Abstract
We suggest an annotation strategy for genes encoded by retroviruses and transposable elements (RETRA genes) based on a set of marker protein domains. Usually RETRA genes are masked in vertebrate genomes prior to the application of automated gene prediction pipelines under the assumption that they provide no selective advantage to the host. Yet, we show that about 1000 genes in four vertebrate gene sets analyzed contain at least one RETRA gene marker domain. Using the conservation of genomic neighborhood (synteny), we were able to discriminate between RETRA genes with putative functionality in the vertebrates and those that probably function only in the context of mobile elements. We identified 35 such genes in human, along with their corresponding mouse and rat orthologs; which included almost all known human genes with similarity to mobile elements. The results also imply that the vast majority of the remaining RETRA genes in current gene sets are unlikely to encode vertebrate functions. To automatically annotate RETRA genes in other vertebrate genomes, we provide as a tool a set of marker protein domains and a manually refined list of domesticated or ancestral RETRA genes for rescuing genes with vertebrate functions.
INTRODUCTION
Preliminary sequence analysis of the draft sequences of the human, mouse and rat genomes (1–3) suggested that less than 5–6% of the genomic sequence appears to be under selective constraint and less than 1–2% is coding for proteins, while most of the genomic sequence comprises neutrally evolving remnants of various transposable elements. Such interspersed repeats are normally assumed not to have any host-specific functionality and are therefore commonly omitted from functional analysis, e.g. by applying the RepeatMasker program (Smit & Green, http://repeatmasker.org) prior to gene prediction (4). However some repetitive elements do encode proteins, and a considerable number of genes predicted in these genomes are similar to Retroviral or Transposon-associated (RETRA) genes. Indeed fragments of transposable elements (TEs) have been found to insert into vertebrate genes, contributing to at least 4% of current coding regions (5,6). Moreover a number of reports demonstrate or propose (7–9) the domestication of genes from TEs by vertebrate genomes. Well-characterized examples include the major centromere-binding protein CENP-B, which is related to pogo-like DNA transposases (9) and telomerase, a reverse transcriptase related to non-LTR retrotransposons (10). Yet in many cases there does not seem to be any relationship between the sizes of protein families with similarity to RETRA genes and the number of well-characterized family members with known functions in the vertebrate genomes. For example as many as 307 human and 244 mouse reverse transcriptases had been predicted in the respective landmark genome sequence papers [see table 25 in (1) and table 11 in (2)] although to our knowledge only one well-characterized vertebrate member, telomerase (11), has been described so far. The inconsistent inclusion of RETRA genes into gene sets can result in misleading comparative analysis due to artificially inflated sizes of RETRA gene families. Therefore, there is a need for reliable identification and annotation of such genes, particularly if they contribute to vertebrate function.
To get an overview of the coding potential of RETRA genes we compiled a list of known characteristic protein domains. We then applied these domains to evaluate the instances of RETRA genes included into several frequently used gene prediction sets derived from four completely sequenced vertebrate genomes (human, mouse, rat and puffer fish), and developed a strategy to discriminate those with likely vertebrate function. For all the candidate RETRA genes in three mammalian genomes, we measured selective constraints to identify genes with a function in the host genome. It has been shown that 97% of human and rat orthologous genes are retained in orthologous genomic regions (3). Hence if a RETRA gene has been preserved in synteny in either rodent or human, we then assume that it is performing a vertebrate function because otherwise purifying selection would have led to the elimination of the gene. This is a much more rigorous criterion than the requirement of a supporting expressed sequence tag (EST), which has been previously used to identify 34 RETRA genes (annotated in the current Ensembl gene set build 34) with putative functionality in human (1), as ESTs can also be derived from pseudogenes or other non-coding regions (12,13). If a RETRA gene is not in synteny, it may either have recently acquired a vertebrate function or, much more likely, it functions only in the context of retroviral or transposon activity. Although the procedure to identify RETRA genes with vertebrate functionality outlined above can be applied in principle automatically, it depends on derived data (e.g. gene predictions) and there are inherent limitations in the methods used (e.g. use of best-reciprocal hits for orthology detection), hence we did a manual refinement of the results. Therefore, the curated data sets obtained, in combination with the marker domains, should result in a reliable automatic method for RETRA gene detection.
TEs and retroviruses with coding potential
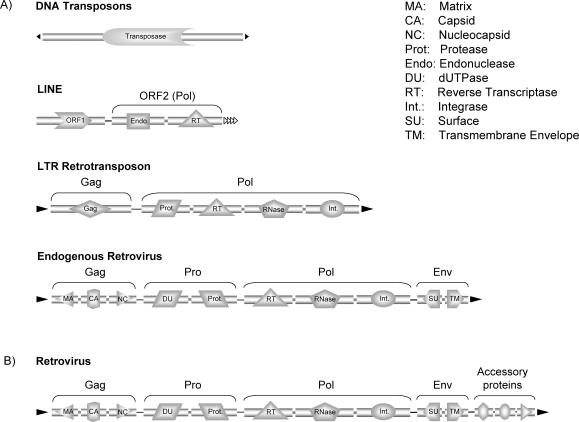
Transposable elements are repetitive mobile sequences that are dispersed throughout the genome. In vertebrates, the content and diversity of these elements varies considerably. In mammalian genomes, the recognizable copies of these elements are estimated to cover 40–50% of their DNA content (1–3), whereas in the more compact vertebrate genome of puffer fish (fugu) the fraction is only 2.7% of the genome (14). TEs can be classified into class I and class II depending on whether their transposition intermediate is RNA or DNA respectively. Each class can be subdivided into elements that code for genes that catalyze transposition (autonomous TEs) (Figure 1) and those that do not contain such genes (non-autonomous TEs).
Figure 1.
