Abstract
Accurate prediction of promoters is fundamental to understanding gene expression patterns, where confidence estimation is one of the main requirements. Using recently developed transductive confidence machine (TCM) techniques, we developed a new program TSSP-TCM for the prediction of plant promoters that also provides confidence of the prediction. The program was trained on 132 and 104 sequences and tested on 40 and 25 sequences (containing TATA and TATA-less promoters, respectively) with known transcription start sites (TSSs). As negative training samples for TCM learning we used coding and intron sequences of plant genes annotated in the GenBank. In the test set of TATA promoters, the program correctly predicted TSS for 35 out of 40 (87.5%) genes with a median deviation of several base pairs from the true site location. For 25 TATA-less promoters, TSSs were predicted for 21 out of 25 (84%) genes, including 14 cases of 5 bp distance between annotated and predicted TSSs. Using TSSP-TCM program we annotated promoters in the whole Arabidopsis genome. The predicted promoters were in good agreement with the start position of known Arabidopsis mRNAs. Thus, TCM technique has produced a plant-oriented promoter prediction tool of high accuracy. TSSP-TCM program and annotated promoters are available at http://mendel.cs.rhul.ac.uk/mendel.php?topic=fgen.
INTRODUCTION
The RNA polymerase II (Pol II) promoter is a key region that regulates differential transcription of protein coding genes. The gene-specific architecture of promoter sequences makes it extremely difficult to devise a general strategy for predicting promoters. Promoter 5′-flanking regions are especially poorly described and understood. They may contain dozens of short motifs (5–10 bases) that serve as recognition sites for proteins involved in transcription initiation, and specific regulation of gene expression. Each promoter has a unique selection and arrangement of such elements generating a unique pattern of gene expression. Several reviews of promoter prediction approaches have been published recently (1–5).
The core promoter is a minimum promoter region that is capable of initiating basal transcription. It contains the transcription start site (TSS) and typically spans from −60 to +40 relative to the TSS. Approximately 30–50% of all known promoters contain a TATA-box located from 45 to 25 bp upstream of the TSS. The TATA-box is apparently the most conserved functional signal in eukaryotic promoters and in some cases can direct accurate transcription initiation by Pol II, even in the absence of other control elements. Many highly expressed genes contain a strong TATA-box in their core promoter. However, in some large groups of genes, like housekeeping and photosynthesis genes, the TATA-box is often absent, and the corresponding promoters are referred to as TATA-less promoters. In these promoters, the exact position of the transcription start point may be controlled by the nucleotide sequence of the transcription initiation region (Inr) or the recently found downstream promoter element (DPE), which is typically observed 30 bp downstream of the TSS (3,6).
The region 200–300 bp immediately upstream of the core promoter constitutes the proximal promoter. The proximal promoter usually contains multiple transcription factor binding sites, which are responsible for specific transcription regulation. The distal part of the promoter (usually known as enhancer/silencer elements) is located further upstream and may also include transcription factor binding sites (1,2,4).
The first comprehensive review of the performance of many general-purpose promoter prediction programs has been presented by Fickett and Hatzigeorgiou (7). Although their relatively small test set (18 sequences) had several problems (8), the results demonstrated that the tested programs can recognize just ∼50% of the promoters with a false positive rate of ∼1 per 700–1000 bp [for more recent related reviews, see (2,8,9)]. Ohler et al. (8) used interpolated Markov chains in their approach and demonstrated slightly improved promoter prediction results, although they identified the same 50% of promoters from the dataset analyzed by Fickett and Hatzigeorgiou (7), while having one false positive prediction for every 849 bp. Later, to further improve the accuracy of eukaryotic promoter recognition, Ohler et al. (10) applied an approach integrating some physical properties of DNA (DNA bendability and GC-content) into their probabilistic promoter recognition system McPromoter and achieved a reduction of ∼30% of false positives, compared with a model solely based on sequence likelihoods. The initial version of our promoter predictor TSSW (11) had an accuracy of 42% with a false positive rate 1/789 bp. Another promoter identification program, Promoter 2.0, was designed by Knudsen (12) applying a combination of neural networks and genetic algorithms. Promoter 2.0 was tested on recognizing promoters in the complete Adenovirus genome (35 937 bp). The program predicted all the 5 known promoter sites on the plus strand and 30 false positive promoters. The average distance between the actual and the closest predicted promoter was ∼115 bp. The TSSW program with the threshold to predict all the 5 promoters produced 35 false positives, but its average distance between predicted and known TSS was just 4 bp (2 promoters predicted exactly, 1 with 1 bp shift, 1 with 5 bp shift and the weakest promoter was predicted with 15 bp shift).
