Abstract
The human prothrombin G20210A polymorphism located at the 3′ cleavage site of the mRNA results in elevated plasma prothrombin levels and increased risk of venous thrombosis. This polymorphism has been shown to directly influence a variety of processes related to prothrombin mRNA metabolism. We have constructed plasmids that express the full-length prothrombin mRNA that is polyadenylated at its natural site. The A allele prothrombin variant was more efficient than the G allele at promoting cleavage at this site in the presence of a competing poly (A) sequence. In the absence of competition, both allelic variants give rise to a similar level of cleavage site heterogeneity. An upstream sequence element (USE) was also identified within the prothrombin 3′-UTR. When placed upstream of two competing poly (A) sites, the USE directed cleavage preferentially to the proximal poly (A) site. In the absence of competition, the USE had no effect on cleavage site selection. This study suggests that the basis for the increase in prothrombin expression in A allele carriers is not due to allelic changes in cleavage site selection per se. In addition, the functionality of USEs needs to be considered within the context of endogenous sequence architecture.
INTRODUCTION
Prothrombin is a central regulatory component of the coagulation cascade which in its active form, thrombin, performs both procoagulant and anticoagulant roles (1). The prothrombin G20210A polymorphism is a single G to A nucleotide transition at the 3′ cleavage site of prothrombin pre-mRNA and is associated with increased levels of plasma prothrombin (2–6). Individuals heterozygous for this polymorphism have a three-fold increased risk of developing venous thrombosis (2). The G20210A polymorphism is functional (7), however, the consequences of this polymorphism on prothrombin gene expression are controversial. Increased plasma prothrombin levels caused by the mutant (A allele) variant have been associated with changes in prothrombin pre-mRNA cleavage and polyadenylation, mRNA stability and translation (8–11). Over the last few years there has been increased emphasis on the functional role of this polymorphism at the level of prothrombin mRNA 3′-end formation.
Specific sequence elements within the pre-mRNA direct the binding of the 3′ pre-mRNA processing complex to the pre-mRNA, which in turn are necessary for cleavage and polyadenylation (reviewed in 12,13). Typically, mammalian 3′ pre-mRNA processing requires the conserved polyadenylation signal (5′-AAUAAA-3′) located 10–30 nt upstream of the cleavage site and a second less conserved element referred to as the downstream sequence element (DSE), usually located within 30 nt downstream of the cleavage site (12). Cleavage does not occur at a consensus site, however a CA dinucleotide is frequently found at the site of cleavage (14,15). A selected number of pre-mRNAs contain additional elements that influence 3′ pre-mRNA processing efficiency besides AAUAAA and DSE. One such element is located upstream of cleavage sites and is referred to as the upstream sequence element (USE). USEs are described as U-rich in sequence (12) and have been predominately characterized in viruses such as SV40, adenovirus, hepatitis B and HIV-1 pre-mRNAs (16–19). More recently, functional USEs have been described in cellular genes including complement factor C2, lamin B2, collagen and 2′-5′-oligoadenylate synthetase enzyme (20–23).
The efficiency and accuracy of an individual poly (A) site is determined by the polyadenylation signal itself and the relative location of auxiliary sequences upstream and downstream of the cleavage site. Bioinformatic analyses have revealed that the efficiency of 3′ pre-mRNA processing is not exclusively mediated by the polyadenylation signal and DSE alone. Previous studies of polyadenylation sites revealed that ∼29% of human genes have alternative poly (A) sites within one gene and the usage of an alternative site is related to biological conditions as opposed to being a random event (24,25). Several studies on core elements suggested that while most (50–60%) human polyadenylation sites contain the canonical AAUAAA element, a large portion of genes have its single-nucleotide variants, with AUUAAA as the most common (24,26,27). DSEs do not have a consensus sequence and are poorly characterized primarily due to the lack of cleavage-site information. Graber and co-workers (26) identified U-rich/UG-rich elements as DSEs, using about 4000 human and 6000 mouse expressed sequence tags. Zarudnaya et al. (28) analysed 244 randomly selected human genes and proposed the existence of auxiliary downstream elements that form G-quadruplex structures in the SV40 late pre-mRNA polyadenylation site as well as in a number of human genes. Recently, Legendre and Gautheret (29) suggested that the DSEs can be characterized as U-rich and probably do not have any specific sequence elements. In their study, the USE was also found to be U-rich; thus the cleavage site is flanked by U-rich sequences. Hence, the relative contribution of the cis-elements involved in cleavage and polyadenylation needs to be analysed on an individual transcript basis.
Recently, it has been demonstrated using a reporter gene assay that the DSE of prothrombin is weak and it overcomes this weakness with a functional USE (30). We have investigated the role of the prothrombin USE in mRNA 3′-end formation using full-length prothrombin mRNA expression vectors. These vectors contain the full-length prothrombin cDNA and downstream genomic sequence (DGS) to facilitate natural prothrombin cleavage and polyadenylation. This model system was used to assess the role of the USE in both a competitive and non-competitive environment for mRNA 3′-end formation. If the USE was placed upstream of two competing poly (A) sites, the USE acted to promote cleavage at the site nearest the USE. Hence, the frequency of mRNA 3′-end formation at a given poly (A) sequence is enhanced in the presence of a functional USE. In the absence of competition, the prothrombin USE had no effect on pre-mRNA 3′-end formation at the level of cleavage site selection but was shown to slightly reduce mRNA expression levels when mutated. Our results also show that the G20210A polymorphism does not significantly influence cleavage site selection. Hence, the basis for the increase in plasma prothrombin expression in A allele prothrombin carriers appears to be unrelated to the selection of the cleavage site per se, but related to other aspects of prothrombin mRNA metabolism.
MATERIALS AND METHODS
Construction of prothrombin expression plasmids containing mutated SV40 polyadenylation signal
To assess the functional role of the G20210A polymorphism we designed constructs that generated the full-length prothrombin mRNA using downstream prothrombin genomic sequences to direct natural 3′-end formation. This was performed using plasmid pCMV-PT-WT (9) as a template. This construct contains the full-length wild-type (G allele) prothrombin cDNA under the control of the CMV promoter and the mRNA derived from this vector is 3′ processed using the SV40 polyadenylation sequence (9). The SV40 polyadenylation signal AAUAAA was mutated to GCGCGA by overlap extension mutagenesis PCR (31) using primers (SJS 009, SJS 014, SJS 015 and SJS 016; see Table 1) with the relevant base substitutions and restriction enzyme sites (HpaI and NotI located within the vector sequence).
Table 1.
