution scores reflect a standardized evolutionary model and introduce inconsistencies when applied to non-standard cases (10). As a result, although the similarity detection between closely related sequences is mostly unaffected by these inconsistencies and produces high-confidence alignments, sequences in the so-called ‘twilight zone’ (<30% sequence identity) are extremely hard to align. This is because the evolutionary scenario relating them becomes virtually undetectable due to the noise introduced by the extent of mutational change that has occurred (11).
Improvements to the alignment of distant sequences have been achieved using several approaches. The evolutionary model describing the relation of a set of sequences can be re-adjusted to fit the sequence set and not an extrapolated generic model. Recently, Yu et al. (10) showed that the use of organism-specific or alignment-set-specific background frequencies for contextual re-adjustment of the standard amino acid exchange weights provides a more sensitive and biologically accurate way to align sequences. Alternatively, structural or homologous sequence information can be incorporated into the alignment process to help identify the distant relations between sequences. The benefits of using related sequence information have been shown in numerous profile–profile alignment methods that apply different profile-scoring schemes (12–28). Many of these scoring schemes have been assessed in recent comparison studies and have shown little significant difference in their respective performances (29,30). However, most of the profile–profile alignment approaches to date have been used mainly for sequence database searching (local pairwise alignment). Multiple alignment methods that use profile information can be separated into two main groups: (i) methods that are given a set of more than two sequences and return these sequences in aligned form; and (ii) methods that take a single sequence as input and collect related sequences by aligning them to that sequence (profile-building). The DbClustal method (31) belongs to the second group because it takes a single sequence as input and uses database-searching to collect homologous sequences for that single sequence. This newly built multiple alignment profile is then used to derive ‘anchor’ points to guide the realignment of the query and homologous sequences using ClustalW (32). Conversely, the profile pre-processing strategy of the PRALINE alignment method (33) belongs to the first group, as it creates pre-alignment profiles for each sequence in a given set by adding information from all other sequences in the set. The method we present in this paper also belongs to the first group of multiple alignment methods. It takes two or more sequences as input, for each of which profiles are generated by database searching and then these profiles are used as starting input for progressive multiple alignment. To our knowledge, this application of profile–profile alignment is yet unexplored. Other methods incorporate structural-based information because structure is more conserved than sequence (34) and, therefore, it remains relatively unchanged through evolution, despite the mutational changes of the residues. Structural input has been used in the form of derived or predicted secondary structure (15,22,33,35,36) and more recently in the form of side-chain contact information derived from tertiary protein structures, by using contact mutation probability matrices (37) in contact-based alignment (38).
In this paper, we present an application of profile–profile alignment for progressive multiple alignment, implemented in PRALINEPSI. Pre-alignment profiles (pre-profiles) are generated using each sequence in a set as a PSI-BLAST (39,40) query. The resulting PSI-BLAST local alignments are filtered for redundancy and converted to PRALINE pre-profiles, which replace the single sequence input that would otherwise be used for the alignment. For further details on the PRALINE alignment algorithm see (33,35,41). This extension of the pre-profile information beyond the sequences in the given set increases the information in the pre-profiles, and the new homologous sequences that are detected act as intermediary steps in the evolutionary paths that connect the sequences in the set. As a result, the increased sensitivity of our method in detecting similarities becomes more evident, the more distant the sequence pairs become (or sequence-profile and profile–profile pairs in multiple sequence alignment).
MATERIALS AND METHODS
PRALINEPSI is written in the ‘ANSI C’ programming language. All programs were run on using locally installed versions of PSI-BLAST (39,40), PRALINE (33,41), ALICAO (38), T-COFFEE v2.03 (42) and MUSCLE v3.51 (17).
The PRALINEPSI algorithm
Here, we concentrate on the PRALINEPSI-related features of the PRALINE multiple sequence alignment tool (Figure 1). Further details on PRALINE and what options it provides can be found in (1,33,35,41).
Figure 1.
The schematic representation of the PRALINEPSI strategy. Each sequence is submitted as a PSI-BLAST query to a database of choice. The resulting local alignments are filtered for redundancy and if no hits are found or all hits are redundant, the search is re-run using a new E-value threshold 10 times less stringent. The final local alignments for each sequence are converted to a pre-profile and given to the PRALINE alignment algorithm.
Generating PSI-BLAST pre-profiles
Each member of a sequence set is successively submitted as a query to a protein sequence database of choice, using PSI-BLAST. The iteration number and E-value cut-off threshold for PSI-BLAST can be manually set to any real number and are part of the quality-control of the hits that will be included in the pre-alignment profiles (pre-profiles). If the E-value threshold is too stringent and returns no hits or only redundant hits, PSI-BLAST is automatically restarted with a higher E-value tolerance in 10-fold increments (e.g. from 10−6 to 10−5, etc.). Each resulting PSI-BLAST local alignment is filtered for redundant hits (100% sequence identity) and converted into a PRALINE pre-profile. The pre-profiles replace the single-sequence input of the basic PRALINE strategy (PRALINEBASIC) (33).
To test the sensitivity of PRALINEPSI to the content of the pre-profiles, we run PSI-BLAST with fixed E-value thresholds 0, 10−6, 10−3, 10−2, 1, 5 and 10. Note that for this test the automatic E-value threshold increments were switched off to allow meaningful comparison between the results of each fixed threshold benchmark.