The current draft of the human genome sequence provides the basis for several annotations of genes, both known and predicted. These annotations, however, do not include promoters. Mapping known expressed sequence tag (EST) and mRNAs does not help: these sequences are usually 5′ incomplete. The first attempt to map promoter locations to the chromosome 22 sequence was based on the PromoterInspector program (13). The program can identify ∼50% of known promoters as genomic regions up to 1 kb in length by discriminating them from the exon, intron and 3′-untranslated region (3′-UTR) sequences.
Recently, Bajic et al. (14) reported the Dragon Promoter Finder (DBF) program, which uses sensors for three functional regions: promoters, exons and introns, and an artificial neural network. Judging by the authors' estimates, that approach has a higher accuracy than three other promoter finding programs which it was compared: NNPP2.1 (15) (http://www.fruitfly.org./seq_tools/promoter.html), Promoter2.0 (12) and PromoterInspector (16). Another tool developed by Down and Hubbard (17) reported a novel hybrid machine-learning method capable of predicting >50% of human TSS with a specificity of >70%.
The methods applied to the long chromosome sequence (mentioned above) report potential starts of transcription as regions of ∼1–2 kb. It is much less precise than the methods that usually identify promoters within 100 bp and tested on sequences (∼3–10 kb), but the later methods clearly have a much higher false positive rate when applied to long genomic sequences. However, we can apply the ‘precise’ approaches to identify promoters in chromosome sequences if we exclude a lot of non-promoter sequences, such as known repeats and internal gene regions (exons and introns). These regions are identified in genome annotation projects by mapping repeats and available EST sequences onto a genome or applying computation gene-finding approaches. After that we can search for promoters (using precise methods) in sequences just before the mapped EST or predicted coding regions (within intergenic space). This avoids scanning the whole-genome sequence and as a result we achieve an acceptable level of false positive predictions.
Wasserman et al. (18) have shown that 98% of experimentally defined transcriptional factor binding sites in the human and mouse orthologous genes, upregulated in the skeletal muscle, are located in the most conservative regions confined to 19% of human DNA sequences. We used several types of conservative blocks to enhance the sensitivity and specificity of the TSSW algorithm, providing pairs of aligned orthologous genomic sequences as input data. Recently, the draft sequence of the mouse genome (19) and a gene expression map of human chromosome 21 orthologs in the mouse (20) have been reported. By exploiting conservative elements in pairs of orthologous genes of human and related species, the PromH program was developed previously (21). The program correctly predicted TSS for all 21 genes of the TATA-promoter test set with a median deviation of 2 bp from true site location. Only for two genes, there was a significant (46 and 105 bp) discrepancy between predicted and annotated TSS positions. For 38 TATA-less promoters from the second test set, TSS was predicted for 27 genes, in 14 cases within 10 bp distance from the annotated TSS, and in 21 cases within 100 bp distance. While requiring the input of pairs of orthologous sequences, such an approach demonstrated better accuracy (on rather limited test data) than currently available promoter predictors annotating single sequences.
However, it is important to improve the accuracy of promoter prediction on single sequences (due to the frequent lack of information about sequences of orthologous genes). Moreover, no promoter prediction tool has been trained and adapted for plants. In a recent review on in silico promoter identification (16), the authors investigated the possibility of predicting promoters based on the detection of CpG/CpNpG islands in the Arabidopsis genome. They conclude that such islands do not provide a straightforward indicator of promoter location, but such features can be used as a component of a more sophisticated promoter predictor. Here, we investigate a new learning and discriminative technique called the transductive confidence machine (TCM), which has been trained and tested on independent sets of well-known promoters. The method presented in the paper allows us not just to make predictions, but more importantly, it also gives valid measures of confidence in the predictions for each individual example in the test set. Validity in our method means that if we set up a confidence level, say, 95%, then we can guarantee that we are not going to have more than 5 errors out of 100 examples. Moreover, the method is flexible in the sense that it can be used with almost all known classifiers, such as support vector machine (SVM), Decision Trees and others. The accuracy of the prediction depends on how good the ‘underlying’ classifier is. The method can be applied to high-dimensional data and requires just one assumption: the examples are assumed to be independent and identically distributed (the iid assumption). Some characteristics of promoters such as the density of functional motifs do not follow normal distribution, which was a limitation of the discriminant analysis approach we used for promoter prediction in previous works (11,21).