Schematic overview of autonomous TEs and retroviruses and their protein content. The highlighted domains are characteristic for these elements and correspond to marker domains used in this study. (A) Protein content of most common vertebrate retrotransposons and DNA transposons. The single ORF of DNA transposons encoding a transposase is depicted. LINEs contain two open reading frames (ORF1 and ORF2): encoding an RNA-binding protein and a pol gene with homology to reverse transcriptases and endonucleases. LTR-retrotransposons can include ORFs such as (i) gag, encoding a protein that forms the structural component of a cytoplasmic particle within which reverse transcription reaction takes place and (ii) pol, which encodes in most elements an aspartic protease (Pro), a reverse transcriptase (RT), a ribonuclease H (RNase H) and an integrase (Int). These elements can also include an env protein and, thus, show a protein content very similar to the endogenous retrovirus. (B) Protein content of retroviruses. In comparison to an endogenous retrovirus, a retrovirus possesses additional proteins that are usually not recognizable in endogenous retrovirus.
Class I elements or retrotransposons replicate through a reverse transcription mechanism and the most common elements of this class are the non-LTR retrotransposon short (SINE) and long (LINE) interspersed nuclear elements, LTR-retrotransposons and endogenous retroviruses. While SINEs have no open reading frames (ORFs) and are therefore always non-autonomous, all other class I elements encode a number of proteins. When retroviruses occasionally insert into the genome of a germ line cell they can become endogenous (Figure 1) and for this reason we also considered retroviruses in this study.
Class II elements or DNA transposons excise and reinsert as DNA. The autonomous DNA transposons usually contain only a single gene encoding a transposase. Vertebrate genomes contain only a few copies of autonomous full-length TEs together with numerous fragmented copies. Taken together, both TEs and Retroviruses have coding potential and we thus derive marker domains of the characteristic ORFs from these elements.
METHODS
Data sets
The analysis was based on publicly available gene prediction sets and genomic sequences as of October 9, 2003 (Supplementary Table 2). Together with the final sets of genes provided by NCBI (http://www.ncbi.nlm.nih.gov) and Ensembl (15) for human, rat and mouse genomes, and by JGI (http://www.jgi.doe.gov) for the fugu genome, we also used gene prediction sets directly produced by automatic gene calling methods, such as GeneScan (16) used in Ensembl and JGI pipelines and Gnomon (http://www.ncbi.nlm.nih.gov/genome/guide/gnomon.html) at NCBI. A comparative gene prediction method, Twinscan (17), was also included.
RETRA protein domain signatures
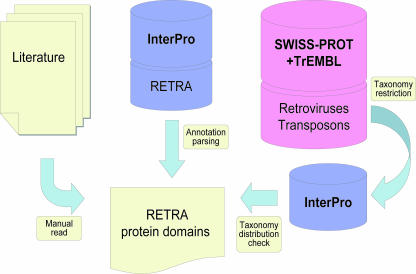
We decided to use a set of protein domains characteristic of genes encoded by transposable elements and retroviruses as a discriminator of such genes. Since protein coding signal is more conserved through evolution, this approach is more sensitive than DNA-based analysis. Protein domain signatures associated with retrotransposons, DNA transposons and retroviruses were identified on the basis of literature survey, InterPro domain annotation and annotation of proteins in SWISS-PROT and TrEMBL databases as follows: (i) we selected all InterPro protein signatures annotated with any of the following keywords (substrings): ‘transposable’, ‘transposase’, ‘transposon’, ‘retroelement’, ‘retroid’, ‘retrotransposon’, ‘retroviral’, ‘retrovirus’, and (ii) we considered all HMM-based InterPro domains that are over-represented (100-fold) in vertebrate proteins annotated in SWISS-PROT and TrEMBL with a keyword (substring) ‘transposa’ in the description or keyword lines with respect to the rest of the vertebrate proteins, or that are over-represented (10 fold) in retroviral proteins with respect to the rest of proteins in the database. The ratios were based on the fraction of the total number of retroviral proteins in the databases. A manual inspection refined this list to a total of 85 RETRA Pfam HMM profiles that we consider as being RETRA gene specific. As an example, this manual refinement excluded from the list two profiles of CCHC Zn-finger (IPR001878) and Endonuclease (IPR005135) domains that are also found in variety of non-RETRA proteins. The list contains a number of profiles characteristic to RETRA genes even though no endogenous genes with the domains have yet been detected.
We used only corresponding signatures from the Pfam database to simplify the surveying procedure. Although not all protein domains are characterized and as Pfam does not have complete coverage (providing profiles for only about 75% of known proteins (18)) the use of HMM profiles gives significant advantages in terms of both sensitivity and specificity. For searching the gene sets using HMMER2, both ls (global) and fs (local) modes were considered yielding practically the same results and the fs mode was selected for further use as it is more tolerant to gene prediction errors (such as gene truncation). The results were filtered using family specific ‘gathering’ cut-offs specified in the HMM model descriptions (18). The selected HMMs (Supplementary Table 1) were retrieved from the Pfam (v.10) database and were scanned against the predicted proteomes (Supplementary Table 2). Parsed results were loaded and analyzed in a PostgreSQL (http://www.postgresql.org) database.
Estimating the number of RETRA genes in vertebrate genomes
Since scanning HMM profiles directly against genomic sequences is extremely CPU intensive, in Supplementary Table 3 we report the number of matches found by TBlastN (24) for sample protein fragments, extracted from the corresponding PfamA seed alignments, in non-masked vertebrate genomic sequences with E-value less than 0.001.
