Abstract
Mulberry (Morus spp.), being an economically important tree, is cultivated in China, India, Thailand, Brazil, Uzbekistan and other Countries across the globe, for its leaves to feed monophagous mulberry silkworm (Bombyx mori). The sustainability of silk industry is directly correlated with the production and continuous supply of high-quality mulberry leaves. In India, it is cultivated on large scale in tropical, sub-tropical and temperate regions under irrigated conditions for silkworm rearing. Drought, low temperature, high salinity and alkalinity, being experienced in widespread areas, are the major abiotic stresses, causing reduction in its potential foliage yield and quality. Further, climate change effects may worsen the productivity of mulberry in near future, not only in India but also across the globe. Although traditional breeding methods contributed immensely towards the development of abiotic stress-tolerant mulberry varieties, still there is lot of scope for implementation of modern genomic and molecular biology tools for accelerating mulberry genetic improvement programmes. This review discusses omics approaches, molecular breeding, plant tissue culture and genetic engineering techniques exploited for mulberry genetic improvement for abiotic stress tolerance. However, high-throughput biotechnological tools such as RNA interference, virus-induced gene silencing, epigenomics and genome editing tools need to be utilized in mulberry to accelerate the progress of functional genomics. The application of genomic tools such as genetic engineering, marker-assisted selection and genomic selection in breeding programmes can hasten the development of climate resilient and productive mulberry varieties leading to the vertical and horizontal expansion for quality silk production.
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0829-z) contains supplementary material, which is available to authorized users.
Keywords: Transgenic mulberry, Marker-assisted selection, Functional genomics, Climate resilience, Genomic tool
Introduction
Mulberry (Morus spp.; Family: Moraceae) is a fast-growing, cross-pollinated, perennial, dioecious and hardy tree (2n = 28–308). Globally, it occurs in warm and moist climates between latitudes of 50°N and 10°S which include Southeastern Asia, Europe, North and South America. Although, mulberry leaves are the sole source of food for the mulberry silkworm (Bombyx mori), its leaf is also used as fodder for life stock. Its fruit, due to ample nutritive values, is used for human consummation. In addition, mulberry tree produces valuable products like timber and pharmaceutically important chemicals which make its widespread occurrence across the World. As mulberry tree is highly heterozygous and characterized by long juvenile period, it is propagated by vegetative means such as stem cuttings and bud grafting for commercial cultivation. Globally, 68 mulberry species have been reported and the majority of the species occur in Asia. China and Japan house 24 and 19 species, respectively, while four mulberry species (M. indica, M. alba, M. laevigata, and M. serrata) are reported from India. Mulberry occurs in nature with greater variation at ploidy level and is cultivated under irrigation system in India. Approximately, 1200 mulberry accessions including indigenous and exotic accessions are being conserved at Central Sericultural Germplasm Resources Centre, India. China stands first in mulberry raw silk production. India is the second largest producer of mulberry raw silk and stands first for raw silk consumption. Mulberry is cultivated in 2.20 lakhs ha in India (Central Silk Board 2015) and the sustainability of Indian silk industry is directly correlated with the production and supply of quality mulberry leaves. However, there is burgeoning demand for quality silk at domestic and international markets, which cannot be met by horizontal expansion of mulberry cultivation in traditional agricultural land, due to competition with other food and cash crops. Hence, it is imperative to utilize marginal, problematic soils and non-traditional areas affected by various abiotic stresses such as alkalinity, salinity and moisture deficit for mulberry cultivation. Abiotic stress tolerance in plant system is a polygenic trait and involves interaction among several genes through signal transduction pathways (Sarkar et al. 2014; Liu et al. 2015). However, drought or soil moisture deficit stress is the detrimental abiotic factor and causes fall of mulberry foliage production immensely. It is projected that about 20% of land surface is under drought across the globe at a given point of time (Sarkar et al. 2016). To combat drought stress, a range of drought adaptation traits such as efficient water conservation, wider and deeper root system, improved photosynthetic yield, water use efficiency (WUE) and cellular tolerance need to be introgressed in mulberry. Other abiotic stresses such as salinity, alkalinity and frost are also detrimental to productivity in various parts of India. In India, salinity-affected area is estimated at 7.61 million ha (Sarkar et al. 2014). The traditional breeding practices, relying on morphological and physiological based phenotyping, have been successfully utilized in developing drought, alkalinity, salinity and frost-tolerant productive mulberry varieties (Susheelamma et al. 1992; Mogili et al. 2008; Vijayan et al. 2009a; Doss et al. 2012). Wild mulberry species such as M. serrata, M. laveigata and germplasm resources have vigorous growth pattern, wider adaptability to harsh environments and palatability to silkworm (Saeed et al. 2016a). However, the abiotic stress adaptive traits of wild species are yet to be introgressed into cultivated mulberry variety for commercial exploitation. In this background, the present review discusses omics approach, molecular breeding, plant tissue culture and genetic engineering techniques exploited for mulberry genetic improvement, especially for abiotic stress tolerance.
Marker-assisted breeding in accelerating mulberry genetic improvement programmes
Molecular markers found its applications in studies on genetic diversity, molecular characterization of germplasm and varieties, development of linkage and QTL map, association mapping, parental selection schemes and marker-assisted selection (MAS) of mulberry progenies (Khurana and Checker 2011; Vijayan et al. 2014). RAPD was the first molecular marker used in mulberry (M. alba) for molecular systematics (Xiang et al. 1995). Subsequently, this marker has been used in various studies for analyzing the extent of genetic diversity and selection of parental lines for trait improvement in mulberry (Feng et al. 1996; Lou et al. 1998; Bhattacharya and Ranade 2001; Naik et al. 2002, 2013, 2015; Zhao and Pan 2002; Awasthi et al. 2004; Mishra et al. 2013; Banerjee et al. 2016). Dominant markers such as ISSR (Vijayan et al. 2004a, 2006; Zhao et al. 2006, 2007a; Kar et al. 2008), DAMD (Bhattacharya and Ranade 2001), AFLP (Sharma et al. 2000; Wang and Yu 2001; Kafkas et al. 2008) were also used for phylogenetic analysis, germplasm characterization, DNA fingerprinting, analysis of genetic fidelity of in vitro regenerated plants and determining genetic variations across the mulberry genotypes (Vijayan et al. 2014; Saha et al. 2016). Important co-dominant markers such as genic-SSR (Krishnan et al. 2013; Arora et al. 2014; Thumilan et al. 2016), genomic-SSR (Aggarwal et al. 2004; Zhao et al. 2005, 2007a; Pinto et al. 2012; Thumilan et al. 2013; Krishnan et al. 2013), SRAP (Zhao et al. 2009) have been used not only to analyze the extent of genetic diversity in cultivated and wild mulberry spp., but also to identify its route of introduction and spreading in India. Very often, combination of multiple marker types such as RAPD and ISSR, or dominant and co-dominant markers have been used in germplasm characterization for better coverage of mulberry genome as compared to using single marker type (Vijayan et al. 2004a; Zhao et al. 2007a) (Table 1). With the advancement of molecular biology and genomic tools, the genomic- and genic-SSR (microsatellite) markers have been isolated from genomic clones and EST sequences of mulberry and these markers also showed transferability to other related species belonging to the family Moraceae, such as fig, ficus and jackfruit (Thumilan et al. 2013, 2016). The dominant (RAPD and ISSR) and co-dominant (SSR) markers have been exploited for the development of linkage maps, QTL maps and marker trait association analysis in mulberry (Mishra and Dandin 2010; Naik et al. 2014).
Table 1.
