Abstract
We have used photo-crosslinking to investigate the structure and dynamics of four-way junction hairpin ribozyme constructs. Four phenylazide photo-crosslinkers were coupled to 2′-NH2-modified U+2 in the substrate and irradiated at different Mg2+ concentrations and temperatures. Consistent with the role of divalent metal ions in hairpin ribozyme folding, we observed more interdomain crosslinks in the presence of Mg2+ than in its absence. In general, we observed intradomain crosslinks to nucleotides 2–11 and interdomain crosslinks to the U1A binding loop. Crosslinks to A26 and G36 in domain B were also observed when crosslinking was carried out at −78°C. In contrast to crosslinking results at higher temperatures (0, 25 and 37°C), similar crosslinks were obtained in the presence and absence of Mg2+ at −78°C, suggesting Mg2+ stabilizes a low-energy hairpin ribozyme conformation. We also evaluated the effects of photo-crosslinker structure and mechanism on crosslinks. First, most crosslinks were to unpaired nucleotides. Second, shorter and longer photo-crosslinkers formed crosslinks to intradomain locations nearer to and farther from photo-crosslinker modification, respectively. Finally, fluorine substitutions on the phenylazide ring did not change the locations of crosslinks, but rather decreased crosslinking efficiency. These findings have implications for the use of phenylazide photo-crosslinkers in structural studies of RNA.
INTRODUCTION
The hairpin ribozyme is a small catalytic RNA motif which facilitates RNA processing during the life cycle of the tobacco ringspot virus (1). Like most small ribozymes, the hairpin catalyzes the sequence-specific cleavage of an RNA substrate via nucleophilic attack of a 2′ hydroxyl group on an adjacent phosphorus atom, resulting in a 2′,3′-cyclic phosphate and a 5′-hydroxyl terminus [reviewed in (2,3)]. The minimum catalytic structure of the hairpin ribozyme is composed of two domains, A and B, each consisting of helices interrupted by loops (Figure 1A). The inclusion of two more helices through a four-way junction, although not necessary for catalysis, has been shown to facilitate folding (4) and is also present in the natural form of the hairpin ribozyme. Most of the nucleotides in loop A and loop B have been shown to be necessary for catalysis (5), suggesting that the two loops interact to form the ribozyme's active structure. For example, the crystal structure (Figure 1B) shows that these loops are stitched together via a ribose-zipper interaction in which the 2′-hydroxyls of nucleotides 10, 11, 24 and 25 hydrogen bond with the N3 atoms of A10 and A24 (6). In addition, G+1 of the substrate forms a base pair with C25 in domain B while U42 extrudes from the helical stack in stem B and forms hydrogen bonds to nucleotides in both domains.
Figure 1.
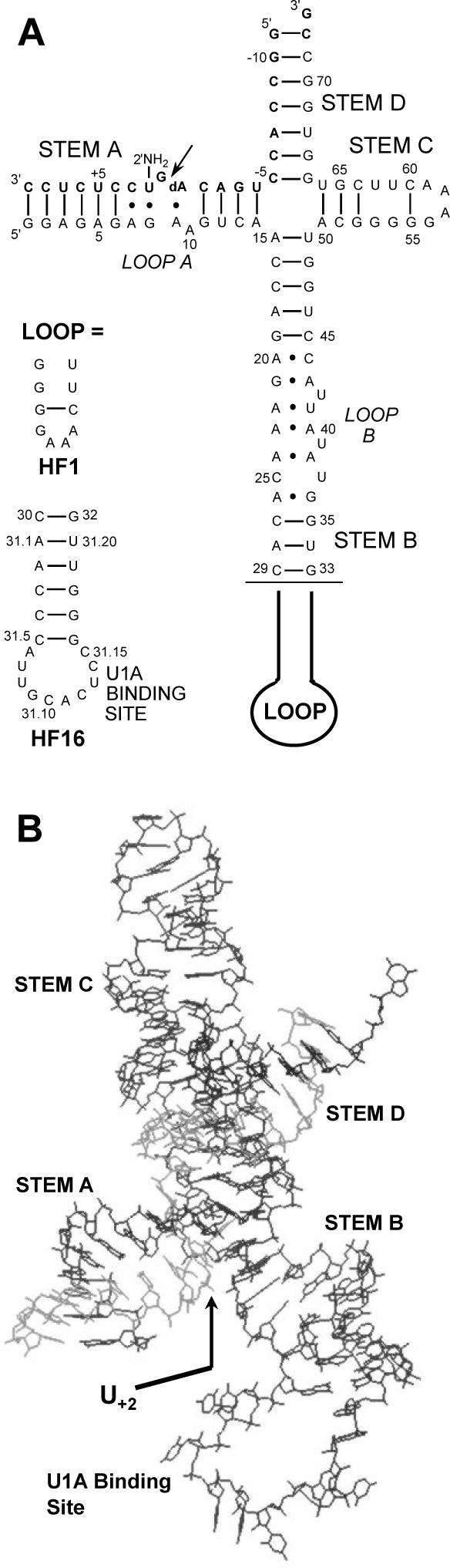
The hairpin ribozyme constructs used in this study. (A) Secondary structure of the HF1 and HF16 hairpin ribozyme constructs, with the 2′-NH2 modified inhibitory substrate strand shown in bold. The arrow indicates the substrate cleavage site. (B) Crystal structure of the HF16 hairpin ribozyme construct (6). The bold arrow indicates the location of the 2′-NH2 modification.
Catalysis by the hairpin ribozyme has been shown to require the presence of metal ions. However, research suggests that this metal ion dependence originates solely in the folding and stabilization of the hairpin's active structure rather than in direct involvement of the metal ions in catalysis itself (7–9). Divalent metal ion-facilitated folding of the hairpin ribozyme into its active structure involves a docking event that has been the subject of intensive study. Folding of a four-way junction hairpin ribozyme construct into its docked form has been shown by ensemble (10) and single molecule (11,12) Fluorescence Resonance Energy Transfer (FRET) experiments to be a two-step process. The first step of folding requires only the diffuse binding of mono- or divalent metal ions, while the second requires site binding of at least one divalent metal ion. A more recent FRET study of conformational distributions of individual hairpin ribozyme molecules in solution showed that the four-way junction was able to promote folding into a docked conformation without specific interactions between loops A and B, but only at higher concentrations of Mg2+ (13). Results from single molecule FRET studies of hairpin ribozymes tethered to a surface suggest that after the first step of docking, the two loops are proximal but are not locked in place by specific interactions between nucleotides in the two loops (14). Because of the transient nature of this intermediate, its structure has not been fully characterized. Loops A and B undergo a significant structural rearrangement upon folding into the fully docked structure from the unfolded conformation, as evidenced by comparison of the independent NMR structures of domains A (15) and B (16) with the crystal structure (6) and Nucleotide Analog Interference Mapping (NAIM) data (17). For example, U+2 is flipped out of the helix in the NMR structure, but pairs with G8 in the crystal structure, while G+1 adopts a 2′-endo sugar pucker in order to base pair with C25 in loop B (6).
Crosslinking experiments have also shown the proximity of domains A and B in the active structure of the hairpin ribozyme (18,19). For example, Burke and coworkers linked phenylazide photo-crosslinkers to phosphorothioates incorporated at specific sites within domains A and B of a two-way junction ribozyme, which gave a number of interdomain crosslinks upon irradiation (19). Here, we describe incorporation of four different photo-crosslinkers into domain A of the natural four-way junction form of the hairpin ribozyme, utilizing the selective reaction of 2′-NH2 groups in RNA with aliphatic isocyanates. The effects of Mg2+ concentration and temperature on the folding and flexibility of the hairpin ribozyme were subsequently studied through photochemical crosslinking. Consistent with the role of divalent metal ions in the folding of the hairpin ribozyme, we observed more interdomain crosslinks in the presence of Mg2+ than in its absence. However, crosslinks to domain B (A26 and G36) were observed only at low temperatures (−78°C). This result indicates that at lower temperatures, the motion between loops A and B is slowed enough to allow for the formation of crosslinks between the two loops. Finally, at −78°C similar crosslinks were obtained in the presence and absence of Mg2+, suggesting that the hairpin prefers the same fold in the presence of Mg2+ as in its absence at this temperature.
A second goal of this study was to evaluate the effects of tether length and chemical reactivity of phenylazide photo-crosslinkers on the crosslinks they form. To this end, we employed four phenylazide crosslinkers differing in both length and photoreactive mechanism. We have found that fluorine substitution on the phenyl ring of a photo-crosslinker lowers its crosslinking efficiency, but does not change the locations to which it crosslinks. However, varying the tether length of the photo-crosslinker resulted in a variety of crosslinks to different locations in the ribozyme.
