Abstract
Background
Generalized pustular psoriasis (GPP) is a rare, debilitating, and often life-threatening inflammatory disease characterized by episodic infiltration of neutrophils into the skin, pustule development, and systemic inflammation, which can manifest in the presence or absence of chronic plaque psoriasis (PV). Current treatments are unsatisfactory warranting a better understanding of GPP pathogenesis.
Objective
To understand better the disease mechanism of GPP to allow improved targeted therapies.
Methods
We performed a gene expression study on formalin-fixed paraffin-embedded GPP (n=28) and PV (n=12) lesional biopsies and healthy control (n=20) skin. Differential gene expression was analyzed using gene ontology and enrichment analysis. Gene expression was validated with qRT-PCR and immunohistochemistry, and a potential disease mechanism investigated using primary human cell culture.
Results
Compared with healthy skin, GPP lesions yielded 479 and PV 854 differentially expressed genes respectively, with 184 upregulated in both diseases. We detected significant contributions of IL-17A, TNF, IL-1, IL-36 and interferons in both diseases; although GPP lesions furnished higher IL-1 and IL-36 and lower IL-17A and interferon-γ mRNA expression than PV. We detected prominent IL-36 expression by keratinocytes proximal to neutrophilic pustules and show that both neutrophils and neutrophil proteases activate IL-36. Suggesting another mechanism regulating IL-36 activity, the protease inhibitors serpin A1 and A3, which inhibit elastase and cathepsin G respectively, were upregulated in both diseases and inhibited activation of IL-36.
Conclusions
Our data indicate sustained activation of IL-1 and IL-36 in GPP, inducing neutrophil chemokine expression, infiltration and pustule formation, suggesting that the IL-1/IL-36 inflammatory axis is a potent driver of disease pathology in GPP.
Keywords: Psoriasis, generalized pustular psoriasis, DITRA, Inflammation, Interleukin
CAPSULE SUMMARY
Current treatments for generalized pustular psoriasis are unsatisfactory. We applied recently-developed techniques for transcriptomic analysis of archived FFPE biopsies revealing pro-inflammatory IL-1, IL-17, TNF and IL-36 activity, which provides a rationale for biologic targeting of these cytokines.
INTRODUCTION
Generalized pustular psoriasis (GPP, OMIM 614204) is a rare, debilitating and life-threatening disease, characterized by episodic infiltration of neutrophils into the skin, pustule development, generalized erythema and desquamation. The acute onset of GPP is frequently accompanied by chills, high-grade fever, fatigue and neutrophilia which can be potentially life-threatening and require hospitalization (1). Cases of GPP can occur either as a distinct entity or preceded by, concurrent with, or followed by chronic plaque psoriasis (psoriasis vulgaris, PV, OMIM 177900) which has complicated the study of the disease. GPP is often classified as a variant of PV yet striking clinical, histological and genetic differences suggest that the two diseases have distinct pathogenic mechanisms (1–5).
Many treatments for PV such as acitretin, cyclosporine A, and anti-TNF biologics are in common use for GPP but typically do not completely control the disease (6). This shortfall in the efficacy of these treatments likely stems from an incomplete overlap of the pathobiology of the two diseases. Resistance to existing treatments and disease recurrence are common with GPP thus there is a critical need to understand better the disease mechanism to develop new effective treatments.
To investigate the pathogenic mechanisms driving GPP, we harnessed a recently-developed technique to perform transcriptional profiling on archived formalin-fixed paraffin-embedded (FFPE) biopsies (7) of GPP skin to assess the transcriptome of GPP lesions, detect differentially expressed genes and identify which inflammatory pathways were activated with a view to the development of new therapeutic interventions tailored to GPP.
Herein we report that an IL36-chemokine-neutrophil axis appears to be central to the pathogenesis of GPP, with a greater utilization of innate immune mechanisms, preferential expression of IL-36 cytokines, KC expression of neutrophil chemokines, IL-36 activation by neutrophil proteases and induction of inflammatory keratinocyte responses. Our data provide a rational basis for targeted anti-cytokine biologic therapy for GPP.
METHODS
Subject recruitment
Archived formalin fixed paraffin embedded tissue was identified by search in our pathology database. Cases with diagnosis of pustular psoriasis were identified and diagnosis was verified by chart review. Healthy controls and patients with chronic plaque psoriasis were identified in our clinic and biopsy was obtained for formalin fixation and paraffin embedding prior to processing and analyses. Healthy volunteers were recruited for blood draws for neutrophil isolation after providing written informed consent. All protocols were approved by the institutional review board of the University of Michigan, Ann Arbor, and the study was carried out in accordance with the Declaration of Helsinki principles.
Microarray analysis
RNA extraction was performed with an E.Z.N.A. FFPE RNA isolation kit (Omega Bio-tek, Norcross, GA) using the xylene-based extraction method as specified by manufacturer and five 20µm thick FFPE sections of biopsy. RNA was eluted into water and stored at −80°C until analyzed. Affymetrix Human Gene ST 2.1 microarrays (Affymetrix, Santa Clara, CA) were processed at the University of Michigan Microarray Core Facility according to the manufacturer’s protocol. Raw microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO series accession number GSE79704. The raw microarray data (.CEL files) were processed in the publicly available software R (www.r-project.org) using a modified version of the “affy” package and Human Entrez Gene custom CDF annotation version 19 (8) (http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp) using the Robust Multichip Average (RMA) method (9). Post-hybridization quality control checks were performed using RNA degradation score, relative log expression (RLE), and normalized unscaled standard errors (NUSE). Data were batch and age-corrected using an implementation of ComBat v3 (10) within the GenePattern pipeline (http://www.GenePattern.org). To remove background, we calculated the median values of all probe sets and removed those probes with expression values below the lowest median value + 1 standard deviation using a custom Perl script. This yielded 25,561 probe sets upon which subsequent analyses were based. Statistical analyses were performed using Significance Analysis of Microarrays method implemented in the MultiExperiment Viewer application (11). Hierarchical clustering and principal components analysis (PCA) were performed on the adjusted expression data using R and gene list comparisons done in Genomatix (www.genomatix.de).
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed as described (12) with the following modifications. The top 250 genes expressed by KC, CD4+ T-cells, CD8+ T-cells, monocytes, neutrophils and dendritic cells (DC) as listed previously (13) or cytokine-treated KC (14) were matched against a ranked list of DEGs in averaged GPP and PV samples. The rank order of matching genes was used to construct a cumulative curve. Effect size was estimated by calculating the area under the curve and normalizing to the maximum possible effect size for each 250 member gene set. Statistical significance was determined by using a phenotype-based permutation test, which preserves the complex co-expression relationships of gene expression data (12). Gene ontology (GO) analysis was performed using the GO_ImmuneSystemProcesses ontology set using ClueGo v2.2.4 in the Cytoscape v.3.2.1 environment (15). A kappa score threshold of 0.4 was used to build networks and a two-sided hypergeometric test with Bonferroni step down correction to determine statistical significance.
Quantitative PCR
Real time RT-PCR was performed after reverse transcribing RNA (High Capacity cDNA Transcription kit; Applied Biosystems, Foster City, CA) quantifying transcripts using a 7900HT Fast Real-Time PCR system (Applied Biosystems) using TaqMan primer sets purchased from Applied Biosystems. All values were normalized to the expression of the housekeeping gene RPLP0. Statistical significance between healthy, PV and GPP samples was determined using a Kruskal-Wallace test with Dunn’s multiple comparison test in GraphPad Prism v6.
Immunohistochemistry
Immunohistochemistry was performed on formalin fixed and paraffin embedded human skin biopsies sectioned at 5 µm, and then immunohistochemically stained after antigen retrieval using polyclonal goat anti-IL-36α (1µg/ml, AF1078, R&D Systems), IL-36γ (2.5µg/ml, rat, Y-12, Santa Cruz Biotech), polyclonal goat anti-CXCL8 (5µg/ml, AF2008, R&D Systems), polyclonal rabbit anti-cathepsin G (5µg/ml, NBP233498, Novus Biologicals), neutrophil elastase (1µg/ml, mouse, NP57, Dako), proteinase 3 (1µg/ml, EPR6277, LSBio), and matched isotype control antibodies. Immunoreactivity was visualized with 3, 3’-diaminobenzidine and counterstained with hematoxylin.