List of molecular markers used for marker-assisted breeding and clonal propagation in Morus spp. during last 10 years
| Sl. no. | Marker type | Purpose | Country | References |
|---|---|---|---|---|
| 1. | RAPD | Genetic variability and phylogenetic relationship among 15 white mulberry genotypes | Turkey | Orhan et al. (2007) |
| 2. | ISSR | Genetic diversity among 66 local varieties belonging to 8 populations | China | Zhao et al. (2007b) |
| 3. | ISSR and SSR | Genetic diversity among 27 mulberry accessions including 19 cultivated and 8 wild accessions | China | Weiguo et al. (2007) |
| 4. | ISSR | Phylogenetic relationship among 18 germplasm collection and association with biochemical parameters | India | Kar et al. (2008) |
| 5. | AFLP | Genetic variability among 43 accessions belonging to M. alba, M. nigra and M. rubra | Turkey | Kafkas et al. (2008) |
| 6. | ISSR | Genetic diversity among ecotypes | China | Zhao et al. (2008) |
| 7. | RAPD | Molecular characterization of inter and intra-specific hybrids | India | Tikader and Dandin (2008) |
| 8. | AFLP | DNA fingerprinting of ten cultivars | China | Huang et al. (2009) |
| 9. | ISSR and RAPD | Association with sprouting and sex expression traits | India | Vijayan et al. (2009b) |
| 10. | SRAP | Genetic diversity among 23 mulberry germplasm accessions from China | Republic of Korea, China | Zhao et al. (2009) |
| 11. | RAPD | Phylogenetic relationship among 47 genotypes | Turkey | Ozrenk et al. (2010) |
| 12. | ISSR | Genetic diversity among 73 local mulberry varieties for development of core collection | China | Lin et al. (2011) |
| 13. | RAPD | Genetic variability among control and ethyl methane sulphonate (EMS) treated clones of mulberry genotype RFS135 | India | Anil Kumar et al. (2012) |
| 14. | RAPD and ISSR | Genetic diversity and phylogenetic relatedness among 20 mulberry varieties | India | Chikkaswamy and Prasad (2012) |
| 15. | RAPD and ISSR | Genetic diversity among 20 mulberry varieties | India | Chikkaswamy et al. (2012) |
| 16. | RAPD and ISSR | Genetic diversity among 21 mulberry genotypes collected from 4 geographic regions of Turkey | Turkey | Ipek et al. (2012) |
| 17. | RAPD | Genetic diversity among 36 genotypes collected from South India | India | Naik et al. (2013) |
| 18. | RAPD, ISSR and SSR | Standardization of novel and efficient DNA extraction protocol | India | Anuradha et al. (2013) |
| 19. | SSR | Genetic diversity among ten accessions belonging to M. alba and M. indica | Kenya | Wangari et al. (2013) |
| 20. | SSR | Phylogenetic relatedness among 17 mulberry genotypes | India | Wani et al. (2013) |
| 21. | RAPD and ISSR | Genetic stability of cryo-preserved dormant buds of different Morus species belonging to indigenous and exotic collection | India | Choudhary et al. (2013) |
| 22. | SSR | Genetic diversity among 36 mulberry genotypes (‘breeders’ collections) | India | Krishnan et al. (2013) |
| 23. | RAPD | Genetic diversity among nine mulberry genotypes with contrasting traits for water use efficiency (WUE) and root | India | Mishra et al. (2013) |
| 24. | RAPD, ISSR and SSR | Genetic diversity among 850 germplasm accessions collected from 23 countries for development of core collection of diverse accessions | India | Guruprasad et al. (2014) |
| 25. | SSR | Genetic diversity among 72 germplasm accessions belonging to two wild species such as M. laevigata and M. serrata collected from different eco-geographic regions of India | India | Naik et al. (2015) |
| 26. | RAPD and ISSR | Genetic fidelity of in vitro regenerated mulberry plants (cv. S1) | India | Saha et al. (2016) |
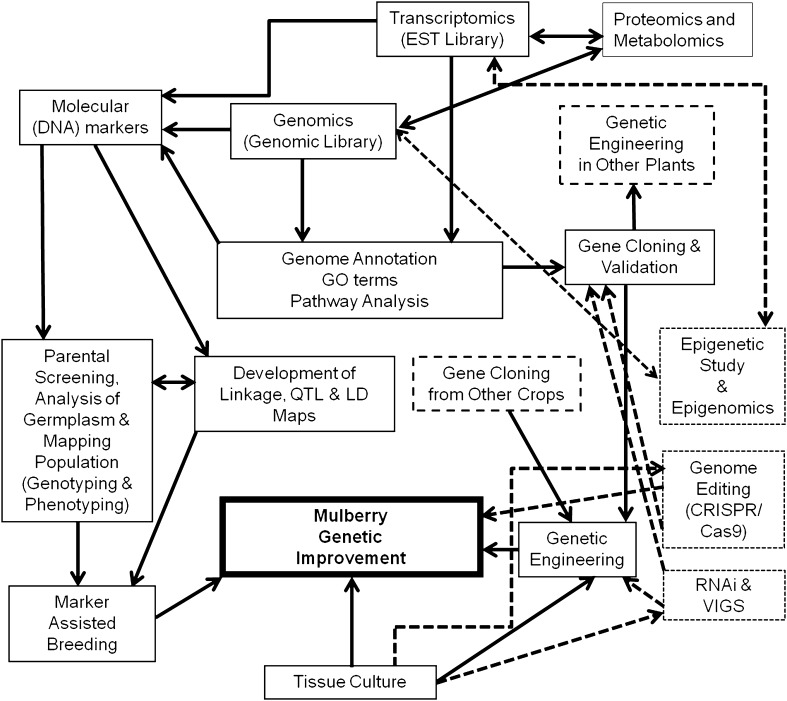
A two-way pseudo-testcross mapping strategy has been adapted in many tree species for construction of genetic linkage maps. This strategy is applicable to the highly heterozygous plant species in which DNA markers segregate in 1:1 ratio in its F1 mapping population, because many alleles are present in only one copy in one parent (Thumilan et al. 2016). The first genetic linkage map was developed in mulberry adapting two-way pseudo-testcross mapping strategy, with 50 F1 full-sib progenies (mapping population) of S36 × V1 cross using RAPD, ISSR and genomic-SSR markers (Venkateswarlu et al. 2006). Attempts were made to develop QTL maps that are specific for important agronomic traits such as WUE, root traits and yield contributing traits in mulberry, using dominant markers such as RAPD and ISSR; few markers showed linkage to the QTLs governing the trait of interest (Naik et al. 2014; Mishra 2014). However, the genetic linkage map needs further saturation for identification of tightly linked markers to the QTLs. Furthermore, introgression lines with improved WUE and root traits have been developed from a cross of Dudia White × MS3 and these lines could be used for validating QTLs associated with drought adaptive traits and as a pre-breeding resource (Mishra 2014). Recently, 134 genomic- and drought adaptive trait-specific genic-SSR markers have been used to develop genetic linkage map using pseudo-testcross mapping strategy in F1 bi-parental mapping population (full-sib, 150 progenies) of Dudia White × UP105 cross; however, linkage between the markers and trait of interest is yet to be determined (Thumilan et al. 2016). The details of molecular maps developed in mulberry are presented in Table 2. Although molecular markers linked to QTLs governing phenotypic traits have been mapped using bi-parental mapping populations, identification of tightly linked markers for the specific traits is still scanty (Mishra 2014; Naik et al. 2014). The linked markers need to be validated in the other unrelated mapping populations to detect accurate association with the particular phenotypic trait. The QTL map further needs to be saturated with most abundant co-dominant marker resources (SNPs) for identification of tightly linked markers to the QTLs responsible for a phenotypic trait variation, and the tightly linked co-dominant markers could be used in MAS in mulberry breeding programmes (Naik et al. 2014). In addition to the saturated linkage map, physical map also needs to be developed for understanding the architecture of mulberry genome, which can open up the avenue for map-based cloning of candidate genes linked to a phenotypic trait of interest. Marker-trait association analysis is an important genomic tool in tree species, which helps in identifying candidate genes responsible for phenotypic trait variations in a natural population. Genome-wide association study (GWAS) or gene scan is an important aspect of association mapping, which surveys genetic variation in whole-genome level to locate candidate genes or narrow genomic regions (QTLs) with significant statistical association with a particular phenotypic trait. SNPs are the most abundant polymorphism present in the genome of mulberry, and their identification and validation help in the analysis of the traits related to abiotic stress tolerance through GWAS and QTL mapping strategies (Vijayan 2010; Khurana and Checker 2011). Genotyping-by-sequencing (GBS) approach is required to be integrated with mulberry genomic research to generate large number of SNPs to be used in QTL and linkage disequilibrium (LD) mapping. Combined approaches of linkage and association mapping can accelerate the progress of QTLs identification and MAS in mulberry genetic improvement programmes (Fig. 1).
Table 2.
Molecular maps developed in mulberry
| Sl. no. | Molecular map | Pedigree of mapping population | Markers | Hereditary nature of marker | Agronomic trait targeted | References |
|---|---|---|---|---|---|---|
| 1. | Genetic linkage map | S36 × V1 | RAPD, ISSR, and SSR | Dominant, co-dominant | No | Venkateswarlu et al. (2006) |
| 2. | QTL map | V1 × Mysore Local | RAPD and ISSR | Dominant | Yield contributing traits | Naik et al. (2014) |
| 3. | QTL map | Himachal Local × MS3 | RAPD and ISSR | Dominant | Water use efficiency | Mishra (2014) |
| 4. | QTL map | Dudia White × UP | RAPD and ISSR | Dominant | Root traits | Mishra (2014) |
| 5. | Genetic linkage map | Dudia White × UP105 | SSR | Co-dominant | No | Thumilan et al. (2016) |
Fig. 1.
Integrated approaches of plant tissue culture, genomic and molecular biology tools for improving abiotic stress tolerance in mulberry
Genetic engineering in mulberry for abiotic stress tolerance
Plant tissue culture technique has numerous applications (Vijayan et al. 2011; Khurana and Checker 2011) in micropropagation (Ohyama and Oka 1976; Kim et al. 1985; Hossain et al. 1992; Bhau and Wakhlu 2003; Chattopadhyay et al. 2011; Zaki et al. 2011), organogenesis through callus phase (Narayan et al. 1989); screening of genotypes for salinity, alkalinity and drought stress tolerance under in vitro condition (Hossain et al. 1991; Tewary et al. 2000; Vijayan et al. 2003, 2004b; Ahmad et al. 2007). Plant tissue culture techniques also helped in the development of triploid mulberry from endosperm culture (Thomas et al. 2000), development of gynogenic plant from ovary (Lakshmi Sita and Ravindran 1991; Thomas et al. 1999), production of somaclonal variants for stress tolerance (cv. SV1) and improved yield contributing traits (Narayan et al. 1989; Susheelamma et al. 1996). Although initial progress has been achieved in androgenesis through anther culture (Shoukang et al. 1987; Jain et al. 1996), but significant success in developing haploid plants has not yet achieved. Similarly, experiments on protoplast culture and somatic hybridization were successful (Ohnishi and Tanabe 1989; Ohnishi and Kiyama 1987; Umate et al. 2005; Umate 2010), but practical realization of these techniques is yet to be achieved.
