Abstract
DNA microarrays were used to evaluate the regulation of the proportion of individual mRNA species in polysomal complexes in leaves of Arabidopsis thaliana under control growth conditions and following a mild dehydration stress (DS). The analysis determined that the percentage of an individual gene transcript in polysomes (ribosome loading) ranged from over 95 to <5%. DS caused a decrease in ribosome loading from 82 to 72%, with maintained polysome association for over 60% of the mRNAs with an increased abundance. To identify sequence features responsible for translational regulation, ribosome loading values and features of full-length mRNA sequences were compared. mRNAs with extreme length or high GU content in the 5′-untranslated regions (5′-UTRs) were generally poorly translated. Under DS, mRNAs with both a high GC content in the 5′-UTR and long open reading frame showed a significant impairment in ribosome loading. Evaluation of initiation A+1UG codon context revealed distinctions in the frequency of adenine in nucleotides −10 to −1 (especially at −4 and −3) in mRNAs with different ribosome loading values. Notably, the mRNA features that contribute to translational regulation could not fully explain the variation in ribosome loading, indicating that additional factors contribute to translational regulation in Arabidopsis.
INTRODUCTION
High-throughput DNA microarray technology has dramatically enhanced the understanding of complicated networks of gene expression. DNA microarrays are routinely used to monitor steady-state transcript abundance, which reflects both transcript synthesis and turnover. However, this technology can also be implemented to measure mRNA turnover (1) and levels of transcripts in messenger ribonucleotide protein particle or polyribosome (polysome) complexes (2–12). We used an oligonucleotide array that monitored 8000 of the ∼28 000 genes of the model plant Arabidopsis thaliana to evaluate the regulation of mRNA translation in rosette leaves (7). This study revealed that the proportion of individual gene transcripts in polysomes varied over a wide range under normal growth conditions, and that mild water deficit stress caused a significant reduction in the level of mRNA in polysomal complexes for the majority of expressed genes. Remarkably, over half of the dehydration-induced mRNAs maintained their association with polysomes under dehydration stress (DS). This and other genome-level surveys of mRNA translation provide a new opportunity to evaluate the features of transcripts that underlie differential mRNA translation.
The analysis of eukaryotic mRNA translation, primarily by use of in vitro systems, has shown that initiation is affected by several features of the 5′-untranslated region (5′-UTR). For example, an extremely short 5′-UTR (<20 nt) inhibited the entry of the 43S pre-initiation complex or recognition of AUG initiation codon (13), whereas a moderately long 5′-UTR promoted initiation (40–100 nt) (14,15). The scanning of the 5′-UTR by the 43S pre-initiation complex was limited by the presence of a strong stem–loop structure, an effect that was dependent on the location and stability of the structure (16). A stem–loop with a predicted free energy value of −20 kcal/mol near to the 5′ end of the mRNA effectively inhibited ribosome entry in vitro; however, a stronger stem–loop structure (−30 kcal/mol) was necessary to abolish ribosome scanning if it was located distant (52 nt) from the 5′ end (17). Although RNA secondary structures have been predicted for plant 5′-UTRs (18), the effect of such structures has not been carefully evaluated. Another factor that contributes to the regulation of initiation of translation in eukaryotes, including plants, is the presence of short uORFs in the 5′ leader that generally impair translation (19–21). The efficiency of initiation is also influenced by the sequences flanking the AUG codon, referred to as the initiation codon context (16). The most frequent nucleotides around the initiation site was reported to be A(A/C)AAA+1UGGC in eudicots and A(A/G)CCA+1UGGC in monocots (22). The nucleotides A−3 and G+4 are the most conserved and are thought to be present in the optimal initiation codon context (14).
Highly abundant mRNAs are typically used as training sets in bioinformatic studies aimed at the determination of sequence features that may contribute to gene regulation, such as nucleotide content and initiation codon context. However, several studies have recognized a discrepancy between the steady-state abundance and translational efficiency of eukaryotic mRNAs (7,23–25). Thus, consideration of the proportion of an individual mRNA species in polysomes may provide a means to identify mRNA features that contribute to translational regulation. Such an evaluation would also require knowledge of the full-length sequence of the mature transcript. There are over 28 000 publicly available full coding-region cDNA sequences for Arabidopsis. These cDNAs provide reliable coding and 3′-UTR sequence information, but may not begin at the 5′ terminus of the mRNA. However, the affinity purification of 5′-7mGppp-capped mRNAs has allowed for the characterization of over 14 000 full-length cDNAs (FL-cDNAs) with 5′-UTR sequences (26–28). These publicly available collections of high quality cDNA and FL-cDNA sequence data provide a valuable resource for bioinformatic characterization of features of the 5′-UTR, coding sequence and 3′-UTR sequences that underlie variation in translational regulation. In this report, a quantitative assessment of the proportion of mRNA in polysomes for over 11 000 genes was used to evaluate the significance of general mRNA sequence features on translational regulation under non-stress (NS) and DS conditions in Arabidopsis.
MATERIALS AND METHODS
Growth conditions and polysome isolation
Plant growth and DS treatment was carried out exactly as described previously (7). Briefly, A.thaliana (Columbia ecotype) plants were grown under short-day conditions (8 h days). Prior to bolting, rosette leaf tissue was harvested from plants grown under well-watered conditions [NS; relative water content (RWC) 81 ± 2.2%] or after 7 days of soil dehydration (DS; RWC, 66 ± 0.1%). The exact procedures (7) were used for the isolation of total cellular RNA and the fractionation of detergent-treated cell extracts into two cellular RNA populations, non-polysomal RNA complexes and polysomal RNA complexes, by centrifugation through 20–60% (w/v) sucrose density gradients.
DNA microarray determination of the proportion of individual mRNAs in polysome complexes
The DNA microarray data were generated with the Affymetrix Arabidopsis whole genome GeneChip (ATH1) exactly as described previously (7) with the only difference in the analysis the version of GeneChip used. Statistical analyses were performed on mRNAs detected as ‘Present’. Briefly, the proportion of mRNA levels in polysomal versus non-polysomal complexes [RL = (expression level in polysomal RNA complexes)/(expression level in non-polysomal RNA complexes)] obtained from the DNA microarray and quantitative real-time RT–PCR (Q-RT–PCR) analyses of 15 genes was compared, as reported previously (7). A high correlation between log2RL values (R = 0.93) was obtained (Supplementary Figure S1). The linear regression equation (log2RLPCR = 2.16 × log2RL + 2.04) was used to convert the RL value obtained by microarray hybridization to that equivalent for Q-RT–PCR under NS and DS conditions. The RL values were normalized to compensate for differences in the quantity of mRNA in the two gradient sub-fractions as well as non-polysomal and polysomal levels under the two conditions. Thus, the normalized ribosome loading (nRL) value is the ratio of quantity of mRNA in the polysomal and non-polysomal fractions for an individual mRNA species. The percentage of an individual mRNA in polysomes [Ribosome loading (%)] was calculated from nRL with the equation:
mRNA sequence dataset
The Institute of Genomic Research (TIGR) Arabidopsis sequences (04/17/03 release, http://www.tigr.org/tdb/e2k1/ath1/) and the Institute of Physical and Chemical Research (RIKEN) Arabidopsis full-length (RAFL) cDNA database (03/12/03 release, http://pfgweb.gsc.riken.go.jp/pub_data/index.html) were used. A database of reliable 5′-UTRs was generated by the identification of cDNAs with identical 5′-UTRs from independent cDNA resources. BLAST [Basic Local Alignment Search Tool (29)] alignment was performed on 28 581 TIGR cDNA sequences against 13 181 RAFL cDNA sequences. BLAST scores over 100 were used to select the RAFL sequences that matched TIGR sequences (15 671 cDNAs). When intron-splicing variants from a single gene in TIGR (∼4.7%, 1267 cDNAs) were identical to a RAFL cDNA, the TIGR cDNA with the highest score was chosen. To exclude TIGR cDNAs that may have been derived from RAFL clones, the TIGR cDNAs with a perfect match in nucleotide identity and length to RAFL cDNAs (1174 cDNAs) were excluded. Finally, TIGR and RAFL cDNAs were compared and the 4151 non-redundant cDNAs that possessed identical 5′ ends in both databases were selected. For mRNA open reading frame (ORF) and 3′-UTR analyses, the TIGR cDNA sequence set was reduced by removal of cDNAs for genes with splicing variants (n = 2678) and cDNAs that lacked a 3′-UTR (n = 10 770). The remaining non-redundant cDNAs (n = 15 133) possessed an ORF and a 3′-UTR. The mRNAs were confirmed to include an initiation codon, continuous ORF and stop codon by use of EditPadPro ver. 4.5.4 (JGsoft, Thailand).