MATERIALS AND METHODS
Training and testing sequences
For training and testing procedures, we used 301 promoters with annotated TSS from PlantProm DB (22). A total of 236 (132 TATA and 104 TATA-less) promoters were taken for learning and 40 TATA promoters (PlantProm DB IDs: PLPR0003, PLPR0010, PLPR0011, PLPR0014, PLPR0015, PLPR0018, PLPR0022, PLPR0024, PLPR0025, PLPR0026, PLPR0034, PLPR0039, PLPR0042, PLPR0043, PLPR0045, PLPR0054, PLPR0057, PLPR0062, PLPR0064, PLPR0065, PLPR0066, PLPR0067, PLPR0071, PLPR0072, PLPR0078, PLPR0087, PLPR0090, PLPR0091, PLPR0111, PLPR0116, PLPR0169, PLPR0171, PLPR0176, PLPR0186, PLPR0202, PLPR0235, PLPR0243, PLPR0253, PLPR0264 and PLPR0286) (PLPR0001, PLPR0009, PLPR0020, PLPR0027, PLPR0037, PLPR0075, PLPR0164 and 25 TATA-less promoters: PLPR0170, PLPR0180, PLPR0182, PLPR0184, PLPR0189, PLPR0190, PLPR0191, PLPR0193, PLPR0199, PLPR0200, PLPR0207, PLPR0239, PLPR0242, PLPR0252, PLPR0254, PLPR0259, PLPR0269 and PLPR0271) were used for testing. As negative samples (non-promoter sequences), 50 000 sequences from CDS and 50 000 sequences from introns of plant genes annotated in GenBank were extracted. The length of all promoter and non-promoter sequences was 351 bp.
Description of the method
Confidence and credibility
Let us assume that we are given a training set of examples (x1, y1),…,(xl, yl), where xi is a vector of attributes (characteristics described below; see Table 1) and yi is a label (promoter or non-promoter), and our goal is to predict the correct label yl+1,…,yl+k for a new (test) set of xl+1,…,xl+k. We make only one assumption about the data generating mechanism: all the examples have been generated independently by some fixed but unknown stochastic mechanism (the iid assumption).
Table 1.
Characteristics of promoter sequences used for TATA and TATA-less promoter recognition and Mahalonobis distance [D2; (32)] showing power of recognition of each characteristic
| Characteristics | D2 for TATA promoters | D2 for TATA-lesspromoters |
|---|---|---|
| Hexaplets −200 : −45 | 2.6 | 1.4 (−100 : −1) |
| TATA box score | 3.4 | 0.9 |
| Triplets around TSS | 4.1 | 0.7 |
| Hexaplets +1: +40 | 0.9 | |
| Sp1-motif content | 0.9 | |
| TATA fixed location | 0.7 | |
| CpG content | 1.4 | 0.7 |
| Similarity −200 : −100 | 0.3 | 0.7 |
| Motif Density (MD) −200 : +1 | 4.5 | 3.2 |
| Direct/Inverted MD −100 : +1 | 4.0 | 3.3 (−100 : −1) |
MD is motif density, computed on known promoters; functional motifs were taken from Plant REGSITE Database (http://softberry.com/berry.phtml?topic=regsitelist).
The method (called TCM or conformal predictors) is based on the recently developed (23–26) computable approximation to algorithmic randomness. To every possible value Y of yl+1, we estimate the ‘randomness level’ (or ‘typicalness’) of the sequence (x1, y1),…,(xl, yl), (xl+1, Y) with respect to the iid model; we can make a confident prediction if and only if exactly one of these two (in the case of binary classifications) sequences is typical. The randomness level is a universal measure of typicalness with respect to the class of iid distributions; if the randomness level is close to 0, it is untypical or strange (27).
Here, the optimal algorithm for making predictions is complemented by some measures of confidence and credibility (24,27):
- Consider all possible values Y1,…,Yk for labels yl+1,…,yl+k and compute the randomness level of every possible completion
Predict the set Y1,…,Yk corresponding to the completion with the largest randomness level.
Output as the confidence in this prediction one minus the second largest randomness level.
Output as the credibility of this prediction the randomness level of the output prediction Y1,…,Yk (i.e. the largest randomness level for all possible predictions).
If the confidence in our prediction exceeds 99% and the prediction is wrong, the actual data sequence belongs to an a priori chosen set of probability <1% (namely, the set of all data sequences with randomness level <1%). Credibility reflects how well our new example fits into our training set. Intuitively, low credibility means that either the training set is non-random or the test examples are not representative of the training set (25).
One of the advantages of this newly developed algorithm is its flexibility: almost all machine learning techniques can be used for prediction. One way to approximate randomness level is to use the SVM (24). Consider, for simplicity, the problem of binary classification with one test example. A SVM maps the original set of vectors into a high-dimensional feature space, and then constructs a linear separating hyperplane (or a linear regression function, in the regression case) in this feature space. According to the SVM approach (28), we should select a separating hyperplane with a small number of errors (or, more generally, a small sum of penalties reflecting the grossness of errors) and a large ‘margin’ (which can be interpreted as the distance from the separating hyperplane to the nearest vectors).
With every possible label Y ∈ {0, 1} (Y = 0 for the positive samples, TATA or TATA-less promoters, and Y = 1 for the negative samples, non-promoter sequences) for xl+1, we associate the SVM optimization problem for the l + 1 examples (the training examples plus the test example labeled with Y). The solutions (Lagrange multipliers) α1, α2,…,αl+1, to this problem reflect the ‘strangeness’ of the examples [αi being the strangeness of (xi, yi), i = 1,…,l and αl+1, being the strangeness of the (xl+1, Y)]. In other words using Lagrange multipliers αi, we can approximate from below the randomness deficiency. This was done in Gammerman et al. (23), where a general function was introduced for the estimation of confidence and credibility, while the SVM is used for prediction.