Assessment of RETRA gene host-specific functionality
To identify RETRA genes that probably encode a host-specific function in mammals, we checked all human genes with the characteristic RETRA domains for conservation of their genomic neighborhood (synteny) in the two rodent genomes. The synteny maps were derived using all genes as orthologous markers as outlined below. Although DNA-level comparison can provide additional details we do not expect many false negatives as it has been estimated that 97% of human and rat orthologous genes are retained in synteny (3). First, we determined putative orthologous genes requiring them to be best reciprocal hits in an inter-species BlastP analysis without low-complexity filtering and using the default E-value cut-off. The synteny of the best reciprocal hits was identified requiring at least two putative orthologous pairs to be nearby on genome but allowing for up to four intervening genes as described before (19) using SyntQL tool (Zdobnov, unpublished). We checked synteny manually for all human genes with RETRA domains for which orthologous genes in mouse or rat were not found automatically. Intrinsic limitations of this approach are discussed in detail in Results and Discussions. In addition, we inspected EST support for the human genes with RETRA domains: all ESTs from dbEST (20) (as of February 2004) were aligned against the human genome (build 34) using stand-alone BLAT (2) and we consider only EST alignments in the genome with a percentage identity greater than 96% and the alignment length greater than 100 bases. If the difference in score between the best hit and second-best hit was less than 10 in a BLAT-like scoring scheme, we considered such an EST alignment as ambiguous.
Data for identifying RETRA genes in vertebrate gene sets
The list of selected 85 RETRA characteristic Pfam HMM models, the models itself from Pfam version 10 and the list of true vertebrate genes with similarity to RETRA genes are available from: http://www.bork.embl-heidelberg.de/Docu/RETRA/.
RESULTS
Marker domains for RETRA genes
To identify characteristic protein signatures that could be used as RETRA gene markers in vertebrate genomes, we surveyed known characteristic domains of RETRA genes as described in the literature (Figure 1) and extended the list by including domains that are clearly annotated as being RETRA in the InterPro database (21) or those that are over-represented in annotated RETRA genes in protein databases (SWISS-PROT and TrEMBL (22); see data flow in Figure 2). As a result we collected a manually curated set of 85 HMM profiles (23) for the domains that can be considered as markers for RETRA genes (Supplementary Table 1, see also Table 3).
Figure 2.
The list of InterPro domain signatures corresponding to genes in DNA transposons, retrotransposons and retroviruses was compiled on the basis of literature survey and two protein and domain annotation-based procedures (for details see Methods).
RETRA genes in vertebrate gene sets
In order to evaluate inconsistencies in RETRA gene inclusion into current vertebrate gene sets, we scanned the 85 marker domains against several popular gene sets (Table 1). The numbers are considerably lower than those in previous releases due to ongoing annotation efforts. For example, we find only 251 candidate RETRA genes in the Ensembl gene set based on human assembly build 33 compared with more than 1000 in the early releases. Despite these considerable improvements there are still as many as 54 predicted genes containing the reverse transcriptase domain [including the well-characterized telomerase and a recently identified LTR retrotransposon element conserved in synteny in human and rodents species (25)] and 127 L1 transposases in this gene set. Given the background of thousands of human reverse transcriptases and L1 transposases in the non-masked human genomic sequence (Supplementary Table 3), gene prediction pipelines already filter out many of the unwanted RETRA genes. Yet Table 1 also indicates that a considerable number of such genes still exist in current annotation schemes, despite manual curation efforts.
Table 1.
RETRA genes of four vertebrates identified in frequently used gene sets
| Ensembl | NCBI | Twinscan (comparative) | |||
|---|---|---|---|---|---|
| Final set v33 (v34) | GeneScan (aut.) | Final set | Gnomon (aut.) | ||
| Human | 251 (230) | 51 | 65 | 60 | 33 |
| Rat | 181 | 479 | 98 | 173 | 190 |
| Mouse | 277 | 475 | 807 | 1195 | 116 |
| Fugu | 679 | 333 | 670 | 220 | na |
| Sum | 1137 | 1338 | 1640 | 1648 | 339a |
(i) The Ensembl (15) pipeline, which is based on (ii) the automatic gene discovery method GeneScan (16), (iii) the NCBI (http://www.ncbi.nlm.nih.gov/genome/guide/build.html) pipeline which is based on (iv) the Gnomon (http://www.ncbi.nlm.nih.gov/genome/guide/gnomon.html) predictor and (v) a comparative gene prediction method, Twinscan (17). All sets correspond to human genome assembly build 33 (see more details in Supplementary Table 2); the reference numbers for the more recent human genome assembly build 34 available from Ensembl are given in brackets. na: twinscan gene predictions for fugu are not available.
aSum is not comparable.
Although the overall number of RETRA genes in vertebrate gene sets appears similar in five popular gene prediction protocols, a more detailed breakdown of the results in different species reveals that different gene prediction pipelines give considerably different results among each other and among different species (Table 2). It indicates that there is still a need for consistent annotation of RETRA genes in gene prediction pipelines. The automatic detection of RETRA genes is complicated by the fact that some RETRA genes encode functionality for the host genome, i.e. are true vertebrate genes. We have thus screened such genes among the RETRA genes recognized by marker domain analysis.
Table 2.