MATERIALS AND METHODS
General
All syntheses involving compounds containing an azide functionality were carried out with care to expose the compounds to a minimal amount of light. TLC analyses were performed on glass plates coated with 250 μm thick silica gel including fluorescent indicator. TLC purifications were performed on glass plates coated with 1.0 mm thick silica gel including fluorescent indicator. Column chromatography was performed with 230–400 mesh silica gel (Merck). All NMR spectra were recorded on a Bruker 500 MHz or 300 MHz spectrometer. 1H spectra were referenced to TMS (0 p.p.m.), CDCl3 (7.26 p.p.m.) or CD3OD (4.87 p.p.m.). 19F spectra were referenced to hexafluorobenzene (−162.9 p.p.m.) which was added as an internal standard. 13C spectra were referenced to CDCl3 (77.36 p.p.m.), CD3OD (49.86 p.p.m.) or C2D5OD (30.60 p.p.m.). Infrared spectra were recorded on a Perkin Elmer 1720 fourier transform infrared spectrophotometer using ZnSe plates. Mass spectra were recorded on a Bruker Esquire Liquid Chromatograph–Ion Trap Mass Spectrometer in CH2Cl2 or CH3OH. Ultraviolet-Visible (UV-Vis) spectra were obtained with a Perkin Elmer Lambda Bio 20 spectrometer. The following extinction coefficients (ε) were used to quantify the oligoribonucleotides in this study: HF16, 921 200 l/mol·cm; HF1, 803 000 l/mol·cm; all hairpin substrate (HPS) constructs, 186 300 l/mol·cm.
Denaturing polyacrylamide gel electrophoresis (DPAGE) gels were prepared with 1× TBE buffer (90 mM Tris–HCl, 90 mM H3BO3, 1 mM Na2EDTA) and 19:1 acrylamide: N,N′-methylene-bis-acrylamide to a final concentration of 12–24%. DPAGE gels were prepared for analysis by autoradiography by first being dried on a Bio-Rad Model 583 gel drier, and then exposed to a Molecular Dynamics Phosphor Screen for 5–48 h. Screens were scanned by a Molecular Dynamics Storm PhosphorImager. Agarose gels were prepared with 1% agarose in 1× TBE buffer (90 mM Tris–HCl, 90 mM H3BO3, 1 mM Na2EDTA). Ethidium bromide was included in the final solution, after heating, to a final concentration of 0.5 μg/ml.
Synthesis of photo-crosslinkers
N-(4-azidobenzoyl)-3-isocyanatopropyl amide (1)
Compound 6 (12.4 mg, 0.057 mmol) was suspended in a solution of N,N,N′,N′-tetramethyl-1,8-naphthalenediamine (Proton Sponge, 49.1 mg, 0.033 mmol) in anhydrous CH2Cl2 (0.25 ml). Sonication was used to ensure a homogeneous suspension. The suspension was added dropwise over ∼5 min to an ice-cold solution of diphosgene (4.0 μl, 0.033 mmol) in anhydrous CH2Cl2 (0.75 ml), resulting in a clear solution. The reaction mixture was stirred at 0°C for 5 min, and the solvent was removed in vacuo. The residue was dissolved in anhydrous CH2Cl2 (1 ml) and washed successively with 1 N HCl (6 × 0.5 ml) and 1 N NaOH (1 × 0.5 ml). The organic layer was dried over MgSO4 and the solvent was removed in vacuo to yield a yellow oil (12.4 mg, 89%). The product was dissolved in anhydrous CDCl3 for NMR analysis. This solution was then aliquotted equally into 24 microtubes (0.52 mg, 2.1 μmol/aliquot) and each aliquot was brought to a total volume of 200 μl with anhydrous CH2Cl2. The solutions were stored in a dessicated container at −20°C until they were used for crosslinking. 1H NMR (CDCl3, 300 MHz): δ 7.67 (d, 2H, J = 8.7 Hz); 6.94 (d, 2H, J = 8.7 Hz); 6.42 (br s, 1H); 3.44 (t, 2H, J = 6.6 Hz); 3.35 (m, 2H, J = 6.6 Hz); 1.82 (m, 2H, J = 6.6 Hz). 13C NMR (CDCl3, 75 mHz): δ 167.02, 143.87, 131.07, 128.98, 119.43, 53.76, 41.23, 37.88, 31.38. IR: ν (cm−1): 2278 (NCO), 2125 (N3), 1637 (CO).
N-(4-azido-2,3,5,6 tetrafluorobenzoyl)-3-isocyanatopropyl amide (2)
Compound 10 (4.1 mg, 14.1 μmol) was suspended in a solution of Proton Sponge (7.0 mg, 32.7 μmol) in anhydrous CH2Cl2 (0.2 ml). Sonication was used to ensure a homogeneous suspension. This suspension was added dropwise over ∼1 min to an ice-cold solution of diphosgene (1.2 μl, 9.87 μmol) in anhydrous CH2Cl2 (0.2 ml), resulting in a clear solution. The reaction mixture was stirred at 0°C for 10 min, after which the solvent was removed in vacuo. The product was dissolved in anhydrous CH2Cl2 (1 ml) and washed with 1 N HCl (6 × 0.2 ml). The organic layer was dried over MgSO4, and the solvent was removed in vacuo, yielding a white solid (2.4 mg, 54%). The product was immediately dissolved in CDCl3 for NMR analysis and then aliquotted equally into 12 microtubes (0.2 mg, 0.63 μmol/aliquot). Each aliquot was brought to a total volume of 200 μl with anhydrous CH2Cl2. The solutions were stored in a dessicated container at −20°C. 1H NMR (CDCl3, 300 MHz): δ 6.18 (br s, 1 H); 3.59 (m, 2H, J = 6.6 Hz); 3.49 (t, 2H, J = 6.6 Hz), 1.93 (m, 2H, J = 6.6 Hz). 19F NMR (CDCl3, 470 MHz): δ −142.10 (2F); −151.42 (2F). IR (cm−1): ν 2287 (NCO), 2128 (N3), 1654 (CO).
4-Azido-2,3,5,6-tetrafluorobenzyl isocyanate (3)
4-Azido-2,3,5,6-tetrafluorobenzyl amine hydrochloride (9.4 mg, 36.6 μmol) was suspended in a solution of Proton Sponge (51.2 mg, 239.0 μmol) in anhydrous CH2Cl2 (0.5 ml). Sonication was used to ensure a homogeneous suspension. This suspension was added dropwise over ∼30 s to an ice-cold solution of diphosgene (4.0 μl, 33.2 μmol) in anhydrous CH2Cl2 (0.5 ml), resulting in a clear solution. The reaction mixture was stirred at 0°C for 10 min, then at room temperature for 5 min, after which the solvent was removed in vacuo. The product was dissolved in anhydrous CH2Cl2 (2 ml) and washed successively with 1 N HCl (6× 1 ml) and 1 N NaOH (1 × 1 ml). The organic layer was dried over MgSO4, and the solvent was removed in vacuo, yielding a white solid (7.6 mg, 84%). The product was immediately dissolved in CDCl3 for NMR analysis. This solution was then aliquotted equally into 22 microtubes (0.35 mg, 1.4 μmol/aliquot) and each aliquot was brought to a total volume of 200 μl with anhydrous CH2Cl2. The solutions were stored in a dessicated container at −20°C. 1H NMR (CDCl3, 500 MHz): δ 5.3 (s). 19F NMR (CDCl3, 470 MHz): δ −144.82 (2F); −152.07 (2F). IR (cm−1): ν 2259 (NCO), 2122 (N3).
p-azidobenzoyl chloride (5)
This compound was synthesized as previously reported (20), with minor changes. p-azidobenzoic acid (4.5 g, 27.6 mmol) and freshly distilled SOCl2 (100 ml, 1.4 mol) were refluxed at 75°C for 30 min. Excess SOCl2 was removed in vacuo to yield a brown solid (5.0 g, 100%), which was used directly in the next step of the synthesis.
N-(4-azidobenzoyl)-1,3-propyldiamine (6)
A solution of 5 (501 mg, 2.76 mmol) in anhydrous ethanol (15 ml) was added dropwise to an ice-cold solution of 1,3-diaminopropane (3.45 ml, 41.3 mmol) in anhydrous ethanol (7.5 ml). The reaction mixture was stirred at 0°C for 4 h, then solid material was removed by filtration and discarded. After removing the solvent in vacuo, 2 M HCl (10 ml) was added to the residue. The resulting white precipitate was removed by filtration and discarded. The filtrate was adjusted to pH 10 with saturated NaOH, and extracted with EtOAc (5 × 2 ml). The organic layer was dried over MgSO4 and the solvent was removed in vacuo. Column chromatography CH3OH:CH2Cl2:Et3N (30:69:1 to 45:54:1) yielded a white solid (49.6 mg, 12%). 1H NMR (CD3OD, 300 MHz): δ 7.86 (d, 2H, J = 8.7 Hz); 7.15 (d, 2H, J = 8.4 Hz); 3.46 (t, 2H, J = 6.6 Hz); 2.76 (t, 2H, J = 6.6 Hz); 1.80 (m, 2H, J = 6.6 Hz). 13C NMR (CD3OD, 75 mHz): δ 170.22, 145.75, 132.97, 131.02, 120.85, 40.36, 39.01, 33.48. IR (cm−1): ν 2128 (N3), 1630 (CO). MS: 220.2 (calcd 220.3 for C10H13ON5).