PCR amplification primers
| Primer | Nucleotide sequence | Orientation | GenBank reference |
|---|---|---|---|
| SJS 009 | 5′-TGAAGAAGTGGATACAGAAGG-3′ | Forward | (1807–1827 PT cDNA) |
| SJS 010 | 5′-CTGGTTACAAGCCTGATGAAG-3′ | Forward | (1632–1653 PT cDNA) |
| SJS 013 | 5′-GGTACGAGCGAAACATTGAAAAGATAT-3′ | Forward | (1278–1305 PT cDNA) |
| SJS 014 | 5′-GGGCGAAAAACCGTCTATCAGGG-3′ | Reverse | (1727–1750 pCI-Neo) |
| SJS 015 | 5′-CCATTATAAGCTGCGCGCGACAAGTTAACAAC-3′ | Forward | (1271–1300 pCI-Neo) |
| SJS 016 | 5′-GTTGTTAACTTGTCGCGCGCAGCTTATAATGG-3′ | Reverse | (1271–1300 pCI-Neo) |
| SJS 017 | 5′-ATAACATGCGGCCGCTTCCTGAGCCCAGAGAGCT-GCCCATGAATAGCACTGGGAGCATTGAGGCTCGCTG-AGAGTCACTTTTATTG-3′ | Reverse | (26764–26844 PT genomic) |
| SJS 018 | 5′-ATAACATGCGGCCGCTTCCTGAGCCCAGAGAGC-TGCCCATGAATAGCACTGGGAGCATTGAGGCTTG-CTGAGAGTCACTTTTATTG-3′ | Reverse | (26764–26844 PT genomic) |
| SJS 025 | 5′-GCAGCATCGAGGCACTAAAACTATGG-3′ | Forward | (1897–1922 PT cDNA) |
| SJS 026 | 5′-AGTGCCTCGATGCTGCTTCTTTCACGG-3′ | Reverse | (1886–1902 PT cDNA) |
| SJS 056 | 5′-TTTATAGCGGCCGCTGCTGAGAGTCACT-3′ | Reverse | (1934–1947 PT cDNA) |
| SJS 069 | 5′-CGGATGGAAGCCGGTCTTGT-3′ | Forward | (2901–2920 pCI-Neo) |
| SJS 089 | 5′-ATAAGAATGCGGCCGCAAGTAAAACCTCTACAAATG-3′ | Forward | (1364–1382 pCI-Neo) |
| SJS 090 | 5′-GTCGGGGCTGGCTTAAC-3′ | Reverse | (3434–3450 pCI-Neo) |
| SJS 125 | 5′-GCGATAGAAGGCGATGCGCT-3′ | Reverse | (3217–3236 pCI-Neo) |
| Nest 13 FNest | 5′-TGCCTGTGAAGGTGACAGTG-3′ | Forward | (1667–1686 PT cDNA) |
| Nest 13F.1 | 5′-ACCCTTTGTCATGAAGAGCC-3′ | Forward | (1690–1710 PT cDNA) |
The name, nucleotide sequence, orientation and GenBank nucleotide reference are provided. The mutations within the sequence are underlined and the restriction enzyme sites are italicized. Prothrombin cDNA (accession no. V00595), prothrombin genomic sequence (accession no. M17262) and pCI-Neo vector (Promega).
To generate the A allele variant of this construct, the G allele prothrombin cDNA was first removed from the SV40-mutated vector by Sal1 and Not1 digestion, and replaced with the A allele variant prothrombin cDNA that had been excised from plasmid pCMV-PT-MUT (9) using the same restriction enzymes. The plasmids generated by this procedure are shown schematically in Figure 1B and are listed below:
PT-G: wild-type (G allele) prothrombin cDNA containing mutated SV40 polyadenylation site.
PT-A: mutant (A allele) prothrombin cDNA containing mutated SV40 polyadenylation site.
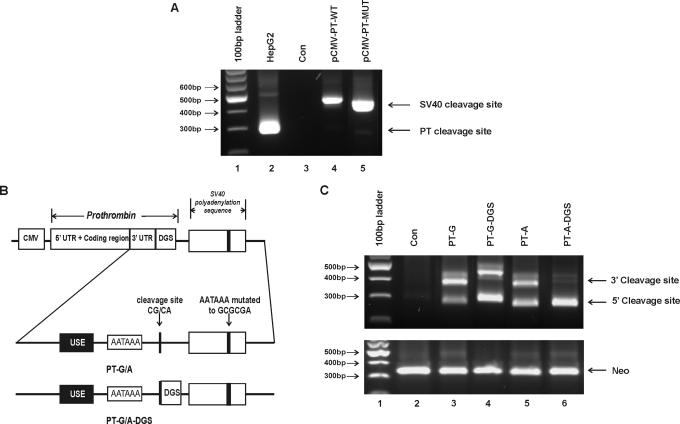
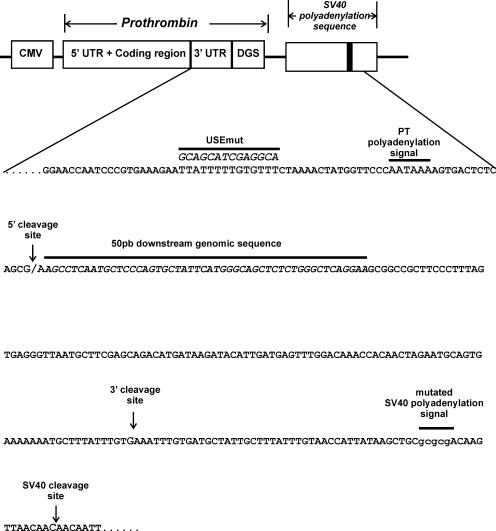
Figure 1.
Prothrombin mRNA is cleaved and polyadenylated in the absence of DGS. (A) 3′RACE results of RNA isolated from cells stably transfected with prothrombin allelic variants. Lane 1, 100 bp DNA ladder; lane 2, Endogenous prothrombin mRNA detected in HepG2 cells; lane 3, HEK-293 cells transfected with the pCI-Neo vector (con); lanes 4 and 5, HEK-293 cells expressing wild-type (pCMV-PT-WT) or mutant (pCMV-PT-MUT) prothrombin variants, respectively, using the SV40 polyadenylation signal. (B) Schematic representation of the constructs expressing prothrombin variants. All constructs express the full-length prothrombin cDNA including the entire 3′-untranslated region (3′-UTR) under the control of the CMV promoter. Cleavage and polyadenylation elements involved in prothrombin 3′-end formation are indicated: USE, upstream sequence element; DGS, 50 bp downstream genomic sequence; AAUAAA, polyadenylation signal; CG/CA, polymorphic cleavage site. All constructs contain mutations within the SV40 polyadenylation signal (AAUAAA mutated to GCGCGA): PT-G/A, allelic prothrombin cDNA with no DGS; PT-G/A-DGS, prothrombin variant cDNA with 50 bp of DGS. (C) 3′RACE results of RNA isolated from HEK-293 cells transiently transfected with prothrombin allelic variants containing mutations within the SV40 poly A signal, with or without the DGS of prothrombin. Lane 1, 100 bp DNA ladder; lane 2, HEK-293 cells transfected with the pCI-Neo vector (con), lane 3, PT-G; lane 4, PT-G-DGS; lane 5, PT-A and lane 6, PT-A-DGS. The 5′ cleavage site represents mRNA produced using the natural prothrombin cleavage site. The slower migrating band (3′ cleavage site) depicts the cryptic downstream cleavage site within the vector sequence. Neomycin phosphotransferase (Neo) mRNA was used as a measure of transfection efficiency (lower panel).