Alignment hierarchy and tree construction
Similarly to the original PRALINE method, the alignment tree is not constructed prior to the progressive steps. First, all pre-profile pairs are scored using their alignment score and the closest two are aligned first. This new profile is then re-aligned to all the remaining pre-profiles and the next highest scoring pair is aligned, whether it is the new profile and a pre-profile or two separate pre-profiles. This continues until all sequences have been aligned and produces the final alignment tree.
Profile alignment
Since all sequence information is in profile form (pre-profiles or profiles), all dynamic programming alignment steps use the profile–profile scoring scheme. We define the score for a profile position (column) pair x and y as the sum of all residue pair scores adjusted according to the residue frequencies of that position:
| 1 |
where αi is the frequency with which residue i appears at position x and βj is the frequency with which residue j appears at position y; pij is the frequency with which residues i and j appear aligned in the dataset used to derive the exchange weights matrix; pi is the background frequency of residue i; and pj is the background frequency of residue j. Commonly, the log() component is simply the exchange weight provided by the selected log-odds substitution matrix (e.g. BLOSUM62).
Alignment method settings for benchmark
For the work described in this paper, we searched a local version of the non-redundant database (NR) (August 2003: 1,428,439 sequences) using PSI-BLAST with three iterations. The benchmarks were carried out using PRALINEPSI with a starting E-value threshold of 10−6. The PRALINEBASIC, profile pre-processing (PRALINEPREPRO) and PRALINEPSI strategies of the PRALINE multiple alignment method were all run using the BLOSUM62 matrix and associated gap penalties 12 (gap-open) and 1 (gap-extension). For better comparison to PRALINEPSI, the PRALINEPREPRO strategy was run so that all sequence set-related information was included in the pre-alignment profiles (pre-processing threshold 0, not optimal). ALICAO (38), T-COFFEE v2.03 (42) and MUSCLE v3.51 (17) were run using their default settings. The ALICAO method was only used in the HOMSTRAD (43) pairwise alignment benchmark because it is not designed for multiple alignment.
The PRALINEPSI strategy has a high computational time compared with the other tested methods. This is due to the time PSI-BLAST needs to search over the NR database, which on a current PC (IBIVU server Xeon 2.4 GHz) averages to ∼60 s per sequence.
Benchmark datasets
HOMSTRAD
We separated the 1032 structure alignments in the HOMSTRAD dataset (36,43) (November 2003) into 633 pairwise and 399 multiple alignment cases. We removed 9 of the pairwise alignments to make the dataset comparable with the published ALICAO benchmark (38). The final pairwise set contained 624 alignments.
BAliBASE
We used reference sets 1–5 of BAliBASE 2.0 (44) to explore the behaviour of PRALINEPSI in different alignment problem cases. Reference 1 is a set of 82 sequence sets that vary in relatedness and length but only contain relatively equidistant sequences. Reference 2 is a set of 23 alignment cases with one orphan sequence (outlier) among a group of related sequences. Reference 3 is a set of 12 alignment cases of two separate groups. References 4 and 5 hold 12 cases each, with N/C-terminal extensions and long internal insertions, respectively. The remaining reference sets 6, 7 and 8 were not used as they represent local alignment problem cases that the methods we are testing are not designed for.
Alignment quality assessment
The quality of the multiple alignments was assessed using both the sum-of-pairs (Q) and column (CS) scores, while the pairwise alignments were assessed only using the sum-of-pairs (Q) score, taking the corresponding reference structure alignments as a standard of truth. For the Q score, all correctly aligned residue pairs are expressed as a percentage of the total number of residue pairs in the alignment (no gapped positions).
For the CS score, all correct alignment positions (all residues of a whole alignment column) are expressed as a percentage of the alignment length.
The BAliBASE alignment cases were assessed using their core block annotations and the software provided by the BAliBASE authors. Some inconsistencies in the software calculations were corrected manually. For all other alignments, we used the VerAlign comparison software, which is available at (http://www.ibivu.cs.vu.nl/programs/veralignwww).
The sequence identities of the pairwise and multiple alignments were calculated as the fraction of aligned identical residue pairs over the total number of aligned residue pairs in the reference structural alignments. The statistical significance of the Q and CS scores for the individual tested methods compared with PRALINEPSI was measured using the Kolmogorov–Smirnov test that has been used in similar assessments (42).
RESULTS
The benchmark assessment presented here has a 2-fold objective. First, we compare the performance of multiple alignment methods in terms of their pairwise and multiple alignment accuracy. Second, we test how the improvements of the pairwise alignments transfer to that of the progressive multiple alignments.
Benchmark on pairwise alignments
We used the 624 HOMSTRAD pairwise alignments as a simple model to illustrate how the homology-extended information in the pre-alignment profiles (pre-profiles) affects similarity detection between sequences of different evolutionary distances.
For a meaningful assessment of PRALINEPSI performance using the incremental strategy from an E-value of 10−6 to a maximum of 10 (PSI-BLAST default setting), we set the alignment quality baseline to that of the basic dynamic programming strategy (sequence-sequence alignment) of PRALINE (PRALINEBASIC) (33) without profile pre-processing and only single-sequence input. To show the performance difference between the PRALINEPSI strategy and that of only using the sequences in a given set for enriching the information for dynamic programming, we aligned the sequence sets using the PRALINE profile pre-processing strategy (profile–profile alignment) (PRALINEPREPRO) (33).