All αi are non-negative and, in practice, only a few of them are different from zero (the support vectors). An easily computable approximation to the randomness level is given by the P-values associated with every completion (x1, y1),…,(xl, yl), (xl+1, Y):
in words, the P-value is the proportion of α's which are at least as large as the last α (23,24). These approximations can be used to assess the randomness level. ‘Randomness’ as a concept only makes sense in connection with a given distribution (e.g. binominal, or iid, etc.). ‘Algorithmic randomness’ is not computable, and therefore requires some approximation, and that is what we use to calculate it. We actually operate not with randomness itself but ‘randomness deficiency’, or ‘strangeness’ and approximate it from below. It is reflected in P-values we compute in the assumption that the data follow iid distribution.
So far, we have assumed that the ‘strangeness values’ αis used to approximate the randomness level are obtained from the SVM algorithm. However, we can get useful αis from many other learning algorithms, such as the nearest neighbors, neural networks, decision trees (24,26).
Features and learning procedures used for recognition
For the characterization of promoter sequences, we use the sequence content and signal features that were found in our previous works as being significantly different in promoter and non-promoter sequences (11,21). The values of Mahalanobis distances (D2) of individual characteristics reflect the power of the feature to separate the signal from non-signal sequences (Table 1). This analysis demonstrated that TATA-less promoters have weaker general features than TATA promoters. Probably TATA-less promoters possess a more gene-specific structure and they will be extremely difficult to predict by any general-purpose methods. Earlier (11,21) discriminant analysis was applied to combine these features in the linear discriminant function. In this work, we applied a more powerful pattern recognition technique that requires just one assumption: the examples are independent and identically distributed. In this study, a package of SVM (freely downloadable at http://www.clrc.rhul.ac.uk/resources/svmdownloadoverview.htm) with dot product kernel has been used to train two classifiers for TATA and TATA-less promoters. We trained our SVM to distinguish between promoter and non-promoter sequences using features discussed above. To estimate the reliablity of the classification produced by SVM, we applied TCM procedure [described above; for details see also (24,26,27) and http://nostradamus.cs.rhul.ac.uk/promoters] to measure the confidence values (P-values) for each of SVM predictions. As predicted promoters, we selected those SVM promoter assignments that have the confidence level ≥0.95. The TCM procedure also provides a credibility measure for different predictions. The measure of credibility provides us with a filter mechanism with which we can ‘reject’ certain predictions. If for any task the consequences of making a wrong prediction are quite severe, we can choose to reject those predictions that have a low credibility value associated with them. The more severe the consequences for making an incorrect prediction are, the higher we can set the rejection threshold. In this study, we accepted as promoters the predictions with the credibility level ≥0.35 for TATA promoters and ≥0.65 for TATA-less promoters.
Algorithm of promoter search
The TSSP-TCM program classifies each position on a given sequence as TSS or non-TSS based on two support vector classifiers (28) (for TATA and TATA-less promoters) with dot product kernel function and eight characteristics calculated in the (−200, +50) region around the current position. If a TATA-box weight matrix gives a score higher than some preliminary defined threshold in the region (−40, −25) from the current position, then the credibility value (23,24) of this position is estimated based on the classifier for TATA promoters; otherwise, it will be estimated by the classifier for TATA-less promoters. Optimal thresholds of credibility value for TATA and TATA-less promoters (0.35 and 0.65, respectively) have been defined on the training dataset. For any pair of predicted TSS, located within 300 bp of each other, only the one with the highest credibility value is retained.
RESULTS AND DISCUSSION
Testing of the method
The learning and testing procedure was repeated 40 times for both TATA and TATA-less promoters (20 computations with negative samples from CDS and 20 computations with negative samples from introns). In every such training and testing procedure, randomly created sets of 1000 non-promoter sequences and the same known 40 TATA and 25 TATA-less promoters were used. Accuracy of recognition is presented in Table 2.
Table 2.
Statistics of testing procedure for 40 TATA and 25 TATA-less promoter sequences of 351 bpa
| Promoter type | Accuracy of discrimination | Negative samples from CDSs (%) | Negative samples from introns (%) |
|---|---|---|---|
| TATA | Mean prediction error for positive samples (%) | 7.4 | 3.5 |
| Mean prediction error for negative samples (%) | 6.0 | 8.7 | |
| TATA-less | Mean prediction error for positive samples (%) | 18.6 | 14.0 |
| Mean prediction error for negative samples (%) | 16.9 | 29.5 |
aA total of 40 various sets of 1000 negative samples of the same length (351 bp), randomly chosen from CDSs (20 sets, totally 20 000 sequences) and introns (20 sets, totally 20 000 samples) of known plant genes. Confidence and credibility levels were ≥0.9 (90%) and ≥0.06 (6%), respectively.