Initiation A+1UG codon context, nucleotide composition and structure analyses
To evaluate the effect of the initiation A+1UG codon context, positions −10 to +5 of the mRNA, the TIGR cDNA sequences with 5′-UTRs ≥10 nt were selected (n = 8232 for NS and n = 8583 for DS for mRNAs detected as ‘Present’ in non-polysomal and polysomal complexes). mRNAs with the highest 1% nRL values under NS (ribosome loading >95%, n = 97) or DS (ribosome loading >91%, n = 113) were designated ‘efficiently translated’. mRNAs with the lowest 5% nRL values under NS (ribosome loading <66%, n = 528) or DS (ribosome loading <50%, n = 550) were designated ‘poorly translated’. The Chi-square test was used to determine the significance of variation in nucleotide frequency. To evaluate the significance of variation in nucleotide composition of this region, the Student's t-test was performed on mRNAs that possessed or lacked the optimal nucleotide between positions −10 and +5 (excluding positions +1 to +3). Similar results were obtained when FL-cDNA sequences (n = 4053) were used for the analysis (data not shown). Nucleotide composition of the 5′- and 3′-UTRs was determined by use of an in-house PERL script. MFOLD (version 3.1) was used to predict RNA free energy and secondary structures for each 5′- and 3′-UTR (30) at the plant growth temperature of 22°C. The 4151 non-redundant FL-cDNAs and 15 133 TIGR cDNAs with a 3′-UTR were used to analyze the predicted RNA secondary structure of 5′- and 3′-UTRs, respectively.
The DNA microarray data, mRNA sequences and mRNA sequence features described in this study are available at (http://bioinfo.ucr.edu/projects/arab_ribosome/search.php). The microarray data are available at the Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/ under GEO accession nos GSM38668, GSM38669, GSM38670 and GSM38671.
RESULTS
Determination of mRNA levels in polysomes by DNA microarray analysis
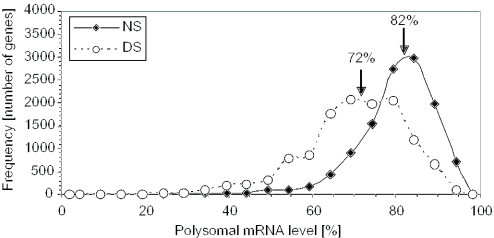
The variation in translation of cellular mRNAs in mature Arabidopsis rosette leaves under NS and DS conditions was determined by use of an oligonucleotide microarray designed to monitor ∼23 000 gene transcripts. Hybridizations were performed with mRNAs from sucrose density gradient fractions that contained non-polysomal and polysomal complexes (≥2 ribosomes per mRNA). The proportion of individual mRNA species in polysomes (ribosome loading) was determined for the genes with transcripts detected in both the non-polysomal and polysomal fractions (Figure 1). The results were consistent with those obtained with a DNA oligonucleotide array that monitored fewer genes (∼8000) (7). In NS leaves, the 5th and 95th percentile values for these genes corresponded to 61.9 and 92.1% of each mRNA species in polysomal complexes, whereas under DS these values fell to 45.9 and 86.8% (Figure 1). The decrease in the average proportion of an mRNA species in polysomes, from 82 to 72%, was significant (P < 0.0001). DS also broadened the modal range of ribosome loading for a large proportion of the mRNAs, indicative of greater constraints on translation. Remarkably, over 50% of the mRNAs with a 2-fold or greater increase in abundance in response to DS showed no decrease in ribosome loading, whereas over 70% of over 11 000 mRNAs monitored showed a significant decrease in ribosome loading (Supplementary Figure S2). This finding indicates that many DS-induced mRNAs can circumvent the global repression in mRNA translation.
Figure 1.
Extensive variability in ribosome loading under NS conditions and global reduction in ribosome loading in response to DS. nRL values were determined for the transcripts detected above background in both non-polysomal and polysomal RNA fractions under at least one condition by DNA microarray hybridization (7). The nRL value for each mRNA was plotted as the percentage of the individual mRNA species in polysomes under NS and DS conditions. Arrows indicate the positions and values of the average nRL (determined with log2-transformed values) for the NS (average log2nRL 2.15 ± 0.87 for n = 11 746) and DS (average log2nRL = 1.3 ± 0.88 for n = 12 310). Averages were statistically different as determined by the Student's t-test on the log2nRL values (P < 10−4).
Effect of the 5′-UTR, coding region and 3′-UTR length on ribosome loading
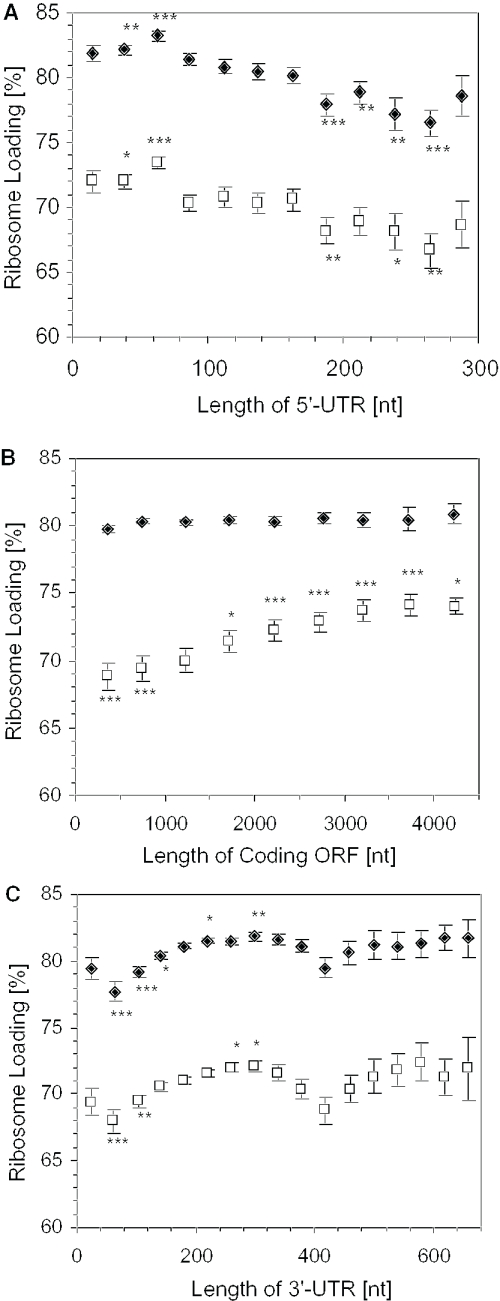
To address the influence of mRNA features on translation, a database was prepared with 5′-UTR, coding and 3′-UTR sequences of full-length and full-coding cDNAs of public collections (See Materials and Methods). Arabidopsis mRNAs ranged in size from 298 to 5754 nt, with an average of 1670 nt (Supplementary Figure S3). The average 5′-UTR length of the FL-cDNAs was 124.7 nt (Supplementary Figure S3), which is also the average 5′-UTR length of human mRNAs (31). The average coding sequence and 3′-UTR length of Arabidopsis mRNAs was 1268 and 248 nt, respectively. Evaluation of the relationship between 5′-UTR and ribosome loading revealed that the optimal 5′-UTR was between 50 and 75 nt under both growth conditions (Table 1 and Figure 2A). mRNAs with extremely short (<25 nt) 5′-UTRs had a lower than average ribosome loading level. The data clearly indicate that mRNAs with long (>175 nt) 5′-UTRs had significantly lower than average ribosome loading. No further decrease in ribosome loading was observed for 5′-UTRs of 175–300 nt. These results demonstrate that the length of the 5′-UTR has a general influence on mRNA translation under NS and DS conditions.