We also compared the quality of the PRALINEPSI alignments with those produced by the contact-based method ALICAO (38) and the latest versions of the top-performing alignment methods T-COFFEE (42) and MUSCLE (17). It is important to clarify that these latter methods use the given sequence information only and are in a strict sense not fairly comparable with the profile–profile methods described above. However, although this is an unfair comparison, results from other MSA methods are essential for our study of how the pairwise accuracy affects that of the progressive multiple alignment. Since there are no multiple alignment programs that use the profile–profile alignment strategy to compare with in the following sections (except PRALINEPREPRO), we chose to compare PRALINEPSI against the best and increasingly popular multiple alignment methods available, namely T-COFFEE and MUSCLE. The comparison is reasonable and interesting since the PRALINEPSI strategy processes additional sequence information obtained via database searches in the background to help align a set of query sequences in the foreground. This effectively means that PRALINEPSI takes a set of unaligned sequences as input and generates a multiple alignment of that same set as output, as do methods, such as T-COFFEE and MUSCLE. In addition, the profile–profile pairwise alignment methods that are currently available are all local alignment programs and therefore cannot be directly compared with our global approach. However, the log-average profile-scoring scheme (19) can be applied to global alignment strategies and is used in MUSCLE as a log-expectation score (17), where position-specific gap penalties are added to the original log-average scoring function.
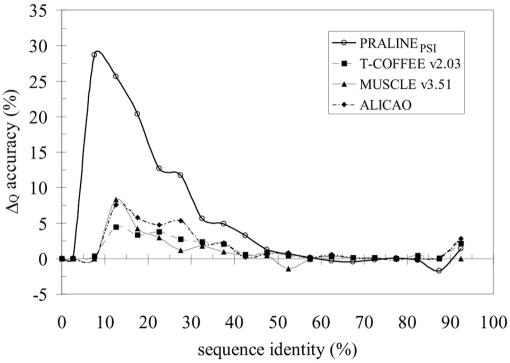
The difference (Δ) in Q scores compared with the PRALINEBASIC strategy is plotted as a function of sequence identity in Figure 2. Owing only to the incorporation of the homology-extended information in the pre-profiles, the difference in alignment quality (ΔQ) is significantly higher compared with the PRALINEBASIC strategy. PRALINEPSI was also significantly better than the other tested methods and improved the most (>65%) and worsened the fewest (<14%) alignment cases, compared with the PRALINEBASIC method (Table 1). By far, the largest improvement was observed in alignment cases with <30% identity (0–30%), although some cases between 30 and 60% were also significantly improved. As could be expected, the alignments above 60% sequence identity (60–100%) were relatively unaffected, albeit the overall quality slightly dropped, but not significantly (∼0.5%).
Figure 2.
Comparison of alignment methods on the 624 HOMSTRAD pairwise alignments (Q score). The difference (Δ) between the average scores of each tested alignment method and that of the PRALINEBASIC method is taken at 5% intervals. The PRALINEPREPRO values for the pairwise alignments are identical to those of PRALINEBASIC and, therefore, they are not included. The PRALINEPSI scores are for the incremental strategy starting with an E-value of 10−6.
Table 1.
The sum-of-pairs (Q) scores of the 624 pairwise alignment HOMSTRAD test cases
| Method | 0–30 (227) (%) | 30–60 (297) (%) | 60–100 (110) (%) | All (624) (%) | Δ Overall (624) (%) | Improved (%) | Worsened (%) | P |
|---|---|---|---|---|---|---|---|---|
| PRALINEBASIC | 57.4 | 89.5 | 98.5 | 79.4 ± 23.2 | – | – | – | <1 × 10−4 |
| PRALINEPSI | 73.2 | 92.7 | 98.0 | 86.6 ± 16.6 | 7.1 | 65.2 | 15.5 | – |
| T-COFFEEv2.03 | 60.8 | 90.8 | 98.7 | 81.3 ± 22.0 | 1.8 | 50.5 | 23.2 | 0.001 |
| MUSCLEv3.51 | 60.6 | 90.1 | 98.6 | 80.9 ± 21.8 | 1.4 | 43.9 | 22.9 | <1 × 10−4 |
| ALICAO | 62.9 | 90.7 | 98.7 | 82.0 ± 21.6 | 2.6 | 51.0 | 18.4 | 0.003 |
Scores are listed separately for sequence identity ranges of 0–30%, 30–60%, 60–100% and the overall scores with their standard deviation (numbers in parentheses are the number of alignments each range contains). The ‘Δ overall’, ‘improved’ and ‘worsened’ columns are with reference to the baseline PRALINEBASIC scores, and the last column ‘P’ shows the statistical significance (P-value from Kolmogorov–Smirnov test) of the overall results of each method compared with those of PRALINEPSI. P-values below 0.05 are underlined. The PRALINEPREPRO scores were not included because due to the lack of extra information (only one extra sequence per profile), they were identical to those of the PRALINEBASIC strategy.
An example of how the extended evolutionary information improves pairwise alignment quality of distant sequences is illustrated in Figure 3. The methyltransferase enzyme alpha chains (HOMSTRAD family ‘SpoU_methylase_N’) from Escherichia coli (top sequence) and Thermus thermophilus share 16.7% sequence identity but have the same α/β knot fold. The very low similarity at the amino acid level causes a register-shift in the alignments of both the single-sequence and contact-based methods. This is entirely prevented by using the homology-extended information in the PRALINEPSI pre-profiles of each sequence and has allowed the correct alignment of the true related regions of these proteins. As a result, the PRALINEBASIC alignment is dramatically improved to over 90% accuracy. The small regions that have been misaligned do not affect the correct alignment of the structural elements of the fold, illustrated by the secondary structure elements of the sequences derived with DSSP (45), in the HOMSTRAD alignment.