We observe very good accuracy of recognition of promoter and non-promoter sequences taking into account that the best promoter recognition programs have an accuracy of ∼50–70%. Interestingly, for TATA and TATA-less promoters the error rate of testing promoter and non-promoter sequences of 351 nt length is higher when using negative samples from introns and CDSs, respectively.
The real task of promoter prediction is slightly different from just discriminating between promoter and non-promoter regions. We should try to identify the most probable promoter location in a long genomic sequence. For testing our recognition function on genomic sequences, we used the same 40 genes with an annotated TATA promoter and 25 genes with an annotated TATA-less promoter by analyzing their known upstream regions. The length of these sequences was 1000 bp or more (including upstream to CDS region plus 30 bp of CDS). The total length of the 40 and 25 genomic sequences mentioned was 75 259 and 42 556 bp, respectively.
TSSW (11) and TSSP-TCM programs classify each position on a given sequence as TSS or non-TSS based on two linear discriminant functions (for TATA and TATA-less promoters) with eight characteristics calculated in the (−200, +50) region around the current position. If the TATA-box weight matrix gives a score higher than some preliminary defined threshold in the region (−40, −25) from the current position, then that position is classified based on the score for TATA promoters, otherwise it will be classified by the score for TATA-less promoters. For any pair of predicted TSS, located within 300 bp of each other, only the one with the highest score is retained, except for one case: if a lower scoring position is predicted as TATA-less promoters near a higher scoring position predicted as TATA promoters, then the first position is also retained as a potential promoter region. In the case of prediction of more than one promoter (TSS) with a high score and if the CDS start is known, the TSS closest to the CDS start was assumed as a predicted promoter. However, of course, it might be the choice of the user.
Testing for genomic sequences was performed by using training with negative samples from both CDSs and introns (Table 3). However, comparing the results of both approaches, we revealed that training on non-promoter sequences from introns gives the best test results in the sense of both false positives and false negatives. It seems reasonable because upstream promoter regions are non-coding DNA that is very similar to intron DNA. Therefore, we will discuss further the use of the training ‘experience’ obtained with intron sequences.
Table 3.
Accuracy of prediction by TSSP-TCM on genomic sequencesa
| Statistic characteristics | For 40 TATA promoters | For 25 TATA-less promoters |
|---|---|---|
| False negatives | 5 | 4 |
| False positives | 14 | 9 |
| False positives' density | 1 per 5375 bp | 1 per ∼4720 bp |
aThe confidence level for the prediction of both promoter classes was 95% or higher. The credibility level was ≥35% for TATA promoters and ≥60% for TATA-less promoters. For every class of promoters only one predicted TSS with the highest credibility level in an interval of 300 bp was taken. TATA and TATA-less promoters predicted were separately estimated by this statistical criterion.
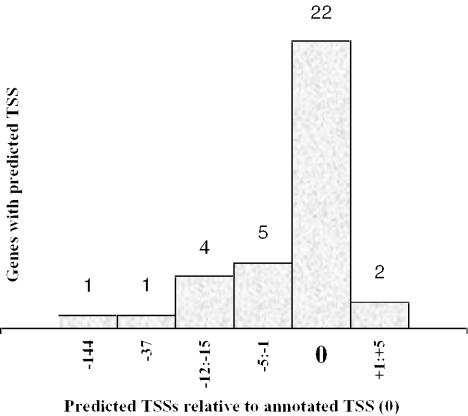
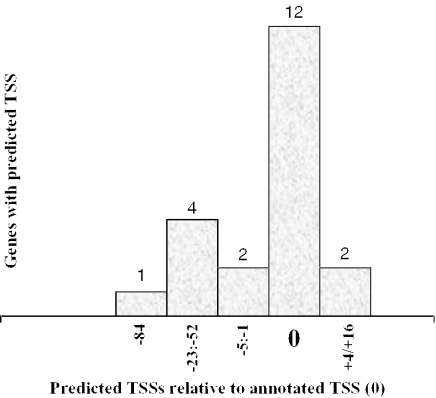
For 35 of the 40 TATA promoters (87.5%) and 21 of the 25 TATA-less promoters (84%), a TSS very close to the known one was predicted (Table 3). For 29 TATA promoter genes of the 35 (72.5%), the distances between the known and nearest predicted TSS were 0–5 bp. The distribution of predicted TSS around real TSS is shown in Figure 1. For 14 TATA-less promoter genes (56%), the distances between the known and nearest predicted TSSs were 0–5 bp (Figure 2). It is interesting that the TSSP-TSM program managed to identify the TATA-less promoter relatively well in spite of the absence of the major TATA-box signal. It is achieved in our model due to significant input in the recognition from many other features of promoter regions. Some of them are specific for TATA-less promoters (such as hexaplet composition of +1 to +40 region and Sp1 motif content) and they probably compensate for the absence of a TATA-box.