IL-36 activation
Neutrophils were prepared from heparinized peripheral blood from healthy volunteers using ficoll and 3% dextran as described (16). NETs were generated on poly-L-lysine (Sigma-Aldrich) coated 12-well cell culture plates (Corning Costar) using 1 million neutrophils stimulated with 20nM PMA for 3 hours at 37°C. NET formation was confirmed by fluorescent microscopy with DAPI staining of DNA. After extensive washing with PBS, 500µl of 0.5µg/ml full-length IL-36α or full-length IL-36γ in PBS were incubated with NETs for 1 hour at 37°C. After which, the processed IL-36 solutions were added to confluent cultures of primary human KC for 24 hours to determine IL-36 activity. Truncated IL-36α (aa 6–158) and IL-36γ (aa 18–169) (R&D Systems) were used at 100ng/ml as positive controls. IL-36 activity was determined from its induction of CXCL1 and CXCL8 mRNA transcripts.
IL-36 activity assays were performed using 100ng/ml full-length IL-36α or full-length IL-36γ (R&D Systems) in unsupplemented medium 154CF (Invitrogen), treated with 10mU neutrophil elastase (Enzo Life Sciences, Farmingdale, NY) or 1mU cathepsin G (Enzo), for 30 minutes at 37°C, then the reactions quenched with either cathepsin G inhibitor I (50µM, EMD), trypsin inhibitor (Sigma-Aldrich, 15µg/ml, inhibiting elastase activity), serpin A1 (1µg/ml, Enzo), or serpin A3 (1µg/ml, Enzo). The mixtures were then transferred onto confluent cultures of primary human KC for 24 hours. IL-36 activity was determined as described above.
RESULTS
Plaque and pustular psoriasis are pathogenically distinct
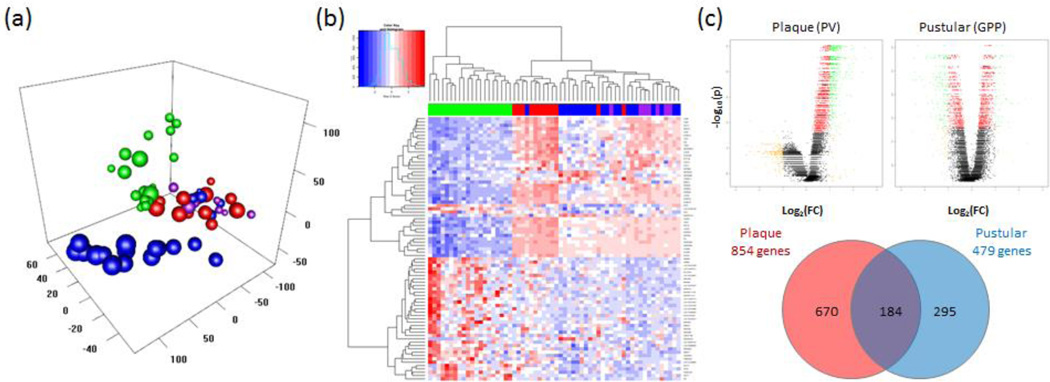
To assess the pathophysiological differences between GPP and chronic plaque psoriasis (psoriasis vulgaris, PV) we analyzed archived FFPE biopsies of confirmed GPP (n=28) and PV (n=12) cases and healthy control (n=20) skin using Affymetrix Human Gene ST 2.1 microarrays. After batch correction, unsupervised principal component analysis (Figure 1a) of the GPP, PV and healthy control samples showed complete separation of the GPP and PV samples from the healthy samples, but there was substantial overlap between the plaque and pustular samples. Unsupervised clustering of transcripts confirmed separation of healthy control and disease samples as well as overlap of GPP and PV transcriptomes (Figure 1b). Interestingly, chart review revealed that 7/28 GPP cases had concurrent PV, and when analyzed by unsupervised clustering analysis, these 7 cases clustered with the GPP-only samples (Figure 1b). Using significance analysis for microarrays, gene expression in GPP and PV lesions were compared with healthy skin, finding 479 differentially expressed genes (DEGs) in GPP and 854 DEGs in PV with at least a 2-fold change in expression (FDR p<0.05) with 184 of these transcripts differentially expressed in both diseases (Figure 1c and Supplemental Table 1).
Figure 1. The transcriptome of generalized pustular psoriasis shares features of plaque psoriasis but is skewed towards innate immune inflammation.
Formalin-fixed paraffin-embedded biopsies of chronic plaque psoriasis (PV, n=12), generalized pustular psoriasis (GPP, n=28) and healthy control skin (NN, n=20) were processed for RNA extraction and analyzed using Affymetrix ST 2.1 arrays. Principal component analysis of the GPP, PV and NN samples showed segregation of the GPP and PV samples from the healthy samples, with some overlap between the plaque and pustular samples. NN, PV, PV+GPP and GPP-only phenotypes are indicated by green, red, purple and blue spheres respectively (a). A heat map generated from unsupervised clustering of differentially expressed transcripts between GPP, PV and NN skin shows heterogeneity of healthy and disease samples. For clarity, the 79 transcripts with 0.25>FC>4.0, and FDR p<0.05 were selected. Blue indicates low expression levels whereas red indicates high expression levels. The green, red, purple and blue bars above the heat map indicate NN, PV, PV+GPP and GPP-only skin samples respectively (b). Volcano plots of gene expression showing roughly equal numbers of up- and down-regulated genes in each disease although many of the down-regulated transcripts in PV did not reach statistical significance. Yellow indicates transcripts with 0.5>FC>2, red denotes FDR p<0.05, and green labels transcripts satisfying both criteria (upper panels, c). Despite overlap of differentially expressed genes between PV and GPP skin lesions, each disease had a set of uniquely-expressed transcripts (lower panel, c).
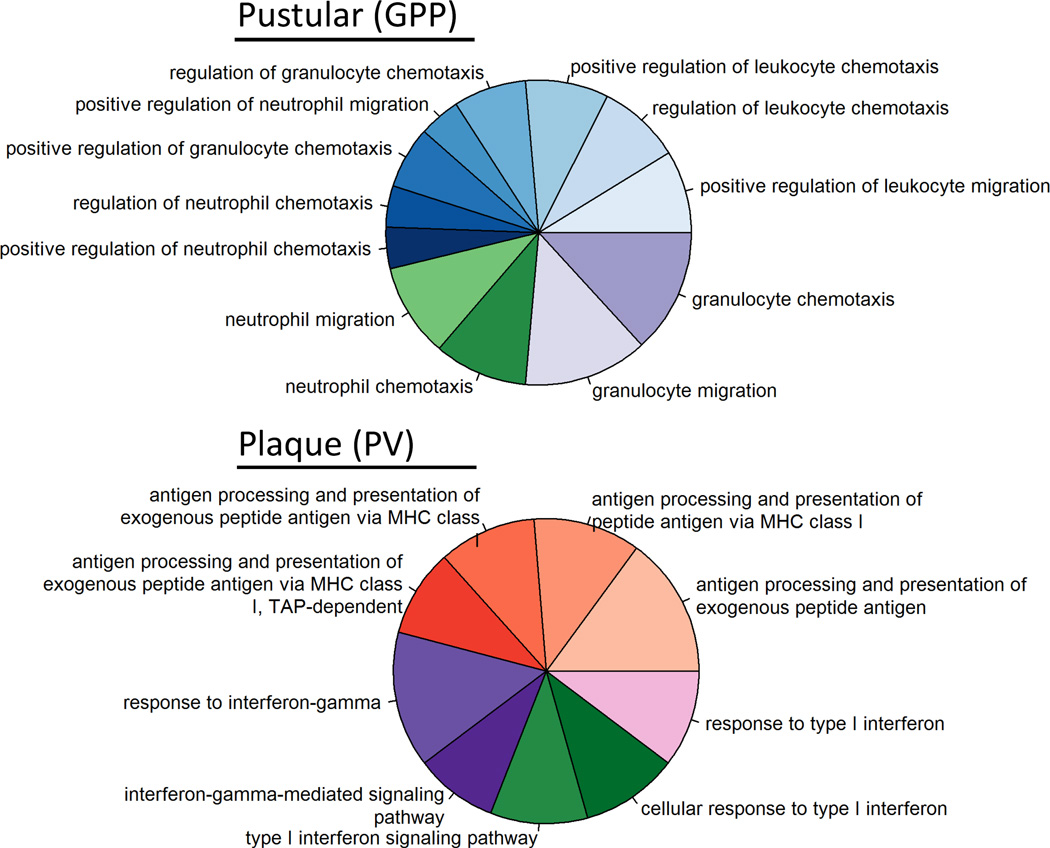
Of the 479 DEGs in GPP vs. healthy skin, 235 were up-regulated and 292 down-regulated. The up-regulated transcripts mapped to 13 gene ontology (GO) Immune System Process terms, with processes related to granulocyte chemotaxis mapping to the vast majority of transcripts (upper panel, Figure 2 and Supplemental Table 2). In contrast, of the 854 DEGs in PV, 169 mapped to 9 GO terms, which included several categories related to the adaptive immune response (lower panel, Figure 2 and Supplemental Table 2).