Figure 3.
Sequence alignments of the protein methyltransferase (HOMSTRAD family ‘SopU_methylase_N’). The numbers in parentheses represent the Q scores of each alignment. The bottom alignment (HOMSTRAD) is the reference alignment derived from structure super-positioning and shows the secondary structures (DSSP-derived). Both the contact-based and the single sequence-based methods show a shift in the matched secondary structure elements, which is entirely prevented by the use of the extended evolutionary information. Correctly aligned residue pairs are denoted by a ‘∧’ sign.
Sensitivity to pre-profile information
We investigated how the stringency of homology-extension balances with the extent of improvement it can provide. PSI-BLAST was invoked using E-value thresholds of 0, 10−6, 10−3, 10−2, 10−1, 1, 5 and 10 (PSI-BLAST default setting) to determine at which point the allowance of false-positive hits in exchange for including more information became detrimental to PRALINEPSI.
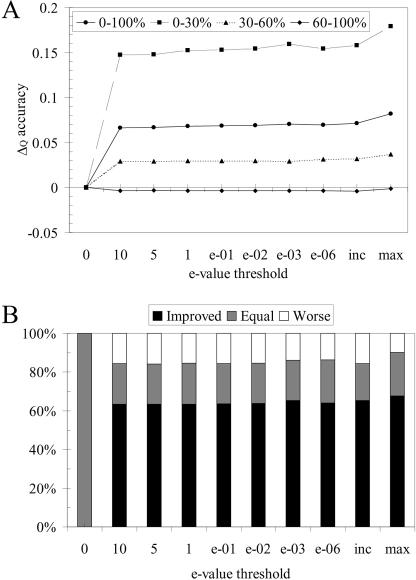
The use of the homology-extended pre-profiles has the same beneficial effect on similarity detection nearly irrespective of the E-value threshold used (Figure 4A). However, although the overall improvement is almost the same for all thresholds tested, the individual correlations of the Q scores of each threshold over the 624 cases show that there is some variation in the results (Table 2). In particular, E-value thresholds 10 and 5 seem to lead to lower alignment quality (Q scores) (Figure 4A). It is clear that the ease of admission of sequences (E-values from 0 to 10) can have an effect on individual cases, although with a minimum correlation coefficient of 0.83, the effect is not dramatic. It is possible that due to the strictness of the threshold, the method would fall back to PRALINEBASIC more often and, as a result, correlate more with the 0 threshold results. However, the overall distribution of improved, unchanged and worsened alignment cases (Figure 4B), in combination with the relatively similar correlation of all thresholds to the PRALINEBASIC scores, is very similar over the E-value thresholds taken. This suggests that a high stringency threshold is adequate to produce good quality alignments and in the cases where no hits or only redundant hits are returned, less stringent thresholds are stable enough to increment too.
Figure 4.
The effects of using E-value thresholds of increasing stringency in PRALINEPSI on the 624 HOMSTRAD pairwise alignments. (A) The difference (Δ) between the average Q scores of PRALINEPSI and the basic PRALINE method, for all cases (0–100% sequence identity) and separately, cases between 0 and 30%, 30 and 60% and 60 and 100% sequence identity. (B) The distributions of improved, equal and worsened cases compared with the basic PRALINE method for each E-value threshold. The ‘inc’ column is the PRALINEPSI incremental strategy starting from a threshold of 10−6, and the ‘max’ column is PRALINEPSI's theoretical upper limit for the tested threshold range.
Table 2.
The correlations between the Q scores of the 624 pairwise alignments of HOMSTRAD aligned by PRALINEPSI using different E-value thresholds
The ‘0’ threshold is equivalent to the PRALINEBASIC strategy.
Next, we re-activated the incrementing of the E-value threshold when no hits or only redundant hits were returned and assessed the quality of the alignments produced by PRALINEPSI with a starting E-value threshold of 10−6 to a maximum of 10 (Figure 4, inc). It is important to note that the ‘inc’ column has no occurrences of non-hit or only redundant PSI-BLAST alignments. Therefore, the percentage of unaffected cases it contains serves as a baseline, further supporting that the distributions of the other thresholds are not greatly biased by the algorithm dropping back to PRALINEBASIC. The incremental strategy covers all alignment cases and shows that the use of the homology-extended information in the pre-profiles greatly improves alignment quality, compared with the basic PRALINE method.
It is understandable that since we have applied a common E-value threshold to all cases, the stringency will cause some sequences to lose useful input and others to incorporate false information. Ideally, one would run each alignment case with its optimum threshold. We investigated the theoretical upper performance limit of PRALINEPSI, by executing each alignment case at its optimum threshold, except 0, and its potential benefits are shown in the ‘max’ dataset results of Figure 4. Although this a priori selection is fictitious, the incremental strategy does not score very far below this upper limit.
Benchmark on multiple alignments
The progressive strategy for multiple alignment is in fact a hierarchical series of pairwise alignments. Therefore, since the incorporation of external information in the form of pre-profiles allows better detection of relations between pairs of distant sequences, it should also produce more accurate multiple alignments. We investigated the effects of using homology-extended information on the 399 HOMSTRAD multiple alignments. PRALINEPSI was run as described for the pairwise alignments above. Alignment quality was assessed using both the Q and CS scores, the latter being the stricter of the two, using the HOMSTRAD structure alignments as a reference. All parameters were kept the same with the only difference being the information content of the pre-alignment profiles.