Mulberry is a highly heterozygous, out-breeding, tree species; its cellular totipotency or in vitro regeneration potential is highly genotype dependent (Raghunath et al. 2013). However, irrigated mulberry genotypes such as Sujanpur5, S13, S799, K2, V1, AR12, DD and S36 were responsive to regeneration and whole plantlets were developed using various type of explants (Vijayan et al. 2000; Kapur et al. 2001; Raghunath et al. 2008, 2013; Rao et al. 2010a; Chitra et al. 2014) (Table 3). The success of genetic transformation is very much dependent on regeneration potential, choice of explant (Sarkar et al. 2016) and Agrobacterium strains to be used in the experiments (Bhatnagar and Khurana 2003). Plant regeneration and genetic transformation have been attempted in mulberry since long time ago (Kim et al. 1985; Machii 1990; Machii et al. 1996; Kapur et al. 2001). However, genetic transformation of mulberry with heterologous genes through various techniques such as particle bombardment, Agrobacterium rhizogenes-mediated, electroporation and in planta has been reported (Sugimura et al. 1999; Oka and Tewary 2000; Machii 1990; Machii et al. 1996; Bhatnagar et al. 2002). Most of these experiments not only failed to regenerate plantlets, but also did not show transgene integration in mulberry genome (Bhatnagar and Khurana 2003). Transgenic mulberry expressing glycinin gene AlaBlb was developed through tissue culture (Jianzhong et al. 2001). In planta, genetic transformation has been successfully attempted in mulberry genetic improvement programme (Ping et al. 2003). Subsequently, an efficient and reproducible protocol with 6% transformation frequency has been used to develop transgenic mulberry cv. K2 through Agrobacterium tumefaciens-mediated genetic transformation. Various explants such as hypocotyl, cotyledon, leaf and leaf callus have been utilized for the transformation experiment, but leaf callus was found to be the choice of explants for regeneration of transgenic plants (Bhatnagar and Khurana 2003). Recently, various abiotic stresses-associated functional genes have been introduced in the mulberry cv. K2 with an appreciable transformation efficiency (20–60%) using cotyledon and hypocotyl explants (Table 4). Heterologous single action genes such as b-carotene hydroxylase1 (bch1), Hva1 (a group 3 late embryogenesis abundant protein) and Osmotin have been introduced in diploid mulberry cv. K2 and the transgenes expression were regulated by constitutive (Actin1, CaMV 35S) and stress inducible (rd29A) promoters (Lal et al. 2008; Das et al. 2011; Checker et al. 2012a; Saeed et al. 2015). Southern blot assay showed one to two copies (s) of heterologous Hva1 gene insertion in mulberry genome and its expression was ascertained by northern blot analysis (Lal et al. 2008), Western blotting and qPCR (Checker et al. 2012a). The Hva1 expression also regulated downstream genes associated with stress tolerance such as chaperone Mi dna J and Mi 2-cysper-oxidin under stress condition. Barley Hva1 expression in transgenic mulberry showed tolerance to drought, salinity and cold stress due to enhanced cellular tolerance, photosynthetic yield, efficient water conservation under stress condition (Lal et al. 2008; Checker et al. 2012a). Bioassay with silkworm indicated normal growth and development of larvae and produced cocoons on par with non-transgenic mulberry after completion of silkworm rearing (Lal et al. 2008). In another study, the expression of tobacco Osmotin gene, driven by CaMV35S promoter, conferred transgenic mulberry with tolerance to both abiotic stress and also biotic challenges (mostly fungal pathogens). Furthermore, when the expression of Osmotin gene was driven by rd29A promoter, transgenic mulberry showed drought and salinity tolerance due to improved membrane stability, osmolytes synthesis and photosynthetic yield (Das et al. 2011). Heterologous expression of SHN1 (wax inducer1/SHINE1) in transgenic mulberry showed dark green shiny appearance of leaf with increased surface wax content. The SHN1transgenic mulberry (cv. M5) showed better leaf moisture retention capacity as compared to wild type (Sajeevan et al. 2017). Overexpression of bch1 gene from M. indica in mulberry cv. K2 showed higher levels of carotenoids and improved oxidative stress (such as high light, heat and UV irradiation) tolerance as compared to non-transgenic plant under non-stress and stress conditions (Saeed et al. 2015) which has paved the way to cisgenic approach for enhancing climate resilience in mulberry (Fig. 1).
Table 3.
List of mulberry (Morus spp.) plantlets regenerated from explants through organogenesis since last decade
| Sl. no. | Explants | Genotype/cultivar | Species | Country | Type of organogenesis | References |
|---|---|---|---|---|---|---|
| 1. | Leaf | S1 | M. alba | India | Direct organogenesis | Vijayan et al. (2000) |
| 2. | Leaf, petiole, intermodal segment | Chinese White, Kokuso27 | M. alba | India | Through callus phase | Bhau and Wakhlu (2001) |
| 3. | Leaf, hypocotyl, cotyledon, petiole, intermodal segment | K2, DD | M. indica | India | Direct organogenesis | Bhatnagar et al. (2001) |
| 4. | Nodal explant | M5, S36, S13, China White |
M. indica
M. alba |
India | Direct organogenesis | Chitra and Padmaja (2002) |
| 5. | Nodal explant | – | M. latifolia | Taiwan | Direct organogenesis | Lu (2002) |
| 6. | Nodal explant, shoot tips | China White, Kokuso27, Ichinose Goshoerami, Rokokuyaso |
M alba
M. multicaulis |
India | Direct organogenesis | Bhau and Wakhlu (2003) |
| 7. | Shoot tip, nodal explant | – | M. alba | India | Direct organogenesis | Anis et al. (2003) |
| 8. | Cotyledon | S36, K2, S1 |
M. indica
M. alba |
India | Direct organogenesis | Thomas (2003) |
| 9. | Leaf | M5, S13, S36 | M. indica | India | Direct organogenesis | Chitra and Padmaja (2005) |
| 10. | Leaf-derived protoplast | S36 | M. indica | India | Through callus phase | Umate et al. (2005) |
| 11. | Apical bud | S54 | M. indica | India | Direct organogenesis | Kavyashree (2007) |
| 12. | Nodal segment | – | M. alba | India | Direct organogenesis | Balakrishnan et al. (2009) |
| 13. | Nodal segment | S36, V1 | M. indica | India | Through callus phase | Rao et al. (2010b) |
| 14. | Nodal segment | – | M. nigra | India | Direct organogenesis | Zaki et al. (2011) |
| 15. | Nodal segment | S1 | M. alba | India | Direct organogenesis | Chattopadhyay et al. (2011) |
| 16. | Nodal segment | V1 | M. indica | India | Direct organogenesis | Sajeevan et al. (2011) |
| 17. | Nodal segment | – | M. macroura | Pakistan | Direct organogenesis | Akram and Aftab (2012) |
| 18. | Nodal segment | S1635 | M. indica | India | Direct organogenesis | Lalitha et al. (2013) |
| 19. | Leaf | V1 | M. indica | India | Direct organogenesis | Raghunath et al. (2013) |
| 20. | Shoot meristem | S36 | M. indica | India | Direct organogenesis | Chitra et al. (2014) |
| 21. | Nodal explant | S1 | M. alba | India | Direct organogenesis | Saha et al. (2016) |
Table 4.
Transgenic mulberry (M. indica cv. K2 and M5) developed for stress tolerance
| Sl. no. | Transgene | Source of transgene | Explant | Performance of transgenic plants | Promoter used for transgene expression | Selectable marker gene | Mode of transformation | A. tumefaciens strain | Plant transformation vector | References |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | Hva1 | Barley | Hypocotyl and cotyledon | Drought and salinity tolerance | Act1 | nptII | Agrobacterium tumefaciens-mediated | LBA4404 | pCAMBIA2301 | Lal et al. (2008) |
| 2. | Osmotin | Tobacco | Hypocotyl and cotyledon | Drought and salinity tolerance; resistance to fungal pathogens (Fusarium pallidoroseum, Colletotrichum gloeosporioides and Colletotrichum dematium) | CaMV35S, rd29A | nptII | Agrobacterium tumefaciens-mediated | Agl1 | pBI121 | Das et al. (2011) |
| 3. | Hva1 | Barley | Hypocotyl and cotyledon | Drought, salinity and cold stress tolerance | rd29A | nptII | Agrobacterium tumefaciens-mediated | Agl1 | pBI121 | Checker et al. (2012a) |
| 4. | bch1 | Mulberry | Hypocotyl and cotyledon | UV, high temperature and high irradiance stress tolerance | CaMV 35S | nptII | Agrobacterium tumefaciens-mediated | – | pCAMBIA2301 | Saeed et al. (2015) |
| 5. | SHN1 | Arabidopsis thaliana | Hypocotyl and cotyledon | Efficient leaf moisture retention capacity | CaMV35S | NptII | Agrobacterium tumefaciens-mediated | EHA105 | pBI121 | Sajeevan et al. (2017) |
Genomics and transcriptomics for identification of novel genes and comprehensive marker resources
High-quality genomic and transcriptomic data generated through next generation sequencing (NGS) platforms provided wealth of information of novel candidate genes and comprehensive molecular markers, thus enhancing the scope for trait-specific genetic improvement in mulberry in terms of productivity, quality and climate resilience. The whole-genome shotgun sequencing of haploid mulberry species (M. notabilis) reported a draft genome of 357 Mb including 128 Mb repetitive sequences with 27,085 high-confidence protein coding loci in tandem with complete gene structure (He et al. 2013). At present, a total of 101,850 nucleotides, 38 whole-genome and transcriptome sequences are available for Morus spp. on the NCBI website (http://www.ncbi.nlm.nih.gov, accessed on April 8, 2017) (Supplementary Table S1).