Introduction of DGS into plasmid pPT-G/A
A 50 bp genomic prothrombin sequence (position 26785–26844 accession no. M17262) downstream of the prothrombin mRNA cleavage site was introduced into the modified allelic prothrombin expression plasmids (above). This was generated by PCR using mega-primers (G and A allele variants) containing the 50 bp genomic sequence (primers SJS 017 and SJS 018, respectively), and an upstream primer (SJS 010) that incorporated the Bcl1 site within the prothrombin 3′-untranslated region (3′-UTR). The mega-primer contained a Not1 site at the 3′ end (the terminal cloning site of prothrombin). Digestion of the PCR product with Bcl1 and Not1 enabled subcloning of 50 bp DGS juxtaposed to the prothrombin cleavage site within PT-G and PT-A. The plasmids generated from these manipulations are shown schematically in Figure 1B and listed below:
PT-G-DGS: wild-type (G allele) prothrombin cDNA containing mutated SV40 polyadenylation signal and 50 bp of DGS.
PT-A-DGS: mutant (A allele) prothrombin cDNA containing mutated SV40 polyadenylation signal and 50 bp of DGS.
Generation of plasmids containing mutations of the prothrombin USE
The prothrombin USE (TTATTTTTGTGTTT, see Results) was mutated to GCAGCATCGAGGCA by overlap mutagenesis PCR using plasmid pPT-G as a template. This was performed using primers harbouring the relevant base substitutions within the USE (SJS 025 and SJS 026) and external primers SJS 013 and SJS 014 that included restriction enzymes EcoRV (located 5′ of the prothrombin USE) and NotI, respectively. The resulting plasmid was referred to as PT-G-USEmut. The A allele prothrombin variant containing a mutated USE(PT-A-USEmut) was generated using primers SJS 013 and SJS 056 (the latter primer introduces the A residue at the poly (A) site and a NotI restriction site at the 3′ end) and PT-G-USEmut as the template. Following this, the DGS was introduced into these plasmids using the strategy outlined above. The plasmids generated from these manipulations are listed below and shown schematically in Figure 4A:
PT-G-USEmut: wild-type (G allele) prothrombin cDNA containing mutated SV40 polyadenylation signal and mutated prothrombin USE.
pPT-A-USEmut: mutant (A allele) prothrombin cDNA containing mutated SV40 polyadenylation signal and mutated prothrombin USE.
PT-G-DGS-USEmut: wild-type (G allele) prothrombin cDNA containing mutated SV40 polyadenylation signal and mutated prothrombin USE with 50 bp of DGS.
PT-A-DGS-USEmut: mutant (A allele) prothrombin cDNA containing mutated SV40 polyadenylation signal and mutated prothrombin USE with 50 bp of DGS.
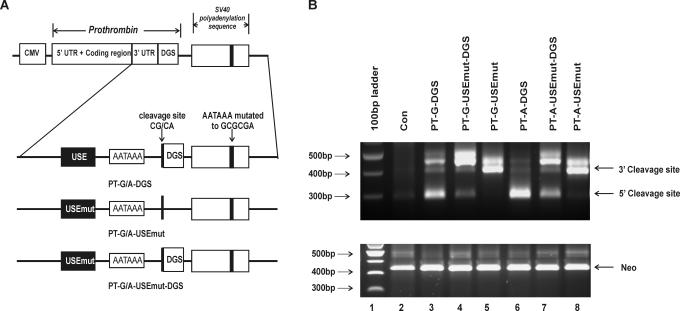
Figure 4.
USE in prothrombin 3′-UTR influences the selection of the natural prothrombin cleavage site. (A) Schematic representation of the constructs expressing prothrombin variants. All constructs express the full-length prothrombin cDNA including the entire 3′-UTR under the control of the CMV promoter. Cleavage and polyadenylation elements involved in prothrombin 3′-end formation are indicated: USE, upstream sequence element; DGS, 50 bp downstream genomic sequences; AAUAAA, polyadenylation signal; CG/CA, polymorphic cleavage site. All constructs contain mutations within the SV40 polyadenylation signal (AAUAAA mutated to GCGCGA): PT-G/A-DGS, prothrombin variant cDNA with 50 bp of DGS; PT-G/A-USEmut, prothrombin variant cDNA with mutated USE; PT-G/A-USEmut-DGS, prothrombin variant cDNA with 50 bp DSG and mutated USE. (B) 3′RACE results of RNA prepared from HEK-293 cells transiently transfected with prothrombin allelic variants containing mutations within the SV40 poly A signal, with or without the DGS and/or USE of prothrombin. Lane 1, 100 bp DNA ladder; lane 2, HEK-293 cells transfected with the pCI-Neo vector (con); lane 3: PT-G-DGS; lane 4: PT-G-USEmut-DGS; lane 5, PT-G-USEmut; lane 6, PT-A-DGS; lane 7, PT-A-USEmut-DGS and lane 8, PT-A-USEmut. The 5′ cleavage site represents mRNA produced using the natural prothrombin cleavage site. The slower migrating band (3′ cleavage site) depicts the cryptic downstream cleavage site within the vector sequence. Neo mRNA was used as a measure of transfection efficiency (lower panel).
Generation of plasmids containing complete deletion of the SV40 poly (A) sequence
To remove the cryptic cleavage site within the SV40 polyadenylation sequence (see Results), the 226 bp late SV40 polyadenylation sequence (1137–1363 bp) within the pCI-Neo vector (Promega) was deleted. The vector was first digested with Not1 and BamH1. This removed a 2.0 kb region that included the SV40 polyadenylation sequence, neomycin resistance gene (neo) and the f1 origin of replication (f1 ori). To replace the region containing Neo and f1 ori, a PCR strategy was adopted. To this end, the region from 1364 to 3350 of pCI-Neo (that excludes the SV40 polyadenylation sequence) was amplified by PCR using upstream primer SJS 089 (located 3′ of the SV40 polyadenylation sequence and incorporating a Not1 site at the 3′end) and downstream primer SJS 090 (located 3′ of BamHI site within the vector). The resulting product was digested with BamHI and NotI and subcloned into the appropriate site within the pCI-Neo vector to create pCIΔSV40. Note the pCIΔSV40 vector retains the pCI-Neo multiple cloning sites. All prothrombin variants were then subcloned into pCIΔSV40 using the SalI and NotI restriction sites within the multiple cloning site. These plasmids are shown schematically in Figure 6A.