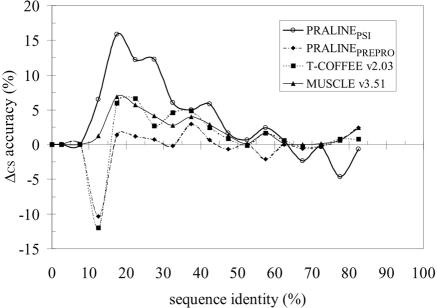
Consistent with the pairwise results, when comparing the quality of the alignments produced by PRALINEPSI with that of the other multiple alignment methods, we observed a similar level of improvement (Figure 5). PRALINEPSI has the highest ratio of improved cases over worsened compared with the PRALINEBASIC strategy. Also, the overall alignment quality is either better than or comparable with the best of the other tested methods throughout all levels of sequence identity (Table 3). This is very interesting because although the T-COFFEE and MUSCLE alignment strategies are different to PRALINE and produce better alignments compared with the PRALINEBASIC strategy, they base their alignment only on the given sequence-set-specific information. Conversely, PRALINEPSI is exactly the same algorithm as PRALINEBASIC, the only difference being the use of the homology-extended information.
Figure 5.
Comparison of alignment methods on the 399 HOMSTRAD multiple alignments (CS score). The difference (Δ) between the average scores of each tested alignment method and that of the PRALINEBASIC method is taken at 5% intervals. The PRALINEPSI scores are for the incremental strategy starting with an E-value of 10−6.
Table 3.
The column (CS) and sum-of-pairs (Q) scores of the 399 mutiple alignment HOMSTRAD test cases
| Method | 0–30 (121) (%) | 30–60 (241) (%) | 60–100 (37) (%) | All (399) (%) | Δ Overall (399) (%) | Improved (%) | Worsened (%) | P |
|---|---|---|---|---|---|---|---|---|
| Column scores (CS) | ||||||||
| PRALINEBASIC | 49.8 | 77.2 | 97.4 | 70.7 ± 22.1 | – | – | – | <1 × 10−4 |
| PRALINEPREPRO | 50.2 | 77.6 | 97.5 | 71.1 ± 22.3 | 0.4 | 46.1 | 31.8 | <1 × 10−4 |
| PRALINEPSI | 62.5 | 81.3 | 96.4 | 77.0 ± 19.6 | 6.3 | 70.2 | 17.0 | – |
| T-COFFEEv2.03 | 53.7 | 79.9 | 97.6 | 73.6 ± 20.9 | 2.9 | 62.2 | 25.6 | 0.041 |
| MUSCLEv3.51 | 54.9 | 79.5 | 97.8 | 73.7 ± 20.8 | 3.0 | 62.4 | 23.1 | 0.027 |
| Sum-of-pairs scores (Q) | ||||||||
| PRALINEBASIC | 60.4 | 85.4 | 98.4 | 79.0 ± 19.2 | – | – | – | <1 × 10−4 |
| PRALINEPREPRO | 61.3 | 85.5 | 98.5 | 79.4 ± 19.6 | 0.3 | 49.1 | 31.6 | 0.003 |
| PRALINEPSI | 72.6 | 88.5 | 97.9 | 84.6 ± 15.7 | 5.5 | 72.4 | 16.3 | – |
| T-COFFEEv2.03 | 64.8 | 87.4 | 98.6 | 81.5 ± 17.8 | 2.5 | 63.7 | 27.3 | 0.050 |
| MUSCLEv3.51 | 65.8 | 87.0 | 98.7 | 81.7 ± 17.4 | 2.6 | 65.2 | 21.8 | 0.034 |
The scores are listed separately for sequence identity ranges of 0–30%, 30–60%, 60–100% and the overall scores with their standard deviation (numbers in parentheses are the number of alignments each range contains). The ‘Δ overall’, ‘improved’ and ‘worsened’ columns are with reference to the baseline PRALINEBASIC scores and the last column ‘P’ shows the statistical significance (P-value from Kolmogorov–Smirnov test) of the overall results of each method compared with those of PRALINEPSI. P-values below 0.05 are underlined.
Similarly to the pairwise benchmark, PRALINEPSI produces better multiple alignments than all other tested methods, especially in the very distant cases. This shows that our initial assumption that the high level of pairwise alignment quality would have a positive effect on multiple alignment was valid. Clearly, the level of improvement in alignment quality is not the same as in the pairwise cases because multiple sequences share more complex inter-relations and the homology-extended information is not always ideal for the sequence or profile pairs. Since the optimization strategies of T-COFFEE and MUSCLE can make very good use of sequence-set-specific information, these methods would largely benefit as well if they would extend likewise the information they use.
Behaviour to specific alignment problems
The HOMSTRAD alignment sets enable us to test the effects of the homology-extended information on alignments of varying difficulty, but the averaged sequence identity values for the multiple alignments did not discern between specific alignment problems biologists and bioinformaticians are faced with, i.e. two sequence sets with low average sequence identity could be a closely related group plus one orphan or two distant groups of closely related sequences. Therefore, we used the BAliBASE multiple alignment benchmark set to test how PRALINEPSI performs on specific alignment cases of known composition. Similarly to the HOMSTRAD benchmark, the BAliBASE sets were aligned with and without homology-extended information and the PRALINEPSI alignments were also compared with results from T-COFFEE and MUSCLE that are to date the highest scoring methods on the BAliBASE reference alignment sets.