Transcriptome resources from the cultivated, wild and haploid mulberry species could be used in genes and markers identification. The 21 Gb RNA-seq data from five tissues (root, bark, winter bud, male flower and leaf) of M. notabilis depicted 5833 unique ESTs (expressed sequence tag) for further gene model prediction and validation. Five mulberry miRNAs are found in hemolymph and silk gland of silkworm that indicate the plant–herbivore relationship at molecular level (He et al. 2013). Morus Genome Database (MorusDB), a web-based, open-access database and also a workbench that is assigned to enable access to large-scale genomic sequences, transcriptomic data, predicted genes and unigenes, functional annotation, ESTs, and transposable elements. This database also sheds light on horizontal genes transfer between mulberry and silkworm, orthologous and paralogous genes (Li et al. 2014; Krishnan et al. 2014). A total of 217, 312 and 961 microsatellite (SSR) were mined from whole-genome sequence and EST sequences of Morus notabilis that could find applications in molecular breeding programmes (Krishnan et al. 2014).
Comparative transcriptomics analysis of M. laevigata and M. serrata leaves showed 24,049 simple sequence repeats (SSRs), 1,201,326 single nucleotide polymorphisms (SNPs) and 67,875 insertion–deletions (InDels). In the light of transcriptome data, comparative expression analysis of stress inducible genes such as Heat Shock Factor 4, Protein HVA22-Like, Aquaporin PIP1; 2 in M. laevigata, M. serrata and M. indica showed genotype-specific differential expression under drought, salinity and cold stress (Saeed et al. 2016a). Transcriptome data of drought-tolerant mulberry genotype Dudia White, upon exposure to drought stress, generated 10,169 EST sequences. A total of 206 SSR markers were developed from these ESTs and validated in 25 mulberry accessions (Thumilan et al. 2016). Similarly, SSR markers have also been identified from ESTs of root and leaf transcriptomes of Morus spp. by in silico approach (Gulyani and Khurana 2011; Checker et al. 2012b; Wang et al. 2014a; Dai et al. 2015). In another study, 222 genomic-SSR and 136 genic-SSR have been identified and characterized from genomic clones and ESTs of M. alba genome (Thumilan et al. 2013). At present, 248 genomic-SSR and 490 genic-SSR (EST-SSR) primers have been designed in mulberry and could be used in marker-assisted breeding programmes (Supplementary Table S2).
Transcriptome data and de novo assembly not only provide large volume of information on protein of known function (PKF) and proteins for secondary metabolic process based on GO (gene ontology) annotation and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping, but also generate data on PUFs, proteins of unknown function (Lal et al. 2009; Gulyani and Khurana 2011; Dai et al. 2015). Leaf transcriptome of drought stress M. alba cv. Dudia White showed large number of PUFs (Dhanyalakshmi et al. 2016). A sequence and structure-based computational analysis model that designated as PUFs Annotation Server (PUFAS) was assigned to function to the PUFs (http://caps.ncbs.res.in/pufas/) that can be used to assign function to PUFs from other plant species. The expression analysis of three annotated PUFs revealed their potential role in stress acclimation pathways (Dhanyalakshmi et al. 2016).
Chloroplast genome analysis helps plant taxonomists in identification of easy-to-use species-specific DNA barcode catalog that paves the way for evolutionary studies in mulberry (Ravi et al. 2008). The complete chloroplast genome of M. indica cv. K2 was sequenced, and comparative genome analysis showed its phylogetic relatedness to Cucumis and Lotus spp. (Ravi et al. 2006), while the chloroplast genome of M. notabilis showed similarity to M. indica and M. mongolica (Chen et al. 2015).
Generation of ESTs and molecular characterization of functional genes
Genomic and molecular biology tools generate valuable information and help in identification of potential ESTs, candidate genes and molecular markers based on molecular mechanism of stress tolerance through GO terms and KEGG pathway mapping. This genomic toolbox is considered as an entry point for targeted genetic manipulation by incorporation of heterologous genes and trait-specific introgression of beneficial QTLs conferring stress tolerance in mulberry (Fig. 1). The ESTs developed from cDNA clones and unigenes, derived from transcriptome of Morus spp., were functionally categorized based on GO annotation and KEGG pathway mapping into protein metabolism, transport, secondary metabolite synthesis, response to abiotic stress, energy metabolism, photosynthesis, disease and pest resistance (Lal et al. 2009; He et al. 2013; Wang et al. 2014a; Dai et al. 2015). Suppression subtractive hybridization of drought susceptible mulberry cv. K2 and tolerant cv. AR12 led to the identification of drought-regulated genes and generation of large number of ESTs (Gulyani and Khurana 2011). The sets of genes coding for abiotic stress-related and wax biosynthesis-associated proteins were functionally validated by expression studies through qPCR, Northern Blotting and cDNA macroarray (Gulyani and Khurana 2011; Das et al. 2013; Wang et al. 2014b; Mamrutha et al. 2017).
Genome-wide scanning of gene super-family helps in identification and functional annotation of its components and in establishing evolutionary relationship (Baranwal et al. 2016). Fifty-four genes with conserved WRKY motifs and 197 lectin genes have been identified in M. notabilis and these genes showed their preferential expression in various tissues (Baranwal et al. 2016; Saeed et al. 2016b). The WRKYs in mulberry species showed genotype-specific expression in response to various stress treatments. Molecular cloning and characterization of Helix–Loop–Helix-144 (bHLH144), Remorin (REM), Early Responsive to Dehydration15 (ERD15) and Nitrite Reductase (NiR) genes have assigned their functions in abiotic stress tolerance and in vitro regeneration potential (Checker and Khurana 2013; Wang et al. 2014b; Sajeevan and Nataraja 2016; Saeed and Khurana 2016). Cloning and molecular characterization (gain-of-function) of Mibch1 gene in mulberry lead to the development of abiotic stress-tolerant cisgenic plants (Lal et al. 2009; Saeed et al. 2015). DREB4A gene that codes for a transcription factor has been isolated from M. notabilis genome. Heterologous expression of MnDREB4A in transgenic tobacco conferred tolerance to high temperature, cold, drought and salt stresses (Liu et al. 2015). Hence, novel genes from mulberry could be used for genetic manipulation in other crops for multiple stress tolerance and trait improvement (Fig. 1).
Conclusions and future perspectives
Mulberry is an economically important plant due to its role in the World economy particularly silk industry. Traditional breeding methods are time-consuming and long-drawn programmes, however, contributed significantly for the development of superior mulberry varieties with improved foliage yield, quality and stress tolerance. However, abiotic stresses form major constraints for vertical and horizontal expansion of mulberry cultivation not only in India but also across the globe. Emerging genomic and molecular biology tools such as genetic engineering, functional genomics, and marker-assisted breeding have been attempted to accelerate the breeding process, but their full potential is yet to be materialized. Each abiotic stress tolerance is polygenic in nature and regulated by signal transduction pathway, so molecular mechanism of targeted abiotic stress tolerance needs to be dissected utilizing high-throughput reverse genetic approaches such as RNA interference (RNAi) and virus-induced gene silencing (VIGS) (Fig. 1). Recently, genome editing tool such as clustered regulatory interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9) systems (CRISPER/Cas9) has been used to introduce targeted mutagenesis in the genome and functional characterization of candidate genes in various crops (Rani et al. 2016). This genome editing system can be explored in mulberry to unravel molecular basis of abiotic stress tolerance. Although, researches on metabolome and proteome are in their infancy for mulberry, these omic approaches could be integrated with transcriptomic and genomic studies to identity and characterize novel genes involved in abiotic stress signaling and secondary metabolite biosynthesis pathways (Khurana and Checker 2011). Furthermore, epigenomic study in mulberry could shed light on epigenetic mechanism of gene expression, regulated by DNA methylation, histone post-translational modifications and RNAi pathway (small non-coding RNAs). The epigenetic mechanism involves in various aspects of life in plant including agronomically important traits and responses to abiotic stress (Schmitz and Zhang 2011).
Mulberry germplasm collection including wild spp. shows extensive genotypic and phenotypic variations for important agronomic traits and could be utilized in crop breeding programmes, thus forming valuable genetic resources for trait pyramiding with the help of tightly linked molecular markers. Transgenic mulberry lines for various abiotic stress tolerance traits have been developed and need to be evaluated under real field conditions for further exploitation, which might be fraught with regulatory issues. However, there is a need to simplify the process of biosafety regulations for urgent realization of transgenic crops under field conditions (Mishra et al. 2017). The physical and genetic linkage maps along with key QTLs for important agronomic traits need to be analyzed further. Morus genomic and transcriptomic data, genic- and genomic-SSRs, SNPs and InDel markers will fuel the endeavors of mapping of expression QTLs (eQTLs) and genes to linkage maps and LD maps (Khurana and Checker 2011). The identification of linked markers or candidate genes will facilitate MAS in breeding programmes for genetic improvement. Transcriptome analysis deciphers the role of candidate genes in stress tolerance and could trigger the impetus for developing improved stress-tolerant variety by genetic engineering. Integrated approaches utilizing advanced genomic tools, with more funds in tandem with conventional breeding methods, could be the necessary steps towards mulberry genetic improvement programmes to enhance its productivity and adaptability to climate change for sustaining silk industry.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- RAPD
Random amplified polymorphic DNA
- DAMD
Directed amplification of minisatellite DNA
- AFLP
Amplified fragment length polymorphisms
- ISSR
Inter-simple sequence repeat (ISSR)
- SRAP
Sequence-related amplified polymorphism
- SSR
Simple sequence repeat
- NCBI
National Center for Biotechnology Information
Author contributions
Conceived the idea of the review paper: TS, VS, TM. Wrote the review paper: TS, TM. Edited the manuscript: VS, TS, TM.