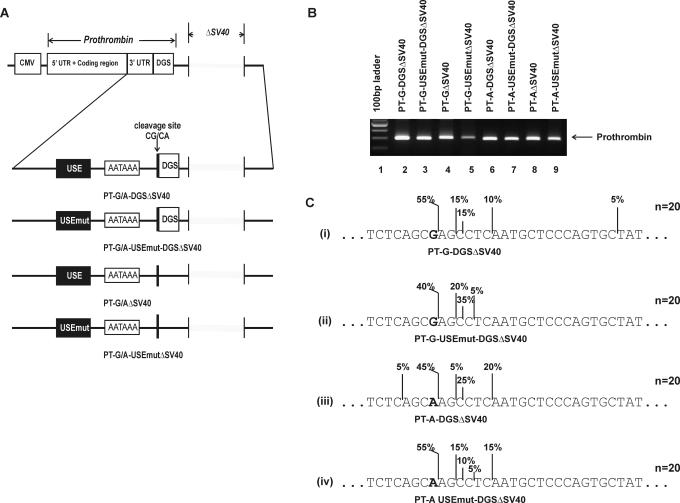
Figure 6.
The prothrombin G20210A and the USE do not impact on cleavage site selection in the absence of competition. (A) Schematic representation of the constructs expressing prothrombin variants. All constructs express the full-length prothrombin cDNA including the entire 3′-UTR under the control of the CMV promoter. Cleavage and polyadenylation elements involved in prothrombin 3′-end formation are indicated: USE, upstream sequence element; DGS, 50 bp downstream genomic sequences; AAUAAA, polyadenylation signal; CG/CA, polymorphic cleavage site. The SV40 polyadenylation sequence has been removed from all constructs (ΔSV40): PT-G/A-DGS, prothrombin variant cDNA with 50 bp of DGS; PT-G/A-USEmut-DGS, prothrombin variant cDNA with 50 bp DSG and mutated USE; PT-G/A, allelic prothrombin cDNA with no DGS; PT-G/A-USEmut, prothrombin variant cDNA with mutated USE. (B) 3′RACE results of RNA prepared from HEK-293 cells transiently transfected with constructs expressing prothrombin allelic variants (with and without DGS and/USE) using vectors that had the SV40 polyadenylation sequence removed. Lane 1, 100 bp DNA ladder; lane 2, PT-G-DGS; lane 3, PT-G-USEmut-DGS; lane 4, PT-G; lane 5, PT-G-USEmut; lane 6, PT-A-DGS; lane 7, PT-A-USEmut-DGS; lane 8, PT-A and lane 9, PT-A-USEmut. These vectors generate a single 3′RACE product representing prothrombin mRNA being cleaved at its natural position. (C) Schematic representation of the percentage usage of prothrombin cleavage sites (determined by sequencing) generated by the above constructs; (i) PT-G-DGSΔSV40 (n = 20), (ii) PT-G-USEmut-DGS ΔSV40 (n = 20), (iii) PT-A-DGS ΔSV40 (n = 20) and (iv) PT-A-USEmut-DGS ΔSV40 (n = 20). The polymorphic residue at position 20210 is shown in bold.
Generation of plasmids containing tandem prothrombin polyadenylation sequences
The pCIΔSV40 vector was used to generate a prothrombin expression plasmid containing tandem copies of the prothrombin polyadenylation sequence (that included the DGS). The prothrombin cDNA (harbouring one copy of the 3′-UTR and DGS) was initially subcloned into the SalI and SmaI sites of pCIΔSV40. The second copy of the prothrombin polyadenylation sequence was introduced into the above construct using SmaI and NotI restriction sites. The tandem arrangement of the polyadenylation sequences in plasmid pCIΔSV40 is shown schematically in Figure 5A.
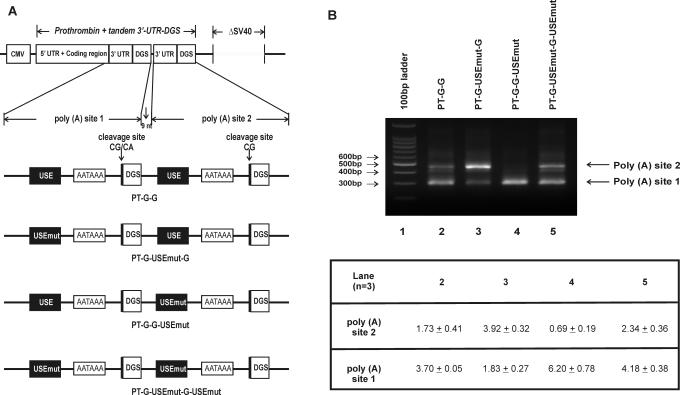
Figure 5.
The USE is functional in the presence of tandem prothrombin poly (A) signals. (A) Schematic representation of constructs expressing the full-length wild-type (G allele) prothrombin mRNA with tandem arrangement of the 3′-UTR and DGS indicated as poly (A) site 1 and poly (A) site 2. All constructs contain a 9 nt space between the tandem repeats. Cleavage and polyadenylation elements involved in prothrombin 3′-end formation are indicated: USE, upstream sequence element; DGS, 50 bp downstream genomic sequences; AAUAAA, polyadenylation signal; CG, wild-type cleavage site. The SV40 polyadenylation sequence (ΔSV40) has been removed from all constructs: PT-G-G, prothrombin cDNA with tandem poly (A) sites (poly A sites 1 and 2); PT-G-USEmut-G, contains mutation of USE within poly (A) site 1; PT-G-G-USEmut, contains mutation of USE within poly (A) site 2; PT-G-USEmut-G-USEmut-DGS, contains mutation of the USE within poly A sites 1 and 2. (B) 3′RACE results of RNA from cells transfected with the tandem prothrombin polyadenylation sequence (3′-UTR + DGS). Lane 1, 100 bp DNA ladder; lane 2, PT-G-G; lane 3, PT-G-USEmut-G; lane 4, PT-G-G-USEmut and lane 5, PT-G-USEmut-G-USEmut. The table below represents the quantitation (Image Quant) of the bands generated by prothrombin mRNA expression using poly (A) site 1 (5′) and 2 (3′) ± standard error of the mean (n = 3). The quantitation was corrected for fragment size.
Cell culture and transfection
Human HEK-293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen), supplemented with 10% (v/v) heat-inactivated foetal bovine serum (HI-FBS), 2mM glutamine, and streptomycin and penicillin, in a humidified atmosphere at 37°C with 5% CO2. Constructs were transfected transiently by calcium phosphate precipitation using 5 μg plasmid DNA in a 60 mm dish as previously described (32), except the transfection mixture was incubated with the cells for 16 h and the glycerol shock was omitted (33). Fresh DMEM containing 10% HI-FBS was then added and cells were maintained for a further 48 h. Total RNA was extracted for further analysis. Stable cell lines were generated by selecting with 1000 μg/ml G418 (Life Technologies, Inc.) and pooling resistant clones.