Table 1.
Effect of mRNA features on ribosome loading under NS and DS conditions
| mRNA feature | Overall range | Average | Maximum RL (%) | Minimum RL (%) | % Contribution | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| NS | DS | Range | NS | DS | Range | NS | DS | |||
| 5′-UTR length (nt) | 0 to 300 | 124.7 | 83.2 | 73.4 | 50 to 70 | 76.5 | 66.7 | 250 to 275 | 6.7 | 6.7 |
| ORF length (nt) | 100 to 4500 | 1268.2 | 80.9 | 74 | 4000 to 4500 | 79.8 | 68.8 | 0 to 500 | 1.1 | 5.2 |
| 3′-UTR length (nt) | 0 to 680 | 247.8 | 81.8 | 72.1 | 280 to 320 | 77.7 | 68 | 40 to 80 | 4.1 | 4.1 |
| 5′-UTR ΔG (kcal/mol) | −145 to 5 | −35 | 83.1 | 73.7 | −10 to 5 | 76.2 | 67.4 | −70 to −55 | 6.9 | 6.3 |
| 3′-UTR ΔG (kcal/mol) | −160 to 5 | −69 | 81.9 | 72.3 | −85 to −70 | 78.6 | 68.8 | −25 to −10 | 3.3 | 3.5 |
| 5′-UTR GC content (%) | 20 to 60 | 39 | 85.9 | 78.3 | 20 to 25 | 75.9 | 60.9 | 55 to 60 | 10.0 | 17.4 |
| 3′-UTR GC content (%) | 20 to 45 | 32 | 81.3 | 71.3 | 30 to 35 | 79.6 | 69.3 | 40 to 45 | 1.7 | 2.0 |
Percentage contribution of each mRNA feature was calculated as maximum percentage ribosome loading − minimum percentage ribosome loading.
Figure 2.
Effect of mRNA 5′-UTR, coding region and 3′-UTR length on ribosome loading. FL-cDNA sequences were grouped based on the length of 5′-UTR, length of coding ORF and 3′-UTR with a window of 25 nt (5′-UTR), 500 nt (ORF) and 40 nt (3′-UTR). The average length of each region within each group was plotted against the average ribosome loading (percentage of an individual mRNA in the polysomes) under NS (closed diamond) or DS (open square). mRNAs that were detected in non-polysomal and polysomal fractions under at least one of the conditions were used for the analysis (A) 5′-UTR, n = 2724, NS; n = 2845, DS; (B) ORF, n = 10 628, for NS; n = 11 056, for DS; and (C) 3′-UTR, n = 9018, for NS; n = 9405, for DS. Error bars indicate S.E. Statistical significance of ribosome loading in each range was determined for log2nRL values against the average log2nRL value for each condition by the Student's t-test (*, P < 0.05, **, P < 0.01, ***, P < 0.001).
The comparison of coding sequence length and ribosome loading showed no over-representation of mRNAs that encode high molecular mass polypeptides in the polysome fraction under NS conditions (Figure 2B). However, mRNAs with long ORFs showed a steady increase in ribosome loading under DS; this increase was significant for ORFs of >1500 nt. The increase in mRNAs with long coding sequences in polysomes under DS most likely indicates a general decrease in translational elongation and/or termination. Although there was no apparent increase in the average size of polysomes in response to DS, based on the analysis of the absorbance profiles of sucrose density gradient fractionated polysomes (7), this analysis suggests there is a global reduction in polypeptide chain elongation under DS. Overall, ORF length contributed 5% of the variation in ribosome loading under DS (Table 2).
Table 2.
Correlation coefficient (R) determined from the comparison of FL-cDNA 5′-UTR nucleotide content and ribosome loading under NS (n = 2716) and DS (n = 2833) conditions
| Nucleotide(s) | Non-stress R | Dehydration stress R |
|---|---|---|
| A | 0.20 | 0.22 |
| U | −0.11 | −0.07 |
| G | −0.16 | −0.20 |
| C | 0.04 | −0.07 |
| AU | 0.12 | 0.21 |
| GC | −0.12 | −0.21 |
| CU | −0.06 | −0.06 |
| AG | 0.06 | 0.06 |
| GU | −0.24 | −0.22 |
| AC | 0.24 | 0.22 |
Mono- and di-nucleotide content reflects the sum of the frequency of the 2 nt.
The average length determined for Arabidopsis 3′-UTRs (247.8 nt) was slightly longer than previously reported for plants (∼200 nt) (32), and significantly longer than the average 5′-UTR length (P < 0.0001) (Supplementary Figure S3). Evaluation of the range in 3′-UTR length on ribosome loading indicated that 280–320 nt was an optimal length under both NS and DS conditions (Figure 2C). We found that mRNAs with a short (40–120 nt) or long (>380 nt) 3′-UTR had a significantly reduced ribosome loading under both conditions. Overall, the 3′-UTR length contributed 4.1% of the variation in ribosome loading, whereas 5′-UTR length accounted for 6.7% of the variation (Table 1).
Effect of RNA free energy and nucleotide composition on ribosome loading
Initiation of translation is an ATP- and GTP-dependent process that begins with the recruitment of eIF4F and eIF3 to the mRNA and capture of the 43S pre-initiation complex, which scans in the 5′ to 3′ direction until an AUG initiation codon is recognized (33,34). Thermodynamically stable RNA secondary structures present in the 5′-UTR impair scanning in vitro (14). To evaluate the contribution of potential secondary structure to polysome association, the free energy (ΔG) of FL-cDNA 5′-UTRs was predicted by use of MFOLD (30,35) and compared with ribosome loading values (Figure 3A). This comparison revealed that mRNAs with weak potential secondary structure in the 5′-UTR (>−20 kcal/mol) had an advantage and mRNAs with a higher potential for secondary structure (<−55 kcal/mol) had a significant disadvantage in ribosome recruitment, respectively. Despite demonstrations by others that the effect of moderately strong stem–loop structures (−20 to −30 kcal/mol) within the 5′-UTR is position dependent (17,36), we failed to detect an effect of moderate ΔG values in the different regions of native 5′-UTRs (Supplementary Figure S4).