4-Azido-2,3,5,6-tetrafluorobenzoyl chloride (7)
This compound was synthesized as previously reported (21), with minor changes. p-Azido-2,3,5,6-tetrafluorobenzoic acid (198.5 mg, 782.9 mmol), which was prepared in two steps from methyl pentafluorobenzoate (21), and freshly distilled SOCl2 (2 ml, 27.4 mmol) were refluxed at 75–85°C for 3 h 15 min. Excess SOCl2 was removed in vacuo to yield a yellow oil (196.4 mg, 92% yield) which was used directly in the next step of the synthesis.
N-triphenylmethyl-1,3-diaminopropane (8)
1,3-diamino propane (4.5 ml, 54 mmol), triphenylmethyl chloride (10.0 g, 36 mmol), and diisopropylethyl amine (9.4 ml, 54 mmol) were dissolved in anhydrous CH2Cl2 (144 ml). The reaction mixture was stirred at room temperature for 4 h, after which it was diluted with CH2Cl2 (65 ml) and washed with 0.1 M Et3NHOAc (3 × 80 ml). The organic layer was dried over MgSO4 and the solvent was removed in vacuo. Column chromatography CH3OH:CH2Cl2:Et3N (0:99:1 to 10:89:1) yielded white crystals (1.7 g, 15%). 1H NMR (CDCl3, 500 MHz): δ 7.60 (d, 6H, J = 7.5 Hz); 7.31 (t, 6H, J = 7.5 Hz); 7.20 (t, 3H, J = 7.5 Hz); 2.75 (t, 2H, J = 7.0 Hz); 2.29 (t, 2H, J = 7.0 Hz); 1.64 (m, 2H, J = 7.0 Hz), 1.49 (br s). 13C NMR (CDCl3, 126 mHz): δ 145.84, 128.25, 127.42, 125.85, 70.55, 41.13, 40.06, 34.10.
N-(4-azido-2,3,5,6 tetrafluorobenzoyl)-3-triphenylmethyl-1,3-diaminopropyl amide (9)
To an ice-cold solution of 7 (70.0 mg, 0.28 mmol) and Proton Sponge (252.5 mg, 1.2 mmol) in anhydrous CH2Cl2 (3.4 ml) was added 8 (189.4 mg, 0.60 mmol). The resulting suspension was sonicated and then stirred at 0°C for 30 min. Excess solid 8 was removed by filtration, and the filtrate was purified by TLC (2% CH3OH/1% Et3N/CH2Cl2) to yield a yellow oil (138.2 mg, 94%). 1H NMR (CDCl3, 500 MHz): δ 7.42 (d, 6H, J = 7.5 Hz); 7.28 (t, 6H, J = 7.5 Hz); 7.22 (t, 3H, J = 7.5 Hz); 3.63 (m, 2H, J = 6.0 Hz); 2.38 (br t, 2H), 1.79 (m, 2H, J = 6.0 Hz). 19F NMR (CDCl3, 470 MHz): δ −141.85 (2F); −151.53 (2F). 13C NMR (CDCl3, 75 mHz): δ 155.55, 145.93, 142.51, 139.04, 128.81, 128.26, 126.81, 121.98, 112.80, 71.50, 42.26, 39.79, 29.78. IR (cm−1): ν 2126 (N3), 1651 (CO).
N-(4-azido-2,3,5,6 tetrafluorobenzoyl) 1,3-diaminopropyl amide (10)
To a solution of 9 (138.2 mg, 0.26 mmol) in CH2Cl2 (8 ml) was added 3% Cl3COOH/CH2Cl2 (8 ml). The reaction mixture was stirred at room temperature for 12.5 h, then extracted with H2O (3× 10 ml). The aqueous layer was adjusted to pH ≥ 10 with 20% NaOH, then extracted with EtOAc (6 × 20 ml). The organic layer was dried over MgSO4 and the solvent was removed in vacuo to yield a white solid (13.2 mg, 18%). 1H NMR (CD3OD, 300 MHz): δ 3.45 (t, 2H, J = 6.9 Hz); 2.74 (t, 2H, J = 6.9 Hz), 1.76 (m, 2H, J = 6.9 Hz). 19F NMR (CD3OD, 470 MHz): δ −142.52 (2F); −151.13 (2F). 13C NMR (CD3OD, 75 mHz): δ 161.06, 147.46, 144.48, 141.20, 123.91, 114.26, 40.48, 39.27, 33.95. IR (cm−1): ν 2129 (N3), 1651 (CO). MS: 292.2 (calcd 292.3 for C10H9ON5F4).
Preparation of ribozyme constructs
The hairpin ribozyme constructs HF1 and HF16 were prepared by in vitro transcription. Transformed cells containing plasmids pHF1, pHF16 and pAVA were generously donated by A. Ferré-D'Amaré (Fred Hutchison Cancer Research Center, Seattle, WA). The plasmid pAVA encodes for a ribozyme engineered to cleave after G92 of the HF16 transcript in order to eliminate 3′ heterogeneity, facilitating subsequent purification (22). Plasmids were expressed in bacterial colonies and linearized as in (6). In vitro transcription was optimized and carried out on a preparatory scale as in (23), using RNA polymerase prepared as in (24). The transcripts were purified by DPAGE as described in (25) to yield 0.36 nmol HF1 and 0.73 nmol HF16. Each purified transcript (10 μM, 50 μl) was dephosphorylated with calf intestinal alkaline phosphatase, then precipitated by addition of H2O (62.5 μl), NaOAc (3 M, 12.5 μl) and ethanol (375 μl, −20°C).
Preparation of photo-crosslinker-modified RNA
Reaction mixtures containing the 2′-NH2-modified RNA constructs were prepared as previously described (26) except formamide was included in the reaction mixtures to a final concentration of 27% in order to denature the RNA. A solution of 1, 2, 3 or 4 in anhydrous CH2Cl2 (2.5 mg/ml, 200 μl) was then dried by vacuum centrifugation and dissolved in anhydrous DMF (−5°C, 5 μl). An aliquot of the resulting solution (0.5 μl) was immediately added to the RNA solution. The mixture was manually mixed using a pipetman, then incubated at −5°C for 30–45 min. Compound 1, 2, 3 or 4 was added 5 more times, each after a 30–45 min incubation period, using the same procedure and a fresh aliquot of isocyanate or isothiocyanates each time. After the final addition, the reaction mixture was incubated at −5°C for 30–45 min, then precipitated by addition of NaOAc (3 M, 25 μl) and ethanol (−20°C, 750 μl). The extent of modification was determined by DPAGE: 1, 84%; 2, 75%; 3, 100%; 4, 100%.
Radiolabeling of oligonucleotides
A solution of oligoribonucleotide (1 μl, 10 μM) was mixed with T4 polynucleotide kinase (PNK 0.5 μl, 5 U, NEN), [γ-32P]ATP (10 μCi/μl, 1.25 μl, NEN), 10× PNK buffer (NEN) and H2O (8.5 μl). Reaction mixtures were incubated at 37°C for 1 h then at 65°C for 10 min, after which they were precipitated by addition of H2O (2.5 μl), NH3OAc (5 M, 60 μl) and EtOH (187.5 μl, −20°C).
Photo-crosslinking
Either the ribozyme or the substrate strand was radiolabeled in all photo-crosslinking experiments. HF1 or HF16 (10 μM, 0.5 μl) was combined with photo-crosslinker-modified hairpin substrate (HPS-1, 2, 3 or 4, 10 μM, 1 μl), buffer containing 10 mM Na2HPO4/NaH2PO4, pH 7.0, 100 mM NaCl and 0.1 mM EDTA (2 μl) and H2O (0.5 μl) and heated at 90°C for 2 min, 60°C, 50°C and 40°C for 5 min each and 22°C for 15 min, to anneal the RNA–substrate constructs. H2O or MgCl2 (50 mM, 1 μl) was added and solutions were equilibrated at room temperature for 15 min. Final concentrations: 2 μM ribozyme, 4 μM substrate, 40 mM NaHPO4/H2PO4, 40 mM NaCl, 0.04 mM EDTA and 0 or 10 mM MgCl2. Crosslinking experiments were also carried out on an analytical scale in 50 mM Tris–HCl, pH 7.5, as in (19). The same crosslinks were formed in each case, in similar yields to crosslinks generated in the phosphate buffer. For temperature-dependence studies, glycerol (10 μl) was mixed with the solutions before irradiation. Tubes were immersed in ice, or, for temperature-dependence studies, isopropanol/dry-ice (−78°C), ice (0°C), trays (25°C) or heating blocks (37°C) and covered with half of a Petri dish to filter out low-wavelength UV light. A handheld 302-nM lamp was rested directly on top of the Petri dish. Solutions were irradiated for 30 min.