RNA extraction and 3′RACE
Total RNA was extracted from cells as described previously (34). The quality of each RNA sample was confirmed by agarose-gel (1%) electrophoresis. cDNA synthesis was performed by 3′ rapid amplification of cDNA ends (3′RACE) (Invitrogen Life Technologies). 2 μg of RNA was used for first strand cDNA synthesis. A nested PCR method was performed to determine the cleavage/polyadenylation site of prothrombin mRNA. 2 μl of the first strand cDNA synthesis was used to amplify prothrombin cDNA sequences by PCR using the Ex 13F Nest primer (Table 1) and reverse primer AUAP (3′RACE kit). Hot start PCR was performed in which Taq polymerase was added after incubation of the reaction for 5 min at 94°C. Thermal cycling was carried out after preincubation at 94°C for 3 min, 25 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s and extension at 72°C for 1 min and a final step at 5 min at 72°C. About 2 μl of 1 in 100 dilution of the above PCR reaction was used as a template for the nested PCR. The second PCR was performed using primer Ex 13F.1 Nest (Table 1) and the same reverse primer as used for the first PCR. Thermal cycling was carried out in a similar fashion to the first PCR except 35 cycles were performed with an annealing temperature of 65°C. Neomycin phosphotransferase (Neo) mRNA was detected using 2 μl of first stand reaction and primers SJS 069 and SJS 125. Thermal cycling was performed as above (25 cycles) with an annealing temperature of 65°C. PCR products were resolved on 2% agarose gels and visualized on a transilluminator using ethidium bromide. To assess PCR products for specificity and to evaluate position of the polyadenylation site, the products were excised from the gel and cloned into pGEM-T Easy vector (Promega). Positive clones were sequenced using the universal T7 primer.
Ribonuclease protection assay
The ribonuclease (RNase) protection assay was carried out using the RPA III kit (Ambion) with 10 μg of total RNA prepared from transiently transfected HEK-293 cells. RNA samples were subjected to DNase-1 digestion (Promega) prior to use. The prothrombin RNA probe was 250 nt in length directed against region 1643–1889 within the prothrombin cDNA (accession no. V00595). The Neo probe was 200 nt in length directed towards position 2472–2680 within the pCI-Neo vector. To generate these RNA probes, prothrombin and Neo cDNA fragments were first subcloned into the pGEM-T Easy vector and then linearized with SacII. The probes were in vitro transcribed using SP6 RNA polymerase. The products were resolved on a denaturing gel (7M urea/5% polyacrylamide gel) and visualized by autoradiography (Kodak BioMax film). Signals were quantitated using ImageQuant Version 5 (Amersham Biosciences) and the results presented in graphical form after correcting for changes in Neo mRNA expression between samples.
Statistical analyses
Results are expressed as the mean ± standard error of the mean (SEM) and analyses were performed by two-sample one tailed Students t-test. P-value < 0.05 was considered as significant.
RESULTS
Prothrombin mRNA is able to cleave and polyadenylate at its natural position in the absence of DGS
Previously, Carter and co-workers (9) generated the full-length prothrombin mRNA using the vector derived SV40 polyadenylation signal (pCMV-PT-WT/MUT). To confirm the position of the 3′ cleavage site, these constructs were first transfected into HEK-293 cells and the 3′ cleavage site of prothrombin mRNA was determined by 3′RACE and sequencing. As shown in Figure 1A (lanes 4 and 5), the G and A allelic prothrombin transcripts generated using the SV40 polyadenylation signal (pCMV-PT-WT/MUT) produce a single product that is ∼200 nt longer than the endogenous prothrombin expressed in Hep G2 cells (Figure 1A, lane 2). Sequence analysis confirmed that cleavage occurred at the SV40 polyadenylation site. This difference, as expected, is fully accounted for by vector-derived sequences.
To assess the functional role of the G20210A polymorphism at the level of 3′ pre-mRNA processing, we designed constructs that generated full-length prothrombin mRNA that was cleaved and polyadenylated at its natural site. To achieve this, the highly efficient SV40 polyadenylation signal within pCMV-PT-WT/MUT vectors was inactivated via mutation (Figure 1B, PT-G/A). In addition to the polyadenylation signal, 3′ pre-mRNA processing also requires DSEs usually located within 30 nt of the cleavage site (12). Therefore, 50 bp of prothrombin DGS was introduced into the vectors containing the mutated SV40 polyadenylation signal to direct natural prothrombin mRNA 3′-end formation (Figure 1B). These prothrombin constructs were transfected into HEK-293 cells and prothrombin mRNA expression and the 3′ cleavage site selection was analysed by 3′RACE and DNA sequencing.
3′RACE results of RNA from HEK-293 cells transfected with the prothrombin expression constructs containing the mutated SV40 polyadenylation signal are shown in Figure 1C. In the absence of both the SV40 polyadenylation signal and downstream prothrombin genomic sequences, both allelic prothrombin expression plasmids (PT-G/A) were able to generate transcripts with cleavage occurring at two sites. The identical pattern was seen for both the A and G allele prothrombin variants (Figure 1C, lanes 3 and 5). Sequencing analysis revealed that the 5′ cleavage site was occurring at the natural prothrombin cleavage site, while the unexpected 3′ site was located within the vector derived-sequence, upstream of the mutated SV40 polyadenylation signal (Figure 2). The faint larger product just above the 3′ cleavage site is the result of the low level of cleavage at the SV40 site (Figure 3C). In principle, the mutated SV40 signal is deactivated, however, we cannot exclude the occurrence of rare cleavage events that are detected by the sensitivity of the PCR technique.
Figure 2.
Schematic representation and nucleotide sequence of prothrombin variants expressed using the vectors harbouring the mutated SV40 polyadenylation signal. Indicated in the figure: the mutated prothrombin upstream sequence element (USEmut); AAUAAA, prothrombin (PT) polyadenylation signal; CG/CA, prothrombin polymorphic cleavage site; 50 bp prothrombin downstream genomic sequence (DGS); cryptic 3′ cleavage site; mutated SV40 polyadenylation signal (gcgcgA); SV40 cleavage site, AC. Due to the presence of adenine residues downstream of the indicated cryptic (1–3 nt) and SV40 (1–2 nt) sites, it is not possible to ascertain the exact site of cleavage by sequencing due to the presence of the poly (A) tail. For simplicity only the last non-adenine residue is indicated as the position of the cryptic and SV40 cleavage sites.
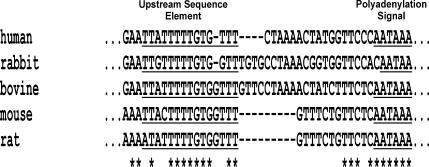
Figure 3.
Prothrombin USE is highly conserved in sequences derived from other mammalian species. Comparison of the prothrombin USE within mammalian species; human (accession no. V00595), mouse (accession no. M81394), rat (accession no. M81397), rabbit (accession no. M81396) and bovine (accession no. NM 173877). Asterisks indicate the conserved nucleotides between the mammalian species.
The molecular basis for the selection of the cryptic 3′ cleavage site is presently unknown but may possibly be due to a variant polyadenylation signal or secondary structure. However, the fact that some cleavage was occurring at the natural position even in the absence of downstream sequences suggests that the prothrombin transcript contains functionally relevant cis-acting elements upstream of the cleavage site to direct the cleavage machinery to the correct location.