Figure 3.
Effect of potential RNA secondary structure formation and GC content in the 5′-UTR on ribosome loading. mRNAs were grouped based on the predicted free energy (ΔG kcal/mol) or the GC content with a 15 kcal/mol (ΔG) or 5% (GC content) window, respectively. (A) The average predicted free energy of the 5′-UTRs in each group under NS (n = 2641, closed diamond) and DS (n = 2758, open square) was plotted. (B) Effect of 5′-UTR secondary structure for the mRNA with 5′-UTR length of 80–180 nt. mRNA sequences from FL-cDNA are grouped based on the predicted free energy (ΔG kcal/mol). (C) The average GC content of the 5′-UTR of each group under NS (closed diamond) and DS (open square) was plotted. (D) The average change in ribosome loading in response to DS was plotted against the average change in GC content of the 5′-UTR of each group in response to DS. Statistical significance of ribosome loading or change in ribosome loading was determined against the average value under each condition by Student's t-test (*, P < 0.05, **, P < 0.01, ***, P < 0.001). Error bars indicate S.E.
The potential for secondary structure in the 5′-UTR accounted for ∼6.5% of the variation in ribosome loading under both NS and DS conditions, similar to the variation caused by 5′-UTR length (Table 1). The strong negative correlation between ΔG and 5′-UTR length (R = −0.97) (data not shown) suggests that as the 5′-UTR increases in length secondary structure potential also increases. Thus, the contribution of 5′-UTR length and ΔG to ribosome loading may not necessarily be additive. However, the evaluation of mRNAs with similar 5′-UTR length (80–180 nt) but varying predicted ΔG indicated that potential secondary structure can independently influence ribosome recruitment (Figure 3B).
The investigation of the influence of 5′-UTR mono- and di-nucleotide composition on ribosome loading revealed that adenine (A) content was positively correlated, whereas guanine (G) content was negatively correlated with ribosome loading under both growth conditions. Uracil (U) and cytosine (C) content had a lesser effect (Table 2). Consistent with the mono-nucleotide influence, the frequency of A and U (AU content) and A and C (AC content) was positively correlated with ribosome loading under both conditions. GU content had a strong negative effect on translation under both NS and DS. In contrast, GC content has a negative effect on translation that was exacerbated by DS (Table 2 and Figure 3C). High GC content (NS: >50% and DS: >42.5%) in the 5′-UTR significantly reduced ribosome loading, whereas low GC content (NS: <32.5% and DS: <37.5%) significantly promoted ribosome loading, as compared with the average GC content under both conditions (39%). Interestingly, cytosolic ribosomal protein mRNAs showed a dramatic reduction in ribosome loading under DS [Supplementary Figure S2, (7)]. These mRNAs have a significantly higher GC content in the 5′-UTR (45.2 ± 5.56%, n = 169, P < 0.0001) than the average. The contribution of GC content to variation in ribosome loading was considerably higher under DS (∼17%) than NS (∼10%) (Table 1). In conclusion, translation of mRNAs with 5′-UTRs with a GC content above the average (39%) is impaired as a consequence of DS (Figure 3D).
The effect of GC content and potential secondary structures in the 3′-UTR on ribosome loading was also considered. The average GC content (32%) and ΔG value (−69 kcal/mol) for the 3′-UTR was significantly different from that of the 5′-UTR (Table 1; Supplementary Figure S5, P < 0.0001). The GC content of the 3′-UTR appeared to have little influence on ribosome loading. Interestingly, a high ΔG ≥ −25 kcal/mol was correlated with reduced ribosome loading (Table 1 and Supplementary Figure S5); however, the poor loading of these mRNAs was frequently correlated with a short 3′-UTR. These results support the notion that re-initiation is impaired by an extremely short 3′-UTR but is generally insensitive to secondary structure formation in the 3′ end.
Effect of upstream AUGs and uORFs on ribosome loading
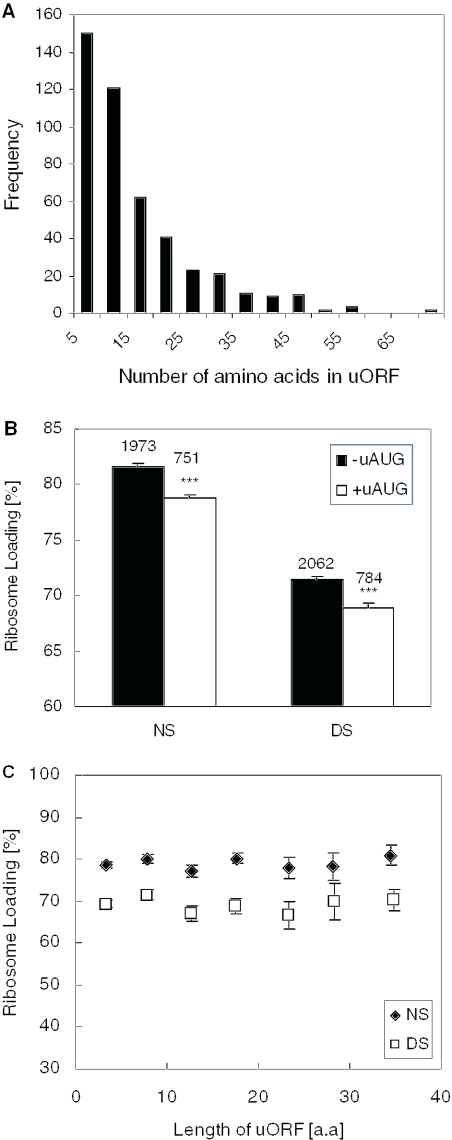
A considerable number of eukaryotic mRNAs contain a 5′AUG triplet upstream of the Met codon that commences the protein coding ORF (uAUG) (37). A previous survey of 1023 Arabidopsis mRNAs found that 22% contained one or more uAUG (19). In our dataset of 4151 FL-cDNA sequences, 73% had no uAUG, 15% had one and the remaining 12% had more than one uAUG. Interestingly, ∼28% of genes with an uAUG encode proteins involved in signal transduction and/or gene regulation (data not shown). We found that the uAUGs initiated uORFs that ranged from 2 to >50 codons in length (Figure 4A), and that the presence of an uAUG(s) significantly impaired in ribosome loading under both growth conditions (Figure 4B). The negative effect was similar over a wide range of uORF lengths (2–40 amino acids) (Figure 4C). Since the average 5′-UTR with one or more uAUG was 226 nt (n = 1132 FL-cDNAs), whereas the average 5′-UTR that lacked an uAUG was 87 nt (n = 3019 FL-cDNAs), the poor translation of these mRNAs could reflect both the presence of an uAUG and/or the ramifications of a long 5′-UTR. It was shown previously that Arabidopsis mRNAs that possess an uAUG typically have an AUG at the start of the coding sequence that is in poor context (19). Thus, some cellular mRNAs may be burdened with multiple features that impair translation.
Figure 4.