Isolation of crosslinks by DPAGE
Crosslinked RNA mixtures were separated by DPAGE (3000 V, 24 h). Bands representing crosslinks were excised, crushed and eluted in 10 mM Tris–HCl, 1 mM Na2EDTA and 250 mM NaCl, pH 7.5 overnight at 4°C. Supernatants were removed by centrifugation and were adjusted to a total volume of 225 μl with H2O. The RNA was precipitated by addition of 3 M NaOAc (23 M, 5 μl), ethanol (−20°C, 750 μl) and glycogen (10 mg/ml, 0.5 μl).
Mapping of crosslinks by limited alkaline hydrolysis and T1 RNase digestion
Each pellet containing crosslinked RNA was dissolved in H2O (3.0 μl) and mixed with unlabeled 5S RNA (2.5 mg/ml, 1 μl) and NaHCO3 (250 mM, 0.5 μl). The solutions were incubated at 90°C for 2.5 min and then quick-frozen in dry ice. This procedure was also performed on a sample of uncrosslinked, radiolabeled RNA to provide a control ladder. Samples were mixed with 8 M urea (2.5 μl) and analyzed by 20% DPAGE (2.5 h, 3000 V).
To prepare the T1 ladders, solutions containing trace amounts (≤1 pmol) of radiolabeled RNA were mixed with buffer containing 8.75 M urea, 1.25 mM Na2EDTA and 31.25 mM sodium-citrate, pH 5.0 (8 μl) and T1 RNase (0.6 U/μl, 1.0 μl, Gibco BRL). Reaction mixtures were incubated at 37°C for 15 min and then quick-frozen in dry ice. T1 RNase digestions were thawed and loaded directly onto gels alongside the limited alkaline hydrolysis digestions.
Hairpin ribozyme activity assays
HF16 (10 μM, 7.2 μl) was mixed with 250 mM Tris–HCl, pH 7.5 (14.4 μl) and H2O (7.2 μl). In separate tubes, 5′-32P radiolabeled cleavable HPS, HPS-2′NH2 or HPS-1 (1 μM, 3.0 μl) was mixed with buffer containing 250 mM Tris–HCl and 0.5 mM Na2EDTA, pH 7.5 (6.0 μl) and H2O (3.0 μl). All RNA solutions were incubated at 90°C for 1 min, then at room temperature for 15 min. To each solution was added 50 mM MgCl2 (7.2 μl to the HF16 solution and 3.0 μl to each of the HPS solutions), which were then equilibrated at room temperature for 15 min. An aliquot of each of the HPS solutions (3 μl) was mixed with stop-mix (3.5 M urea, 25 mM Na2EDTA and 0.025% each of bromophenol blue and xylene cyanole, 6 μl) for t0 timepoints. Reactions were initiated by addition of HF16 solution (12 μl) to each of the remaining HPS solutions (12 μl). Final concentrations (24 μl): 1 μM ribozyme, 100 nM substrate, 50 mM Tris–HCl, 0.1 mM Na2EDTA and 10 mM MgCl. Reactions were carried out at 25°C and time points were taken by combining an aliquot of each reaction mixture (5 μl) with stop-mix (10 μl). Time points were analyzed by 20% DPAGE (1.5 h, 500 V).
RESULTS
Synthesis of photo-crosslinkers
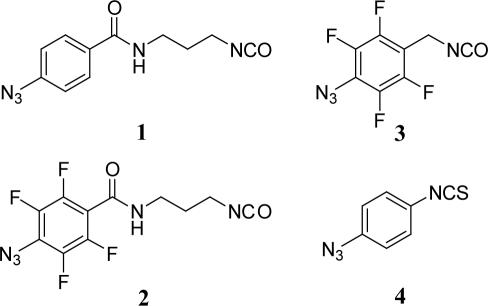
In an effort to explore the effects of tether length and photo-crosslinking chemistry on crosslink yields and locations, we have used four crosslinkers, N-(4-azidobenzoyl)-3-isocyanatopropyl amide (1), N-(4-azido-2,3,5,6-tetrafluorobenzoyl)-3-isocyanatopropyl amide (2), 4-azido-2,3,5,6-tetrafluorobenzyl isocyanate (3) and 4-azidoisothiocyanate (4) (Figure 2). Upon irradiation, the phenylazide groups of crosslinkers 1 and 4 are expected to undergo rearrangement to a highly electrophilic dehydroazepine ring and react with nucleophilic atoms in the RNA (27,28), while the fluorinated crosslinkers 2 and 3 are expected to react as the singlet nitrene intermediate, primarily via C–H bond insertion (29–31). These four photo-crosslinkers also represent different molecular ‘rulers’. Measured from the reactive nitrogen in the azide group to the 2′-NH2 group in the RNA, the crosslinkers are 15.2 (1 and 2), 10.7 (3) and 9.5 Å (4) long. Photo-crosslinker 2 is the same length as 1 and was included in order to directly compare the effects of the different arylazide chemistry pathways on quantity and locations of crosslinks.
Figure 2.
The four photo-crosslinking agents 1–4 used in this study that span distances of 15.2, 15.2, 10.7 and 9.5 Å, respectively.
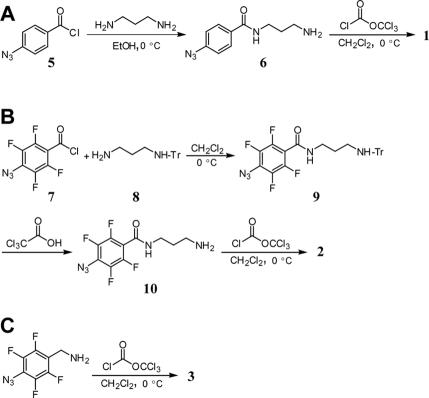
The synthetic routes for crosslinkers 1–3 are shown in Figure 3. With the exception of 4, which was purchased as an isothiocyanate, each photo-crosslinker was synthesized from its corresponding primary amine by treatment with diphosgene in the presence of N,N,N′,N′-tetramethyl-1,8-naphthalenediamine (Proton Sponge) (32). The compound 4-azidobenzoyl chloride (5) (21) was treated with 1,3-diaminopropane in a simple displacement reaction generating 6 in 12% yield. An attempt was made to prepare amine 10 in the same manner as 6 by reaction of acid chloride 7 (21) with a large excess of 1,3-diaminopropane. However, IR, NMR and mass spectral data of the purified product indicated that the azide had been reduced to the amine during this step. This result was corroborated by a very low level of crosslinking when it was irradiated in a hairpin ribozyme/substrate complex. The reduction of the azide was circumvented by using monotritylated 1,3-diaminopropane (8). Compound 8 was prepared in 15% yield by reaction of 1,3-diaminopropane with triphenylmethyl (trityl) chloride. Reaction of 8 with 7 yielded protected amine 9 in 94% yield. The trityl group was removed by incubation of 9 in trichloroacetic acid to form 10 in 18% yield. Finally, 10 was converted to isocyanate 2 as described for 1, in 54% yield. The amine precursor to 3 was commercially available and was converted to the isocyanate 3 in 84% yield.
Figure 3.
Synthetic routes for crosslinkers 1 (A), 2 (B) and 3 (C).
Incorporation of photo-crosslinkers into RNA
The substrate strand was chosen for photo-crosslinker modification because of its short length, allowing us to use synthetic 2′-NH2 modified oligonucleotides, and the U+2 position was chosen because of its proximity to domain B in the crystal structure. We expected to observe crosslinks to A26, C27 and A28, which span distances of 5.1, 4.9 and 5.9 Å, respectively, from the 2′-NH2 group of U+2. In order to avoid complication in analysis of photo-crosslinking data due to substrate cleavage by the ribozyme, the substrate contained a deoxyadenosine nucleotide at the cleavage site (Figure 1). The crosslinkers were incorporated into the hairpin ribozyme by reaction of a 2′-NH2 modification at the U+2 position in the substrate strand with their isocyanate or isothiocyanate functionalities, which react specifically with aliphatic amines over hydroxyl groups and aromatic amino groups in RNA (26). Photo-crosslinkers 1–4 in DMF were added to solutions of hairpin substrate in borate buffer and formamide at −4°C, resulting in 75–100% coupling.