Introduction of 50 bp of DGS resulted in an allele-specific alteration in the cleavage site selection (Figure 1C, lanes 4 and 6). Plasmids expressing the wild-type (G allele) prothrombin variant produced transcripts that utilized the same two cleavage sites, one occurring at the natural position, and the other at the cryptic site. In contrast, plasmids expressing the mutant (A allele) prothrombin produced transcripts that were cleaved at the natural position, with only a small percentage of transcripts being cleaved at the cryptic 3′ position. Hence, the A allele variant is stronger than the G allele at maintaining cleavage at the prothrombin poly (A) sequence.
An upstream element in the prothrombin 3′-UTR influences selection of the 3′ cleavage site
Results presented in Figure 1C (lanes 3 and 5) provide evidence that prothrombin mRNA contains a functional upstream sequence that influences the selection of the cleavage site. Such USEs within the 3′-UTR have been described in only a few mammalian genes, and in these cases, the identified USE was shown to be highly U-rich. Close analysis of the prothrombin 3′-UTR revealed the presence of a putative U-rich USE located 17 nt from the natural prothrombin polyadenylation signal (Figure 2). Comparison of the human prothrombin transcript with the prothrombin sequence derived from rabbit, bovine, mouse and rat revealed that this USE was highly conserved (Figure 3) which is suggestive of functional relevance. To determine if the USE plays a role in prothrombin 3′-end formation, a 14 bp substitution (Figure 2) was introduced into this region within the SV40 polyadenylation signal-inactivated plasmids (Figure 4A). 3′RACE analysis of HEK-293 cells transfected with these expression constructs revealed that the natural cleavage site was rarely used and the cleavage site was moved to the downstream cryptic position for both allelic constructs (Figure 4B, lanes 4 and 7). Furthermore, removal of the genomic downstream sequences from these constructs resulted in all transcripts being 3′cleaved at the cryptic site within the vector (Figure 4B, lanes 5 and 8). Taken together, these data indicate that a functional USE within the prothrombin 3′-UTR influences the selection of the 3′ cleavage site.
The USE is functional in the presence of tandem prothrombin poly (A) signals
The constructs containing an inactivated SV40 polyadenylation signal possessed a cryptic cleavage site within the SV40 poly (A) sequence which competed with the natural prothrombin cleavage site (Figure 1C). To assess the role of the USE in an environment in which the competing poly(A) signals are of similar strength, we removed the cryptic 3′ cleavage site by deleting the entire 226 bp SV40 polyadenylation sequence (i.e. ΔSV40) and incorporated a tandem copy of the full-length G allele variant of the prothrombin 3′-UTR and DGS (Figure 5A). These plasmids were transfected into HEK-293 cells and 3′RACE was performed to assess the preferred sites of cleavage. As shown in Figure 5B, both cleavage sites are utilized in the presence of tandem repeat sequences with unaltered USE and DSE, with cleavage favouring the poly (A) site within the first repeat [i.e. poly (A) site 1] (lane 2). When the USE was selectively mutated within poly (A) site 1 of the 3′-UTR, poly (A) site 2 was preferred (lane 3). In the reverse experiment, mutation of the USE within poly (A) site 2 forced cleavage to occur at poly (A) site 1 (lane 4). When the USE was mutated within both poly (A) sites, cleavage appeared to occur favourably at poly (A) site 1 (lane 5), similar to the pattern seen for the wild-type tandem transcript (lane 2). These results demonstrate that frequency of 3′-end formation of a given poly (A) signal within a competitive environment is enhanced in the presence of a functional USE.
The prothrombin G20201A and the USE do not impact on cleavage site selection in the absence of competition
It was important to assess the role of the USE in a non-competitive environment. To assess this, prothrombin expression constructs containing a single copy of the prothrombin 3′-UTR and DGS (i.e. the native configuration) were expressed using vectors (pPT-G/A-ΔSV40, Figure 6A). The ΔSV40 constructs were transiently transfected into HEK-293 cells and the cleavage site was determined by 3′RACE and sequencing. The precise position of the cleavage site of the prothrombin mRNA cannot be accurately determined after the addition of a poly (A) tail since an A residue is present at position 20211 within the gene. Hence, the cleavage site for the G allele prothrombin mRNA variant can occur at 20210G or 20211A, whereas the A allele variant cleavage could occur at 20209C, 20210A or 20211A. Here, we have defined the CG (G allele) or CA (A allele) dinucleotide preceding the poly (A) tail as being cleaved at position 20210. In the absence of any competing downstream cleavage site, both prothrombin variants (G and A allele) produced a single transcript with a size consistent with the natural cleavage site (Figure 6B). Indeed, mutagenesis of the USE had little effect on prothrombin mRNA production (Figure 6B, compare lanes 2 with 3 and 6 with 7) and did not influence cleavage site selection. All transcripts containing either the G or A allelic variant with or without an intact USE produced a similar heterogeneous population of transcripts (Figure 6C). This data indicates that neither the G20210A polymorphism nor the USE influence selection of the cleavage site in the absence of competition in this model system.
The influence of the USE and DGS on prothrombin mRNA expression levels
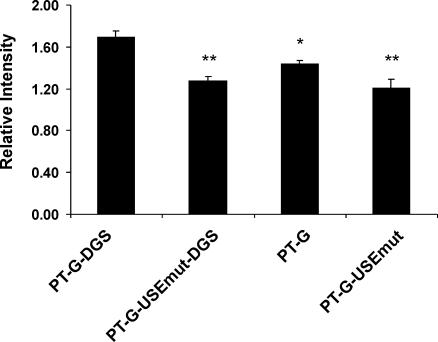
To determine whether the USE either alone or in combination with the DGS could affect amount of prothrombin mRNA production, RNase protection assays were undertaken on total RNA from cells transfected with prothrombin G allele variants containing mutations/deletions in either or in both elements. Mutagenesis of the USE or the removal of DGS (either alone or in combination) resulted in a reduction in levels of prothrombin mRNA (Figure 7). However, since the majority of prothrombin mRNA expression is maintained in the absence of both the USE and the DGS, it implies that other elements within the prothrombin 3′-UTR are critical for maximal mRNA production.
Figure 7.
The influence of the USE and DGS on prothrombin mRNA expression levels. Graphical representation of prothrombin mRNA production determined by RNase protection assays. HEK-293 cells were transiently transfected with the wild-type prothrombin variant (PT-G-DGS) or the same plasmid containing either a mutation of the USE (PT-G-USEmut-DGS), removal of the DGS (PT-G) or both mutation of the USE and removal of the DGS (PT-G-USEmut). The signals for PT and Neo were quantitated using ImageQuant. The bar diagram indicates the level of prothrombin mRNA expression ± SEM after normalization for transfection efficiency with neo mRNA. Asterisks indicate significant difference in mRNA expression compared to PT-G-DGS control (*P < 0.05, **P < 0.02; n = 3).
DISCUSSION
The molecular function of the prothrombin G20210A polymorphism has been associated with various aspects of gene regulation including mRNA 3′-end formation, stability and translation (8–11,35). In general, the polymorphism appears to have a critical effect at the level of mRNA 3′-end formation. Using a reporter gene system it was demonstrated that the A allele transcript of prothrombin promotes increased efficiency of cleavage and polyadenylation compared to the G allele (8,10). Additionally, the A allele of prothrombin has been found to give rise to a homogeneous population of transcripts in contrast to the G allele in vivo (using liver tissue) and in vitro (using a reporter gene system) (11,35). Here, we have assessed the role of the G20210A polymorphism and 3′-end formation within the context of the full-length prothrombin mRNA.