Detailed breakdown of RETRA domain occurrences in public gene sets
| Pfam | InterPro description | Ensembl | NCBI | TwinScan | ||
|---|---|---|---|---|---|---|
| Final set v33 (v34) | GeneScan (aut.) | Final set | Gnomon (aut.) | |||
| PF02994 | (IPR004244) L1 transposable element | homo: 127(131) | homo: 0 | homo: 2 | homo: 3 | homo: 0 |
| rat: 48 | rat: 0 | rat: 25 | rat: 26 | rat: 1 | ||
| mouse: 1 | mouse: 0 | mouse: 45 | mouse: 52 | mouse: 0 | ||
| fugu: 1 | fugu: 1 | fugu: 1 | fugu: 2 | fugu: na | ||
| PF00078 | (IPR000477) RNA-directed DNA polymerase (Reverse transcriptase) | homo: 54 (41) | homo: 3 | homo: 12 | homo: 8 | homo: 2 |
| rat: 59 | rat: 3 | rat: 5 | rat: 7 | rat: 99 | ||
| mouse: 6 | mouse: 3 | mouse: 19 | mouse: 20 | mouse: 3 | ||
| fugu: 368 | fugu: 209 | fugu: 364 | fugu: 120 | fugu: na | ||
| PF00429 | (IPR002050) ENV polyprotein (coat polyprotein) | homo: 13 (9) | homo: 0 | homo: 2 | homo: 1 | homo: 0 |
| rat: 5 | rat: 94 | rat: 31 | rat: 55 | rat: 25 | ||
| mouse: 40 | mouse: 64 | mouse: 209 | mouse: 297 | mouse: 11 | ||
| fugu: 0 | fugu: 0 | fugu: 0 | fugu: 0 | fugu: na | ||
| PF03184 | (IPR004875) CENP-B protein | homo: 13 (12) | homo: 12 | homo: 14 | homo: 12 | homo: 9 |
| rat: 8 | rat: 7 | rat: 8 | rat: 9 | rat: 9 | ||
| mouse: 10 | mouse: 9 | mouse: 8 | mouse: 8 | mouse: 9 | ||
| fugu: 18 | fugu: 23 | fugu: 18 | fugu: 12 | fugu: na | ||
| PF00665 | (IPR001584) Integrase, catalytic domain | homo: 10 (10) | homo: 7 | homo: 7 | homo: 9 | homo: 3 |
| rat: 4 | rat: 7 | rat: 3 | rat: 3 | rat: 4 | ||
| mouse: 21 | mouse: 13 | mouse: 50 | mouse: 74 | mouse: 3 | ||
| fugu: 108 | fugu: 41 | fugu: 108 | fugu: 61 | fugu: na | ||
| PF05699 | (IPR008906) HAT dimerisation | homo: 6 (8) | homo: 7 | homo: 8 | homo: 7 | homo: 7 |
| rat: 3 | rat: 4 | rat: 6 | rat: 7 | rat: 7 | ||
| mouse: 4 | mouse: 7 | mouse: 3 | mouse: 6 | mouse: 7 | ||
| fugu: 64 | fugu: 18 | fugu: 61 | fugu: 9 | fugu: na | ||
| PF00692 | (IPR008180) DeoxyUTP pyrophosphatase | homo: 5 (3) | homo: 4 | homo: 3 | homo: 4 | homo: 1 |
| rat: 6 | rat: 24 | rat: 2 | rat: 3 | rat: 3 | ||
| mouse: 58 | mouse: 93 | mouse: 152 | mouse: 237 | mouse: 19 | ||
| fugu: 1 | fugu: 0 | fugu: 1 | fugu: 1 | fugu: na | ||
| PF02337 | (IPR003322) Retroviral GAG p10 protein | homo: 4 (2) | homo: 2 | homo: 2 | homo: 4 | homo: 1 |
| rat: 34 | rat: 249 | rat: 3 | rat: 31 | rat: 9 | ||
| mouse: 61 | mouse: 93 | mouse: 58 | mouse: 239 | mouse: 10 | ||
| fugu: 0 | fugu: 0 | fugu: 0 | fugu: 0 | fugu: na | ||
| PF03732 | (IPR005162) Retrotransposon gag protein | homo: 4 (4) | homo: 5 | homo: 4 | homo: 5 | homo: 5 |
| rat: 1 | rat: 1 | rat: 3 | rat: 3 | rat: 2 | ||
| mouse: 4 | mouse: 6 | mouse: 4 | mouse: 5 | mouse: 4 | ||
| fugu: 40 | fugu: 24 | fugu: 40 | fugu: 13 | fugu: na | ||
| PF00075 | (IPR002156) RNase H | homo: 3 (6) | homo: 5 | homo: 5 | homo: 5 | homo: 3 |
| rat: 4 | rat: 22 | rat: 4 | rat: 4 | rat: 1 | ||
| mouse: 8 | mouse: 13 | mouse: 30 | mouse: 43 | mouse: 7 | ||
| fugu: 34 | fugu: 12 | fugu: 33 | fugu: 9 | fugu: na | ||
| PF00077 | (IPR001995) Peptidase A2A, retrovirus | homo: 3 (1) | homo: 0 | homo: 1 | homo: 0 | homo: 0 |
| rat: 4 | rat: 14 | rat: 1 | rat: 2 | rat: 0 | ||
| mouse: 44 | mouse: 55 | mouse: 99 | mouse: 145 | mouse: 9 | ||
| fugu: 11 | fugu: 4 | fugu: 11 | fugu: 1 | fugu: na | ||
| PF00607 | (IPR000721) Retroviral nucleocapsid protein Gag | homo: 3 (2) | homo: 0 | homo: 1 | homo: 1 | homo: 1 |
| rat: 3 | rat: 69 | rat: 13 | rat: 45 | rat: 36 | ||
| mouse: 48 | mouse: 140 | mouse: 102 | mouse: 268 | mouse: 53 | ||
| fugu: 0 | fugu: 0 | fugu: 0 | fugu: 0 | fugu: na | ||
| PF00552 | (IPR001037) Retroviral integrase, C-terminal | homo: 1 (4) | homo: 0 | homo: 2 | homo: 0 | homo: 0 |
| rat: 0 | rat: 5 | rat: 1 | rat: 1 | rat: 1 | ||
| mouse: 28 | mouse: 28 | mouse: 180 | mouse: 43 | mouse: 0 | ||
| fugu: 0 | fugu: 0 | fugu: 0 | fugu: 0 | fugu: na | ||
| PF01498 | (IPR002492) Transposase, Tc1/Tc3 | homo: 0 (0) | homo: 0 | homo: 0 | homo: 0 | homo: 0 |
| rat: 0 | rat: 0 | rat: 0 | rat: 0 | rat: 0 | ||
| mouse: 0 | mouse: 0 | mouse: 0 | mouse: 0 | mouse: 0 | ||
| fugu: 42 | fugu: 14 | fugu: 41 | fugu: 5 | fugu: na | ||
| PF05380 | (IPR008042) Retrotransposon, Pao | homo: 0 (0) | homo: 0 | homo: 0 | homo: 0 | homo: 0 |
| rat: 0 | rat: 0 | rat: 0 | rat: 0 | rat: 0 | ||
| mouse: 0 | mouse: 0 | mouse: 0 | mouse: 0 | mouse: 0 | ||
| fugu: 13 | fugu: 7 | fugu: 13 | fugu: 12 | fugu: na | ||
Only domains that match at least 10 proteins in any of the Ensembl sets are shown. At the time of analysis only an Ensembl gene set based on human genome assembly build 34 was available, shown in brackets.