Effect of presence of an AUG or ORF upstream of initiation codon on ribosome loading. mRNAs in FL-cDNA sequence dataset with or without an uAUG (an AUG upstream of the initiation codon) were selected. (A) Frequency distribution of uORF length. (B) Ribosome loading values under NS and DS conditions were compared for mRNAs that lacked (black bar) or possessed one or more uAUG(s) (white bar). Statistical significance of ribosome loading values (log2nRL) of mRNAs with an uAUG as compared with an average log2nRL of mRNAs without uAUG was determined under NS or DS conditions by Student's t-test (***P < 0.0001). Error bars indicate S.E. Values above bars indicate the sample size. (C) Ribosome loading values under NS and DS conditions for mRNAs with a uORF of varying length.
Initiation codon context and ribosome loading
The initiation codon consensus can be determined by use of the 50/75 consensus rule (the most frequent nucleotides that surround the AUG are indicated in uppercase if the relative frequency of a nucleotide is ≥50% and at least twice as frequent as the second most frequent nucleotide, if not, the sum of the frequency of 2 nt is ≥75% as a co-consensus in uppercase, and in lowercase if neither of the former conditions are met) (38). The initiation codon consensus sequence derived for all genes with transcripts detected under both growth conditions was a−10aaaaaaA/GaaA+1UGGc+4 (Figure 5A). The preference for a purine in the −3 and +4 positions was consistent with earlier determinations for eudicots (38). For genes that were highly loaded under NS or DS conditions (approximately the highest 1%), the most frequent nucleotides were AAAAAAAAAAA+1UGGC (Figure 5B and D). Notably, the occurrence of A−4, A−3, A−1 and C+5 was significantly higher in these mRNAs than the average or in the poorly loaded mRNAs under NS conditions (Figure 5C). Moreover, the analysis showed that mRNAs with the highest proportion in polysomes under DS had a significantly higher frequency of A−10, A−9 and A−6 than the highly loaded mRNAs under NS conditions (Figure 5D). As observed under NS conditions, the mRNAs that were poorly loaded under DS had a much lower frequency of A in these positions (Figure 5E).
Figure 5.
Evaluation of frequency of nucleotides surrounding the initiation codon AUG in mRNAs. TIGR cDNA sequences with 10 or greater nucleotides were selected for the determination of nucleotide frequency at positions between −10 and +5 (initiation codon AUG corresponds to position +1 to +3). (A) All genes with mRNAs detected under NS and DS conditions (n = 7870). mRNAs with very high (approximately highest 1% ribosome loading values) (B and D) and very low (approximately lowest 5% ribosome loading values) nRL (C and E) under NS and DS conditions, respectively. The statistical significance of the frequency of the nucleotide with the highest value at each position was determined by Chi-square test against the combined frequency of other nucleotides of all mRNAs under the same condition; asterisks in parenthesis indicate the comparison against mRNAs with very low nRL, *, P < 0.05, **, P < 0.01, ***, P < 0.001. Gene numbers (n) in samples indicated.
To shed additional light on the significance of nucleotides flanking the initiation codon, we monitored the difference between the average ribosome loading of mRNAs with and without the most frequent nucleotide at each position (Figure 6). This analysis showed that residues that significantly affect ribosome loading were distinct under the two conditions: NS, AAAaaaAAaAA+1UGGC and DS, AAAaAaAAaAA+1UGGC (residues shown in bold uppercase correlated with significantly increased ribosome loading). The most critical residues were A−4, A−3, A−1, G+4 and C+5 under both conditions. The largest decrease in average ribosome loading was observed when C+5 was absent. The only difference between the two conditions was at position −6, where an A had a significant positive effect under DS. Strikingly, the high frequency of A−2 in mRNAs with high ribosome loading under NS or DS did not significantly affect ribosome loading.
Figure 6.
Effect of most frequent nucleotide at position −10 to +5 on dehydration-induced change in ribosome loading. Difference between average ribosome loading values of mRNAs with and without the most frequent nucleotide determined from the mRNA set with highest 1% ribosome loading under NS and DS at each position between −10 and +5. Statistical significance of the difference was determined by Student's t-test for the average log2nRL value of mRNAs with and without the corresponding nucleotide (*, P < 0.05, **, P < 0.01, ***, P < 0.001).
DISCUSSION
The recruitment of the 43S pre-initiation complex to an mRNA is a competitive process that is determined by features of the mRNA, the cellular mileu and the activity of the translational machinery (23). The mechanism of initiation of translation in plants and other eukaryotes involves a pseudo-circularized mRNA formed by interactions between the 5′-cap and 3′-tail which involve eIF4G, eIF4B and poly(A) binding protein (20,34,39). The mRNA cap–tail interaction promotes the scanning of the 43S pre-initiation complex and secondary re-initiation events. In plants, discrimination between mRNAs in translational initiation was reported for a number of mRNAs in focused studies [reviewed in (24,40)]. More recently, differential mRNA translation in Arabidopsis leaves was confirmed at the global level by quantitative assessment of the accumulation of mRNAs in polysomal complexes under both NS and DS conditions (Figure 1) (7). Here, we expanded the comparison of steady-state total and polysomal mRNA abundance of Arabidopsis genes and identified several mRNA features that contribute to translational regulation under these two growth conditions. The results indicate that features of the 5′-UTR, ORF and 3′-UTR play a general role in the regulation of translation of individual gene transcripts (Table 1). Furthermore, our findings provide support to the hypothesis that the nucleotides that surround the initiation codon influence translational efficiency.
The role of the 5′-UTR in translational regulation
The 5′-UTR had the greatest effect on ribosome loading due to the influence of nucleotide composition, length, potential secondary structure and the presence of uAUGs. mRNAs with high ribosome loading generally had a 5′-UTR with high A content, whereas poorly loaded mRNAs generally had 5′-UTRs with elevated G, U and GU contents (Table 2). In addition, a 5′-UTR, which was long, had a ΔG prediction of <−55 kcal/mol and/or contained uAUGs significantly impaired ribosome loading under both growth conditions. Strikingly, of the 5′-UTR features evaluated, only high GC content clearly contributed to the differential reduction of ribosome loading under DS (Figure 3B and C). This observation could reflect a higher requirement for ATP-dependent RNA helicase activity for the initiation of mRNA with a high GC content in the leader sequence. These results suggest that under DS, when nucleotide triphosphate levels are reduced, ribosome loading is significantly modulated by the requirement for ATP consumption in the scanning process (41,42).
The initiation codon context and translation
The results presented here provide evidence that the context surrounding the initiation codon contributes to control of translational initiation under NS conditions, and differentially influences translation under DS. mRNAs that were highly loaded under both NS and DS conditions had a slightly modified consensus sequence than predicted from the survey of all Arabidopsis genes. The modified consensus sequence was characterized by a preference for A from position −10 to −1. Our finding that A−3 is favored in highly translated mRNAs is consistent with the analyses of AUG context in stably transformed tobacco cells (43,44). However, another study observed that reporter gene mRNAs with an A-rich (AAACAAUGG) initiation codon context were expressed at similar levels as those with a less A-rich (CCACCAUGG) region in transiently transformed tobacco protoplasts (45).
The significantly higher frequency of A at positions −7, −9 and −10 in the highly translated mRNAs under DS further indicates that the modulation of expression involves constraints within a larger region initiation codon context region than previously considered by mutational analyses. The A-rich initiation codon context could be favorable due to minimization of secondary structure formation or increased interaction with eIF1. Remarkably, the optimal sequence (a−10aaaaaaA/GaaA+1UGGc+4) based on nucleotide frequency in the efficiently translated mRNAs did not occur in any of the highly loaded mRNAs. Moreover, many mRNAs with extremely high ribosome loading under NS or DS had several substitutions in the critical positions around the AUG. These results appear to indicate that a general reduction in the capacity for secondary structure formation around the AUG may be beneficial, whereas poly(A) tracts in this region may be unfavorable.