It is important to note that BAliBASE is critically small and as the P-values from the Kolmogorov–Smirnov test show, the statistical significance of most of the results on BAliBASE presented here are too low to allow confident conclusions to be drawn (Table 4).
Table 4.
The column (CS) and sum-of-pair (Q) scores of the BAliBASE test cases in references 1–5
| Method | REF 1 (82) (%) | P | REF 2 (23) (%) | P | REF 3 (12) (%) | P | REF 4 (12) (%) | P | REF 5 (12) (%) | P | Weighted average (%) | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Column scores (CS) | ||||||||||||
| PRALINEBASIC | 76.9 | 0.425 | 51.0 | 0.593 | 54.0 | 0.786 | 38.5 | 0.991 | 59.8 | 0.786 | 66.0 | 0.187 |
| PRALINEPREPRO | 78.4 | 0.425 | 56.2 | 0.842 | 50.8 | 0.786 | 30.7 | 0.991 | 77.1 | 0.786 | 68.3 | 0.949 |
| PRALINEPSI | 83.9 | – | 61.0 | – | 55.8 | – | 53.9 | – | 68.6 | – | 73.9 | – |
| T-COFFEEv2.03 | 78.9 | 0.548 | 58.5 | 0.593 | 54.8 | 0.786 | 70.8 | 0.186 | 86.1 | 0.186 | 73.4 | 0.768 |
| MUSCLEv3.51 | 79.9 | 0.914 | 60.2 | 0.842 | 58.3 | 0.786 | 63.3 | 0.186 | 91.4 | 0.066 | 74.4 | 0.858 |
| Sum-of-pairs scores (Q) | ||||||||||||
| PRALINEBASIC | 85.0 | 0.319 | 91.0 | 0.017 | 77.1 | 0.991 | 73.2 | 0.991 | 82.5 | 0.786 | 84.1 | 0.030 |
| PRALINEPREPRO | 86.0 | 0.425 | 93.1 | 0.593 | 77.9 | 0.991 | 74.1 | 0.991 | 88.9 | 0.991 | 85.7 | 0.858 |
| PRALINEPSI | 90.4 | – | 94.0 | – | 76.4 | – | 79.9 | – | 81.8 | – | 88.2 | – |
| T-COFFEEv2.03 | 86.2 | 0.425 | 93.9 | 0.842 | 76.7 | 0.786 | 88.3 | 0.433 | 94.6 | 0.186 | 87.5 | 0.858 |
| MUSCLEv3.51 | 87.0 | 0.914 | 93.7 | 0.842 | 79.6 | 0.433 | 88.9 | 0.186 | 97.8 | 0.019 | 88.5 | 0.928 |
The scores are listed separately for each reference set and the overall average, weighted relative to the number of alignments in each reference set (numbers in parentheses are the number of alignments each set contains). The ‘P’ columns show the statistical significance (P-value from Kolmogorov–Smirnov test) of the results of each method compared with PRALINEPSI. P-values below 0.05 are underlined.
Overall, the alignment cases of reference 1, 2 and 3 comprise over 80% of the alignment cases in BAliBASE and contain most of the distantly related sequences (based on average sequence identity). Our results show that the use of the homology-extended information in these distant sequence cases (>100 alignments) consistently improves the alignment quality compared with the basic PRALINE method, albeit the improvement is not as high as that of T-COFFEE and MUSCLE in the 24 alignment cases in references 4 and 5 (Table 4). Considering the alignment cases of the two latter sets (long insertions and terminal extensions), the differences in the improvement levels are mainly results of the distinct gap weighting of the individual alignment methods. Nonetheless, such alignment cases can be easily detected by the difference in sequence lengths and, therefore, a user would be encouraged to use the MUSCLE or T-COFFEE methods when aligning such sequence sets.
DISCUSSION
The use of profiles to store evolutionary information improves alignment quality and has been known for some time now. One of the most famous examples has been the transition of BLAST to the more accurate PSI-BLAST database-searching tool and more recently to numerous database-search tools that use profile–profile alignment strategies. However, although this highly successful technique allowed the correct detection of very distant homologues, it is not included in top-performing multiple alignment methods. In this paper, we have shown that the dramatic benefits of using homology-extended information for pairwise alignment are stably sustained through the progressive steps of multiple alignment. This suggests that there is information to be extracted from residue sequences before extending to structure, for which the available data remain limiting.
The PRALINEPSI strategy can positively affect the field of database searching, which is one of the most important computational areas in biological research. With PRALINEPSI, we are able to detect similarities between distant sequences with a higher accuracy, but we also use database searching as our means of collecting the extended information. In iterative alignment-based search tools, such as QUEST (46,47), this introduces an optimization scenario that allows the use of the search hits for better alignment before they are used for the next step.
The PRALINEPSI strategy does not intervene with further alignment optimizations such as the re-adjustment of amino acid substitution matrices (10), profile–profile scoring techniques (12,16,18–20,24,25,28,48) and the incorporation of contact or structural information (15,22). Since the extended information is in the form of a profile, contact and structural information can be readily incorporated to further enrich the position-specific information for the alignment. Furthermore, the alignment routine still uses substitution matrices and, therefore, the re-adjustment strategies are applicable. Finally, all pairwise alignments in both pairwise and multiple alignment cases are in the profile–profile form, allowing for any profile-scoring technique to be applied. Therefore, homology-extended sequence alignment should be used together with the aforementioned alignment optimizations in current and future multiple alignment methods.