Compliance with compliance standards
Conflict of interest
All the authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-017-0829-z) contains supplementary material, which is available to authorized users.
References
- Aggarwal RK, Udaykumar D, Hender PS, Sarkar A, Singh L. Isolation and characterization of six novel microsatellite markers for mulberry (Morus indica) Mol Ecol Notes. 2004;4:477–479. doi: 10.1111/j.1471-8286.2004.00718.x. [DOI] [Google Scholar]
- Ahmad P, Sharma S, Srivastava PS. In vitro selection of NaHCO3 tolerant cultivars of Morus alba (Local and Sujanpuri) in response to morphological and biochemical parameters. Hort Sci (Prague) 2007;34(3):114–122. [Google Scholar]
- Akram M, Aftab F. Efficient micropropagation and rooting of king white mulberry (Morus macroura Miq.) var. laevigata from nodal explants of mature tree. Pak J Bot. 2012;44:285–289. [Google Scholar]
- Anil Kumar HV, Muralidhar TS, Munirajappa R. RAPD analysis of EMS mutagenised mulberry genotype RFS135. Schol J Biotech. 2012;1:1–7. [Google Scholar]
- Anis M, Faisal M, Singh SK. Micropropagation of mulberry (Morus alba L.) through in vitro culture of shoot tip and nodal explants. Plant Tiss Cult. 2003;13:47–51. [Google Scholar]
- Anuradha JH, Vijayan K, Nair CV, Manjula A. A novel and efficient protocol for the isolation of genomic DNA from mulberry (Morus L.) Emir J Food Agr. 2013;25:124–131. doi: 10.9755/ejfa.v25i2.11660. [DOI] [Google Scholar]
- Arora V, Ghosh MK, Gangopadhyay G. SSR markers for assessing the hybrid nature of two high yielding mulberry varieties. Int J Genet Eng Biotechnol. 2014;52:191–196. [Google Scholar]
- Awasthi AK, Nagaraja GM, Naik GV, Kanginakudru S, Thangavelu K, Nagaraju J. Genetic diversity and relationships in mulberry (genus Morus) as revealed by RAPD and ISSR marker assays. BMC Genet. 2004;5:1. doi: 10.1186/1471-2156-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan V, Latha MR, Ravindran KC, Robinson JP. Clonal propagation of Morus alba L. through nodal and axillary bud explants. Bot Res Int. 2009;2:42–49. [Google Scholar]
- Banerjee R, Chattopadhyay S, Saha AK. Genetic diversity and relationship of mulberry genotypes revealed by RAPD and ISSR markers. J Crop Improv. 2016 [Google Scholar]
- Baranwal VK, Negi N, Khurana P. Genome-wide identification and structural, functional and evolutionary analysis of WRKY components of mulberry. Sci Rep. 2016;6:30794. doi: 10.1038/srep30794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar S, Khurana P. Agrobacterium tumefaciens-mediated transformation of Indian mulberry, Morus indica cv. K-2: a time-phased screening strategy. Plant Cell Rep. 2003;21(7):669–675. doi: 10.1007/s00299-003-0572-2. [DOI] [PubMed] [Google Scholar]
- Bhatnagar S, Kapur A, Khurana P. TDZ mediated differentiation in commercially valuable Indian mulberry, Morus indica cultivars K2 and DD. Plant Biotechnol. 2001;18:61–65. doi: 10.5511/plantbiotechnology.18.61. [DOI] [Google Scholar]
- Bhatnagar S, Kapur A, Khurana P. Evaluation of parameters for high efficiency gene transfer via particle bombardment in Indian mulberry. Indian J Exp Biol. 2002;40:1387–1393. [PubMed] [Google Scholar]
- Bhattacharya E, Ranade SA. Molecular distinction among varieties of mulberry using RAPD and DAMD profiles. BMC Plant Biol. 2001;1:3. doi: 10.1186/1471-2229-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhau BS, Wakhlu AA. Effect of genotype, explant type and growth regulators on organogenesis in Morus alba. Plant Cell Tiss Organ Cult. 2001;66:25–29. doi: 10.1023/A:1010617212237. [DOI] [Google Scholar]
- Bhau BS, Wakhlu AK. Rapid micropropagation of five cultivars of mulberry. Biol Plant. 2003;46:349–355. doi: 10.1023/A:1024313832737. [DOI] [Google Scholar]
- Central Silk Board (2015). Annual report 2014–15. Retrieved from http://www.csb.gov.in/assets/Uploads/pdf-files/Graph-English-1415.pdf
- Chattopadhyay S, Doss SG, Halder S, Ali AK, Bajpai AK. Comparative micropropagation efficiency of diploid and triploid mulberry (Morus alba cv. S1) from axillary bud explants. Afr J Biotechnol. 2011;10(79):18153–18159. doi: 10.5897/AJB10.1474. [DOI] [Google Scholar]
- Checker VG, Khurana P. Molecular and functional characterization of mulberry EST encoding remorin (MiREM) involved in abiotic stress. Plant Cell Rep. 2013;32:1729–1741. doi: 10.1007/s00299-013-1483-5. [DOI] [PubMed] [Google Scholar]
- Checker VG, Chibbar AK, Khurana P. Stress-inducible expression of barley hva1 gene in transgenic mulberry displays enhanced tolerance against drought, salinity and cold stress. Transgenic Res. 2012;21(5):939–957. doi: 10.1007/s11248-011-9577-8. [DOI] [PubMed] [Google Scholar]
- Checker VG, Saeed B, Khurana P. Analysis of expressed sequence tags from mulberry (Morus indica) roots and implications for comparative transcriptomics and marker identification. Tree Genet Genomes. 2012;8:1437–1450. doi: 10.1007/s11295-012-0531-6. [DOI] [Google Scholar]
- Chen C, Zhou W, Huang Y, Wang ZZ. The complete chloroplast genome sequence of the mulberry Morus notabilis (Moreae), mitochondrial DNA. J DNA Mapp Seq Anal. 2015 doi: 10.3109/19401736.2015.1053127. [DOI] [PubMed] [Google Scholar]
- Chikkaswamy BK, Prasad MP. Evaluation of genetic diversity and relationships in mulberry varieties using RAPD and ISSR molecular markers. Int J Mol Biol. 2012;3:2–70. [Google Scholar]
- Chikkaswamy BK, Paramanik RC, Debnath A, Sadana MS. Evaluation of genetic diversity in mulberry varieties using molecular markers. Nature Sci. 2012;10:45–60. [Google Scholar]
- Chitra DSV, Padmaja G. Seasonal influence on axillary bud sprouting and micropropagation of elite cultivars of mulberry. Sci Hort. 2002;92:55–68. doi: 10.1016/S0304-4238(01)00279-5. [DOI] [Google Scholar]
- Chitra DSV, Padmaja G. Shoot regeneration via direct organogenesis from in vitro derived leaves of mulberry using thidiazuron and 6-benzylaminopurine. Sci Hort. 2005;106:593–602. doi: 10.1016/j.scienta.2005.05.008. [DOI] [Google Scholar]
- Chitra DSV, Chinthapalli B, Padmaja G. Efficient regeneration system for genetic transformation of mulberry (Morus indica L. Cultivar S-36) using in vitro derived shoot meristems. Am J Plant Sci. 2014;5:1–6. doi: 10.4236/ajps.2014.51001. [DOI] [Google Scholar]
- Choudhary R, Chaudhury R, Malik SK, Kumar S, Pal D. Genetic stability of mulberry germplasm after cryopreservation by two-step freezing technique. Afr J Biotechnol. 2013;12(41):5983–5993. doi: 10.5897/AJB2013.12916. [DOI] [Google Scholar]
- Dai F, Tang C, Wang Z, Luo G, He L, Yao L. De novo assembly, gene annotation, and marker development of mulberry (Morus atropurpurea) transcriptome. Tree Genet Genomes. 2015;11:26. doi: 10.1007/s11295-015-0851-4. [DOI] [Google Scholar]
- Das M, Chauhan H, Chhibbar A, Haq QMR, Khurana P. High-efficiency transformation and selective tolerance against biotic and abiotic stress in mulberry, Morus indica cv. K-2, by constitutive and inducible expression of tobacco Osmotin. Transgenic Res. 2011;20(2):231–246. doi: 10.1007/s11248-010-9405-6. [DOI] [PubMed] [Google Scholar]
- Das M, Tetoriya M, Haq QMR, Khurana P. Screening and expression analysis of hal3a, dehydrin and nhx1 in ten genotypes of mulberry for abiotic stress tolerance. Sericologia. 2013;53(2):1–10. [Google Scholar]
- Dhanyalakshmi KH, Naika MBN, Sajeevan RS, Mathew OK, Shafi KM, Sowdhamini R, Nataraja KN. An approach to function annotation for proteins of unknown function (PUFs) in the transcriptome of Indian mulberry. PLoS One. 2016;11(3):e0151323. doi: 10.1371/journal.pone.0151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss SG, Chakraborti SP, Roychowdhuri S, Das NK, Vijayan K, Ghosh PD. Development of mulberry varieties for sustainable growth and leaf yield in temperate and subtropical regions of India. Euphytica. 2012;185(2):215–225. doi: 10.1007/s10681-011-0523-x. [DOI] [Google Scholar]