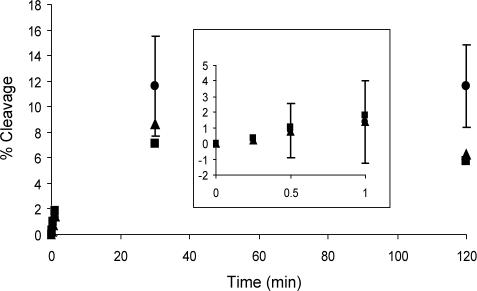
Two different hairpin ribozyme constructs, HF1 and HF16 (Figure 1), were used in this study. HF16 contains a loop onto which the U1A protein RNA binding domain was grafted to facilitate crystallization (6). In HF1, the U1A binding domain is replaced with a smaller stem–loop, but this construct is otherwise identical to HF16. In order to determine whether the 2′-NH2 or photo-crosslinker modifications on the substrate affect the ability of the ribozyme to adopt its active conformation, both constructs were incubated with a cleavable substrate bearing no modification, 2′-NH2 or 2′-1 at the U+2 position. A time course for the cleavage showed that all three substrates were cleaved to a similar extent, suggesting these modifications do not alter the tertiary structure of the ribozyme (Figure 4). The equilibrium ratio of cleaved to intact HPS, HPS-2′NH2 and HPS-1 was 0.12, 0.07 and 0.09, respectively. In comparison, the internal equilibrium of substrate cleavage by a four-way junction hairpin ribozyme was determined to be between 0.03 and 0.43 (33). Thus, 2′-NH2 and 2′-1 modifications do not significantly alter the activity of the hairpin ribozyme.
Figure 4.
Time course of HF16 hairpin ribozyme-catalyzed cleavage of unmodified (circles), 2′-NH2-modified (squares) and photo-crosslinker 1-modified (triangles) substrate. Insert shows an expansion of early time points. SD values were 2, 2, 3 and 4% for 2′-NH2-modified HF16 activity and 0.4, 2, 3 and 1% for 1-modified HF16 activity at 0.5, 1, 30 and 120 min, respectively.
Photo-crosslinking efficiency
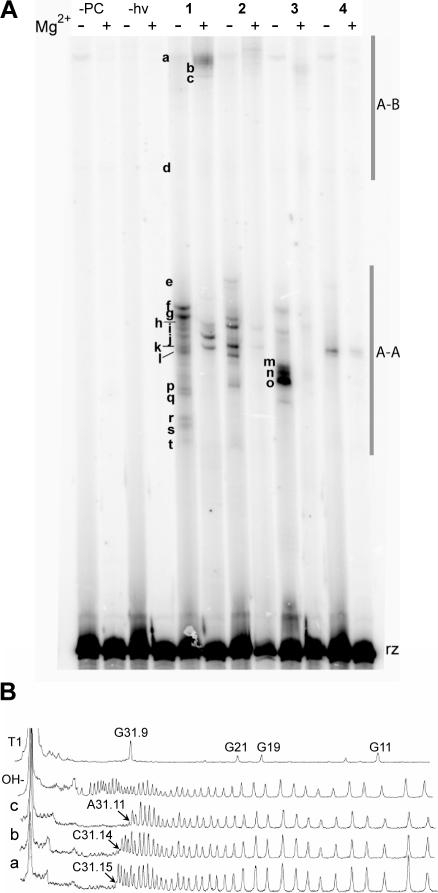
Photo-crosslinker-modified substrates were annealed to each 5′-32P radiolabeled ribozyme construct and irradiated with 302 nm light at 0°C, both in the presence and absence of Mg2+ (10 mM). Crosslinked ribozyme–substrate complexes were separated from uncrosslinked ribozyme by DPAGE (Figure 5), and quantified. The total crosslink yields and the number of crosslinks decreased with decreasing tether length and with fluorine substitution, both with and without Mg2+ (Table 1).
Figure 5.
(A) DPAGE analysis of crosslinks formed by all four photo-crosslinkers in the absence and presence of Mg2+ (10 mM). Bands representing crosslinks are designated with lowercase letters. The bands marked A–A represent intradomain crosslinks; those marked A–B represent interdomain crosslinks. ‘rz’ indicates uncrosslinked hairpin ribozyme. (B) Sample densitometry traces of bands a, b and c. Arrows point to the first absent band from the 5′ end of the sequence, which indicates the location of the crosslink.
Table 1.
Summary of the locations of crosslinks and their distances from U+2
| Crosslink site | Band # | Distance (Å) | 1 (15.2 Å) | 2 (15.2 Å) | 3 (10.7 Å) | 4 (9.5 Å) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Temperature (°C) | −78 | 0 | 25 | 37 | 0 | 0 | 0 | ||||||||||
| Mg2+ | − | + | − | + | − | + | − | + | − | + | − | + | − | + | |||
| G2 | t | 13.6 | 1 | ||||||||||||||
| A3 | r–s | 16.8 | 2 | 0.3 | 3 | 0.7 | 3 | ||||||||||
| G4 | p–q | 19.9 | 2 | 1 | 4 | 0.4 | |||||||||||
| G6 | o | 13.1 | 1 | ||||||||||||||
| A7 | m–n | 11.3 | 8 | 0.3 | |||||||||||||
| G8 | 2 | 5 | |||||||||||||||
| A9 | i–l | 10 | 3 | 4 | 17 | 6 | 15 | 1 | 17 | 2 | 11 | 2 | 7 | 0.1 | 4 | 0.4 | |
| A10 | e–h | 10.8 | 3 | 2 | 9 | 2 | 10 | 9 | 14 | 11 | 6 | 2 | 2 | 0.4 | |||
| G11 | 0.1 | ||||||||||||||||
| G21 | d | 24.5 | 0.6 | 6 | 1 | 0.1 | |||||||||||
| A26 | 0.7 | 2 | |||||||||||||||
| A31.11* | c | 26.5 | 0.3 | ||||||||||||||
| C31.14* | b | 14 | 6 | 0.2 | |||||||||||||
| C31.15* | a | 12.5 | 0.4 | 2 | 0.6 | 0.5 | 6 | 23 | 3 | 20 | 0.3 | 0.1 | |||||
| G36 | 0.2 | 0.5 | |||||||||||||||
| Total crosslink yield | 10 | 16 | 38 | 15 | 32 | 33 | 41 | 34 | 18 | 4 | 21 | 1.3 | 4.1 | 0.8 | |||
Crosslink yields are reported as percentages of uncrosslinked ribozyme. Underlined and bold numbers indicate crosslinks that were only observed in HF1 or HF16, respectively. All other crosslinks at 0°C were observed in both hairpin constructs. Data at −78, 25 and 37°C reflect crosslinks in HF16 only. Bold nucleotides were only observed at −78°C. The distances to nucleotides marked with an asterisk are those reported in the crystal structure with the U1A protein bound to the loop. SD values ranged from 0.1 to 0.4% for all crosslink yields except those for A9 and A10, which reflected larger SD values of 2–5%.
Mapping of crosslinks
Two distinct sets of crosslinks were consistently observed. Although all crosslinks had lower gel mobility than uncrosslinked ribozyme, crosslinks a–d had significantly lower mobility than crosslinks e–t (Figure 5). Crosslinks were purified by DPAGE, subjected to limited alkaline hydrolysis and run alongside hydrolysis and T1 RNase ladders of unmodified ribozyme. Gaps in the ladders were compared to the sequencing lanes and the locations of crosslinks were identified. Five crosslinks (A3, G4, A7, A9 and A10) were represented by more than one band (r–s, p–q, m–n, i–l and e–h, respectively), which is probably due to crosslinking to different atoms in the same nucleotide. We found that high-mobility crosslinks e–t represent intradomain crosslinks to nucleotides in domain A and low-mobility crosslinks a–d represent interdomain crosslinks to nucleotides in loop B. Table 1 summarizes the mapping data (the locations of crosslinked nucleotides and their distances to U+2, the site of crosslinker incorporation) for crosslinkers 1–4. Crosslinks to A9 and A10 were the most reproducible intradomain crosslinks under all conditions. In the absence of Mg2+, crosslinks were also formed to nucleotides in the 3′ direction of these sites (G2–A7). We were not able to map crosslinks generated by 4 because of the low level of crosslinking, but we were able to compare the mobility of the crosslinks generated by 4 with those formed by 1–3. The mobility of crosslinks produced by 4 indicates that this photo-crosslinker forms crosslinks to A9 and C31.15 both in the absence and presence of Mg2+. Interdomain crosslinks were only observed to G21 and three nucleotides (A31.11, C31.14 and C31.15) in the U1A binding loop of HF16, rather than the expected sites of crosslinking in domain B (A26–A28), when the experiments were carried out at 0°C. However, at −78°C, interdomain crosslinks were observed at A26 and G36, as discussed below.