Identification of mammalian genes with similarity to RETRA genes
Under the assumption that synteny between species as divergent as human and rodents should be a sufficient denominator for host-specific functionality, we analyzed all identified RETRA genes in mammals for this feature. In brief, our synteny analysis requires the conservation of local genomic neighborhood of putative orthologs (19), operationally defined by best reciprocal hits in an inter-species BlastP analysis. Since the method relies on existing genome annotation, the analysis was complemented by manual inspection of all human genes with RETRA domains for which no putative mouse or rat orthologs were found in synteny automatically. As orthology and synteny identification methods are sensitive to genome sequence completeness and quality, the fugu genome was not included in this analysis.
The majority of the RETRA domain families contain none or one gene in synteny, the latter being often a known human gene with similarity to RETRA genes (see Table 3 and below). Other families of RETRA genes contain a few genes in synteny, the majority of which have not been noted before. Examples are genes with HAT dimerization (InterPro family identifier: IPR008906), Integrase (IPR001584) and BED finger (IPR003656) domains (Table 3). Surprisingly, in a small number of families (almost) all genes were found in synteny between human and rodents, suggesting that the members of these families perform host-specific functions in vertebrates.
Table 3.
Detailed analysis of putative RETRA genes in human Ensembl gene set (corresponding to genome assembly build 34)
| InterPro (Pfam) | RETRA domain description | Total | EST | BRH | Synteny | Known |
|---|---|---|---|---|---|---|
| IPR004244 (PF02994) | L1 transposable element | 127 | 7(1) | 3 | 0(1) | |
| IPR000477 (PF00078) | RNA-directed DNA polymerase (Reverse transcriptase) | 54 | 9(1) | 5 | 1(2) | Telomerase (O14746) (36,37) Hur1 (25) |
| IPR004875 (PF03184) | CENP-B protein | 14 | 14(12) | 9 | 9(12) | CENP-B (P07199) (29) |
| Jerky (Q60976) (30) | ||||||
| TIGD2, TIGD3, TIGD6 and TIGD7 (31) | ||||||
| IPR006695 (PF04218) | CENP-B, N-terminal DNA-binding | YCE7_HUMANa (Q9Y3E5) (38) | ||||
| IPR002050 (PF00429) | ENV polyprotein (coat polyprotein) | 13 | 6(0) | 2 | 0(0) | Syncytin 1+ and 2+ (Q9NZG3, P60508) (33,35) |
| IPR001584 (PF00665) | Integrase, catalytic domain | 10 | 9(6) | 4 | 3(6) | Gin-1 (NM_017676) (39) |
| IPR008906 (PF05699) | HAT dimerisation | 6 | 6(5) | 3 | 3(5) | P52rIPK (O43422)a (40) |
| ZBED1+ (NM_004729) (34) | ||||||
| IPR008180 (PF00692) | DeoxyUTP pyrophosphatase | 5 | 3(1) | 2 | 1(1) | dUTP pyrophosphatase (P33316) (41) |
| IPR004295 (PF03056) | Env gp36 protein, HERV | 4 | 2(0) | 1 | 0(0) | |
| IPR005162 (PF03732) | Retrotransposon gag protein | 4 | 4(4) | 4 | 4(4) | PEG10(Q9UPV1) (42)PNMA2 (O94959) (26) |
| IPR003322 (PF02337) | Retroviral GAG p10 protein | 4 | 1(0) | 2 | 0(0) | |
| IPR003656 (PF02892) | BED finger | 4 | 4(3) | 1 | 2(3) | ZBED1+ (NM_004729) (34) |
| IPR001995 (PF00077) | Peptidase A2A, retrovirus | 3 | 1(0) | 1 | 0(0) | |
| IPR000721 (PF00607) | Retroviral nucleocapsid protein Gag | 3 | 2(0) | 2 | 0(0) | |
| IPR002156 (PF00075) | Rnase H | 3 | 2(1) | 2 | 1(1) | RNase H (O60930) (43) |
| IPR004191 (PF02920) | Tn916 integrase, N-terminal DNA binding | 2 | 2(2) | 1 | 2(2) | Liprin-beta 1a (Q9ULJ0) and Liprin-beta 2 (Q8ND30) (44) |
| IPR003036 (PF02093) | Core shell protein Gag P30 | 2 | 1(0) | 1 | 0(0) | |
| IPR001888 (PF01359) | Transposase, type 1 | 2 | 1(1) | 1 | 1(1) | SETMAR (NM_006515) (45) |
| IPR001037 (PF00552) | Retroviral integrase, C-terminal | 1 | 1(0) | 1 | 0(0) | |
| IPR003308 (PF02022) | Integrase, N-terminal zinc-binding | 1 | 0(0) | 1 | 0(0) | |
| IPR002514 (PF01527) | Transposase IS3 | 1 | 1(1) | 0 | 1(1) |
We identified 35 RETRA genes in synteny between humans and rodents (some genes have more than one domain listed in the table). Of them, 21 have been reported in literature, and 18 of which were detected by our approach (with exception of Syncytin1, Syncytin2 and ZBED1 labeled with +). Detailed list of human genes, and corresponding orthologs in other species, containing RETRA domains but retained in synteny is provided in supplementary material (Supplementary Table 5). Total: total number of proteins with corresponding RETRA domain (some genes may have more than one domain); EST: number of genes confirmed by at least one EST or mRNA (in brackets the number of genes in synteny with matched ESTs is shown); BRH: number of human genes with a putative ortholog in the mouse or rat genomes identified by best reciprocal hit in the current gene sets; Synteny: number of putative orthologs found in synteny identified by an automatic procedure (with manual inspection shown in brackets); Known: previously known human genes with these domains reported in literature. These domains can be used as RETRA markers in annotation pipelines provided a rescue procedure for functional genes is used.