Our study also confirmed that there is a preference for G+4 and C+5 in mRNAs with extremely high ribosome loading under both conditions. These nucleotides would result in an Ala residue at the second position of the polypeptide, as noted previously (22). Consistent with our observations, a previous study (43) showed that the base substitution of G+4 significantly reduced the reporter gene expression. It should also be considered that nucleotide variation at position +4 and/or +5 may have ramifications on protein stability according to the N-end rule, where Ala is fairly stable residue in Escherichia coli and yeast (44,46).
The role of the 3′-UTR in translational regulation
The effect of the length and other features of the 3′-UTR on translation has not been extensively studied in plants. Tanguay and Gallie (47) reported that the extension of the 3′-UTR length from 4 to 104 nt increased the translational efficiency of non-polyadenylated mRNAs in a transient expression system using Chinese hamster ovary cells. However, polyadenylated reporter gene mRNAs showed little increase in translation when the 3′-UTR was extended from 27 to 161 nt. We found that the average 3′-UTR length for Arabidopsis mRNAs was 248 nt. In contrast, the 3′-UTR of mammalian mRNAs is generally longer (>400 nt) (32). The survey of 3′-UTR features and polysome association presented here indicated that mRNAs with a short 3′-UTR (40–120 nt) were translated at significantly reduced levels in Arabidopsis leaves. From these results, it can be speculated that a minimal distance between the stop codon and poly(A) tail is critical for the re-initiation process. A long 3′-UTR (>300 nt) may not adversely affect this process, although the increased variation in ribosome loading of mRNAs with long 3′-UTRs is noteworthy. This might reflect the presence of additional features in long 3′-UTRs that play a role in other processes, such as differential regulation of polyadenylation site selection, mRNA stability, transport and the subcellular location of translation (32,48,49).
Evidence of increased number of ribosomes per mRNA under DS
Several studies have noted a shift of plant mRNAs to larger polysomes in response to environmental stimuli (40). We found here that mRNAs with a long ORF (>1500 nt) have significantly higher ribosome loading values under DS, indicating a greater number of ribosomes per mRNA. This could reflect an enhancement of initiation/re-initiation or reduced elongation/termination. It is well established that initiation is reduced under DS. Therefore, these results may indicate that initiation is not the sole limiting factor and that elongation/termination is also decreased. This could be a consequence of reduced availability of GTP. The contribution of ORF length to polysome association under DS was significant over the entire range of coding sequence length, consistent with the conclusion that elongation/termination was globally repressed irrespective of mRNA length under DS.
Mechanism of selective mRNA translation under DS
About 50% of the genes that were highly induced at the level of mRNA abundance by DS showed little reduction in ribosome loading, whereas the majority of mRNAs showed reduced translation (Supplementary Figure S2) (7). A survey of the genes that displayed maintained ribosome loading in response to DS indicated that only ∼9% of genes represented in the FL-cDNA set (32/349 genes) had a 5′-UTR of the optimal length (30–70 nt) and GC content (<40%). Moreover, a significant number of mRNAs with optimal 5′-UTR length and GC content displayed reduced ribosome loading under DS conditions (193/349 mRNAs with ΔnRL < 0, P < 0.01). This finding indicates that although the mRNA features identified in this analysis generally contribute to translational regulation, other factors are likely to be responsible for the maintained translation of a subset of cellular mRNAs under DS. Our analysis considered that specific mRNA sequences might be present in the mRNAs that are efficiently or poorly translated under DS. Such sequences might promote assembly of the initiation complex or reduce dependency on cap–tail interactions, thereby allowing certain mRNAs to be efficiently translated under DS. This escape of translational repression might be facilitated directly by the RNA sequence or indirectly though interaction with RNA-binding proteins that enhance the cap–tail interaction (20,39). However, despite considerable effort, we were unable to identify motifs that augment or impair ribosome loading under DS using publicly available motif explore programs [e.g. MEME (50), GPRM (51) and SLASH (52)] (data not shown). This leads us to speculate that mRNA sequences per se may not be solely responsible for differential mRNA translation.
The finding that the majority of the DS-induced mRNAs also showed maintained translation (Supplementary Figure S2) raises the possibility that transcriptional induction is coupled with efficient translation during DS. The coupling of nuclear and cytoplasmic regulatory mechanisms was reported in response to heat stress in yeast, where induced mRNAs also showed efficient association with polysomes (23,53). The mechanisms that co-regulate transcription/splicing/export events with translation remain to be elucidated. Several recent studies indicate that transcriptional activity is linked to splicing, polyadenylation and turnover; all of these processes have been shown to influence translation (54–57). The coupling of transcription to post-transcriptional events most likely involves heterogeneous nuclear ribonucleoproteins (hnRNPs) and factors involved in post-transcriptional processes that are transferred from the polymerase II transcription complex to the hnRNA (58). The transferred proteins have been shown to interact with splicing, export and translation factors (53).
An alternative explanation for the apparent absence of sequence motifs that regulate differential mRNA translation is that the exact leader sequence might be regulated by the use of alternative transcription start sites and/or removal of 5′ intron(s). It is clear that a long structured 5′-UTR is likely to cause inefficient initiation, thus removal of such a structure or uAUGs by alternative transcription (59–61) or splicing (62–64) could lead to a higher rate of initiation. In humans, 35% of mRNAs undergo alternative splicing, occurring mostly in the 5′-UTR (65). Additional features/mechanisms including IRES, initiation without t-RNAMet or mRNA–rRNA pairing might also contribute to translational regulation in response to DS (66).
In conclusion, this study provides strong evidence that multiple mRNA sequence features contribute to the regulation of mRNA translation under standard growth conditions and during DS. Although 5′-UTR GC content, initiation codon context and ORF length generally contribute to the differential translation of mRNAs under DS, these features do not appear to be solely responsible for the observed dynamics in mRNA translation. Moreover, our failure to identify the presence of mRNA sequence motifs that correlate with maintenance versus impairment of translation of individual mRNAs under DS (data not shown) leads to the suggestion that translational regulation may involve aspects of gene regulation that have yet to be appreciated. In light of recent genetic analyses that have identified several nuclear mRNA-binding proteins and export factors that play a role in gene expression in response to DS and abscisic acid (67,68), we propose that the evaluation of the coupling of nuclear and cytoplasmic gene regulation deserves additional attention. Future studies that make use of DNA microarrays to distinguish transcriptional activity, nuclear RNA populations, splicing variants and polysomal mRNAs could provide greater understanding of the nucleotide elements that are critical to the continuum of cellular events that may underlie differential mRNA translation.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Dr Thomas Girke for his assistance in the preparation of the mRNA sequence databases and Dr Elizabeth Bray for many insightful discussions. This research was supported by a grant from the National Science Foundation (DBI 0211857) to J.B.-S. Funding to pay the Open Access publication charges for this article was provided by University of California Riverside Agricultural Experiment Station Funds.