Mg2+ dependence
The presence of Mg2+ had three distinct effects on both crosslinking efficiency and locations of crosslinks. First, total crosslink yields were lower by 60–94% in the presence of Mg2+ than in its absence. Second, the ratio of the yields of interdomain crosslinks to intradomain crosslinks was higher in the presence of Mg2+ than in its absence. Third, more crosslinks inconsistent with the crystal structure were formed in the absence of Mg2+ than in its presence (Table 1). It should be noted that the distances reported in Table 1 are those between the 2′-N of U+2 and the nearest atom on the crosslinked nucleoside. Thus, the reported distances are only approximate since the atomic connectivity of the crosslinks is not known.
Temperature dependence
In order to observe any potential effects of ribozyme flexibility on crosslink locations, we performed crosslinking experiments with 1 at varying temperatures. Prior to irradiation, hairpin ribozyme/photo-crosslinker-modified substrate complexes were mixed with glycerol to a final concentration of 67% to prevent freezing. These samples were irradiated at −78, 0, 25 and 37°C for 30 min with 302 nm light, as above. Comparison of crosslinking at 0°C for 30 min containing glycerol with those containing no glycerol revealed that the inclusion of glycerol in the reaction mixture did not affect either crosslinking yields or the identity of the crosslinks, suggesting that the ribozyme adopts the same tertiary structure and dynamics under both conditions.
Two temperature-dependent effects were observed in these experiments (Figure 6). First, photo-crosslinking efficiency increased with increasing temperatures (Table 1). These results are consistent with temperature-dependent laser flash photolysis studies, which suggest that the dehydroazepine intermediate is more reactive at higher temperatures (34). Second, two new intradomain crosslinks were observed, to G8 and G11, and two new interdomain crosslinks were observed, to A26 and G36 at −78°C (Figure 6). Third, the crosslinks to G2–A7, A31.11 and C31.14 were not observed at −78°C. We also observed that most crosslinks that were produced by 1 in the absence of Mg2+ were also formed in its presence at −78°C, with the exception of crosslinks to G11 and G21, which were observed only in the absence of Mg2+. Finally, fewer crosslinks inconsistent with the crystal structure formed at −78°C, compared to 0°C (Table 1).
Figure 6.
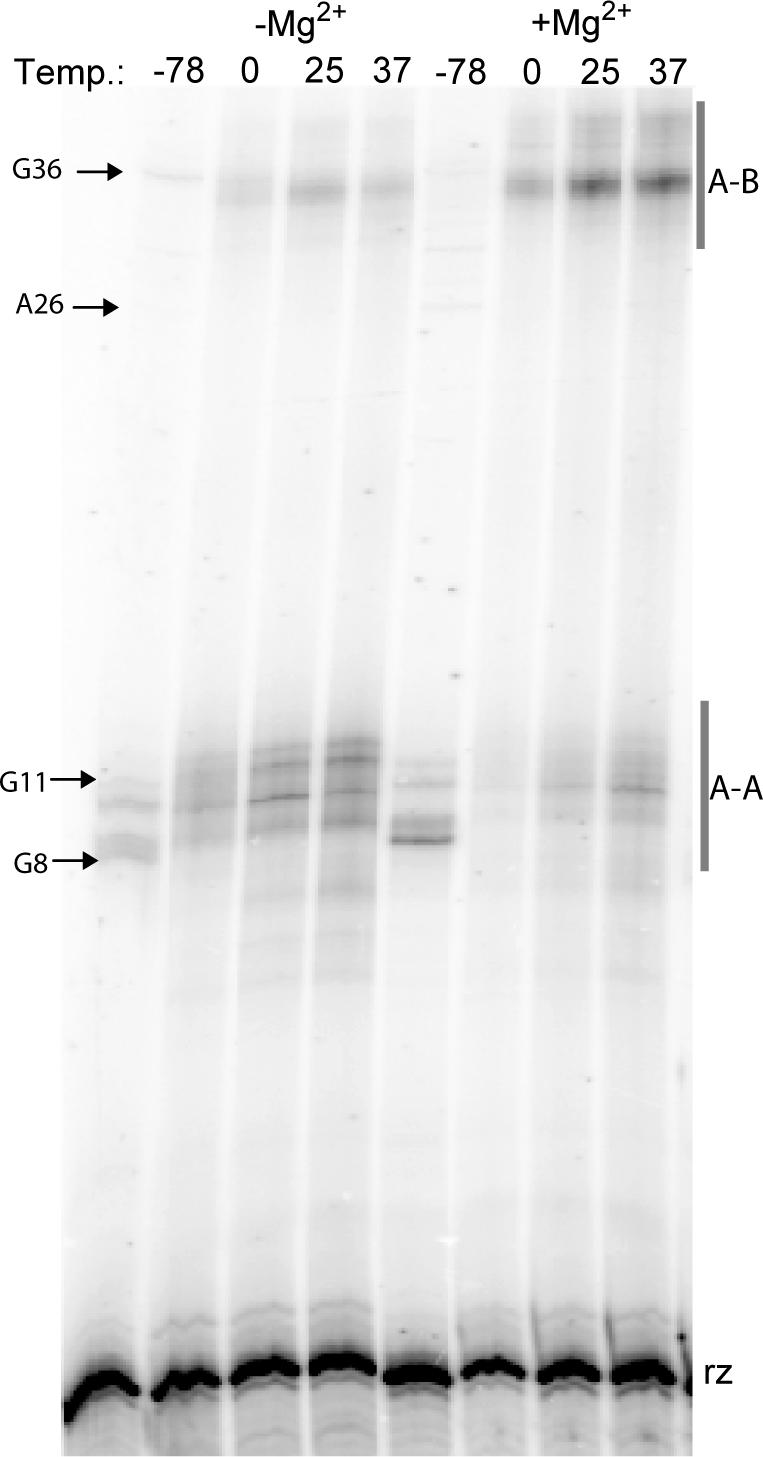
DPAGE analysis of temperature-dependent photo-crosslinking in the absence and presence of Mg2+ (10 mM). Bands representing new crosslinks are marked accordingly, except G11 which was only observed under the larger-scale conditions of preparatory experiments. The bands marked A–A represent intradomain crosslinks; those marked A–B represent interdomain crosslinks. ‘rz’ indicates uncrosslinked hairpin ribozyme. Temperature units are °C.
DISCUSSION
The goals of this study were 2-fold. First, by performing crosslinking experiments in the absence and presence of Mg2+ and at different temperatures, we studied the folding and flexibility of the hairpin ribozyme. Second, we compared the crosslinks formed by four different photo-crosslinking agents of different lengths and chemical reactivity under various conditions, in order to observe the effects of tether length and chemistry on crosslinks. To our knowledge, this is the first example of such a comparative study within the same RNA.
Mg2+ dependence
Comparison of photo-crosslinks in the absence and presence of 10 mM Mg2+ has allowed us to monitor divalent metal-ion dependent flexibility and folding of the hairpin ribozyme. First, we observed a higher ratio of inter- to intradomain crosslinks in the presence of Mg2+ than in its absence. This difference is evidence for a higher ratio of docked to undocked ribozyme in the presence of divalent metal ions, which has previously been demonstrated in ensemble FRET studies of a hairpin ribozyme four-way junction (12). Second, we observed a higher total yield of crosslinks in the absence of Mg2+ than in its presence, primarily due to a higher number and yield of intradomain crosslinks. Third, we observed a higher number of crosslinks in the presence of Mg2+, which were consistent with the crystal structure, suggesting that the hairpin is folded into a tertiary conformation that is similar to the crystal structure at 10 mM Mg2+. All of these data are consistent with previous findings that Mg2+ concentrations in the millimolar range are required for full docking of the two domains (10).
Temperature dependence
Crosslinks to expected sites in domain B of the hairpin ribozyme (A26–A28) were not observed at 0°C. Thus, we performed crosslinking experiments with 1 at −78°C in order to investigate whether flexibility of the ribozyme itself could be contributing to the lack of crosslinks to the expected region of the ribozyme. We found that crosslinks formed at this temperature were more consistent with the crystal structure of the ribozyme than those formed at higher temperatures in particular to A26 and G36. These crosslinks were not observed at 0–37°C, but are consistent with the crystal structure (they are 5.0 and 9.5 Å, respectively, from U+2), implying that the two domains are docked. The observation of more crosslinks, which are consistent with the crystal structure at −78°C could be explained by a more static structure, while crosslinking data at higher temperatures reflects a more dynamic structure.