Figure 2. The transcriptome of pustular psoriasis lesions are enriched for innate immune genes.
Gene ontology analysis of DEGs in GPP and PV using the GO_ImmuneSystemProcess category. GO analysis highlighted an over-representation of innate immune genes in GPP (A) contrasting with more prominent expression of genes involved in acquired immunity in PV (B). Categories shown are those reaching statistical significance using a two-sided hypergeometric test. Details of individual GO terms are listed in Supplemental Table 2.
Pustular psoriasis lesions have heightened IL-1/36 cytokine activity
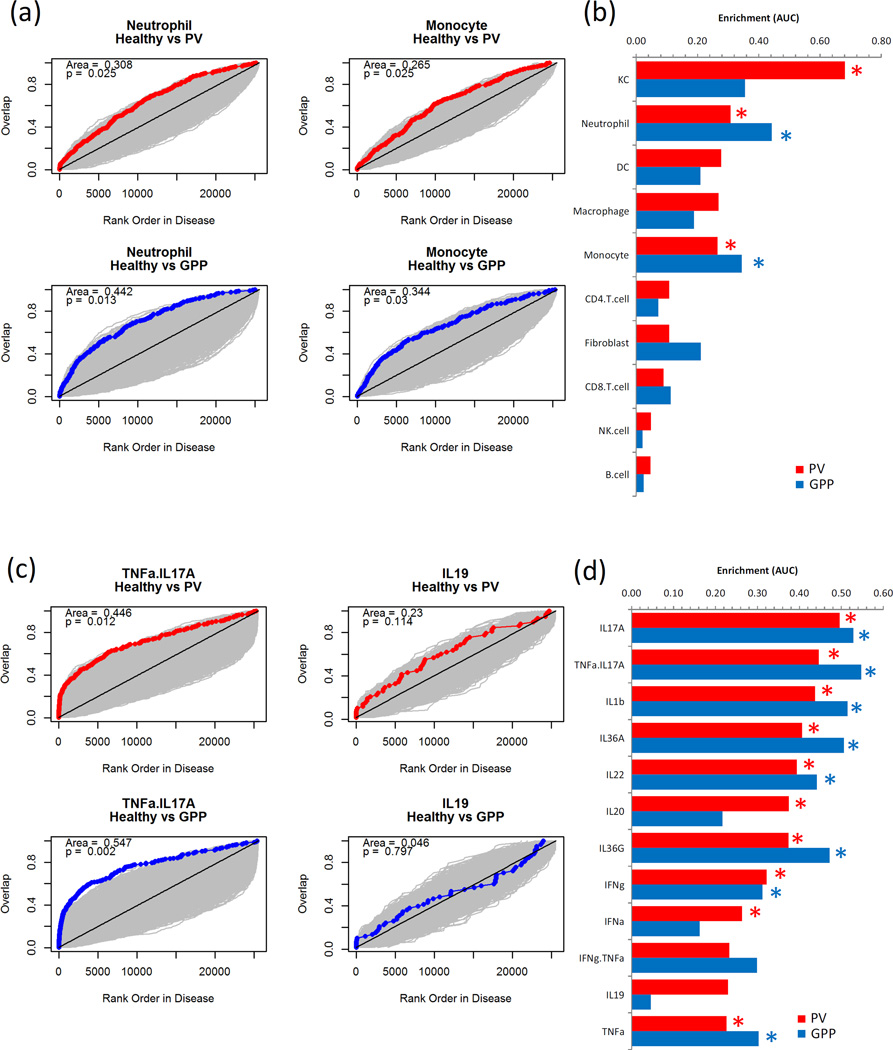
DEGs in PV and GPP were interrogated using gene set enrichment analysis (GSEA) using gene sets comprised of the 250 most abundantly expressed genes in keratinocytes (KC), CD4+ T-cells, CD8+ T-cells, monocytes, neutrophils and dendritic cells (DC) (13). This revealed that a greater abundance of genes expressed by neutrophils and monocytes were detected in GPP compared with PV biopsies (Figure 3a and b). We extended this approach by using gene sets generated from KC cultures treated with a panel of recombinant cytokines to discern cytokine activity in the biopsies. We found evidence of IL-17A, IL-1β, IL-36α, IL-36γ, IL-22, TNF-α, and IFN-γ activity in the tissues, with much of this cytokine activity more prominent in pustular lesions than plaque biopsies (Figure 3c and d).
Figure 3. Neutrophil and monocyte transcripts show greater enrichment in pustular psoriasis (GPP) lesions than plaque psoriasis (PV), whereas robust cytokine gene induction is evident in both diseases.
Microarray datasets for KC, CD4+ T-cells, CD8+ T-cells, monocytes, neutrophils and dendritic cells (DC) were compiled and used to generate signature gene lists (13). DEGs in PV vs NN and GPP vs NN comparisons were ordered by fold-change and the rank position each of the cell signature genes noted and cumulative overlap between the sets plotted (red/blue line), showing a greater up-regulation of neutrophil-specific genes in GPP than PV lesions. Grey areas indicate space generated by 1000 random permutations of NN, PV and GPP datasets from which statistical significance is derived (a). Data from each of the cell signature comparisons is summarized, red bars PV, blue bars GPP, statistical significance indicated by *p<0.05 permutation test (b). Likewise gene signatures for cytokine activity on KC were generated and used to discern cytokine activity within the PV and GPP samples, showing prominent and significant presence of genes induced by TNF-α+IL17A in PV and GPP lesions, contrasted by the lack of enrichment in genes induced by IL-19 (c and d).