Figure 1.
Location of the predicted nearest TSS in relationship with the known TSSs for 35 out of 40 genes with annotated TATA promoters.
Figure 2.
Location of the predicted nearest TSSs in relationship with the known TSS for 21 out of 25 genes with annotated TATA-less promoters.
Our results indicate that the TCM technique can be successfully applied to combine complex features of promoter sequences and to design an accurate promoter identification program. According to our estimations based on the latest (May 2003) annotation of the Arabidopsis genome (GenBank accession nos NC_003070, NC_003071, NC_003074, NC_003075 and NC_003076), a gene density in the genome is ∼4.4 kb per gene for 16 811 genes supported by cDNAs and EST, and ∼7.1 kb per gene for all 27 128 genes. Therefore, upstream regions of genes should be ∼1–4 kb and the developed approach, having a true prediction rate ∼85% and one false positive prediction in ∼4000–5000 bp, can be used for the annotation of promoter regions in plant genomes. Promoter candidates generated in this way can be further verified experimentally. Taking into account that currently we have just several hundred experimentally tested promoter sequences, the TSSP-TCM program may be of considerable value for molecular biologists in deciphering the regulation of genes encoded in sequenced genomes or in interpreting the results of expression profiling.
The search for promoters in the genome of Arabidopsis thaliana
Annotations of five chromosomes of A.thaliana (May 2003; GenBank accession nos NC_003070, NC_003071, NC_003074, NC_003075 and NC_003076) include 27 128 genes, where 16 811 of them have known cDNA/mRNA. For testing the TSSP-TCM program on the genome level, we selected 13 350 (out of 16 811) genes that have known mRNA with 5′-UTR ≥ 20 bp. For every such gene, a region (P − L, P + 30 bp) around the annotated CDS (started from position P) was taken into search for promoters. L was the distance to the previous gene or 5000 bp, if that gene is located further than 5000 bp. If the length of upstream region was <350 bp, than L = 350 bp was taken. The summary of promoter search results is presented in Table 4.
Table 4.
Summary of promoter prediction for 13 350 mRNA supported genes of A.thaliana
| Analyzed | 13 350 genes |
| At least, 1 promoter found | 9653 (72.3%) genes |
| At least, 1 TATA promoter found | 6141 (46.0%) genes |
| At least, 1 TATA-less promoter found | 6717 (50.3%) genes |
| The predicted TATA promoter is the closest to the annotated mRNA start | 4465 (46.3%) genes |
| The predicted TATA-less promoter is the closest to the annotated mRNA start | 5188 (53.7%) genes |
| Total length of analyzed sequences | 27 709 288 bp |
| Total number promoters predicted | 17 717 |
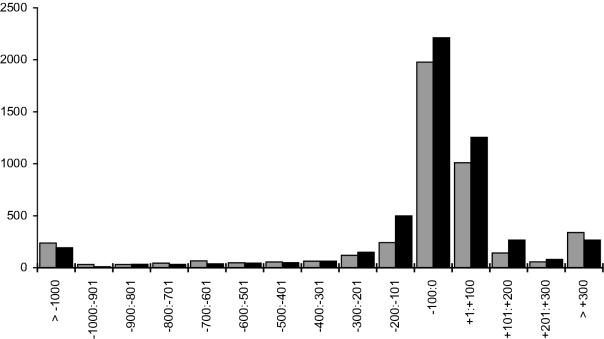
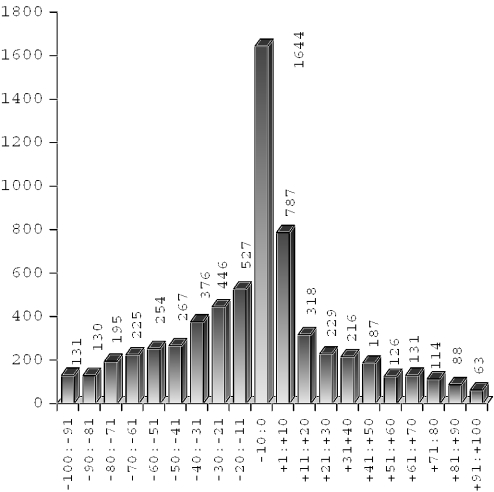
By analyzing the location of TSS positions predicted by TSSP-TCM (Figure 3), we could observe a very profound peak near the start positions of known transcripts. Some discrepancies here might appear due to non-complete sequencing of mRNA 5′ ends. It often happens that some number of nucleotides is absent on both 5′ and 3′ ends of mRNA generated by cDNA sequencing. If we consider shorter intervals (Figure 4), then we can see that the most often predicted promoters are located within 1–10 bp of the putative mRNA start. Taking this into account, we have observed a very good agreement between the predicted locations and the locations of TSSs supported by mRNA. It prompted us to annotate the promoters in the whole genome of Arabidopsis using regions upstream of the annotated first coding exons.