REFERENCES
- 1.Gutierrez R.A., Ewing R.M., Cherry J.M., Green P.J. Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc. Natl Acad. Sci. USA. 2002;99:11513–11518. doi: 10.1073/pnas.152204099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arava Y., Wang Y., Storey J.D., Liu C.L., Brown P.O., Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter M.S., Kuhn K.M., Sarnow P. Cellular internal ribosome entry site elements and the use of cDNA microarrays in their investigation. In: Sonenberg N., Hershey J.W.B., Mathews M.B., editors. Translational Control of Gene Expression. NY: Cold Spring Harbor Laboratory Press; 2000. pp. 615–635. [Google Scholar]
- 4.Diehn M., Eisen M.B., Botstein D., Brown P.O. Large-scale identification of secreted and membrane-associated gene products using DNA microarrays. Nature Genet. 2000;25:58–62. doi: 10.1038/75603. [DOI] [PubMed] [Google Scholar]
- 5.Grolleau A., Bowman J., Pradet-Balade B., Puravs E., Hanash S., Garcia-Sanz J.A., Beretta L. Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics. J. Biol. Chem. 2002;277:22175–22184. doi: 10.1074/jbc.M202014200. [DOI] [PubMed] [Google Scholar]
- 6.Johannes G., Carter M.S., Eisen M.B., Brown P.O., Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl Acad. Sci. USA. 1999;9:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawaguchi R., Girke T., Bray E.A., Bailey-Serres J. Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J. 2004;38:823–839. doi: 10.1111/j.1365-313X.2004.02090.x. [DOI] [PubMed] [Google Scholar]
- 8.Kuhn K.M., DeRisi J.L., Brown P.O., Sarnow P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell. Biol. 2001;21:916–927. doi: 10.1128/MCB.21.3.916-927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pradet-Balade B., Boulme F., Beug H., Mullner E.W., Garcia-Sanz J.A. Translation control: bridging the gap between genomics and proteomics? Trends Biochem. Sci. 2001;26:225–229. doi: 10.1016/s0968-0004(00)01776-x. [DOI] [PubMed] [Google Scholar]
- 10.Zong Q., Schummer M., Hood L., Morris D.R. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc. Natl Acad. Sci. USA. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacKay V.L., Li X., Flory M.R., Turcott E., Law G.L., Serikawa K.A., Xu X.L., Lee H., Goodlett D.R., Aebersold R., Zhao L.P., Morris D.R. Gene expression analyzed by high-resolution state array analysis and quantitative proteomics: response of yeast to mating pheromone. Mol. Cell. Proteomics. 2004;3:478–489. doi: 10.1074/mcp.M300129-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Serikawa K.A., Xu X.L., MacKay V.L., Law G.L., Zong Q., Zhao L.P., Bumgarner R., Morris D.R. The transcriptome and its translation during recovery from cell cycle arrest in Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2003;2:191–204. doi: 10.1074/mcp.D200002-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Kozak M. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991;1:111–115. [PMC free article] [PubMed] [Google Scholar]
- 14.Kozak M. Determinants of translational fidelity and efficiency in vertebrate mRNAs. Biochimie. 1994;76:815–821. doi: 10.1016/0300-9084(94)90182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gayen A.K., Peffley D.M. The length of 5′-untranslated leader sequences influences distribution of 3-hydroxy-3-methylglutaryl-coenzyme A reductase mRNA in polysomes: effects of lovastatin, oxysterols, and mevalonate. Arch. Biochem. Biophys. 1995;322:475–485. doi: 10.1006/abbi.1995.1491. [DOI] [PubMed] [Google Scholar]
- 16.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eukaryotic mRNAs. Mol. Cell. Biol. 1989;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaff P., Riesner D., Steger G. RNA structure and the regulation of gene expression. Plant Mol. Biol. 1996;32:89–106. doi: 10.1007/BF00039379. [DOI] [PubMed] [Google Scholar]
- 19.Rogozin I.B., Kochetov A.V., Kondrashov F.A., Koonin E.V., Milanesi L. Presence of ATG triplets in 5′ untranslated regions of eukaryotic cDNAs correlates with a ‘weak’ context of the start codon. Bioinformatics. 2001;17:890–900. doi: 10.1093/bioinformatics/17.10.890. [DOI] [PubMed] [Google Scholar]
- 20.Kawaguchi R., Bailey-Serres J. Regulation of translational initiation in plants. Curr. Opin. Plant Biol. 2002;5:460–465. doi: 10.1016/s1369-5266(02)00290-x. [DOI] [PubMed] [Google Scholar]
- 21.Vilela C., McCarthy J.E. Regulation of fungal gene expression via short open reading frames in the mRNA 5′untranslated region. Mol. Microbiol. 2003;49:859–867. doi: 10.1046/j.1365-2958.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- 22.Joshi C.P., Zhou H., Huang X., Chiang V.L. Context sequences of translation initiation codon in plants. Plant Mol. Biol. 1997;35:993–1001. doi: 10.1023/a:1005816823636. [DOI] [PubMed] [Google Scholar]
- 23.Preiss T., Baron-Benhamou J., Ansorge W., Hentze M.W. Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nature Struct. Biol. 2003;10:1039–1047. doi: 10.1038/nsb1015. [DOI] [PubMed] [Google Scholar]
- 24.Kawaguchi R., Bray E.A., Bailey-Serres J. Water-deficit-induced translational control in Nicotiana tabacum. Plant Cell Environ. 2003;26:221–229. [Google Scholar]
- 25.Zhu J., Spencer E.D., Kaspar R.L. Differential translation of TOP mRNAs in rapamycin-treated human B lymphocytes. Biochim. Biophys. Acta. 2003;1628:50–55. doi: 10.1016/s0167-4781(03)00117-9. [DOI] [PubMed] [Google Scholar]
- 26.Carninci P., Waki K., Shiraki T., Konno H., Shibata K., Itoh M., Aizawa K., Arakawa T., Ishii Y., Sasaki D., et al. Targeting a complex transcriptome: the construction of the mouse full-length cDNA encyclopedia. Genome Res. 2003;13:1273–1289. doi: 10.1101/gr.1119703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas B.J., Volfovsky N., Town C.D., Troukhan M., Alexandrov N., Feldmann K.A., Flavell R.B., White O., Salzberg S.L. Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol. 2002;3:1–12. doi: 10.1186/gb-2002-3-6-research0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seki M., Narusaka M., Kamiya A., Ishida J., Satou M., Sakurai T., Nakajima M., Enju A., Akiyama K., Oono Y., et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- 29.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki Y., Yoshitomo-Nakagawa K., Maruyama K., Suyama A., Sugano S. Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene. 2000;200:149–156. doi: 10.1016/s0378-1119(97)00411-3. [DOI] [PubMed] [Google Scholar]
- 32.Mazumder B., Seshadri V., Fox P.L. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 33.Kozak M. Evaluation of the “scanning model” for initiation of protein synthesis in eucaryotes. Cell. 1980;22:7–8. doi: 10.1016/0092-8674(80)90148-8. [DOI] [PubMed] [Google Scholar]
- 34.Preiss T., Hentze M.W. Starting the protein synthesis machine: eukaryotic translation initiation. BioEssays. 2003;25:1201–1211. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- 35.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 36.Gallie D.R., Ling J., Niepel M., Morley S.J., Pain V.M. The role of 5′-leader length, secondary structure and PABP concentration on cap and poly(A) tail function during translation in Xenopus oocytes. Nucleic Acids Res. 2000;28:2943–2953. doi: 10.1093/nar/28.15.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peri S., Pandey A. A reassessment of the translation initiation codon in vertebrates. Trends Genet. 2001;17:685–687. doi: 10.1016/s0168-9525(01)02493-3. [DOI] [PubMed] [Google Scholar]