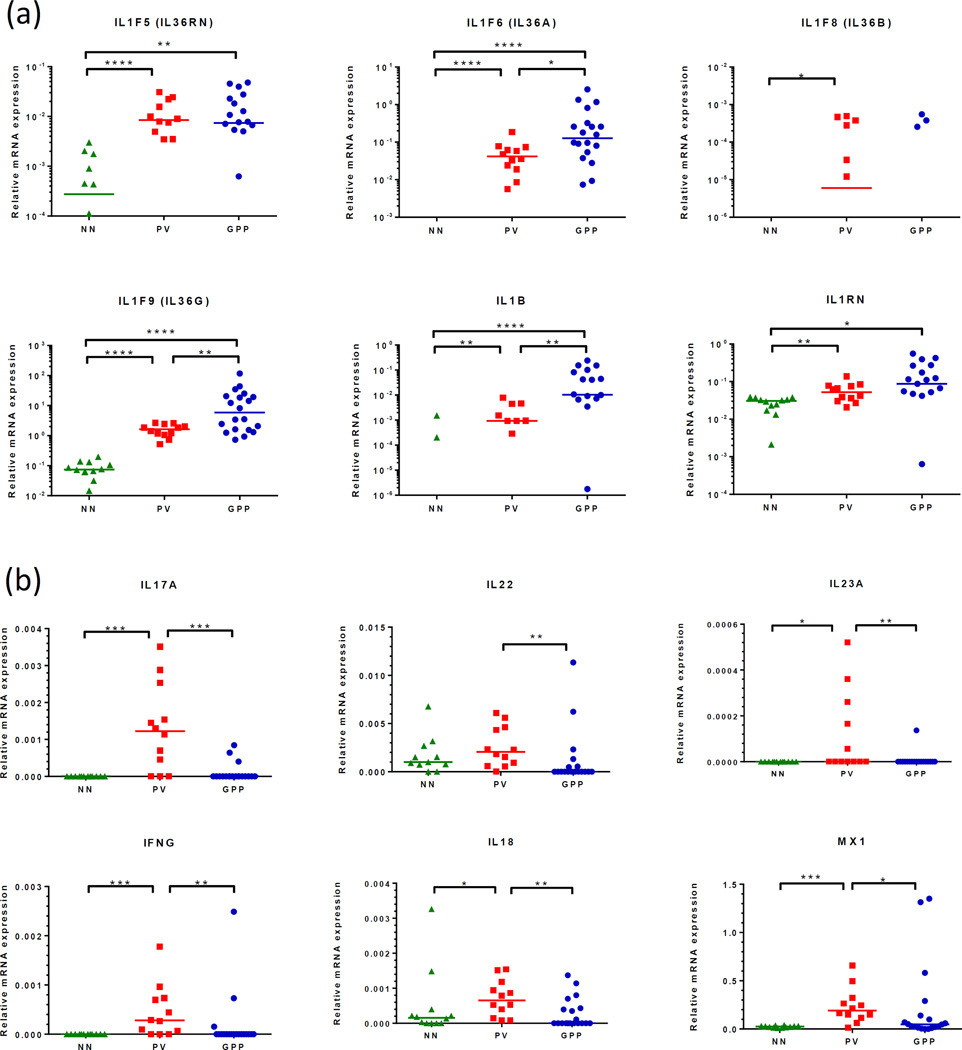
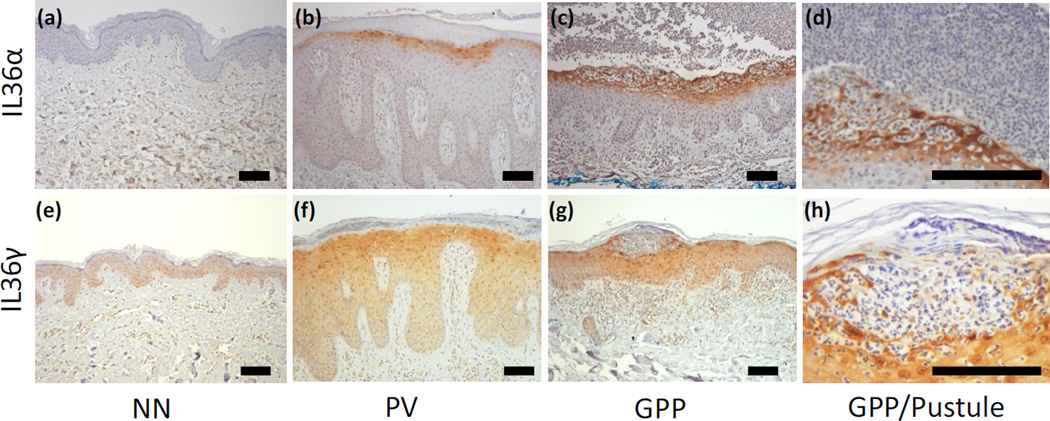
To confirm the increased cytokine activity in GPP skin biopsies, we performed qRT-PCR and found that while we could confirm increased expression of IL-1β, IL-1RA, IL-36α, IL-36β, IL-36γ and IL-36RA in plaque psoriasis skin, we found a significantly greater abundance of transcripts for IL-1β, IL-36α and IL-36γ in GPP versus PV lesions, with no significant changes in the expression levels of the respective receptor antagonists (Figure 4a). Conversely, and suggesting a departure from typical Th1/Th17 pathophysiology, we found that transcripts for IL-17A, IL-22, IL-23p19, IFN-γ, IL-18 and myxovirus resistance 1 (MX-1, an interferon response gene) were all significantly more abundant in plaques compared with pustular lesions (Figure 4b). The elevated IL-36α and IL-36γ expression in GPP was confirmed at the level of protein expression by immunohistochemistry, with an increased intensity of IL-36α and IL-36γ expression, particularly by the KC surrounding the neutrophilic pustule, but not the neutrophils themselves (Figure 5).
Figure 4. GPP lesions have a heightened IL-1/IL-36 cytokine axis but less pronounced Th1/Th17 gene expression than PV.
Quantitative RT-PCR revealed more abundant expression of IL36A, IL36G and IL1B transcripts in GPP compared with PV biopsies, this difference was not seen with respect to the receptor antagonists IL1RN and IL36RN (a). Suggesting a departure from typical Th1/Th17 pathophysiology, IL23A, IL17A, IFNG, CXCL9, CXCL10 and MX1 transcript expression was found to be significantly lower in GPP compared to PV lesions (b). Statistical significance indicated *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Kruskal-Wallace test with Dunn’s multiple comparison test.
Figure 5. Immunohistochemical detection of IL-36α and IL-36γ in skin confirms elevated expression of IL-36 in GPP and localizes IL-36 expression to keratinocytes surrounding the neutrophilic pustule.
IL-36α was not expressed by healthy skin (a), found in the cytoplasm of the uppermost layers of viable KC in PV lesions (b) and expressed at a higher intensity by a wider band of KC in GPP lesions (c). IL-36α expression was strongest in the LC proximal to neutrophilic pustules, but not the neutrophils themselves (d). Faint expression of IL-36γ could be detected in the cytoplasm, but was most apparent in the nuclei of healthy skin KC (e). IL-36γ was intensified in PV lesions (f) yet this was more intense in GPP lesions (g), most pronounced in the KC adjacent to pustules (h). Representative images from 6 GPP, PV and NN donors used (Supplemental Figure 1). DAB with hematoxylin counterstain. Scale bar 100µm.