Figure 3.
Distribution of distances (D) between the predicted TSS and the annotated start of mRNA (gray, 4465 TATA promoters; black, 5188 TATA-less promoters).
Figure 4.
Distribution of distances (D) of predicted (6454 TATA and TATA-less promoters) closer than 100 bp to the annotated start of mRNA.
The set of predicted promoters for 18 601 genes (out of 27 128 annotated ones) in the whole genome is presented at our plant genomic server (http://mendel.cs.rhul.ac.uk/) and might be used for further verification and experimental studies of the regulation of plant genes. To date Arabidopsis [(29); GenBank accession nos NC_003070, NC_003071, NC_003074, NC_003075 and NC_003076] and rice (30,31) genome sequences are available. Besides, several new plant genome sequencing projects (such as Populus trichocarpa, Lycopersicon esculentum, Zea mays and Medicago truncatula) are underway. It is impossible in the near future to investigate their genes and promoters by experimental techniques. The computational methods help to speed up this process. Promoter prediction generates a set of probable candidates, making it possible to avoid the initial testing of thousands of potential genomic fragments. Having cDNA libraries an alternative strategy to identify promoters would be to align full-length cDNA sequences to the genome sequence. However, most cDNA clones do not extend to the TSS. It is estimated that only 50–80% of cDNA extend to the TSS, making it unreliable to base conclusions on individual cDNA alignments (10). TSSs can be located thousands of bases upstream of CDS regions of a gene. Therefore, approximate gene location by prediction of coding exons or mapping known cDNA sequences does not provide enough precise localization of TSS for experimental study. However, having annotated coding exons we can limit promoter search to intergenic regions and apply our promoter prediction software to annotate promoters in the whole genome. Owing to the much better prediction accuracy of coding genes (compared with promoter prediction accuracy) we believe that using the initial placement of coding gene regions is a good strategy to reduce promoter search space. With an average intergenic region size ∼2000–3000 bp and false positive rate 1/5000 bp, we will not generate as many false positives as when running promoter prediction on whole-genomic sequences. We understand that finding promoters in genomic sequences is far from being a trivial problem. There are now <1000 experimentally identified functional motifs of plant origin available for the development of promoter recognition functions. More accurate approaches will require knowledge of a significantly larger set of plant regulatory motifs and probably their patterns. Another problem that limits the development of more accurate algorithms is the very small number of experimentally verified plant promoter sequences (∼500). It prevents the discovery of some complex promoter features and their significant combinations.
Our current predictions can be considered as the first draft of promoter annotation and as the next step in the characterization of genomes using computational tools (in addition to the sets of computationally derived gene structures that are still changing, but serve as important resources in genome studies). Recently, we have developed a new promoter identification program, PromH, which using pairs of orthologous gene sequences (human and mouse) significantly improves the quality of promoter prediction (21). Having several annotated plant genomes we plan to select their orthologous genes and apply a plant-specific version of the PromHP program to further improve promoter annotation.
Acknowledgments
PlantProm Database is designed and maintained at Royal Holloway, University of London in collaboration with Softberry Inc. (USA). This work was partially supported by BBSRC grant 111/BIO14428 ‘Pattern recognition techniques for gene and promoter identification and classification in plant genomic sequences’. Funding to pay the Open Access publication charges for this article was provided by JICS, UK.