The most striking observation from the low-temperature photo-crosslinking studies was that most crosslinks in both constructs that were formed at −78°C with 1 were observed both in the presence and absence of Mg2+. In contrast, at 0°C, many crosslinks generated by 1 and 3 were formed under only one or the other condition. Comparison of crosslinks generated by 1 at −78°C with those formed by 1 at 0°C implies that Mg2+ stabilizes an already preferred fold, which is consistent with the crystal structure of the ribozyme.
Intra- versus interdomain crosslinks
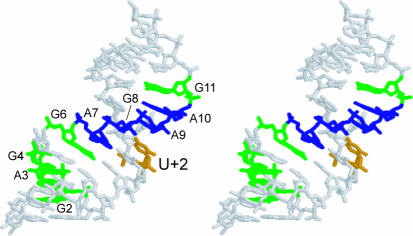
Intradomain crosslinks were the strongest, most reproducible crosslinks. Figure 7 shows the locations of these intradomain crosslinks in the crystal structure (6) relative to the site of crosslinker incorporation (U+2). Although a few crosslinks to nucleotides out of theoretical range of the crosslinker that formed them were observed in the absence of Mg2+, none which spanned distances more than 1.3 Å longer than the crosslinker in the crystal structure were observed in the presence of Mg2+ (Table 1). These data suggest that unfolded or misfolded conformations of the hairpin ribozyme exist in the absence of Mg2+, or that the hairpin ribozyme is simply more flexible under those conditions.
Figure 7.
Stereoview of domain A of the hairpin ribozyme showing all intradomain crosslinks observed in this study. The site of photo-crosslinker incorporation (U+2) is shown in orange. Green nucleotides represent crosslinks formed only in the absence of Mg2+ and blue nucleotides represent crosslinks formed both in the absence and presence of Mg2+.
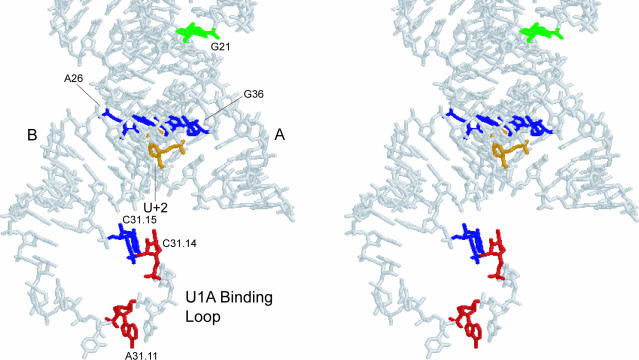
Figure 8 shows the locations of all interdomain crosslinks in three-dimensional space. Most interdomain crosslinks at 0°C were to the U1A protein binding domain, instead of domain B. Both photo-crosslinkers 1 and 3 formed crosslinks to A31.11, C31.14 and C31.15 in this loop. This is a nonconserved, flexible loop which probably inhabits several locations relative to the rest of the ribozyme, making it accessible to the photo-crosslinkers. Therefore, crosslinks to this loop are to be expected. The lack of crosslinks to the nucleotides A26–A28 of domain B, which are 4.9–5.9 Å from U+2, was somewhat surprising at first. Two possible explanations exist for this phenomenon. First, the U1A loop may be physically in the way of the expected area of crosslinking due to its size and flexibility. However, crosslinking with the HF1 construct that lacks the loop does not yield crosslinks to A26–A28 at 0°C. The second, and more likely explanation, is that the U1A binding loop presents more reactive functional groups to the crosslinkers than domain B because of its lack of secondary or tertiary structure. Indeed, four of the five crosslinks observed in Burke and coworkers' crosslinking study of the hairpin ribozyme were to nucleotides in loops A and B (19), which do not form canonical base pairs in the crystal structure (6). Likewise, the majority of crosslinks in a study of the ribosome which employed a phenylazide crosslinker were to unpaired nucleotides (35). Previous studies have shown that dehydroazepine rings, which are the likely rearrangement product of unsubstituted phenylazide rings, react preferably with nitrogen of the nucleoside bases (27,28), which would only be exposed in unpaired nucleotides. Therefore, the lack of crosslinks to A26–A28 seems to reflect the reactivity of these nucleotides toward aryl nitrenes, rather than poor placement of the crosslinking agents.
Figure 8.
Stereoview of domains A and B of the hairpin ribozyme showing all interdomain crosslinks observed in this study. The site of photo-crosslinker incorporation (U+2) is shown in orange. Green nucleotides represent crosslinks formed only in the absence of Mg2+, red nucleotides represent crosslinks formed only in the presence of Mg2+ and blue nucleotides represent crosslinks formed under both conditions.
Reactivity of photo-crosslinkers
In addition to allowing us to explore structural and dynamic properties of the hairpin ribozyme in aqueous solutions, our data provide information about the reactive properties of the photo-crosslinkers themselves. We found that decreasing tether length results in fewer crosslinks and lower yields. ‘Zero-length’ photo-crosslinkers such as 4-thio uridine, have been used extensively in investigations of the structure of RNA (36–39). Although fewer crosslinks are obtained, they give higher resolution structural data because the distance constraints they provide can be confined to within 4 Å (38). Similarly, we observe fewer crosslinks with the shorter crosslinkers, but the distance constraints they provide are confined to a smaller range.
Fluorine substitution decreases crosslinking yields. Phenylazide 2, which has the same structure as 1 except for the fluorine substitutions in the aromatic ring, crosslinks with about an order of magnitude less efficiency than 1. This could be due to higher reactivity of the nitrene formed upon irradiation of 2, which could decrease its lifetime due to reactions with solvent and buffer molecules. In addition, the nitrene generated by photolysis of unsubstituted aryl azides can rearrange to an electrophilic dehydroazepine intermediate, providing a photo-crosslinking mechanism by nucleophilic addition (27,28). An important result from this study is that no new crosslinks were formed by 2 compared to 1, showing that the determinant of photo-crosslinking sites for these phenylazide crosslinkers is the length, rather than the chemistry of the photo-crosslinker.
SUMMARY
In conclusion, photo-crosslinking was used in this study to monitor both structural and dynamic features of RNA. All crosslinks formed in the presence of Mg2+ were consistent with the hairpin's crystal structure. Crosslinks that were out of theoretical range of the crosslinkers formed only in the absence of Mg2+ and can be explained by increased flexibility of the ribozyme in the absence of Mg2+. We have gained further insight into the role of divalent metal ions in the folding of the hairpin ribozyme through temperature-dependent photo-crosslinking experiments. At low temperatures, Mg2+ did not appear to be required for folding, suggesting that the docked form of the hairpin ribozyme four-way junction construct appears to be a low-energy structure that is stabilized by Mg2+. Photo-crosslinkers with longer tether lengths generated a higher number of crosslinks to nucleotides farther away from the site of photo-crosslinker incorporation, than those with shorter tether lengths. Inclusion of fluorine substitutions into the phenylazide ring did not change the locations to which the crosslinks formed, but only decreased the number and yields of crosslinks, showing that unsubstituted phenylazide crosslinkers are better suited for studying RNA structure. The crosslinking data show that a major limitation of photo-crosslinking approaches with phenylazide crosslinkers is that they preferably react with unpaired nucleotides. Nonetheless, phenylazide photo-crosslinkers are useful structural reporters that can provide insights into RNA structure and dynamics, especially when used in conjunction with other structural data.
Acknowledgments
We thank members of the Sigurdsson Research Group for critical review of the manuscript, and Dr Adrian Ferré-D'Amaré for his help with preparation of the plasmids for transcription of the hairpin ribozyme constructs used in this study. This work was supported by a grant from the National Institutes of Health (GM56947). Funding to pay the Open Access publication charges for this article was provided by the University of Washington.