The neutrophilic environment of GPP lesions activates IL-36α and IL-36γ
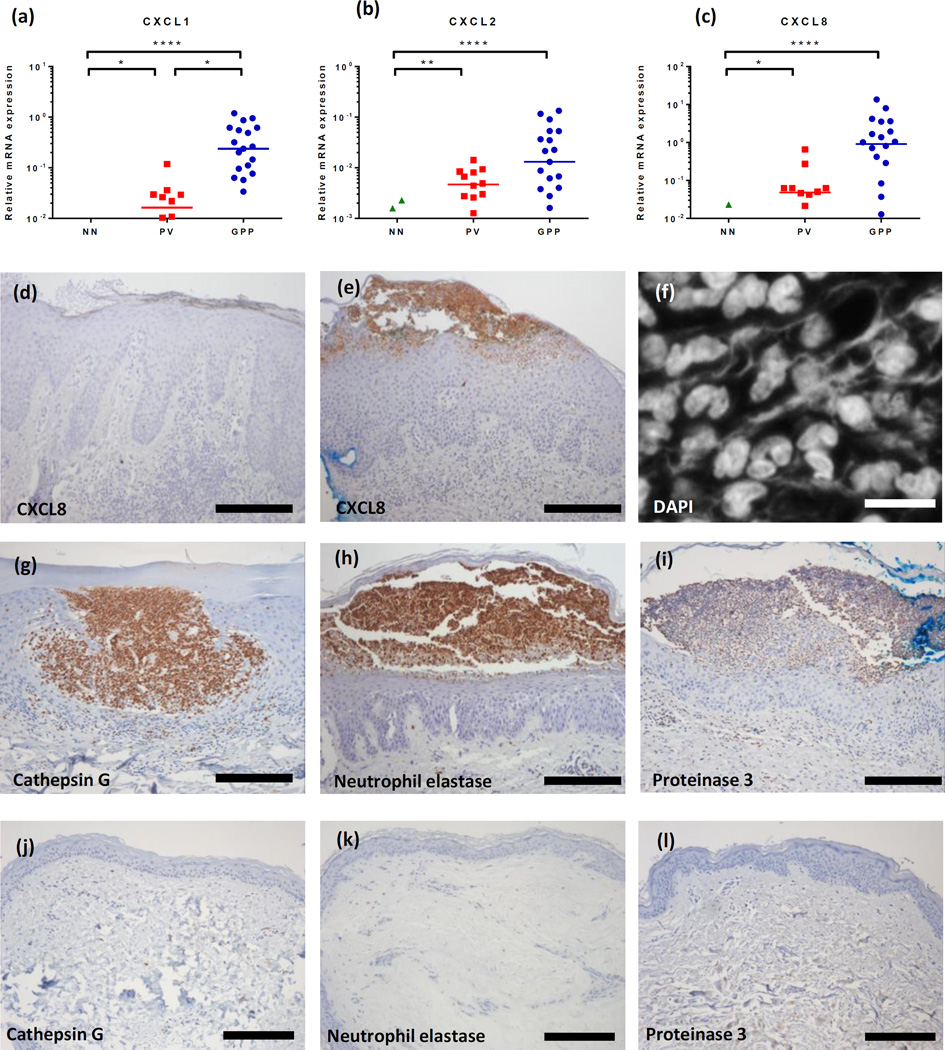
GPP is characterized by periodic neutrophil infiltration into the skin and development of pustules. In concordance with this we observed strongly enhanced expression of the neutrophil chemokines CXCL1, CXCL2 and CXCL8 (IL8) in GPP, likely the factors driving the neutrophilic skin infiltration. Compared with plaque lesions, we found 15-, 5.8-, and 20-fold more transcripts for CXCL1, CXCL2 and CXCL8 in GPP lesions respectively (Figure 6a–c). The enhanced expression of CXCL8 was particularly striking also at the level of protein expression (Figure 6d and e). Given that we witnessed significantly more IL-36 expression in GPP compared with PV lesions and that neutrophils have been suggested to be a source of enzymes that are capable of processing IL-36 isoforms (17, 18), we assessed the expression of neutrophilic proteases in GPP, as well as the presence of networks of extracellular DNA suggestive of neutrophil extracellular traps (NETs), structures resulting from the programmed death of neutrophils (netosis) which expose the neutrophil proteases elastase, cathepsin G and proteinase 3 to the extracellular milieu. NETs were detected in both PV and GPP skin (Figure 6f) and immunohistochemistry confirmed the expression of cathepsin G, elastase and proteinase 3 by neutrophils in GPP skin (Figure 6g–i).
Figure 6. The neutrophil chemokines CXCL1, CXCL2 and CXCL8 are more strongly expressed in GPP than PV lesions.
Using qRT-PCR we detected significantly higher expression of CXCL1, CXCL2, and IL8 transcripts in GPP (n=20) compared with plaque psoriasis (n=12) or healthy control skin (NN, n=12) (a–c). CXCL8 immunoreactivity was barely evident in PV (d) but very strong in and around neutrophilic pustules in GPP biopsies (e). NET formation was prominent in GPP lesions as visualized with DAPI (f) as was the expression of the neutrophil proteases cathepsin G, elastase, and proteinase 3 in GPP (g, h, i) but not healthy control skin (j, k, l). Representative images of at least 6 GPP, PV, NN donors used (Supplemental Figure 1). Scale bar, d- e, and g-l: 200µm, f: 10µm. Statistical significance indicated *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Kruskal-Wallace test with Dunn’s multiple comparison test.
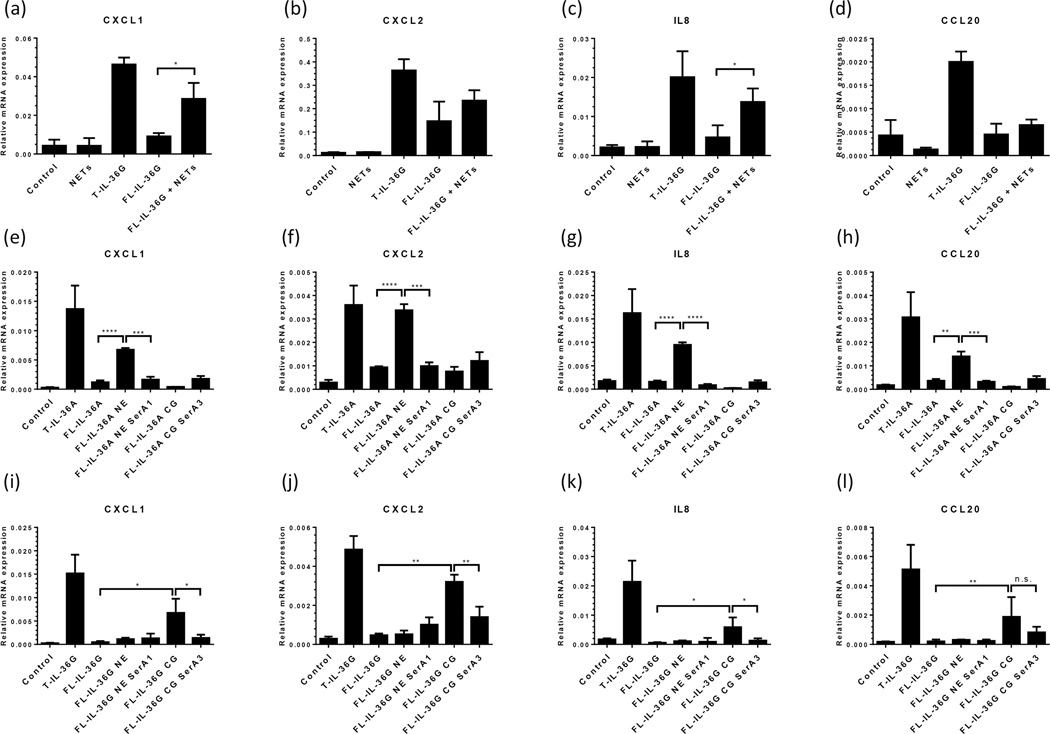
Given the close juxtaposition of heightened levels of IL-36 cytokines and the proteases implicated in their proteolytic activation, we then asked whether neutrophil NETs were capable of activating IL-36α and IL-36γ in vitro. Full-length (FL)-IL-36α and FL-IL-36γ were treated with NETs generated from PMA-activated neutrophils for 1 hour then exposed to cultures of primary human KC to assay their activity with respect to the induction of KC inflammatory responses. Exposure to NETs significantly increased the ability of FL-IL-36γ to induce CXCL1 and CXCL8 expression by KC more than 3-fold compared with untreated FL-IL-36γ (Figure 7a and c), suggesting that enzyme activity within the NETs caused FL-IL-36γ activation. Next we approached the IL-36 activation question using purified neutrophil elastase and cathepsin G to examine their influence on the activity of FL-IL-36α and FL-IL-36γ. Treatment of FL-IL-36α (1µg/ml) with 10mU neutrophil elastase before addition to cultures of primary KC lead to enhanced KC production of CXCL1, CXCL2, IL8 and CCL20 transcripts (Figure 7e–h). However, treatment of FL-IL-36α with cathepsin G did not increase the cytokine’s activity. On the other hand, no increase in FL-IL-36γ activity was seen with elastase treatment, but incubation of FL-IL-36γ with 1mU cathepsin G lead to a significant increase in its ability to induce chemokine transcript production by KC (Figure 7i–l)
Figure 7. Neutrophil extracellular traps (NETs) and neutrophil proteases activate IL-36.
Exposure to NETs for 1 hour (a–d) significantly increased the ability of full-length (FL)-IL-36γ to induce CXCL1 and IL8 expression by KC more than 3-fold compared with untreated FL-IL-36γ (a and c), suggesting that enzyme activity within the NETs caused FL-IL-36γ activation. Truncated (T)-IL-36 as positive control for IL-36 activity. Purified neutrophil elastase (NE, e–h) and cathepsin G (CG, i–l) activated FL-IL-36α and FL-IL-36γ respectively which was inhibited by the addition of specific inhibitors of elastase (serpin A1) or cathepsin G (serpin A3). Statistical significance determined with Student’s t-test and indicated *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, n.s. non-significant.