- Feng LC, Guangwei Y, Maode Y, Yifu K, Chenjun J, Zhonghuai Y. Studies on the genetic identities and relationships of mulberry cultivated species (Morus L.) by a random amplified polymorphic DNA assay. Acta Sericol Sin. 1996;22:135–139. [Google Scholar]
- Gulyani V, Khurana P. Identification and expression profiling of drought-regulated genes in mulberry (Morus sp.) by suppression subtractive hybridization of susceptible and tolerant cultivars. Tree Genet Genomes. 2011;7:725–738. doi: 10.1007/s11295-011-0369-3. [DOI] [Google Scholar]
- Guruprasad Krishnan RR, Dandin SB, Naik VG. Groupwise sampling: a strategy to sample core entries from RAPD marker data with application to mulberry. Trees. 2014;28:723–731. doi: 10.1007/s00468-014-0984-3. [DOI] [Google Scholar]
- He N, Zhang C, Qi X, Zhao S, Tao Y, Yang G, Lee TH, Wang X, Cai Q, Li D, Lu M, Liao S, Luo G, He R, Tan X, Xu Y, Li T, Zhao A, Jia L, Fu Q, Zeng Q, Gao C, Ma B, Liang J, Wang X, Shang J, Song P, Wu H, Fan L, Wang Q, Shuai Q, Zhu J, Wei C, Zhu-Salzman K, Jin D, Wang J, Liu T, Yu M, Tang C, Wang Z, Dai F, Chen J, Liu Y, Zhao S, Lin T, Zhang S, Wang J, Wang J, Yang H, Yang G, Wang J, Paterson AH, Xia Q, Ji D, Xiang Z. Draft genome sequence of the mulberry tree Morus notabilis. Nature Commun. 2013;4:2445. doi: 10.1038/ncomms3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M, Rahama SM, Jorder OI. Isolation of sodium chloride resistant genotypes in mulberry cultivars. Bull Sericult Res. 1991;2:67–73. [Google Scholar]
- Hossain M, Rahman SM, Zaman A, Jorder OI, Islam R. Micropropagation of M. laevigata Wall. from matured trees. Plant Cell Rep. 1992;11:522–527. doi: 10.1007/BF00236269. [DOI] [PubMed] [Google Scholar]
- Huang R-Z, Yan X-P, Li J, Zhang X-W. AFLP finger print analysis for 10 mulberry cultivars in Hunan province. Sci Seric. 2009;35:837–841. [Google Scholar]
- Ipek M, Pirlak L, Kafkas S. Molecular characterization of mulberry (Morus spp.) genotypes via RAPD and ISSR. J Sci Food Agr. 2012;92:1633–1637. doi: 10.1002/jsfa.4749. [DOI] [PubMed] [Google Scholar]
- Jain AK, Sarkar A, Datta RK. Induction of haploid callus and embryogenesis in in vitro cultured anthers of mulberry (Morus indica) Plant Cell Tiss Organ Cult. 1996;44:143–147. doi: 10.1007/BF00048192. [DOI] [Google Scholar]
- Jianzhong T, Chengfu L, Hongli W, Mingqi C. Transgenic plants via transformation of glycinin gene to mulberry. J Agr Biotechnol. 2001;9(4):400–402. [Google Scholar]
- Kafkas S, Ozgen M, Dogan Y, Ozgen B, Ercisli S, Serce S. Molecular characterization of mulberry accessions in Turkey by AFLP markers. J Am Soc Hort Sci. 2008;133:593–597. [Google Scholar]
- Kapur A, Bhatnagar S, Khurana P. Efficient regeneration from mature leaf explants of Indian mulberry via organogenesis. Sericologia. 2001;41:207–214. [Google Scholar]
- Kar P, Srivastava PP, Awasthi AK, Urs SR. Genetic variability and association of ISSR markers with some biochemical traits in mulberry (Morus spp.) genetic resources available in India. Tree Genet Genomes. 2008;4:75–83. doi: 10.1007/s11295-007-0089-x. [DOI] [Google Scholar]
- Kavyashree R. A repeatable protocol for in vitro micropropagation of mulberry variety S54. Indian J Biotechnol. 2007;6:385–388. [Google Scholar]
- Khurana P, Checker VG. The advent of genomics in mulberry and perspectives for productivity enhancement. Plant Cell Rep. 2011;30:825–838. doi: 10.1007/s00299-011-1059-1. [DOI] [PubMed] [Google Scholar]
- Kim HR, Patel KR, Thorpe TA. Regeneration of mulberry plants through tissue culture. Bot Gazette. 1985;146:335–340. doi: 10.1086/337533. [DOI] [Google Scholar]
- Krishnan RR, Naik VG, Ramesh SR, Qadri SMH. Microsatellite marker analysis reveals the events of the introduction and spread of cultivated mulberry in the Indian subcontinent. Plant Genet Resour. 2013;12(1):129–139. doi: 10.1017/S1479262113000415. [DOI] [Google Scholar]
- Krishnan RR, Sumathy R, Bindroo BB, Naik VG. MulSatDB: a first online database for mulberry microsatellites. Trees. 2014;28:1793–1799. doi: 10.1007/s00468-014-1086-y. [DOI] [Google Scholar]
- Lakshmi Sita G, Ravindran S. Gynogenic plants from ovary cultures of mulberry (Morus indica) In: Prakash J, Pierik KLM, editors. Horticulture new techniques and applications. London: Kluwer; 1991. pp. 225–229. [Google Scholar]
- Lal S, Gulyani V, Khurana P. Overexpression of hva1 gene from barley generates tolerance to salinity and water stress in transgenic mulberry (Morus indica) Transgenic Res. 2008;17:651–663. doi: 10.1007/s11248-007-9145-4. [DOI] [PubMed] [Google Scholar]
- Lal S, Ravi V, Khurana JP, Khurana P. Repertoire of leaf expressed sequence tags (ESTs) and partial characterization of stress-related and membrane transporter genes from mulberry (Morus indica L.) Tree Genet Genomes. 2009;5(2):359–374. doi: 10.1007/s11295-008-0192-7. [DOI] [Google Scholar]
- Lalitha N, Kih S, Banerjee R, Chattopadhya S, Saha AK, Bindroo BB. High frequency multiple shoot induction and in vitro regeneration of mulberry (Morus indica L. cv. S-1635) Int J Adv Res. 2013;1:22–26. [Google Scholar]
- Li T, Qi X, Zeng Q, Xiang Z, He N (2014) MorusDB: a resource for mulberry genomics and genome biology. Database, 2014, p.bau054 [DOI] [PMC free article] [PubMed]
- Lin Z, Weiguo Z, Junbai C, Yong H, Xing JS, Liu L, Qiang S. Analysis of genetic diversity and construction of core collection of local mulberry varieties from Shanxi Province based on ISSR marker. Afr J Biotech. 2011;10:7756–7765. doi: 10.5897/AJB11.172. [DOI] [Google Scholar]
- Liu XQ, Liu CY, Guo Q, Zhang M, Cao BN, Xiang ZH, Zhao AC. Mulberry transcription factor MnDREB4A confers tolerance to multiple abiotic stresses in transgenic tobacco. PLoS One. 2015;10(12):e0145619. doi: 10.1371/journal.pone.0145619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou CF, Zhang YZ, Zhou JM. Polymorphisms of genomic DNA in parents and their resulting hybrids in mulberry (Morus) Sericologia. 1998;38:437–445. [Google Scholar]
- Lu M-C. Micropropagation of Morus latifolia Poilet using axillary buds from mature trees. Sci Hort. 2002;96:329–341. doi: 10.1016/S0304-4238(02)00120-6. [DOI] [Google Scholar]
- Machii M. Leaf disc transformation of mulberry plant (Morus alba L.) by Agrobacterium Ti plasmid. J Sericult Sci Jpn. 1990;59:105–110. [Google Scholar]
- Machii M, Sung GB, Yamanuchi H, Koyama A. Transient expression of GUS gene introduced into mulberry plant by particle bombardment. J Sericult Sci Jpn. 1996;65:503–506. [Google Scholar]
- Mamrutha HM, Nataraja KN, Rama N, Kosma DK, Mogili T, Lakshmi KJ, Kumar MD, Jenks MA. Leaf surface wax composition of genetically diverse mulberry (Morus sp.) genotypes and its close association with expression of genes involved in wax metabolism. Curr Sci. 2017;112(4):759–766. doi: 10.18520/cs/v112/i04/759-766. [DOI] [Google Scholar]
- Mishra S (2014) Genetic analysis of traits controlling water use efficiency and rooting in mulberry (Morus spp.) by molecular markers. Ph.D. Thesis, University of Mysore, Mysuru, India
- Mishra S, Dandin SB. Molecular characterization of mulberry genotypes in relation to photosynthetic efficiency. Indian J Seric. 2010;49(1):50–57. [Google Scholar]
- Mishra S, Naik VG, Sukumar M, Pinto MV, Sheshashayee MS, Dandin SB. Genetic analysis of parental genotypes for mapping of water use efficiency and root traits in mulberry. Indian J Genet Plant Breed (The) 2013;73(4):405–410. doi: 10.5958/j.0975-6906.73.4.061. [DOI] [Google Scholar]
- Mishra GP, Singh B, Seth T, Singh AK, Halder J, Krishnan N, Tiwari SK, Singh PM. Biotechnological advancements and begomovirus management in okra (Abelmoschus esculentus L.): status and perspectives. Front Plant Sci. 2017;8:360. doi: 10.3389/fpls.2017.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogili T, Rajashekar K, Tripathi PM, Sathyanarayana K, Balakrishna R, Reddy MM. Screening mulberry genotypes for tolerance to alkalinity stress. Adv Plant Sci. 2008;2:621–629. [Google Scholar]
