Abstract
Background
Somatic mutations of BRAF or NRAS activating the MAP kinase cell signaling pathway are present in 70% of cutaneous melanomas. The mutant allele frequency of BRAF V600E (M%BRAF) was recently shown to be highly heterogeneous in melanomas. The present study focuses on the NRAS Q61 mutant allele frequency (M%NRAS).
Methods
Retrospective quantitative analyze of 104 NRAS mutated melanomas was performed using pyrosequencing. Mechanisms of M%NRAS imbalance were studied by fluorescence in situ hybridization (FISH) and microsatellite analysis.
Results
M%NRAS was increased in 27.9% of cases. FISH revealed that chromosome 1 instability was the predominant mechanism of M%NRAS increase, with chromosome 1 polysomy observed in 28.6% of cases and intra-tumor cellular heterogeneity with copy number variations of chromosome 1/NRAS in 23.8%. Acquired copy-neutral loss of heterozygosity (LOH) was less frequent (19%). However, most samples with high M%NRAS had only one copy of NRAS locus surrounding regions suggesting a WT allele loss. Clinical characteristics and survival of patients with either <60% or ≥60% of M%NRAS were not different.
Conclusion
As recently shown for M%BRAF, M%NRAS is highly heterogeneous. The clinical impacts of high M%NRAS should be investigated in a larger series of patients.
Electronic supplementary material
The online version of this article (doi:10.1186/s12895-017-0061-x) contains supplementary material, which is available to authorized users.
Keywords: Melanoma, M%NRAS, Imbalance, Pyrosequencing, WT allele loss
Background
Cutaneous melanoma is a highly aggressive and treatment-resistant human cancer. The most frequent genetic alterations involve genes of the MAP kinase signaling pathway [1–3]. Activating hot-spot mutations are mainly found in BRAF (codon V600) and in NRAS (codon Q61, and less frequently in the codons G12 and G13) genes, in 35–50% and 15–25% of cutaneous melanoma, respectively [4, 5]. Among BRAF alterations, the BRAF V600E mutation in exon 15 is predominant (85%) and due to a substitution of a valine to a glutamic acid (c.1799 T > A, p.V600E) [6, 7]. BRAF and NRAS mutations are almost always mutually exclusive [8, 9].
Mutant NRAS melanomas have been reported to have more aggressive clinical features than other subtypes, with thicker lesions, elevated mitotic activity, and higher rates of lymph node metastasis [10–12]. Additionally, NRAS mutation status was reported as a predictor of poorer outcomes with lower median survival compared to non-NRAS mutated melanoma [10, 13].
The discovery of BRAF mutations led to the development of targeted treatments [14, 15]. However despite major clinical benefit in melanomas with BRAF mutation, secondary resistance occurs in most patients during the first year of treatment. Thus combinations of BRAF and MEK inhibitors have been developed, and were shown to induce longer progression free survivals (PFS) of patients with BRAF mutated melanomas [16–18]. By contrast, targeted treatment of patients with NRAS mutated melanomas is still a challenge, although an international phase 3 prospective study with the MEK inhibitor binimetinib recently provided promising results [19].
We recently studied the frequency of BRAF mutant alleles (M%BRAF) and showed that M%BRAF is highly heterogeneous and frequently increased in BRAF mutated melanomas [20]. Interestingly, a recent clinical study showed that the increased BRAF V600 mutation level was significantly associated with a better response rate to vemurafenib during the first 10 months of treatment [21]. These observations highlighted the importance of quantitative evaluation of BRAF mutation before melanoma treatment.
Although biological and clinical implication of the frequency of mutant alleles of BRAF in melanomas are currently under investigation, no data are available concerning the variation of M%NRAS. Accordingly, we conducted this study to investigate NRAS Q61 mutations and M%NRAS in a series of 199 melanomas wild type for BRAF V600. The mechanisms of the M%NRAS variations were then studied by fluorescence in situ hybridization (FISH) and by amplified fragment length polymorphism (AFLP).
Methods
Patients and samples
Melanoma samples were obtained from the bank of biological resources of Ambroise Paré Hospital in Boulogne-Billancourt. The research was performed in compliance with the ethical principles of the Helsinki Declaration (1964) and with the French ethics laws. Patients were informed and approved the use of their samples for research purpose. Tumor samples collection was declared to the French Ministry of Research (DC 2009–933). Melan-Cohort study was approved by CPP IDF 8 Ethics committee (030209) and registered with https://www.clinicaltrials.gov/ct2/search (NCT00839410). Clinical and survival data were collected from clinical records of the Dermatology Department of Ambroise Paré Hospital.
The frequency of NRAS Q61 mutations was evaluated in a consecutive series of melanomas received for diagnosis from March 2013 until May 2015. Additionally, a second series of patients, whose samples were received earlier to March 2013, mutated for NRAS were also included for the evaluation of M%NRAS. In our previous paper concerning the BRAF mutant allele frequency in melanoma [20], we observed a distinct distribution of the percentage of mutated allele according to the percentage of tumor cells. However, the inter-pathologist reproducibility for the evaluation of tumor cell content was substantial for the 80% cut-off (κ = 0.79). Therefore, we excluded samples with less than 80% of melanoma cells from further NRAS molecular analysis.