aIn the human proteins P52rIPK (O43422), Liprin-beta 1 and YCE7_HUMAN found in synteny with rodents we detected RETRA domains only in the human lineage suggesting the recent acquisition of the domains in these proteins. Yet, YCE7_HUMAN gene seems to acquire only N-terminal CENP-B domain.
+Known RETRA genes recently acquired a host function missed by our approach.
For example, an entire family of gag-like proteins (IPR005162) (at least four paralogs in mammals) appears to play a role in mammalian biology. One member of this family has been detected as an antigen in patients with testicular cancer (26) and another, PEG10, is probably a regulator of transcription (27). We can only speculate about the mammalian functionality of the two other members of this gag-like family, but the recent discovery of the cellular interaction partner of the homologous viral gag protein of the Moloney murine leukemia virus, endophilin 2 (28), might give a first hint.
Another family for which host-specific functionality in mammals was discovered using our procedure contains homologs of the centromeric protein CENP-B (IPR004875). Out of 13 CENP-B family members that were detected (not counting YCE7_HUMAN gene, see Table 3), 11 were found to be in synteny in mammals including two experimentally proven human genes, CENP-B itself and jerky (29,30). The mammalian function of the remaining nine genes is unknown, although all of them with one exception have already been either predicted based on EST support (1) or based on orthology in rodents (31). CENP-B binds to alpha-repetitive sequences at the centromere of autosomes and the X chromosome (29), therefore the presence of homologs with slightly different binding specificities might explain the inability of CENP-B to bind Y chromosome centromeric regions.
In total, 35 human genes with similarity to RETRA genes were found in synteny; of which only 27 were recognized automatically as best-reciprocal hits in synteny with rodents, and the remainder through manual analysis of all other candidates detected by the marker domains.
Comparison to known human genes with similarity to RETRA genes
To get an overview of how many known human genes with similarity to RETRA our procedure identified, we surveyed the literature for the respective reports. We retrieved 21 genes with a proven human function (Table 3), of which 18 were recovered by our procedure as likely having a host function (in 3 of these 18 cases synteny was detected only by manual refinement). Of the three genes that we did not detect using synteny, Syncytin1 and Syncytin2, were previously reported as primate-specific acquisitions (32,33) and the third, human transcription factor ZBED1 with HAT and BED finger domains, was described before as a homolog of the Drosophila DREF transcription regulator (34).
In addition to the recovery of 18 out of 21 known mammalian RETRA genes, our procedure led to the identification of an additional 17 expressed human genes with similarity to RETRA genes (Table 3); they all have retained their genomic neighborhood in rodents and human, and therefore being under selective constraint they are likely to have a specific function in mammals. We combined these two sets and recorded the respective vertebrate orthologs to derive a list of vertebrate genes with similarity to RETRA (‘rescue list’). Despite additional manual effort to compile this list, it can now be used automatically in conjunction with the set of marker domains for the annotation of RETRA genes in forthcoming vertebrate genomes. This concept can also be extended to other metazoan genomes.
DISCUSSION
Although most RETRA genes are commonly filtered out prior to the application of automated gene prediction pipelines (4) as elements without any selective advantage for the host, we find that current gene prediction pipelines still include a considerable number of RETRA genes (Table 1), and detect inconsistencies not only between methods applied to the same genome but also between applications of each method to different genomes. These discrepancies cannot be explained merely by the different time points at which the analyses were done or by the amount of manual work invested for a particular genome. They are most likely due to the absence of an established criterion for the annotation of RETRA genes, which can easily lead to erroneous conclusions in comparative analyses. We therefore propose that, as is attempted by the repeat masking process, RETRA elements should be specifically identified and excluded from gene sets unless evidence for host-specific functionality is found.
For this reason we have developed a procedure for the identification of RETRA genes and suggest a criterion based on genomic neighborhood conservation to distinguish between those with a host-specific function and those that function only in the context of mobile elements. By applying this methodology we have identified the vast majority of described RETRA genes with functionality in mammals (e.g. Telomerase, CENP-B, RNAse H etc.) and have revealed additional functional genes, all with EST or mRNA support. In contrast, only 15% of the non-syntenic RETRA genes are supported by uniquely mapped ESTs or mRNAs (Supplementary Table 4), although this is likely to be a significant underestimate due to high level of sequence similarity within families of repetitive elements. The sensitivity of the method could in principle be further increased as the orthology identification is far from being perfect and the domain detection also has its limits. For instance, the human transcription factor ZBED1 (34), whose homolog can be found in Drosophila but not in rodents. Yet synteny will not be able to identify all RETRA genes with functionality in the host genomes as it is possible that true orthologous genes have been translocated, lost or even acquired and domesticated, since the divergence of rodents and human approximately 75 MYA ago (2). This seems to be the case for Syncytin1 and Syncytin2 in primates (32,33). Apparently both proteins have been acquired from human endogenous retroviruses (HERV-W and HERV-FRD, respectively) envelope proteins and now potentially have a role in placenta formation (32,33,35). Although this is a clear example of the domestication of a RETRA gene, analysis of the phylogenetic trees of other RETRA gene families does not give a clear picture of the origin of these genes i.e. in most cases we were not able to distinguish whether they have been domesticated by the vertebrates or have been picked up by the mobile elements (data not shown). As the method relies in part on existing genome annotation (e.g. gene sets in different vertebrates), the analysis might have to be extended to the whole genome to allow a more comprehensive overview of RETRA genes.