- 38.Joshi C.P. An inspection of the domain between putative TATA box and translational start site in 79 plant genes. Nucleic Acids Res. 1987;16:6643–6653. doi: 10.1093/nar/15.16.6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallie D.R. Protein–protein interactions required during translation. Plant Mol. Biol. 2002;50:949–970. doi: 10.1023/a:1021220910664. [DOI] [PubMed] [Google Scholar]
- 40.Bailey-Serres J. Selective translation of cytoplasmic mRNAs in plants. Trends Plant Sci. 1999;4:142–148. doi: 10.1016/s1360-1385(99)01386-2. [DOI] [PubMed] [Google Scholar]
- 41.Flexas J., Medrano H. Drought-inhibition of photosynthesis in C3 plants: stomatal and non-stomatal limitations revisited. Ann. Bot. 2002;89:183–189. doi: 10.1093/aob/mcf027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gimenez C., Mitchell V.J., Lawlor D.W. Regulation of photosynthetic rate of low sunflower hybrids under water stress. Plant Physiol. 1992;98:516–524. doi: 10.1104/pp.98.2.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lukaszewicz M., Feuermannl M., Jerouville B., Stas A., Boutry M. In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Sci. 2000;154:89–98. doi: 10.1016/s0168-9452(00)00195-3. [DOI] [PubMed] [Google Scholar]
- 44.Taylor J.L., Jones J.D.G., Sandler S., Mueller G.M., Bedbrook J., Dunsmuir P. Optimizing the expression of chimeric genes in plant cells. Mol. Gen. Genet. 1987;210:572–577. [Google Scholar]
- 45.Guerineau F., Lucy A., Mullineaux P. Effect of two consensus sequences preceding the translational initiator codon on gene expression in plant protoplasts. Plant Mol. Biol. 1992;18:815–818. doi: 10.1007/BF00020027. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida S., Ito M., Callis J., Nishida I., Watanabe A. A delayed leaf senescence mutant is defective in arginyl-tRNA:protein arginyltransferase, a component of the N-end rule pathway in Arabidopsis. Plant J. 2002;32:129–137. doi: 10.1046/j.1365-313x.2002.01407.x. [DOI] [PubMed] [Google Scholar]
- 47.Tanguay R.L., Gallie D.R. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 1996;16:146–156. doi: 10.1128/mcb.16.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Derrigo M., Cestelli A., Savettieri G., Di Liegro I. RNA–protein interactions in the control of stability and localization of messenger RNA. Int. J. Mol. Med. 2000;5:111–123. [PubMed] [Google Scholar]
- 49.Macdonald P. Diversity in translational regulation. Curr. Opin. Cell Biol. 2001;13:326–331. doi: 10.1016/s0955-0674(00)00215-5. [DOI] [PubMed] [Google Scholar]
- 50.Bailey T.L., Elkan C. The value of prior knowledge in discovering motifs with MEME. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1995;3:21–29. [PubMed] [Google Scholar]
- 51.Hu Y.J. GPRM: A genetic programming approach to finding common RNA secondary structure elements. Nucleic Acids Res. 2003;31:3446–3449. doi: 10.1093/nar/gkg521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gorodkin J., Stricklin S.L., Stormo G.D. Discovering common stem–loop motifs in unaligned RNA sequences. Nucleic Acids Res. 2001;29:2135–2144. doi: 10.1093/nar/29.10.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolffe A.P., Meric F. Coupling transcription to translation: a novel site for the regulation of eukaryotic gene expression. Int. J. Biochem. Cell. Biol. 1996;28:247–257. doi: 10.1016/1357-2725(95)00141-7. [DOI] [PubMed] [Google Scholar]
- 54.Hirose Y., Manley J.L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 55.Mazan-Mamczarz K., Galban S., Lopez de Silanes I., Martindale J.L., Atasoy U., Keene J.D., Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl Acad. Sci. USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Proudfoot N.J., Furger A., Dye M.J. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 57.Rosonina E., Bakowski M.A., McCracken S., Blencowe B.J. Transcriptional activators control splicing and 3′-end cleavage levels. J. Biol. Chem. 2003;278:43034–43040. doi: 10.1074/jbc.M307289200. [DOI] [PubMed] [Google Scholar]
- 58.Manley J.L. Nuclear coupling: RNA processing reaches back to transcription. Nature Struct. Biol. 2002;9:790–791. doi: 10.1038/nsb1102-790. [DOI] [PubMed] [Google Scholar]
- 59.Wang Y., Newton D.C., Robb G.B., Kau C.L., Miller T.L., Cheung A.H., Hall A.V., VanDamme S., Wilcox J.N., Marsden P.A. RNA diversity has profound effects on the translation of neuronal nitric oxide synthase. Proc. Natl Acad. Sci. USA. 1999;96:12150–12155. doi: 10.1073/pnas.96.21.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pozner A., Goldenberg D., Negreanu V., Le S.Y., Elroy-Stein O., Levanon D., Groner Y. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol. Cell. Biol. 2000;20:2297–2307. doi: 10.1128/mcb.20.7.2297-2307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tanaka M., Ito S., Kiuchi K. The 5′-untranslated region of the mouse glial cell line-derived neurotrophic factor gene regulates expression at both the transcriptional and translational levels. Brain Res. Mol. Brain Res. 2001;91:81–95. doi: 10.1016/s0169-328x(01)00125-5. [DOI] [PubMed] [Google Scholar]
- 62.Gayen A.K., Peffley D.M. The length of 5′-untranslated leader sequences influences distribution of 3-hydroxy-3-methylglutaryl-coenzyme A reductase mRNA in polysomes: effects of lovastatin, oxysterols, and mevalonate. Arch. Biochem. Biophys. 1995;322:475–485. doi: 10.1006/abbi.1995.1491. [DOI] [PubMed] [Google Scholar]
- 63.Larsen L.K., Amri E.Z., Mandrup S., Pacot C., Kristiansen K. Genomic organization of the mouse peroxisome proliferator-activated receptor beta/delta gene: alternative promoter usage and splicing yield transcripts exhibiting differential translational efficiency. Biochem. J. 2002;366:767–775. doi: 10.1042/BJ20011821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arora S., Chauhan S.S. Identification and characterization of a novel human cathepsin L splice variant. Gene. 2002;293:123–131. doi: 10.1016/s0378-1119(02)00700-x. [DOI] [PubMed] [Google Scholar]
- 65.Mironov A.A., Fickett J.W., Gelfand M.S. Frequent alternative splicing of human genes. Genome Res. 1999;9:1288–1293. doi: 10.1101/gr.9.12.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kozak M. New ways of initiating translation in eukaryotes? Mol. Cell. Biol. 2001;21:1899–1907. doi: 10.1128/MCB.21.6.1899-1907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li J., Kinoshita T., Pandey S., Ng C.K., Gygi S.P., Shimazaki K., Assmann S.M. Modulation of an RNA-binding protein by abscisic-acid-activated protein kinase. Nature. 2002;418:793–797. doi: 10.1038/nature00936. [DOI] [PubMed] [Google Scholar]
- 68.Hugouvieux V., Kwak J.M., Schroeder J.I. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.