Physiologic protease inhibitors serpin A1 and A3 regulate IL-36 activation
Serpin A1 and A3 are extracellular protease inhibitors with activity against neutrophil elastase and cathepsin G respectively (19). Here we found that the induction of FL-IL-36α activity was prevented by addition of serpin A1 (Figure 7e–h) and FL-IL-36γ activity prevented by treatment with the specific cathepsin G inhibitor serpin A3 (Figure 7i–l). Suggestive of attempts, albeit insufficient, to regulate IL-36 activity in skin, both of these protease inhibitors were found to be upregulated in our microarray dataset: serpin A1 was 1.86-fold (q=0.0048) and 1.78-fold (q<0.001) upregulated in PV and GPP lesions respectively, compared with healthy skin; whereas serpin A3 expression was increased 1.94-fold (q=0.020) and 2.2-fold (q=0.035) in PV and GPP lesions respectively.
DISCUSSION
The adaptive immune system, particularly CD4+ and CD8+ T cells, has a critical, if not causal, role in the pathogenesis of PV and as such, agents specifically targeting elements of adaptive immunity have had great success in treating plaque psoriasis (20, 21). These therapies are generally less effective for GPP (6), suggesting a divergent pathogenic mechanism in the pustular disease. Using archived FFPE skin biopsy material, microarrays and a bioinformatics approach we have revealed that although the transcriptome of GPP overlaps with PV (Figure 1), the pustular disease has a much more pronounced utilization of the innate immune system than PV (Figure 2 and Supplemental Table 2). Our data suggest that GPP is a disease primarily involving the activities of KC, neutrophils and monocytes (Figure 3a), with inflammatory processes mainly driven by IL-36, IL-1, TNF-α/IL-17A (Figure 3b). Moreover, the increased expression (Figures 4 and 5), processing and activity (Figure 7) of IL-36 cytokines might be a central mechanism that promotes neutrophil accumulation in the epidermis.
The importance of IL-36 in pustular skin disease was recently highlighted when missense mutations in IL36RN, which affect the structure and function of the IL-36 receptor antagonist (IL-36RA), were found to be associated with GPP (3, 22). Loss of IL-36RA function leads to unrestrained IL-36 activity, as evidenced by induction of IL-1β, IL-6, and IL-8 production, and neutrophil infiltration. GPP can present either alone or preceded by, concurrent with, or followed by PV (1). Mutations in IL36RN have been shown to account for 46–82% of cases of GPP without associated PV (5, 23). The proportion of IL36RN mutant carriage is much lower (10–17%) in cases of GPP associated with PV (2), supporting the notion of divergent pathogenic mechanisms in the two diseases. A recent meta-analysis of 233 GPP cases revealed that carriage of 1 or 2 IL36RN mutant alleles conferred a more severe clinical phenotype with an earlier age of onset and increased risk of systemic inflammation than non-carriage (2). A gene-dosage effect was also apparent in that homozygous carriers had an earlier age of onset than heterozygotes. In addition, mutations in AP1S3 (24) and CARD14 (25) affecting the structure and function of the AP-1 complex subunit σ1C, and the NF-κB activator CARD14 respectively, have also been associated with GPP, giving further insight into the mechanisms that may underlie this disease.
The three isoforms of IL-36, IL-36α, β and γ, form a trio of pro-inflammatory IL-1 family cytokines which we (26) and others (27, 28) have shown are overexpressed in plaque psoriasis skin where they may drive keratinocyte inflammatory responses (26), synergize with other pro-inflammatory cytokines (26, 29), and promote dendritic cell activation (30, 31). The effects of IL-36 on skin have been modeled in mice (26, 27). Transgenic expression of IL-36α in murine epidermis lead to epidermal thickening, hyperkeratosis, mixed inflammatory cell infiltrate, and elevated chemokine expression (CCL2, CXCL6, CCL7, and CXCL2) (27). Backcrossing to an IL-36RN-deficient mouse resulted in more severe skin lesions, reflecting the effect of nonsense IL-36RN mutations in GPP. This phenotype was partly reversed by the use of an anti-Gr1 neutrophil-depleting antibody or TNF-neutralizing antibodies, suggesting roles for both neutrophils and TNF in IL-36α-induced skin hyperplasia. IL-36α injected subcutaneously into mice induces acanthosis and a striking infiltration of granulocytes (CD11b+) concomitant with chemokine and growth factor induction (30). IL-36 family expression is also elevated in the skin of psoriasiform KC-Tie2 mice (26) and during imiquimod-induced skin inflammation (26, 32). One of the early features of imiquimod-treated mouse skin is neutrophil infiltration, and the inflammatory phenotype is ablated in IL-36 receptor deficient mice (32) further implicating a neutrophil/IL-36 axis as a central driver in the development of skin inflammation in GPP.
Like their IL-1 family counterparts IL-1β, IL-18 and IL-33 (33–35), the IL-36 cytokines require N-terminal peptide cleavage for activity (36) and it was recently demonstrated that IL-36 activation could be carried out by the neutrophil serine proteases elastase and cathepsin G (17). Here we show that neutrophils activated to form NETs have the ability to activate full-length IL-36γ (Figure 7a–d) and of the three proteases prominently expressed in neutrophilic pustules (Figure 6g–h), elastase could activate IL-36α and cathepsin G was found to activate IL-36γ increasing their ability to induce KC to produce neutrophil chemokines (Figure 7e–l). Interestingly we detected an upregulation of two endogenous protease inhibitors, serpins A1 and A3 in both PV and GPP (Figure 7e–l) and which appear to be labile to induction by inflammatory cytokines (data not shown), suggesting a counter-regulatory mechanism to control IL-36 activity. However, the serpin mechanism of action is one of suicide inhibition and a 1:1 serpin:protease stoichiometry is required (19), thus the approximate 2-fold upregulation of serpins A1 and A3 we detected in lesional skin is unlikely to counter the formidable protease expression afforded by the neutrophilic infiltrate (Figure 6). This imbalance may suggest a yet unexplored therapeutic angle in pustular diseases.
We have shown that GPP utilizes pathways both overlapping and separate from PV, and because of their efficacy in moderate-severe PV, a number of therapies specifically targeting cytokines are in use for GPP. Several reports describe the use of the anakinra, an IL-1 receptor antagonist, to treat GPP (37–39). Anakinra appears to induce a rapid normalization of systemic inflammatory symptoms followed by improvement of the pustular skin eruption. The responses to IL-1 receptor inhibition in the skin tend to be incomplete however, with erythema and hyperkeratosis remaining in some cases, which suggests that IL-1 is not playing a central role in GPP but acts in a positive feedback loop inducing and being induced by IL-36 (26). The relevance of using IL-1 receptor antagonism to tackle a disease which may be primarily driven by deviations in IL-36 signaling, particularly when IL36RN mutations are present, has recently been questioned (40). TNF-α is a central mediator in chronic plaque psoriasis as evidenced by the effectiveness of therapies that block TNF-α activity. Of the three TNF blockers in use for PV, infliximab has most commonly been used for GPP (41, 42) and as such has become one of the recommended treatment options for severe acute GPP despite the lack of adequate clinical trials (6). Infliximab has been reported to have a rapid effect, with systemic inflammation and skin pustules starting to recede in as little as 2 days from the first infusion. In this context, the efficacy of infliximab likely stems from the rapid availability of the drug following infusion, and its inhibition of the synergy between TNF and multiple inflammatory cytokines including IL-36, IL-17A, IL-1β (26, 29, 43, 44). We detected increased IL-17A activity in GPP lesions using GSEA of microarray data (Figure 3) despite not being able to detect a significant increase in IL-17A transcripts by qRT-PCR (Figure 4); this discordance may be the result of the lack of sensitivity of the PCR assay for IL-17A in FFPE samples, contrasting with the ability of GSEA to detect the downstream effects of cytokines on transcriptional networks, or possibly similarities in IL-1/17/36-induced gene sets driving a perceived IL-17A signature in the data. Notwithstanding, biologics targeting IL-17A signaling have been used with success for GPP. An initial case report detailed the rapid improvement of both systemic symptoms and the skin disease of one GPP patient with secukinumab (45), followed by a clinical trial which provided encouraging, but because of its design not conclusive, data from 10 out of 11 GPP patients on secukinumab (46). Likewise brodalumab, a biologic targeting IL-17RA, was shown to induce clinical improvement in 11 out of 12 GPP patients (47).