The full-length prothrombin mRNA was first generated using mammalian expression vectors with the SV40 polyadenylation signal mutated containing the complete prothrombin cDNA and 50 bp of the DGS. The DGS was included to provide prothrombin with its DSE which is necessary for mRNA 3′-end formation. These constructs, with an inactivated SV40 polyadenylation signal generated transcripts with two cleavage sites; one occurring at the natural prothrombin poly (A) site and the other at an unforeseen 3′ distal cleavage site, located within the SV40 vector-derived sequence. The cryptic 3′ distal cleavage site was upstream of the mutated SV40 polyadenylation signal. This phenomenon was not specific for HEK-293 cells as it was reproduced in transfected NIH 3T3 and HeLa cells (data not shown). Secondary structures can influence the ability of DSEs to promote cleavage and polyadenylation (28,36). We suspect that a secondary structure formed within the pre-mRNA generated from these constructs optimally positions the cryptic site for cleavage using the prothrombin polyadenylation signal. However, it is equally likely that cleavage is occurring using an unconventional polyadenylation signal (37). For instance the SV40 polyadenylation signal (AAUAAA) when mutated to AAAAAA retained ∼5% polyadenylation efficiency in vitro (14). We have observed the presence of a stretch of AAAAAA located 13 nt upstream of the cryptic cleavage site which, when combined with the U-rich element (4 out of 5 U tract) located 19 nt downstream (refer to Figure 2 in Results), may facilitate the cleavage at this site. Four out of five U residues located within 6–25 nt downstream of the late SV40 polyadenylation signal were sufficient to mediate 3′ pre-mRNA processing (38).
The production of two cleavage sites by vectors containing the inactivated SV40 polyadenylation signal provided a convenient competition assay to ascertain the cis- elements necessary for cleavage at the natural prothrombin mRNA site. These constructs demonstrated that the CA dinucleotide (A allele) at the cleavage site of prothrombin directs more efficient pre-mRNA 3′-end formation compared to the CG dinucleotide (G allele). This supports the earlier observations from tandem poly (A) signal competition assays (8,10) and extends the evidence towards the functional importance of this polymorphism. This indirect competition assay also revealed that the USE of prothrombin is required for efficient mRNA 3′-end formation at its natural site. The importance of a USE in mRNA 3′-end formation was further evaluated using constructs expressing prothrombin that had the entire SV40 polyadenylation sequence removed. These constructs generated transcripts that cleaved and polyadenylated only at prothrombin's endogenous poly (A) site.
Within this context, both variants of the prothrombin gene produced a similar heterogenous mRNA population in HEK-293 cells. This agrees with the data obtained within the context of a reporter gene system containing the prothrombin 3′-UTR and DGSs (∼70 nt) in Hep G2 cells (10). In contrast, homogenous transcript production by the A allele was previously described in an in vivo study using liver tissue (11) and within the context of the reporter gene system containing the terminal region of the prothrombin gene which included the last two exons, the last intron and the DGS (35). In this study, the expression level of the reporter gene was further increased by incorporating additional DGS (∼1 kb). It cannot be excluded that these additional genomic sequences may be required to promote homogenous prothrombin mRNA population by the A allele. Splicing and polyadenylation events are coupled at the level of terminal intron removal (39–41). Our experimental system did not contain introns and thus 3′ pre-mRNA processing was independent of any splicing events. Therefore, the effect of splicing of the last intron in cleavage site selection remains to be determined.
A recent study using reporter genes also demonstrated that the USE of prothrombin influences the efficiency of cleavage and polyadenylation when in a competitive environment (30). We have demonstrated that the USE is indeed important for prothrombin poly (A) site selection in a competitive environment, but whether competition occurs naturally is unknown. However, it is interesting to note the presence of a potential polyadenylation signal (AAUAAA) located 627 bp downstream of the AAUAAA found within the 3′-UTR of the prothrombin gene. If this downstream site is present within the prothrombin pre-mRNA, the USE within the 3′-UTR may be required to ensure cleavage at the first polyadenylation signal and thus, prevent mRNA 3′-end formation at this downstream site.
Within the context of the full-length prothrombin cDNA, the USE was shown to have a minor effect on levels of mRNA expression as assessed by RNase protection assays, yet had no influence on cleavage site selection. This suggests that the prothrombin gene may have other positional elements within its 3′-UTR that promote efficient pre-mRNA 3′-end formation. The influence of USEs on mRNA 3′-end formation has been often characterized using reporter gene approaches or in vitro assays (20,30,42). Our study demonstrated the importance of characterizing the role of USEs within its natural context to gain a complete understanding of its role in mRNA 3′-end formation.
In conclusion, we have provided novel insights into prothrombin gene expression by utilizing an experimental system that generated full-length prothrombin mRNA which was cleaved and polyadenylated at the natural cleavage site. Within this context the prothrombin G20210A polymorphism does not appear to impact on cleavage site selection. This suggests that the G20210A polymorphism may modulate other aspects of gene regulation besides mRNA 3′-end formation.
Acknowledgments
This work was supported by grants obtained by R.L.M. from the National Health and Medical Research Council of Australia (NH&MRC). Funding to pay the Open Access publication charges for this article was provided by the NH&MRC.