- Naik VG, Sarkar A, Sathyanarayana N. DNA finger printing of Mysore Local and V1 cultivars of mulberry (Morus spp.) with RAPD markers. Indian J Genet. 2002;62:193–196. [Google Scholar]
- Naik VG, Subbulakshmi N, Pinto MV, Mishra S, Guruprasad Qadri SMH. Assessment of genetic diversity among mulberry collections from South India using phenotypic and RAPD markers. Indian J Seric. 2013;52(1):34–43. [Google Scholar]
- Naik VG, Thumilan B, Sarkar A, Dandin SB, Pinto MV, Sivaprasad V. Development of genetic linkage map of mulberry using molecular markers and identification of QTLs linked to yield and yield contributing traits. Sericologia. 2014;54(4):221–229. [Google Scholar]
- Naik VG, Dandin SB, Tikader A, Pinto MV. Molecular diversity of wild mulberry (Morus spp.) of Indian subcontinent. Indian J Biotechnol. 2015;14(3):334–343. [Google Scholar]
- Narayan P, Chakraborty SP, Rao GS. Regeneration of plantlets from the callus of stem segments of mature plants of Morus alba L. Proc Indian Natl Sci Acad B. 1989;55:469–472. [Google Scholar]
- Ohnishi T, Kiyama S. Effects of change in temperature, pH, Ca ion concentration in the solution used for protoplast fusion on the improvement of the fusion ability of mulberry protoplasts. J Sericult Sci Jpn. 1987;56:418–421. [Google Scholar]
- Ohnishi T, Tanabe K. On the protoplast fusion of mulberry and paper mulberry by electrofusion method. J Seri Scricult Jpn. 1989;58:353–354. [Google Scholar]
- Ohyama K, Oka S. Regeneration of whole plants from isolated shoot tips of mulberry. J Sericult Sci Jpn. 1976;45:115–120. [Google Scholar]
- Oka S, Tewary PK. Induction of hairy roots from hypocotyls of mulberry (Morus indica L.) by Japanese wild strains of Agrobacterium rhizogenes. J Sericult Sci Jpn. 2000;69:13–19. [Google Scholar]
- Orhan E, Ercisli S, Yildirim N, Agar G. Genetic variations among mulberry genotypes (Morus alba) as revealed by random amplified polymorphic DNA (RAPD) markers. Plant Syst Evol. 2007;265:251–258. doi: 10.1007/s00606-007-0525-2. [DOI] [Google Scholar]
- Ozrenk K, Sensoy RIG, Erdinc C, Guleryuz M, Aykanat A. Molecular characterization of mulberry germplasm from Eastern Anatolia. Afr J Biotech. 2010;9:1–6. [Google Scholar]
- Ping LX, Nogawa M, Shioiri H, Nozue M, Makita N, Takeda M, Bao L, Kojima M. In planta transformation of mulberry trees (Morus alba L.) by Agrobacterium tumefaciens. J Insect Biotechnol Sericol. 2003;72(3):177–184. [Google Scholar]
- Pinto MV, Naik VG, Qadri SMH. Genetic variability studies in mulberry using microsatellite markers. J Sericult Technol. 2012;3(1–2):38–43. [Google Scholar]
- Raghunath MK, Lal S, Khurana P. In vitro plant regeneration from different explants of elite mulberry (Morus sp.) genotypes AR12, DD and S13. Bangladesh J Seric. 2008;2–3:31–39. [Google Scholar]
- Raghunath MK, Nataraj KN, Meghana JS, Sanjeevan RS, Rajan MV, Qadri SMH. In vitro plant regeneration of Morus indica L. cv. V-1 using leaf explants. Am J Plant Sci. 2013;4(10):2001–2005. doi: 10.4236/ajps.2013.410249. [DOI] [Google Scholar]
- Rani R, Yadav P, Barbadikar KM, Baliyan N, Malhotra EV, Singh BK, Kumar A, Singh D. CRISPR/Cas9: a promising way to exploit genetic variation in plants. Biotechnol Lett. 2016;38(12):1991–2006. doi: 10.1007/s10529-016-2195-z. [DOI] [PubMed] [Google Scholar]
- Rao PJSVVNH, Nuthan D, Krishna KS. A protocol for in vitro regeneration of rainfed mulberry varieties through callus phase. Euro J Biol Sci. 2010;2:80–86. [Google Scholar]
- Rao PJSVVNH, Nuthan D, Krishna KS, Basavaraja MK. In vitro propagation of irrigated mulberry varieties using nodal explants. Curr Biotica. 2010;3:555–564. [Google Scholar]
- Ravi V, Khurana JP, Tyagi AK, Khurana P. The chloroplast genome of mulberry: complete nucleotide sequence, gene organization and comparative analysis. Tree Genet Genomes. 2006;3:49–59. doi: 10.1007/s11295-006-0051-3. [DOI] [Google Scholar]
- Ravi V, Tyagi AK, Khurana JP, Khurana P. An update on chloroplast genomes. Plant Syst Evol. 2008;271:101–122. doi: 10.1007/s00606-007-0608-0. [DOI] [Google Scholar]
- Saeed B, Khurana P. Transcription activation activity of ERD15 protein from Morus indica. Plant Physiol Biochem. 2016 doi: 10.1016/j.plaphy.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Saeed B, Das M, Haq QMR, Khurana P. Overexpression of beta carotene hydroxylase-1 (bch1) in mulberry, Morus indica cv. K-2, confers tolerance against high-temperature and high-irradiance stress induced damage. Plant Cell Tiss Organ Cult. 2015;120(3):1003–1015. doi: 10.1007/s11240-014-0654-6. [DOI] [Google Scholar]
- Saeed B, Baranwal VK, Khurana P. Comparative transcriptomics and comprehensive marker resource development in mulberry. BMC Genomics. 2016;17:98. doi: 10.1186/s12864-016-2417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed B, Baranwal VK, Khurana P (2016b) Identification and expression profiling of Lectin gene superfamily in mulberry. Plant Genome 9(2). doi: 10.3835/plantgenome2015.10.0107 [DOI] [PubMed]
- Saha S, Adhikari S, Dey T, Ghosh P. RAPD and ISSR based evaluation of genetic stability of micropropagated plantlets of Morus alba L. variety S-1. Meta Gene. 2016;7:7–15. doi: 10.1016/j.mgene.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajeevan RS, Nataraja KN. Molecular cloning and characterization of a novel basic helix–loop–helix-144 (bHLH144)-like transcription factor from Morus alba (L.) Plant Gene. 2016;5:109–117. doi: 10.1016/j.plgene.2016.01.004. [DOI] [Google Scholar]
- Sajeevan RS, Singh SJ, Nataraja KN, Shivanna MV. An efficient in vitro protocol for multiple shoot induction in mulberry, Morus alba L variety V1. Intl Res J Plant Sci. 2011;2:254–261. [Google Scholar]
- Sajeevan RS, Nataraja KN, Shivashankara KS, Pallavi N, Gurumurthy DS, Shivanna MB. Expression of Arabidopsis SHN1 in Indian mulberry (Morus indica L.) increases leaf surface wax content and reduces post-harvest water loss. Front Plant Sci. 2017;8:418. doi: 10.3389/fpls.2017.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar T, Radhakrishnan T, Kumar A, Mishra GP, Dobaria JR. Heterologous expression of AtDREB1A gene in transgenic peanut conferred tolerance to drought and salinity stresses. PLoS One. 2014;9(12):e110507. doi: 10.1371/journal.pone.0110507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar T, Radhakrishnan T, Kumar A, Mishra GP, Dobaria JR. Stress inducible expression of AtDREB1A transcription factor in transgenic peanut (Arachis hypogaea L.) crop conferred tolerance to soil-moisture deficit stress. Front Plant Sci. 2016;7:935. doi: 10.3389/fpls.2016.00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RJ, Zhang X. High-throughput approaches for plant epigenomic studies. Curr Opin Plant Biol. 2011;14:130–136. doi: 10.1016/j.pbi.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma AC, Sharma R, Machii H. Assessment of genetic diversity in a Morus germplasm collection using fluorescence-based AFLP markers. Theor Appl Genet. 2000;101:1049–1055. doi: 10.1007/s001220051579. [DOI] [Google Scholar]
- Shoukang L, Dongfeng J, Jun Q. In vitro production of haploid plants from mulberry (Morus) anther culture. Sci Sinica. 1987;30:853–863. [Google Scholar]
- Sugimura Y, Miyazaki J, Yonebayashi K, Kotani E, Furusawa T. Gene transfer by electroporation into protoplasts isolated from mulberry call. J Sericult Sci Jpn. 1999;68:49–53. [Google Scholar]
- Susheelamma BN, Kumar JS, Mogili T, Sengupta K, Padma MN, Suryanarayana N. Evaluation techniques for screening for drought resistance in mulberry. Sericologia. 1992;32:609–614. [Google Scholar]
- Susheelamma BN, Shekhar KR, Sarkar A, Rao MR, Datta RK. Genotypes and hormonal effects on callus formation and regeneration in mulberry. Euphytica. 1996;90:25–29. [Google Scholar]