The above mentioned therapies have largely been tested empirically for GPP, but now our data (Figures 3 and 4) provide evidence for the selection of these treatments as we demonstrate that responses to IL-1, IL-17A and TNF-α are heightened in GPP, with the IL-1 and IL-36 families of cytokines being the most robust. Our data provide a rational basis for the development of therapies specifically targeting these pathways, particularly the IL-36 system in GPP (48, 49).
Supplementary Material
CLINICAL IMPLICATIONS.
This work demonstrates the sustained activity of IL-1, IL-17, TNF and IL-36 cytokines in generalized pustular psoriasis lesions, which provides a rational basis for targeting these cytokines in this disease.
Acknowledgments
We thank the research subjects who participated in this study. We also thank Dr. Stefan W. Stoll for kindly providing primary keratinocyte cultures. We acknowledge Drs. Celine Berthier, Viji Nair (Internal Medicine, University of Michigan, supported by National Institutes of Health (NIH) award DK100845), Philip Stuart and Dr. Lam C. Tsoi (Dermatology) for assistance with biostatistical analyses. AJ is supported by NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) K01 AR064765, the National Psoriasis Foundation USA and the Babcock Memorial Trust. JMK is supported by the NIH NIAMS K08 AR063668 JEG is supported by NIH NIAMS K08 AR060802, R01 AR069071, The A. Alfred Taubman Medical Research Institute as the Kenneth and Frances Eisenberg Emerging Scholar Award, and the Doris Duke Charitable Foundation Grant (#2013106). ZQY is supported by Jiangsu Government Scholarship for Oversea Studies. This work was supported in part by a grant from Novartis to JEG. The funding institutions had no role in the study.
Abbreviations
- GPP
generalized pustular psoriasis
- PV
psoriasis vulgaris
- DEG
differentially expressed gene
- FFPE
formalin-fixed paraffin-embedded
- NET
neutrophil extracellular trap
- GSEA
gene set enrichment analysis
- PCA
principal component analysis
- FL
full-length
- FDR
false discovery rate
REFERENCES
- 1.Gudjonsson JE, Elder JT. Chapter 18. Psoriasis. In: Goldsmith LA, Katz SI, Gilchrest BA, Paller AS, Leffell DJKW, editors. Fitzpatrick’s dermatology in general medicine. 1. 8th. New York: McGraw-Hill Medical; 2012. [Google Scholar]
- 2.Hussain S, Berki DM, Choon SE, Burden AD, Allen MH, Arostegui JI, et al. IL36RN mutations define a severe autoinflammatory phenotype of generalized pustular psoriasis. J Allergy Clin Immunol. 2015;135(4):1067–1070. doi: 10.1016/j.jaci.2014.09.043. e9. [DOI] [PubMed] [Google Scholar]
- 3.Onoufriadis A, Simpson MA, Pink AE, Di Meglio P, Smith CH, Pullabhatla V, et al. Mutations in IL36RN/IL1F5 are associated with the severe episodic inflammatory skin disease known as generalized pustular psoriasis. Am J Hum Genet. 2011;89(3):432–437. doi: 10.1016/j.ajhg.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Setta-Kaffetzi N, Navarini AA, Patel VM, Pullabhatla V, Pink AE, Choon SE, et al. Rare pathogenic variants in IL36RN underlie a spectrum of psoriasis-associated pustular phenotypes. J Invest Dermatol. 2013;133(5):1366–1369. doi: 10.1038/jid.2012.490. [DOI] [PubMed] [Google Scholar]
- 5.Sugiura K, Takemoto A, Yamaguchi M, Takahashi H, Shoda Y, Mitsuma T, et al. The majority of generalized pustular psoriasis without psoriasis vulgaris is caused by deficiency of interleukin-36 receptor antagonist. J Invest Dermatol. 2013;133(11):2514–2521. doi: 10.1038/jid.2013.230. [DOI] [PubMed] [Google Scholar]
- 6.Robinson A, Van Voorhees AS, Hsu S, Korman NJ, Lebwohl MG, Bebo BF, Jr, et al. Treatment of pustular psoriasis: from the Medical Board of the National Psoriasis Foundation. J Am Acad Dermatol. 2012;67(2):279–288. doi: 10.1016/j.jaad.2011.01.032. [DOI] [PubMed] [Google Scholar]
- 7.Hodgin JB, Borczuk AC, Nasr SH, Markowitz GS, Nair V, Martini S, et al. A molecular profile of focal segmental glomerulosclerosis from formalin-fixed, paraffin-embedded tissue. Am J Pathol. 2010;177(4):1674–1686. doi: 10.2353/ajpath.2010.090746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 10.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 11.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swindell WR, Stuart PE, Sarkar MK, Voorhees JJ, Elder JT, Johnston A, et al. Cellular dissection of psoriasis for transcriptome analyses and the post-GWAS era. BMC medical genomics. 2014;7:27. doi: 10.1186/1755-8794-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swindell WR, Johnston A, Voorhees JJ, Elder JT, Gudjonsson JE. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC Genomics. 2013;14:527. doi: 10.1186/1471-2164-14-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clark RA, Nauseef WM. Isolation and functional analysis of neutrophils. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im0723s19. Chapter 7:Unit 7 23. [DOI] [PubMed] [Google Scholar]
- 17.Henry CM, Sullivan GP, Clancy DM, Afonina IS, Kulms D, Martin SJ. Neutrophil-Derived Proteases Escalate Inflammation through Activation of IL-36 Family Cytokines. Cell Rep. 2016;14(4):708–722. doi: 10.1016/j.celrep.2015.12.072. [DOI] [PubMed] [Google Scholar]
- 18.Macleod T, Doble R, McGonagle D, Wasson CW, Alase A, Stacey M, et al. Neutrophil Elastase-mediated proteolysis activates the anti-inflammatory cytokine IL-36 Receptor antagonist. Scientific reports. 2016;6:24880. doi: 10.1038/srep24880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, et al. An overview of the serpin superfamily. Genome Biol. 2006;7(5):216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt J, Rosumeck S, Thomaschewski G, Sporbeck B, Haufe E, Nast A. Efficacy and safety of systemic treatments for moderate-to-severe psoriasis: meta-analysis of randomized controlled trials. Br J Dermatol. 2014;170(2):274–303. doi: 10.1111/bjd.12663. [DOI] [PubMed] [Google Scholar]
- 21.Leonardi CL, Romiti R, Tebbey PW. Ten years on: the impact of biologics on the practice of dermatology. Dermatol Clin. 2015;33(1):111–125. doi: 10.1016/j.det.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Marrakchi S, Guigue P, Renshaw BR, Puel A, Pei XY, Fraitag S, et al. Interleukin-36-receptor antagonist deficiency and generalized pustular psoriasis. N Engl J Med. 2011;365(7):620–628. doi: 10.1056/NEJMoa1013068. [DOI] [PubMed] [Google Scholar]
- 23.Korber A, Mossner R, Renner R, Sticht H, Wilsmann-Theis D, Schulz P, et al. Mutations in IL36RN in patients with generalized pustular psoriasis. J Invest Dermatol. 2013;133(11):2634–2637. doi: 10.1038/jid.2013.214. [DOI] [PubMed] [Google Scholar]