These limitations should not hamper the detection of both RETRA genes and the subset with vertebrate functionality as we not only provide the set of marker domains but also a list of known or identified RETRA genes with functionality in human, mouse and rat. Thus, even without synteny identification, these true mammalian genes together with their orthologs in fugu can be used to rescue genes with similarity to RETRA genes in other vertebrates. When applied to the four vertebrates, over 1000 RETRA genes can be flagged (Table 1), the vast majority of which should probably not be included in the gene sets. In human, 33–251 RETRA genes are found depending on the gene prediction method used (Tables 1 and 2), despite manual curation efforts. We have identified 35 true human genes (none of the methods predicted the complete set of genes in mammals). The number of RETRA genes increases significantly in more automatically annotated organisms (e.g. in the mouse gene sets 116-807 RETRA proteins are found). For other vertebrate genomes to be sequenced, a reproducible automatic pipeline should be applied to separate RETRA genes from host genes and we have taken the first step in this direction. In summary, the proposed use of HMM models of characteristic RETRA protein domains for identification of RETRA genes has greater sensitivity than DNA similarity-based methods. The concept of using conserved synteny in species as divergent as human and rodents to identify RETRA genes with host-specific functionality has been proven to be able to detect the vast majority of such genes, despite the limitations discussed above. The fact that the majority of these genes are also present in fugu genome indicates that the compiled list of such genes is applicable with high confidence to other more distant vertebrate genomes. The automatic procedure proposed here consists of two steps: (i) The identification of RETRA genes using the set of RETRA marker domains we have derived here (this can be done using standard HMM searches (Eddy, http://hmmer.wustl.edu/) and (ii) Detection of genes with vertebrate functions in those sets using the ‘rescue list’ derived here by manually refined synteny analysis.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors are grateful to all members of the P. Bork group and T. Gibson for the useful discussions. M.C. is a recipient of a FEBS long-term Fellowship. E.D.H. is a recipient of an E-STAR fellowship funded by the EC FP6 Marie Curie Host Fellowship for Early Stage Research Training under contract number MEST-CT-2004-504640. Funding to pay the Open Access publication charges for this article was provided by EMBL.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 3.Gibbs R.A., Weinstock G.M., Metzker M.L., Muzny D.M., Sodergren E.J., Scherer S., Scott G., Steffen D., Worley K.C., Burch P.E., et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 4.Curwen V., Eyras E., Andrews T.D., Clarke L., Mongin E., Searle S.M., Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nekrutenko A., Li W.H. Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 2001;17:619–621. doi: 10.1016/s0168-9525(01)02445-3. [DOI] [PubMed] [Google Scholar]
- 6.Li W.H., Gu Z., Wang H., Nekrutenko A. Evolutionary analyses of the human genome. Nature. 2001;409:847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- 7.Smit A.F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 8.Jurka J., Kapitonov V.V. Sectorial mutagenesis by transposable elements. Genetica. 1999;107:239–248. [PubMed] [Google Scholar]
- 9.Kipling D., Warburton P.E. Centromeres, CENP-B and Tigger too. Trends Genet. 1997;13:141–145. doi: 10.1016/s0168-9525(97)01098-6. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura T.M., Cech T.R. Reversing time: origin of telomerase. Cell. 1998;92:587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 11.Harrington L., Zhou W., McPhail T., Oulton R., Yeung D.S., Mar V., Bass M.B., Robinson M.O. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 1997;11:3109–3115. doi: 10.1101/gad.11.23.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mighell A.J., Smith N.R., Robinson P.A., Markham A.F. Vertebrate pseudogenes. FEBS Lett. 2000;468:109–114. doi: 10.1016/s0014-5793(00)01199-6. [DOI] [PubMed] [Google Scholar]
- 13.Okazaki Y., Furuno M., Kasukawa T., Adachi J., Bono H., Kondo S., Nikaido I., Osato N., Saito R., Suzuki H., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 14.Aparicio S., Chapman J., Stupka E., Putnam N., Chia J.M., Dehal P., Christoffels A., Rash S., Hoon S., Smit A., et al. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 15.Hubbard T., Barker D., Birney E., Cameron G., Chen Y., Clark L., Cox T., Cuff J., Curwen V., Down T., et al. The Ensembl genome database project. Nucleic Acids Res. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 17.Korf I., Flicek P., Duan D., Brent M.R. Integrating genomic homology into gene structure prediction. Bioinformatics. 2001;17:S140–148. doi: 10.1093/bioinformatics/17.suppl_1.s140. [DOI] [PubMed] [Google Scholar]
- 18.Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zdobnov E.M., von Mering C., Letunic I., Torrents D., Suyama M., Copley R.R., Christophides G.K., Thomasova D., Holt R.A., Subramanian G.M., et al. Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster. Science. 2002;298:149–159. doi: 10.1126/science.1077061. [DOI] [PubMed] [Google Scholar]