REFERENCES
- 1.Lemon B., Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- 2.Pedersen A.G., Baldi P., Chauvin Y., Brunak S. The biology of eukaryotic promoter prediction—a review. Comput. Chem. 1999;23:191–207. doi: 10.1016/s0097-8485(99)00015-7. [DOI] [PubMed] [Google Scholar]
- 3.Smale S.T. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim. Biophys. Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 4.Smale S.T. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- 5.Werner T. Models for prediction and recognition of eukaryotic promoters. Mamm. Genome. 1999;10:168–175. doi: 10.1007/s003359900963. [DOI] [PubMed] [Google Scholar]
- 6.Burke T.W., Kadonaga J.T. The downstream promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fickett J., Hatzigeorgiou A. Eukaryotic promoter recognition. Genome Res. 1997;7:861–878. doi: 10.1101/gr.7.9.861. [DOI] [PubMed] [Google Scholar]
- 8.Ohler U., Harbeck S., Niemann H., Noth E., Reese M. Interpolated Markov chains for eukaryotic promoter recognition. Bioinformatics. 1999;15:362–369. doi: 10.1093/bioinformatics/15.5.362. [DOI] [PubMed] [Google Scholar]
- 9.Ohler U., Niemann H. Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet. 2001;17:56–60. doi: 10.1016/s0168-9525(00)02174-0. [DOI] [PubMed] [Google Scholar]
- 10.Ohler U., Niemann H., Lia G.-C., Rubin G.M. Joint modeling of DNA sequence and physical properties to improve eukaryotic promoter recognition. Bioinformatics. 2001;17:S199–S206. doi: 10.1093/bioinformatics/17.suppl_1.s199. [DOI] [PubMed] [Google Scholar]
- 11.Solovyev V.V., Salamov A.A. The Gene-Finder computer tools for analysis of human and model organisms genome sequences. In: Rawling C., Clark D., Altman R., Hunter L., Lengauer T., Wodak S., editors. Proceedings of the Fifth International Conference on Intelligent Systems for Molecular Biology; 21–25 June; Halkidiki, Greece. AAAI Press; 1997. pp. 294–302. [PubMed] [Google Scholar]
- 12.Knudsen S. Promoter 2.0: for the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- 13.Scherf M., Klingenhoff A., Frech K., Quandt K., Schneider R., Grote K., Frisch M., Gailus-Durner V., Seidel A., Brack-Werner R., Werner T. First pass annotation of promoters of human chromosome 22. Genome Res. 2001;11:333–340. doi: 10.1101/gr.154601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bajic V.B., Seah S.H., Chong A., Zhang G., Koh J.L.Y., Brusic V. Dragon promoter finder: recognition of vertebrate RNA polymerase II promoters. Bioinformatics. 2002;18:198–199. doi: 10.1093/bioinformatics/18.1.198. [DOI] [PubMed] [Google Scholar]
- 15.Reese M., Harris N.L., Eeckman F.H. Large scale sequencing specific neural networks for promoter and splice site recognition. In: Hunter L., Klein T.E., editors. Biocomputing Proceedings of the 1996 Pacific Symposium; 2–7 January; World Scientific Co., Singapore. 1996. [Google Scholar]
- 16.Scherf M., Klingenhoff A., Werner T. Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J. Mol. Biol. 2000;297:599–606. doi: 10.1006/jmbi.2000.3589. [DOI] [PubMed] [Google Scholar]
- 17.Down T.A., Hubbard T.J. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasserman W.W., Palumbo M., Thompson W., Fickett J.W., Lawrence C.E. Human-mouse genome comparisons to locate regulatory sites. Nature Genet. 2000;26:225–228. doi: 10.1038/79965. [DOI] [PubMed] [Google Scholar]
- 19.Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 20.The HSA21 Expression Map Initiative. A gene expression map of human chromosome 21 orthologues in the mouse. Nature. 2002;420:586–590. doi: 10.1038/nature01270. [DOI] [PubMed] [Google Scholar]
- 21.Solovyev V., Shahmuradov I. PromH: promoters identification using orthologous genomic sequences. Nucleic Acids Res. 2003;31:3540–3545. doi: 10.1093/nar/gkg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shahmuradov I., Gammerman A., Hancock J.M., Bramley P.M., Solovyev V.V. PlantProm: a database of plant promoter sequences. Nucleic Acids Res. 2003;31:114–117. doi: 10.1093/nar/gkg041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gammerman A., Vapnik V.N., Vovk V. Learning by transduction. In: Cooper G.F., Moral S., editors. Proceedings of the Fourteenth Conference on Uncertainty in Artificial Intelligence; 24–27 July; Madison, WI. Morgan Kaufmann, San Francisco, CA. 1998. pp. 148–156. [Google Scholar]
- 24.Gammerman A., Vovk V. Prediction algorithms and confidence measures based on algorithmic randomness theory. Theoret. Comput. Sci. 2002;287:209–217. [Google Scholar]
- 25.Saunders C., Gammerman A., Vovk V. Computationally efficient transductive machines. Proceedings of the Eleventh International Conference on Algorithmic Learning Theory; 11–13 December; Sydney, Australia. Springer-Verlag: Lecture Notes in Artificial Intelligence; 2000. pp. 325–333. [Google Scholar]
- 26.Vovk V., Gammerman A., Shafer G. Algorithmic Learning in a Random World. Springer-Verlag: 2004. in press. [Google Scholar]
- 27.Vovk V., Gammerman A., Saunders C. Machine-learning applications of algorithmic randomness. In: Bratko I., Dzeroski S., editors. Proceedings of the Sixteenth International Conference on Machine Learning; 27–30 June; Bled, Slovenia. Morgan Kaufmann, San Francisco, CA. 1999. pp. 444–453. [Google Scholar]
- 28.Vapnik V.N. Statistical Learning Theory. NY: Wiley; 1998. [DOI] [PubMed] [Google Scholar]
- 29.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 30.Goff S.A., Ricke D., Lan T.H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 31.Yu J., Hu S., Wang J., Wong G.K., Li S., Liu B., Deng Y., Dai L., Zhou Y., Zhang X., Cao M., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 32.Afifi A.A., Azen S.P. Statistical Analysis. A Computer Oriented Approach. NY: Academic Press; 1979. [Google Scholar]