- 24.Setta-Kaffetzi N, Simpson MA, Navarini AA, Patel VM, Lu HC, Allen MH, et al. AP1S3 mutations are associated with pustular psoriasis and impaired Toll-like receptor 3 trafficking. Am J Hum Genet. 2014;94(5):790–797. doi: 10.1016/j.ajhg.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordan CT, Cao L, Roberson ED, Pierson KC, Yang CF, Joyce CE, et al. PSORS2 is due to mutations in CARD14. Am J Hum Genet. 2012;90(5):784–795. doi: 10.1016/j.ajhg.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston A, Xing X, Guzman AM, Riblett M, Loyd CM, Ward NL, et al. IL-1F5, −F6, −F8, and −F9: a novel IL-1 family signaling system that is active in psoriasis and promotes keratinocyte antimicrobial peptide expression. J Immunol. 2011;186(4):2613–2622. doi: 10.4049/jimmunol.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, Weng N, et al. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J Exp Med. 2007;204(11):2603–2614. doi: 10.1084/jem.20070157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debets R, Timans JC, Homey B, Zurawski S, Sana TR, Lo S, et al. Two novel IL-1 family members, IL-1 delta and IL-1 epsilon, function as an antagonist and agonist of NF-kappa B activation through the orphan IL-1 receptor-related protein 2. J Immunol. 2001;167(3):1440–1446. doi: 10.4049/jimmunol.167.3.1440. [DOI] [PubMed] [Google Scholar]
- 29.Carrier Y, Ma HL, Ramon HE, Napierata L, Small C, O’Toole M, et al. Inter-regulation of Th17 cytokines and the IL-36 cytokines in vitro and in vivo: implications in psoriasis pathogenesis. J Invest Dermatol. 2011;131(12):2428–2437. doi: 10.1038/jid.2011.234. [DOI] [PubMed] [Google Scholar]
- 30.Foster AM, Baliwag J, Chen CS, Guzman AM, Stoll SW, Gudjonsson JE, et al. IL-36 promotes myeloid cell infiltration, activation, and inflammatory activity in skin. J Immunol. 2014;192(12):6053–6061. doi: 10.4049/jimmunol.1301481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mutamba S, Allison A, Mahida Y, Barrow P, Foster N. Expression of IL-1Rrp2 by human myelomonocytic cells is unique to DCs and facilitates DC maturation by IL-1F8 and IL-1F9. Eur J Immunol. 2012;42(3):607–617. doi: 10.1002/eji.201142035. [DOI] [PubMed] [Google Scholar]
- 32.Tortola L, Rosenwald E, Abel B, Blumberg H, Schafer M, Coyle AJ, et al. Psoriasiform dermatitis is driven by IL-36-mediated DC-keratinocyte crosstalk. J Clin Invest. 2012;122(11):3965–3976. doi: 10.1172/JCI63451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lefrancais E, Roga S, Gautier V, Gonzalez-de-Peredo A, Monsarrat B, Girard JP, et al. IL-33 is processed into mature bioactive forms by neutrophil elastase and cathepsin G. Proc Natl Acad Sci U S A. 2012;109(5):1673–1678. doi: 10.1073/pnas.1115884109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefrancais E, Duval A, Mirey E, Roga S, Espinosa E, Cayrol C, et al. Central domain of IL-33 is cleaved by mast cell proteases for potent activation of group-2 innate lymphoid cells. Proc Natl Acad Sci U S A. 2014;111(43):15502–15507. doi: 10.1073/pnas.1410700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snelgrove RJ, Gregory LG, Peiro T, Akthar S, Campbell GA, Walker SA, et al. Alternaria-derived serine protease activity drives IL-33-mediated asthma exacerbations. J Allergy Clin Immunol. 2014;134(3):583–592. doi: 10.1016/j.jaci.2014.02.002. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Towne JE, Renshaw BR, Douangpanya J, Lipsky BP, Shen M, Gabel CA, et al. Interleukin-36 (IL-36) ligands require processing for full agonist (IL-36{alpha}, IL-36{beta} and IL-36{gamma}) or antagonist (IL-36Ra) activity. J Biol Chem. 2011;286(49):42594–42602. doi: 10.1074/jbc.M111.267922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rossi-Semerano L, Piram M, Chiaverini C, De Ricaud D, Smahi A, Kone-Paut I. First clinical description of an infant with interleukin-36-receptor antagonist deficiency successfully treated with anakinra. Pediatrics. 2013;132(4):e1043–e1047. doi: 10.1542/peds.2012-3935. [DOI] [PubMed] [Google Scholar]
- 38.Huffmeier U, Watzold M, Mohr J, Schon MP, Mossner R. Successful therapy with anakinra in a patient with generalized pustular psoriasis carrying IL36RN mutations. Br J Dermatol. 2014;170(1):202–204. doi: 10.1111/bjd.12548. [DOI] [PubMed] [Google Scholar]
- 39.Viguier M, Guigue P, Pages C, Smahi A, Bachelez H. Successful treatment of generalized pustular psoriasis with the interleukin-1-receptor antagonist Anakinra: lack of correlation with IL1RN mutations. Ann Intern Med. 2010;153(1):66–67. doi: 10.7326/0003-4819-153-1-201007060-00030. [DOI] [PubMed] [Google Scholar]
- 40.Tauber M, Viguier M, Le Gall C, Smahi A, Bachelez H. Is it relevant to use an interleukin-1-inhibiting strategy for the treatment of patients with deficiency of interleukin-36 receptor antagonist? Br J Dermatol. 2014;170(5):1198–1199. doi: 10.1111/bjd.12805. [DOI] [PubMed] [Google Scholar]
- 41.Elewski BE. Infliximab for the treatment of severe pustular psoriasis. J Am Acad Dermatol. 2002;47(5):796–767. doi: 10.1067/mjd.2002.128382. [DOI] [PubMed] [Google Scholar]
- 42.Newland MR, Weinstein A, Kerdel F. Rapid response to infliximab in severe pustular psoriasis, von Zumbusch type. International journal of dermatology. 2002;41(7):449–452. doi: 10.1046/j.1365-4362.2002.01543.x. [DOI] [PubMed] [Google Scholar]
- 43.Johnston A, Guzman AM, Swindell WR, Wang F, Kang S, Gudjonsson JE. Early tissue responses in psoriasis to the antitumour necrosis factor-alpha biologic etanercept suggest reduced interleukin-17 receptor expression and signalling. Br J Dermatol. 2014;171(1):97–107. doi: 10.1111/bjd.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiricozzi A, Guttman-Yassky E, Suarez-Farinas M, Nograles KE, Tian S, Cardinale I, et al. Integrative responses to IL-17 and TNF-alpha in human keratinocytes account for key inflammatory pathogenic circuits in psoriasis. J Invest Dermatol. 2011;131(3):677–687. doi: 10.1038/jid.2010.340. [DOI] [PubMed] [Google Scholar]
- 45.Bohner A, Roenneberg S, Eyerich K, Eberlein B, Biedermann T. Acute Generalized Pustular Psoriasis Treated With the IL-17A Antibody Secukinumab. JAMA Dermatol. 2015:1–2. doi: 10.1001/jamadermatol.2015.4686. [DOI] [PubMed] [Google Scholar]
- 46.Imafuku S, Honma M, Okubo Y, Komine M, Ohtsuki M, Morita A, et al. Efficacy and safety of secukinumab in patients with generalized pustular psoriasis: A 52-week analysis from phase III open-label multicenter Japanese study. The Journal of dermatology. 2016 doi: 10.1111/1346-8138.13306. [DOI] [PubMed] [Google Scholar]
- 47.Yamasaki K, Nakagawa H, Kubo Y, Ootaki K Japanese brodalumab study g. Efficacy and safety of brodalumab in patients with generalized pustular psoriasis and psoriatic erythroderma: results from a 52-week, open-label study. Br J Dermatol. 2016 doi: 10.1111/bjd.14702. [DOI] [PubMed] [Google Scholar]
- 48.Wolf J, Ferris LK. Anti-IL-36R antibodies, potentially useful for the treatment of psoriasis: a patent evaluation of WO2013074569. Expert Opin Ther Pat. 2014;24(4):477–479. doi: 10.1517/13543776.2014.881473. [DOI] [PubMed] [Google Scholar]
- 49.Gunther S, Sundberg EJ. Molecular determinants of agonist and antagonist signaling through the IL-36 receptor. J Immunol. 2014;193(2):921–930. doi: 10.4049/jimmunol.1400538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.