As would be expected, PRALINEPSI's use of the PSI-BLAST search engine over a database as large as the NR makes its computational time much higher than that of fast methods, such as MUSCLE. However, since the development of software such as IMPALA (49), a sequence can be used to search a position-specific profile database rather than the much larger sequence databases, making the inclusion of appropriate profiles much faster and less CPU intensive. Also, the large size of the pre-profiles that sometimes contain over 1000 sequences creates a bottleneck at the progressive all-against-all alignment steps. Nonetheless, since the PRALINE code has been parallelized (50), the PRALINEPSI strategy computational time can be improved.
More importantly, for fields that rely on very high alignment accuracy, such as comparative modelling, secondary structure prediction, threading and detection of evolutionary relationships, the improvement in alignment accuracy is far more important than the speed at which the alignments are generated. A significantly better alignment of two or more distant sequences can provide answers to questions that do not rely on speedy solutions. Considering the apparent success of using profile–profile alignment beyond the pairwise stage, we expect that more multiple sequence alignment algorithms will employ homology-extended profile information instead of single-sequence input as starting points for the progressive strategy.
AVAILABILITY
The PRALINEPSI strategy is part of the freely available PRALINE WWW Server at http://ibivu.cs.vu.nl/programs/pralinewww/. The PRALINE source code can be made available upon request.
Acknowledgments
We thank the Vrije Universiteit for funding this project. Thanks are also due to the authors of the software and databases we have used for making them freely available online and two anonymous referees for their constructive comments. Funding to pay the Open Access publication charges for this article was provided by The Vrije Universiteit Amsterdam.
REFERENCES
- 1.Simossis V.A., Kleinjung J., Heringa J. In: Current Protocols in Bioinformatics. Baxevanis A.D., editor. NY: John Wiley; 2003. pp. 3.7.1–3.7.25. [DOI] [PubMed] [Google Scholar]
- 2.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 3.Feng D.F., Doolittle R.F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J. Mol. Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- 4.Henikoff S., Henikoff J.G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker W.C., Ketcham L.K., Dayhoff M.O. A comprehensive examination of protein sequences for evidence of internal gene duplication. J. Mol. Evol. 1978;10:265–281. doi: 10.1007/BF01734217. [DOI] [PubMed] [Google Scholar]
- 6.Jones D.T., Taylor W.R., Thornton J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 7.Gonnet G.H., Cohen M.A., Benner S.A. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 8.Muller T., Vingron M. Modeling amino acid replacement. J. Comput. Biol. 2000;7:761–776. doi: 10.1089/10665270050514918. [DOI] [PubMed] [Google Scholar]
- 9.Muller T., Spang R., Vingron M. Estimating amino acid substitution models: a comparison of Dayhoff's estimator, the resolvent approach and a maximum likelihood method. Mol. Biol. Evol. 2002;19:8–13. doi: 10.1093/oxfordjournals.molbev.a003985. [DOI] [PubMed] [Google Scholar]
- 10.Yu Y.K., Wootton J.C., Altschul S.F. The compositional adjustment of amino acid substitution matrices. Proc. Natl Acad. Sci. USA. 2003;100:15688–15693. doi: 10.1073/pnas.2533904100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 12.Wang G., Dunbrack R.L., Jr Scoring profile-to-profile sequence alignments. Protein Sci. 2004;13:1612–1626. doi: 10.1110/ps.03601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomii K., Akiyama Y. FORTE: a profile–profile comparison tool for protein fold recognition. Bioinformatics. 2004;20:594–595. doi: 10.1093/bioinformatics/btg474. [DOI] [PubMed] [Google Scholar]
- 14.von Ohsen N., Sommer I., Zimmer R., Lengauer T. Arby: automatic protein structure prediction using profile–profile alignment and confidence measures. Bioinformatics. 2004;20:2228–2235. doi: 10.1093/bioinformatics/bth232. [DOI] [PubMed] [Google Scholar]
- 15.Ginalski K., von Grotthuss M., Grishin N.V., Rychlewski L. Detecting distant homology with Meta-BASIC. Nucleic Acids Res. 2004;32:W576–W581. doi: 10.1093/nar/gkh370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R.C., Sjolander K. COACH: profile–profile alignment of protein families using hidden Markov models. Bioinformatics. 2004;20:1309–1318. doi: 10.1093/bioinformatics/bth091. [DOI] [PubMed] [Google Scholar]
- 17.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capriotti E., Fariselli P., Rossi I., Casadio R. A Shannon entropy-based filter detects high- quality profile–profile alignments in searches for remote homologues. Proteins. 2004;54:351–360. doi: 10.1002/prot.10564. [DOI] [PubMed] [Google Scholar]
- 19.von Ohsen N., Sommer I., Zimmer R. Pac. Symp. Biocomput. 2003. Profile–profile alignment: a powerful tool for protein structure prediction; pp. 252–263. [PubMed] [Google Scholar]