- Tewary PK, Sharma A, Raghunath MK, Sarkar A. In vitro response of promising mulberry (Morus sp.) genotypes for tolerance to salt and osmotic stresses. Plant Growth Regul. 2000;30(1):17–21. doi: 10.1023/A:1006297830318. [DOI] [Google Scholar]
- Thomas TD. Thidiazuron induced multiple shoot induction and plant regeneration from cotyledonary explants of mulberry. Biol Plant. 2003;46:529–533. doi: 10.1023/A:1024807426591. [DOI] [Google Scholar]
- Thomas TD, Bhatnagar AK, Rajdan MK, Bhojwani SS. A reproducible protocol for the production gynogenic haploids of mulberry, Morus alba L. Euphytica. 1999;110:169–173. doi: 10.1023/A:1003797328246. [DOI] [Google Scholar]
- Thomas TD, Bhatnagar AK, Bhojwani SS. Production of triploid plants of mulberry (Morus alba L.) by endosperm culture. Plant Cell Rep. 2000;19(4):395–399. doi: 10.1007/s002990050746. [DOI] [PubMed] [Google Scholar]
- Thumilan BM, Kadam NN, Biradar J, Sowmya HR, Mahadeva A, Madhura JN, Makarla U, Khurana P, Sreeman SM. Development and characterization of microsatellite markers for Morus spp. and assessment of their transferability to other closely related species. BMC Plant Biol. 2013;13:194. doi: 10.1186/1471-2229-13-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumilan BM, Sajeevan RS, Biradar J, Madhuri T, Nataraja KN, Sreeman SM. Development and characterization of genic SSR markers from Indian mulberry transcriptome and their transferability to related species of Moraceae. PLoS One. 2016;11(9):e0162909. doi: 10.1371/journal.pone.0162909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikader A, Dandin SB. DNA fingerprint of inter and intra-specific hybrids from Morus species using RAPD. Geobios. 2008;35:113–120. [Google Scholar]
- Umate P. Mulberry improvements via plastid transformation and tissue culture engineering. Plant Signal Behav. 2010;5(7):785–787. doi: 10.4161/psb.5.7.12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umate P, Rao KV, Kiranmayee K, Jaya Sree T, Sadanandam A. Plant regeneration of mulberry (Morus indica) from mesophyll-derived protoplasts. Plant Cell Tiss Organ Cult. 2005;82(3):289–293. doi: 10.1007/s11240-005-1541-y. [DOI] [Google Scholar]
- Venkateswarlu M, Raje US, Surendra NB, Shashidhar HE, Maheswaran M, Veeraiah TM, Sabitha MG. A first genetic linkage map of mulberry (Morus spp.) using RAPD, ISSR, and SSR markers and pseudotestcross mapping strategy. Tree Genet Genomes. 2006;3:15–24. doi: 10.1007/s11295-006-0048-y. [DOI] [Google Scholar]
- Vijayan K. The emerging role of genomic tools in mulberry (Morus) genetic improvement. Tree Genet Genomes. 2010;6:613–625. doi: 10.1007/s11295-010-0276-z. [DOI] [Google Scholar]
- Vijayan K, Chakraborti SP, Roy BN. Plant regeneration from leaf explants of mulberry: Influence of sugar, genotype and 6-benzyladenine. Indian J Exp Biol. 2000;38(5):504–508. [PubMed] [Google Scholar]
- Vijayan K, Chakraborti SP, Ghosh PD. In vitro screening of mulberry for salinity tolerance. Plant Cell Rep. 2003;22:350–357. doi: 10.1007/s00299-003-0695-5. [DOI] [PubMed] [Google Scholar]
- Vijayan K, Srivastava PP, Awasthi AK. Analysis of phylogenetic relationship among five mulberry (Morus) species using molecular markers. Genome. 2004;47:439–448. doi: 10.1139/g03-147. [DOI] [PubMed] [Google Scholar]
- Vijayan K, Chakraborti SP, Ghosh PD. Screening of mulberry (Morus spp.) for salinity tolerance through in vitro seed germination. Indian J Biotech. 2004;3:47–51. [Google Scholar]
- Vijayan K, Srivastava PP, Nair CV, Tikader A, Awasthi AK, Urs SR. Molecular characterization and identification of markers associated with leaf yield traits in mulberry using ISSR markers. Plant Breed. 2006;125:298–301. doi: 10.1111/j.1439-0523.2006.01212.x. [DOI] [Google Scholar]
- Vijayan K, Doss SG, Chakraborti SP, Ghosh PD. Breeding for salinity resistance in (Morus spp.) Euphytica. 2009;169(3):403–411. doi: 10.1007/s10681-009-9972-x. [DOI] [Google Scholar]
- Vijayan K, Nair CV, Chatterjee SN. Diversification of mulberry (Morus indica var. S36), a vegetatively propagated tree species. Casp J Env Sci. 2009;7:23–30. [Google Scholar]
- Vijayan K, Tikader A, da Silva JAT. Application of tissue culture techniques propagation and crop improvement in mulberry (Morus spp) Tree For Sci Biotechnol. 2011;5(1):1–13. [Google Scholar]
- Vijayan K, Raju PJ, Tikader A, Saratchnadra B. Biotechnology of mulberry (Morus L.)—a review. Emir J Food Agr. 2014;26(6):472–496. [Google Scholar]
- Wang ZW, Yu MD. AFLP analysis of genetic background of polyploid breeding materials of mulberry. Acta Sericol Sin. 2001;27:170–176. [Google Scholar]
- Wang H, Tong W, Feng L, Jiao Q, Long L, Fang R, Zhao W. De Novo transcriptome analysis of mulberry (Morus L) under drought stress using RNA seq technology. Russ J Bioorg Chem. 2014;40(4):423–432. doi: 10.1134/S1068162014040037. [DOI] [PubMed] [Google Scholar]
- Wang XL, Yu YS, Yang Y, Liu CY, Li J, Yu MD. Molecular cloning and expression of a nitrite reductase gene from mulberry (Morus L.) Plant Cell Tiss Organ Cult. 2014;121(2):301–309. doi: 10.1007/s11240-014-0701-3. [DOI] [Google Scholar]
- Wangari NP, Gacheri KM, Theophilus MM, Lucas N. Use of SSR markers for genetic diversity studies in mulberry accessions grown in Kenya. Int J Biotech Mol Biol Res. 2013;4:38–44. doi: 10.5897/IJBMBR11.057. [DOI] [Google Scholar]
- Wani SA, Bhat MA, Malik GN, Zaki FA, Mir MR, Wani N, Bhat KM. Genetic diversity and relationship assessment among mulberry (Morus spp.) genotypes by simple sequence repeat (SSR) marker profile. Afr J Biotech. 2013;12:3181–3187. [Google Scholar]
- Weiguo Z, Zhihua Z, Xuexia M, Yong Z, Sibao W, Jianhua H, Hui X, Yile P, Yongping H. A comparison of genetic variation among wild and cultivated Morus species (Moraceae: Morus) as revealed by ISSR and SSR markers. Biodiv Conserv. 2007;16:275–290. doi: 10.1007/s10531-005-6973-5. [DOI] [Google Scholar]
- Xiang Z, Zhang Z, Yu M. A preliminary report on the application of RAPD in systematics of Morus alba. Acta Sericol Sin. 1995;21:203–207. [Google Scholar]
- Zaki M, Kaloo ZA, Sofi M. Micropropagation of Morus nigra L. from nodal segments with axillary buds. World J Agr Sci. 2011;7:496–503. [Google Scholar]
- Zhao WG, Pan YL. RAPD analysis for the germplasm resources of genus mulberry. Acta Sericol Sin. 2002;26:1–8. [Google Scholar]
- Zhao W, Miao X, Jia S, Pan Y, Huang Y. Isolation and characterization of microsatellite loci from the mulberry, Morus L. Plant Sci. 2005;168:519–525. doi: 10.1016/j.plantsci.2004.09.020. [DOI] [Google Scholar]
- Zhao WG, Zhou ZH, Miao XX, Wang SB, Zhang L, Pan Y, Huang YP. Genetic relatedness among cultivated and wild mulberry as revealed by inter-simple sequence repeat (ISSR) analysis in China. Can J Plant Sci. 2006;86:251–257. doi: 10.4141/P04-110. [DOI] [Google Scholar]
- Zhao ZZ, Xuexia M, Yong Z, Sibao W, Jianhua H, Hui X, Yile P, Yongping H. A comparison of genetic variation among wild and cultivated Morus species (Moraceae: Morus) as revealed ISSR and SSR markers. Biodiv Conserv. 2007;16:275–290. doi: 10.1007/s10531-005-6973-5. [DOI] [Google Scholar]
- Zhao W, Wang Y, Chen T, Jia G, Wang X, Qi J, Pang Y, Wang S, Li Z, Huang Y, Pan Y, Yang Y-H. Genetic structure of mulberry from different ecotypes revealed by ISSRs in China: an implications for conservation of local mulberry varieties. Sci Hort. 2007;115:47–55. doi: 10.1016/j.scienta.2007.07.017. [DOI] [Google Scholar]
- Zhao WG, Wang W, Yang YH, Huang YP, Pan YL. Genetic diversity of mulberry local varieties from different ecotype as revealed by ISSR analysis in China. Acta Sericol Sin. 2008;34:1–5. [Google Scholar]
- Zhao W, Fang R, Pan Y, Yang Y, Chung JW, Chung IM, Park YJ. Analysis of genetic relationships of mulberry (Morus L.) germplasm using sequence-related amplified polymorphism (SRAP) markers. Afr J Biotechnol. 2009;8:2604–2610. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.