REFERENCES
- 1.Spronk H.M., Govers-Riemslag J.W., ten Cate H. The blood coagulation system as a molecular machine. Bioessays. 2003;25:1220–1228. doi: 10.1002/bies.10360. [DOI] [PubMed] [Google Scholar]
- 2.Poort S.R., Rosendaal F.R., Reitsma P.H., Bertina R.M. A common genetic variation in the 3′-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood. 1996;88:3698–3703. [PubMed] [Google Scholar]
- 3.Brown K., Luddington R., Williamson D., Baker P., Baglin T. Risk of venous thromboembolism associated with a G to A transition at position 20210 in the 3′-untranslated region of the prothrombin gene. Br. J. Haematol. 1997;98:907–909. doi: 10.1046/j.1365-2141.1997.3093130.x. [DOI] [PubMed] [Google Scholar]
- 4.Souto J.C., Coll I., Llobet D., del Rio E., Oliver A., Mateo J., Borrell M., Fontcuberta J. The prothrombin 20210A allele is the most prevalent genetic risk factor for venous thromboembolism in the Spanish population. Thromb. Haemost. 1998;80:366–369. [PubMed] [Google Scholar]
- 5.Margaglione M., Brancaccio V., Giuliani N., D'Andrea G., Cappucci G., Iannaccone L., Vecchione G., Grandone E., Di Minno G. Increased risk for venous thrombosis in carriers of the prothrombin G→A20210 gene variant. Ann. Intern. Med. 1998;129:89–93. doi: 10.7326/0003-4819-129-2-199807150-00003. [DOI] [PubMed] [Google Scholar]
- 6.Makris M., Preston F.E., Beauchamp N.J., Cooper P.C., Daly M.E., Hampton K.K., Bayliss P., Peake I.R., Miller G.J. Co-inheritance of the 20210A allele of the prothrombin gene increases the risk of thrombosis in subjects with familial thrombophilia. Thromb. Haemost. 1997;78:1426–1429. [PubMed] [Google Scholar]
- 7.Soria J.M., Almasy L., Souto J.C., Tirado I., Borell M., Mateo J., Slifer S., Stone W., Blangero J., Fontcuberta J. Linkage analysis demonstrates that the prothrombin G20210A mutation jointly influences plasma prothrombin levels and risk of thrombosis. Blood. 2000;95:2780–2785. [PubMed] [Google Scholar]
- 8.Gehring N.H., Frede U., Neu-Yilik G., Hundsdoerfer P., Vetter B., Hentze M.W., Kulozik A.E. Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nature Genet. 2001;28:389–392. doi: 10.1038/ng578. [DOI] [PubMed] [Google Scholar]
- 9.Carter A.M., Sachchithananthan M., Stasinopoulos S., Maurer F., Medcalf R.L. Prothrombin G20210A is a bifunctional gene polymorphism. Thromb. Haemost. 2002;87:846–853. [PubMed] [Google Scholar]
- 10.Ceelie H., Spaargaren-van Riel C.C., Bertina R.M., Vos H.L. G20210A is a functional mutation in the prothrombin gene; effect on protein levels and 3′-end formation. J. Thromb. Haemost. 2004;2:119–127. doi: 10.1111/j.1538-7836.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 11.Pollak E.S., Lam H.S., Russell J.E. The G20210A mutation does not affect the stability of prothrombin mRNA in vivo. Blood. 2002;100:359–362. doi: 10.1182/blood-2002-02-0412. [DOI] [PubMed] [Google Scholar]
- 12.Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahle E., Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 14.Sheets M.D., Ogg S.C., Wickens M.P. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F., MacDonald C.C., Wilusz J. Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res. 1995;23:2614–2620. doi: 10.1093/nar/23.14.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schek N., Cooke C., Alwine J.C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol. Cell. Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeZazzo J.D., Imperiale M.J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol. Cell. Biol. 1989;9:4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russnak R., Ganem D. Sequences 5′ to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990;4:764–776. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- 19.Gilmartin G.M., Fleming E.S., Oetjen J., Graveley B.R. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 20.Moreira A., Takagaki Y., Brackenridge S., Wollerton M., Manley J.L., Proudfoot N.J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brackenridge S., Ashe H.L., Giacca M., Proudfoot N.J. Transcription and polyadenylation in a short human intergenic region. Nucleic Acids Res. 1997;25:2326–2336. doi: 10.1093/nar/25.12.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Natalizio B.J., Muniz L.C., Arhin G.K., Wilusz J., Lutz C.S. Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J. Biol. Chem. 2002;277:42733–42740. doi: 10.1074/jbc.M208070200. [DOI] [PubMed] [Google Scholar]
- 23.Aissouni Y., Perez C., Calmels B., Benech P.D. The cleavage/polyadenylation activity triggered by a U-rich motif sequence is differently required depending on the poly(A) site location at either the first or last 3′-terminal exon of the 2′-5′ oligo(A) synthetase gene. J. Biol. Chem. 2002;277:35808–35814. doi: 10.1074/jbc.M200540200. [DOI] [PubMed] [Google Scholar]
- 24.Beaudoing E., Freier S., Wyatt J.R., Claverie J.M., Gautheret D. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10:1001–1010. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaudoing E., Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graber J.H., Cantor C.R., Mohr S.C., Smith T.F. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl Acad. Sci. USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabaska J.E., Zhang M.Q. Detection of polyadenylation signals in human DNA sequences. Gene. 1999;231:77–86. doi: 10.1016/s0378-1119(99)00104-3. [DOI] [PubMed] [Google Scholar]
- 28.Zarudnaya M.I., Kolomiets I.M., Potyahaylo A.L., Hovorun D.M. Downstream elements of mammalian pre-mRNA polyadenylation signals: primary, secondary and higher-order structures. Nucleic Acids Res. 2003;31:1375–1386. doi: 10.1093/nar/gkg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legendre M., Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danckwardt S., Gehring N.H., Neu-Yilik G., Hundsdoerfer P., Pforsich M., Hentze M.W., Kulozik A.E. The prothrombin 3′end formation signal reveals a unique architecture that is sensitive to thrombophilic gain-of-function mutations. Blood. 2004;104:428–435. doi: 10.1182/blood-2003-08-2894. [DOI] [PubMed] [Google Scholar]
- 31.Ho S.N., Hunt H.D., Horton R.M., Pullen J.K., Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 32.Costa M., Shen Y., Maurer F., Medcalf R.L. Transcriptional regulation of the tissue-type plasminogen-activator gene in human endothelial cells: identification of nuclear factors that recognise functional elements in the tissue-type plasminogen-activator gene promoter. Eur. J. Biochem. 1998;258:123–131. doi: 10.1046/j.1432-1327.1998.2580123.x. [DOI] [PubMed] [Google Scholar]
- 33.Lai W.S., Carballo E., Strum J.R., Kennington E.A., Phillips R.S., Blackshear P.J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 35.von Ahsen N., Oellerich M. The intronic prothrombin 19911A>G polymorphism influences splicing efficiency and modulates effects of the 20210G>A polymorphism on mRNA amount and expression in a stable reporter gene assay system. Blood. 2004;103:586–593. doi: 10.1182/blood-2003-02-0419. [DOI] [PubMed] [Google Scholar]
- 36.Brown P.H., Tiley L.S., Cullen B.R. Effect of RNA secondary structure on polyadenylation site selection. Genes Dev. 1991;5:1277–1284. doi: 10.1101/gad.5.7.1277. [DOI] [PubMed] [Google Scholar]
- 37.MacDonald C.C., Redondo J.L. Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol. Cell. Endocrinol. 2002;190:1–8. doi: 10.1016/s0303-7207(02)00044-8. [DOI] [PubMed] [Google Scholar]
- 38.Chou Z.F., Chen F., Wilusz J. Sequence and position requirements for uridylate-rich downstream elements of polyadenylation signals. Nucleic Acids Res. 1994;22:2525–2531. doi: 10.1093/nar/22.13.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niwa M., Rose S.D., Berget S.M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 40.Cooke C., Hans H., Alwine J.C. Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 1999;19:4971–4979. doi: 10.1128/mcb.19.7.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooke C., Alwine J.C. Characterization of specific protein-RNA complexes associated with the coupling of polyadenylation and last-intron removal. Mol. Cell. Biol. 2002;22:4579–4586. doi: 10.1128/MCB.22.13.4579-4586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brackenridge S., Proudfoot N.J. Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol. Cell. Biol. 2000;20:2660–2669. doi: 10.1128/mcb.20.8.2660-2669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]