- 20.Boguski M.S., Lowe T.M., Tolstoshev C.M. dbEST—database for “expressed sequence tags”. Nature Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 21.Mulder N.J., Apweiler R., Attwood T.K., Bairoch A., Barrell D., Bateman A., Binns D., Biswas M., Bradley P., Bork P., et al. The InterPro Database, 2003 brings increased coverage and new features. Nucleic Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boeckmann B., Bairoch A., Apweiler R., Blatter M.C., Estreicher A., Gasteiger E., Martin M.J., Michoud K., O'Donovan C., Phan I., et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonnhammer E.L., Eddy S.R., Birney E., Bateman A., Durbin R. Pfam: multiple sequence alignments and HMM-profiles of protein domains. Nucleic Acids Res. 1998;26:320–322. doi: 10.1093/nar/26.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lynch C., Tristem M. A co-opted gypsy-type LTR-retrotransposon is conserved in the genomes of humans, sheep, mice, and rats. Curr. Biol. 2003;13:1518–1523. doi: 10.1016/s0960-9822(03)00618-3. [DOI] [PubMed] [Google Scholar]
- 26.Voltz R., Gultekin S.H., Rosenfeld M.R., Gerstner E., Eichen J., Posner J.B., Dalmau J. A serologic marker of paraneoplastic limbic and brain-stem encephalitis in patients with testicular cancer. N. Engl. J. Med. 1999;340:1788–1795. doi: 10.1056/NEJM199906103402303. [DOI] [PubMed] [Google Scholar]
- 27.Steplewski A., Krynska B., Tretiakova A., Haas S., Khalili K., Amini S. MyEF-3, a developmentally controlled brain-derived nuclear protein which specifically interacts with myelin basic protein proximal regulatory sequences. Biochem. Biophys. Res. Commun. 1998;243:295–301. doi: 10.1006/bbrc.1997.7821. [DOI] [PubMed] [Google Scholar]
- 28.Wang M.Q., Kim W., Gao G., Torrey T.A., Morse H.C., 3rd, De Camilli P., Goff S.P. Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production. J. Biol. 2003;3:4. doi: 10.1186/1475-4924-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masumoto H., Masukata H., Muro Y., Nozaki N., Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W., Seto J., Donovan G., Toth M. Jerky, a protein deficient in a mouse epilepsy model, is associated with translationally inactive mRNA in neurons. J. Neurosci. 2002;22:176–182. doi: 10.1523/JNEUROSCI.22-01-00176.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson H.M. Evolution of DNA Transposons in Eukaryotes. Washington,D.C: ASM Press; 2002. [Google Scholar]
- 32.Mi S., Lee X., Li X., Veldman G.M., Finnerty H., Racie L., LaVallie E., Tang X.Y., Edouard P., Howes S., et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 33.Blaise S., de Parseval N., Benit L., Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl Acad. Sci. USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohshima N., Takahashi M., Hirose F. Identification of a human homologue of the DREF transcription factor with a potential role in regulation of the histone H1 gene. J. Biol. Chem. 2003;278:22928–22938. doi: 10.1074/jbc.M303109200. [DOI] [PubMed] [Google Scholar]
- 35.Mallet F., Bouton O., Prudhomme S., Cheynet V., Oriol G., Bonnaud B., Lucotte G., Duret L., Mandrand B. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl Acad. Sci. USA. 2004;101:1731–1736. doi: 10.1073/pnas.0305763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura T.M., Morin G.B., Chapman K.B., Weinrich S.L., Andrews W.H., Lingner J., Harley C.B., Cech T.R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 37.Meyerson M., Counter C.M., Eaton E.N., Ellisen L.W., Steiner P., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 38.De Pereda J.M., Waas W.F., Jan Y., Ruoslahti E., Schimmel P., Pascual J. Crystal structure of a human peptidyl-tRNA hydrolase reveals a new fold and suggests basis for a bifunctional activity. J. Biol. Chem. 2004;279:8111–8115. doi: 10.1074/jbc.M311449200. [DOI] [PubMed] [Google Scholar]
- 39.Llorens C., Marin I. A mammalian gene evolved from the integrase domain of an LTR retrotransposon. Mol. Biol. Evol. 2001;18:1597–1600. doi: 10.1093/oxfordjournals.molbev.a003947. [DOI] [PubMed] [Google Scholar]
- 40.Gale M., Jr, Blakely C.M., Hopkins D.A., Melville M.W., Wambach M., Romano P.R., Katze M.G. Regulation of interferon-induced protein kinase PKR: modulation of P58IPK inhibitory function by a novel protein, P52rIPK. Mol. Cell Biol. 1998;18:859–871. doi: 10.1128/mcb.18.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McIntosh E.M., Ager D.D., Gadsden M.H., Haynes R.H. Human dUTP pyrophosphatase: cDNA sequence and potential biological importance of the enzyme. Proc. Natl Acad. Sci. USA. 1992;89:8020–8024. doi: 10.1073/pnas.89.17.8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ono R., Kobayashi S., Wagatsuma H., Aisaka K., Kohda T., Kaneko-Ishino T., Ishino F. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics. 2001;73:232–237. doi: 10.1006/geno.2001.6494. [DOI] [PubMed] [Google Scholar]
- 43.Cerritelli S.M., Crouch R.J. Cloning, expression, and mapping of ribonucleases H of human and mouse related to bacterial RNase HI. Genomics. 1998;53:300–307. doi: 10.1006/geno.1998.5497. [DOI] [PubMed] [Google Scholar]
- 44.Serra-Pages C., Medley Q.G., Tang M., Hart A., Streuli M. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J. Biol. Chem. 1998;273:15611–15620. doi: 10.1074/jbc.273.25.15611. [DOI] [PubMed] [Google Scholar]
- 45.Robertson H.M., Zumpano K.L. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene. 1997;205:203–217. doi: 10.1016/s0378-1119(97)00472-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.