- 20.Sadreyev R.I., Baker D., Grishin N.V. Profile–profile comparisons by COMPASS predict intricate homologies between protein families. Protein Sci. 2003;12:2262–2272. doi: 10.1110/ps.03197403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mittelman D., Sadreyev R., Grishin N. Probabilistic scoring measures for profile–profile comparison yield more accurate short seed alignments. Bioinformatics. 2003;19:1531–1539. doi: 10.1093/bioinformatics/btg185. [DOI] [PubMed] [Google Scholar]
- 22.Ginalski K., Pas J., Wyrwicz L.S., von Grotthuss M., Bujnicki J.M., Rychlewski L. ORFeus: detection of distant homology using sequence profiles and predicted secondary structure. Nucleic Acids Res. 2003;31:3804–3807. doi: 10.1093/nar/gkg504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaroszewski L., Rychlewski L., Godzik A. Improving the quality of twilight-zone alignments. Protein Sci. 2000;9:1487–1496. doi: 10.1110/ps.9.8.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rychlewski L., Jaroszewski L., Li W., Godzik A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soding J. Protein homology detection by HMM–HMM comparison. Bioinformatics. 2004 doi: 10.1093/bioinformatics/bti125. doi:10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 26.Sadreyev R., Grishin N. COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance. J. Mol. Biol. 2003;326:317–336. doi: 10.1016/s0022-2836(02)01371-2. [DOI] [PubMed] [Google Scholar]
- 27.Chung R., Yona G. Protein family comparison using statistical models and predicted structural information. BMC Bioinformatics. 2004;5:183. doi: 10.1186/1471-2105-5-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yona G., Levitt M. Within the twilight zone: a sensitive profile–profile comparison tool based on information theory. J. Mol. Biol. 2002;315:1257–1275. doi: 10.1006/jmbi.2001.5293. [DOI] [PubMed] [Google Scholar]
- 29.Edgar R.C., Sjolander K. A comparison of scoring functions for protein sequence profile alignment. Bioinformatics. 2004;20:1301–1308. doi: 10.1093/bioinformatics/bth090. [DOI] [PubMed] [Google Scholar]
- 30.Ohlson T., Wallner B., Elofsson A. Profile–profile methods provide improved fold-recognition: a study of different profile–profile alignment methods. Proteins. 2004;57:188–197. doi: 10.1002/prot.20184. [DOI] [PubMed] [Google Scholar]
- 31.Thompson J.D., Plewniak F., Thierry J., Poch O. DbClustal: rapid and reliable global multiple alignments of protein sequences detected by database searches. Nucleic Acids Res. 2000;28:2919–2926. doi: 10.1093/nar/28.15.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heringa J. Two strategies for sequence comparison: profile-preprocessed and secondary structure-induced multiple alignment. Comput. Chem. 1999;23:341–364. doi: 10.1016/s0097-8485(99)00012-1. [DOI] [PubMed] [Google Scholar]
- 34.Chothia C., Lesk A.M. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heringa J. Computational methods for protein secondary structure prediction using multiple sequence alignments. Curr. Protein Pept. Sci. 2000;1:273–301. doi: 10.2174/1389203003381324. [DOI] [PubMed] [Google Scholar]
- 36.Stebbings L.A., Mizuguchi K. HOMSTRAD: recent developments of the Homologous Protein Structure Alignment Database. Nucleic Acids Res. 2004;32:D203–D207. doi: 10.1093/nar/gkh027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin K., Kleinjung J., Taylor W.R., Heringa J. Testing homology with Contact Accepted mutatiOn (CAO): a contact-based Markov model of protein evolution. Comput. Biol. Chem. 2003;27:93–102. doi: 10.1016/s1476-9271(03)00022-7. [DOI] [PubMed] [Google Scholar]
- 38.Kleinjung J., Romein J., Lin K., Heringa J. Contact-based sequence alignment. Nucleic Acids Res. 2004;32:2464–2473. doi: 10.1093/nar/gkh566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul S.F., Koonin E.V. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 40.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simossis V.A., Heringa J. The PRALINE online server: optimising progressive multiple alignment on the web. Comput. Biol. Chem. 2003;27:511–519. doi: 10.1016/j.compbiolchem.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 43.Mizuguchi K., Deane C.M., Blundell T.L., Overington J.P. HOMSTRAD: a database of protein structure alignments for homologous families. Protein Sci. 1998;7:2469–2471. doi: 10.1002/pro.5560071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bahr A., Thompson J.D., Thierry J.C., Poch O. BAliBASE (Benchmark Alignment dataBASE): enhancements for repeats, transmembrane sequences and circular permutations. Nucleic Acids Res. 2001;29:323–326. doi: 10.1093/nar/29.1.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 46.Taylor W.R. Dynamic sequence databank searching with templates and multiple alignment. J. Mol. Biol. 1998;280:375–406. doi: 10.1006/jmbi.1998.1853. [DOI] [PubMed] [Google Scholar]
- 47.Taylor W.R., Brown N.P. Iterated sequence databank search methods. Comput. Chem. 1999;23:365–385. doi: 10.1016/s0097-8485(99)00017-0. [DOI] [PubMed] [Google Scholar]
- 48.Pei J., Sadreyev R., Grishin N.V. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 49.Schaffer A.A., Wolf Y.I., Ponting C.P., Koonin E.V., Aravind L., Altschul S.F. IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics. 1999;15:1000–1011. doi: 10.1093/bioinformatics/15.12.1000. [DOI] [PubMed] [Google Scholar]
- 50.Kleinjung J., Douglas N., Heringa J. Parallelized multiple alignment. Bioinformatics. 2002;18:1270–1271. doi: 10.1093/bioinformatics/18.9.1270. [DOI] [PubMed] [Google Scholar]