REFERENCES
- 1.Hampel K.J., Walter N.G., Burke J.M. RNA catalytic properties of the minimum (−)sTRSV sequence. Biochemistry. 1989;28:4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- 2.Ferre-D'Amare A.R. The hairpin ribozyme. Biopolymers. 2003;73:71–78. doi: 10.1002/bip.10516. [DOI] [PubMed] [Google Scholar]
- 3.Fedor M. Structure and function of the hairpin ribozyme. J. Mol. Biol. 2000;297:269–291. doi: 10.1006/jmbi.2000.3560. [DOI] [PubMed] [Google Scholar]
- 4.Walter F., Murchie A.I.H., Lilley D.M.J. Folding of the four-way RNA junction of the hairpin ribozyme. Biochemistry. 1998;37:17629–17636. doi: 10.1021/bi9821115. [DOI] [PubMed] [Google Scholar]
- 5.Siwkowski A., Shippy R., Hampel A. Analysis of hairpin ribozyme base mutations in loops 2 and 4 and their effects on cis-cleavage in vitro. Biochemistry. 1997;36:3930–3940. doi: 10.1021/bi9628735. [DOI] [PubMed] [Google Scholar]
- 6.Rupert P.B., Ferre-D'Amare A. Crystal structure of a hairpin ribozyme–inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 7.Hampel A., Cowan J.A. A unique mechanism for RNA catalysis: the role of metal cofactors in hairpin ribozyme cleavage. Chem. Biol. 1997;4:513–517. doi: 10.1016/s1074-5521(97)90323-9. [DOI] [PubMed] [Google Scholar]
- 8.Nesbitt S., Hegg L.A., Fedor M. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- 9.Young K.J., Gill F., Grasby J.A. Metal ions play a passive role in the hairpin ribozyme catalysed reaction. Nucleic Acids Res. 1997;25:3760–3766. doi: 10.1093/nar/25.19.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson T.J., Lilley D.M.J. Metal ion binding and the folding of the hairpin ribozyme. RNA. 2002;8:587–600. doi: 10.1017/s1355838202020514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan E., Wilson T.J., Nahas M., Clegg R.M., Lilley D.M.J., Ha T. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc. Natl Acad. Sci. USA. 2003;100:9308–9313. doi: 10.1073/pnas.1233536100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohng S., Wilson T.J., Tan E., Clegg R.M., Lilley D.M.J., Ha T. Conformational flexibility of four-way junctions in RNA. J. Mol. Biol. 2004;336:69–79. doi: 10.1016/j.jmb.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 13.Pljevaljcic G., Millar D.P., Deniz A.A. Freely diffusing single hairpin ribozymes provide insights into the role of secondary structure and partially folded states in RNA folding. Biophys. J. 2004;87:457–467. doi: 10.1529/biophysj.103.036087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bokinsky G., Rueda D., Misra V.K., Rhodes M.M., Gordus A., Babcock H.P., Walter N.G., Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc. Natl Acad. Sci. USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Z., Tinoco J.Ignacio. Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry. 1996;35:6026–6036. doi: 10.1021/bi952985g. [DOI] [PubMed] [Google Scholar]
- 16.Butcher S.E., Allain F.H.-T., Feigon J. Solution structure of the loop B domain from the hairpin ribozyme. Nature Struct. Biol. 1999;6:212–216. doi: 10.1038/6651. [DOI] [PubMed] [Google Scholar]
- 17.Ryder S.P., Strobel S.A. Comparative analysis of hairpin ribozyme structures and interference data. Nucleic Acids Res. 2002;30:1287–1291. doi: 10.1093/nar/30.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Earnshaw D.J., Masquida B., Muller S., Sigurdsson S.T., Eckstein F., Westhof E., Gait M.J. Inter-domain cross-linking and molecular modelling of the hairpin ribozyme. J. Mol. Biol. 1997;274:197–212. doi: 10.1006/jmbi.1997.1405. [DOI] [PubMed] [Google Scholar]
- 19.Pinard R., Heckman J.E., Burke J.M. Alignment of the two domains of the hairpin ribozyme–substrate complex defined by interdomain photoaffinity crosslinking. J. Mol. Biol. 1999;287:239–251. doi: 10.1006/jmbi.1999.2626. [DOI] [PubMed] [Google Scholar]
- 20.Merrill S.H., Unruh C.C. Photosensitive azide polymers. J. Appl. Polym. Sci. 1963;7:273–279. [Google Scholar]
- 21.Keana J.F.W., Cai S.X. New reagents for photoaffinity labeling: synthesis and photolysis of functionalized perfluorophenyl azides. J. Org. Chem. 1990;55:3640–3647. [Google Scholar]
- 22.Ferre-D'Amare A.R., Doudna J.A. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikonowicz E.P., Sirr A., Legault P., Jucker F.M., Baer L.M., Pardi A. Preparation of 13C and 15N labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res. 1992;20:4507–4513. doi: 10.1093/nar/20.17.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He B., Rong M., Lyakhov D., Gartenstein H., Diaz G., Castagna R., McAllister W.T., Durbin R.K. Rapid mutagenesis and purification of phage RNA polymerases. Protein Expr. Purif. 1997;9:142–151. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- 25.Markley J.C., Godde F., Sigurdsson S.T. Identification and characterization of a divalent metal ion-dependent cleavage site in the hammerhead ribozyme. Biochemistry. 2001;40:13849–13856. doi: 10.1021/bi015634c. [DOI] [PubMed] [Google Scholar]
- 26.Sigurdsson S.T., Eckstein F. Site specific labelling of sugar residues in oligoribonucleotides: reactions of aliphatic isocyanates with 2′ amino groups. Nucleic Acids Res. 1996;24:3129–3133. doi: 10.1093/nar/24.16.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buchmueller K.L., Hill B.T., Platz M.S., Weeks K.M. RNA-tethered phenyl azide photocrosslinking via a short-lived indiscriminant electrophile. J. Am. Chem. Soc. 2003;125:10850–10861. doi: 10.1021/ja035743+. [DOI] [PubMed] [Google Scholar]
- 28.Schrock A.K., Schuster G.B. Photochemistry of phenyl azide: chemical properties of the transient intermediates. J. Am. Chem. Soc. 1984;106:5228–5234. [Google Scholar]
- 29.Gritsan N.P., Gudmundsdottir A.D., Tigelaar D., Zhu Z., Karney W.L., Hadad C.M., Platz M.S. A laser flash photolysis and quantum chemical study of the fluorinated derivatives of singlet phenylnitrene. J. Am. Chem. Soc. 2001;123:1951–1962. doi: 10.1021/ja9944305. [DOI] [PubMed] [Google Scholar]
- 30.Schnapp K.A., Platz M.S. A laser flash photolysis study of di-, tri- and tetrafluorinated phenylnitrenes; implications for photoaffinity labeling. Bioconjug. Chem. 1993;4:178–183. doi: 10.1021/bc00020a011. [DOI] [PubMed] [Google Scholar]
- 31.Schnapp K.A., Poe R., Leyva E., Soundararajan N., Platz M.S. Exploratory photochemistry of fluorinated aryl azides. Implications for the design of photoaffinity labeling reagents. Bioconjug. Chem. 1993;4:172–177. doi: 10.1021/bc00020a010. [DOI] [PubMed] [Google Scholar]
- 32.Sigurdsson S.T., Seeger B., Kutzke U., Eckstein F. A mild and simple method for the preparation of isocyanates from aliphatic amines using trichloromethyl chloroformate. Synthesis of an isocyanate containing an activated disulfide. J. Org. Chem. 1996;61:3883–3884. doi: 10.1021/jo9521920. [DOI] [PubMed] [Google Scholar]
- 33.Fedor M.J. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry. 1999;38:11040–11050. doi: 10.1021/bi991069q. [DOI] [PubMed] [Google Scholar]
- 34.Poe R., Schnapp K.A., Young M.J.T., Grayzar J., Platz M.S. Chemistry and kinetics of singlet (pentafluorophenyl)nitrene. J. Am. Chem. Soc. 1992;114:5054–5067. [Google Scholar]
- 35.Demeshkina N., Repkova M., Ven‘Yaminova A., Graifer D., Karpova G. Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P, and E sites: a crosslinking study with mRNA analogs carrying an aryl azide group at either the uracil or the guanine residue. RNA. 2000;6:1727–1736. doi: 10.1017/s1355838200000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Favre A., Fourrey J.L. Structural probing of small endonucleolytic ribozymes in solution using thio-substituted nucleobases as intrinsic photolabels. Acc. Chem. Res. 1995;28:375–382. [Google Scholar]
- 37.Bravo C., Lescure F., Laugaa P., Fourrey J.L., Favre A. Folding of the HDV antigenomic ribozyme pseudoknot structure deduced from long-range photocrosslinks. Nucleic Acids Res. 1996;24:1351–1359. doi: 10.1093/nar/24.7.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bravo C., Woisard A., Fourrey J.-L., Laugaa P., Favre A. A Y form of hammerhead ribozyme trapped by photo-cross-links retains full cleavage activity. Biochimie. 1999;81:201–212. doi: 10.1016/s0300-9084(99)80053-4. [DOI] [PubMed] [Google Scholar]
- 39.Hiley S.L., Sood V.D., Fan J., Collins R.A. 4-thio-U cross-linking identifies the active site of the VS ribozyme. EMBO J. 2002;21:4691–4698. doi: 10.1093/emboj/cdf462. [DOI] [PMC free article] [PubMed] [Google Scholar]