NRAS mutant allele detection and quantification
Before DNA extraction, HES slides were reviewed to confirm the presence of melanoma cells and to select areas with highest density of tumor cells for macrodissection. For microsatellite analysis, DNA from corresponding normal tissue section was extracted. Genomic DNA was extracted from formalin-fixed and paraffin-embedded (FFPE) fragments of melanoma as previously described [22].
Pyrosequencing was performed as already described [23]. Profiles for different NRAS mutations were established and confirmed by Sanger sequencing method (Additional file 1: Figure S1). Three different assays for detection of NRAS Q61 mutation were designed and primers used for DNA amplification, pyrosequencing, and Sanger method are presented in Table 1. Pyrosequencing Assay 1 allows the quantification of all but one NRAS Q61 mutation (c.183A > T p.Q61H) [24]. Pyrosequencing Assay 2 is an edited version of Assay 1, in which the order of injected nucleotides was modified to allow the quantification of all NRAS Q61 mutations. Pyrosequencing Assay 3 was developed to rescue some cases with a very bad FFPE DNA quality. For patients with several samples available, the M%NRAS used was that obtained from the first metastasis.
Table 1.
Primers and pyrosequencing assay information. Primers sequences used for PCR, Sanger sequencing, pyrosequencing and AFLP technique
| NRAS Pyrosequencing ASSAY 1 | |
| PCR 124 nt | |
| Primer F-Biotine | ACACCCCCAGGATTCTTACAGA |
| Primer R | GCCTGTCCTCATGTATTGGTC |
| Pyrosequencing primer | CATGGCACTGTACTCTTC |
| Nucleotide injection order | GTTACGTCAGCTG |
| NRAS Pyrosequencing ASSAY 2 | |
| PCR 124 nt | |
| Primer F-Biotine | ACACCCCCAGGATTCTTACAGA |
| Primer R | GCCTGTCCTCATGTATTGGTC |
| Pyrosequencing primer | CATGGCACTGTACTCTTC |
| Nucleotide injection order | GCATACGTCAG |
| NRAS Pyrosequencing ASSAY 3 | |
| PCR 90 nt | |
| Primer F-Biotine | ACAAGTGGTTATAGATGGTGA |
| Primer R | ATGTATTGGTCTCTCATGGCA |
| Pyrosequencing primer | CATGGCACTGTACTCTTC |
| Nucleotide injection order | GCATACGTCAGCT |
| Sanger Sequencing | |
| Primer F | ACAAGTGGTTATAGATGGTGA |
| Primer R | ATGTATTGGTCTCTCATGGCA |
| AFLP primers | |
| rs3219599 F | TTCAAGGCTGCAGTGAGCTA |
| rs3219599 R | AGTGGAAGCTAGACACACATTAAGA |
| s3219653 F | CCAGAGAGACAGAACTGAACAAA |
| s3219653 R | CAAATTTTGGACCTGCCATG |
| rs3219587 F | GGGCAAATGGAGGAAAGAGA |
| rs3219587 R | TAAAAATACCCCCACCCCACT |
| rs3220698 F | TTAAAAAACGTACTGCCACATTCA |
| rs3220698 R | GGCAGAAACCAGGAAATGTAGTA |
| rs3220987 F | GGCTTTTAGCTATGATTTGAGA |
| rs3220987 R | GACTCAGGAAATAAACAAGGC |
| rs3220389 F | CGCTGCTCACTCCTCCTCTGA |
| rs3220389 R | AGTGCTGCTCTCAGTGAACTC |
| rs3219612 F | AGCACACAATATACTCTCTCAGA |
| rs3219612 R | ACCTGGGCAAAAGAGTAAGACC |
| rs3219703 F | AACGAAGGTGTACTGGGACTGGT |
| rs3219703 R | ACAGGGATGTGAGGGATTTTTTC |
Microsatellite analysis
Eight microsatellite markers were selected from the NCBI dbSNP short genetic variations database and analyzed by AFLP. Their positions are showed in Fig. 1, and the primers used in Table 1. No highly heterozygous microsatellite was identified within NRAS gene; thus the genetic status of this gene was evaluated on the basis of two microsatellites closed to NRAS locus (rs3220698 and rs3220987). PCR reactions were performed with fluorescent-labeled forward primers and the amplified PCR products were analyzed by capillary array electrophoresis on the ABI PRISM 3100 sequencer (Applied Biosystems, Foster City, USA) and GeneScan software (Applied Biosystems). Detection of loss of heterozygosity was performed as detailed in Loss of Heterozygosity Analysis Getting Started Guide. Two independent experiments were performed to confirm LOH results. The probability of one NRAS allele loss was evaluated as very high if only one allele was detected in each NRAS locus surrounding microsatellites (e.g. Fig. 1, case Y11.136). If only one of NRAS surrounding markers presented one microsatellite allele, this probability was evaluated as mean (e.g. Fig. 1, case Y14.711). The analysis was considered as inconclusive if both NRAS locus surrounding microsatellites gave non informative results (e.g. Fig. 1, case Y10.1471).
Fig. 1.
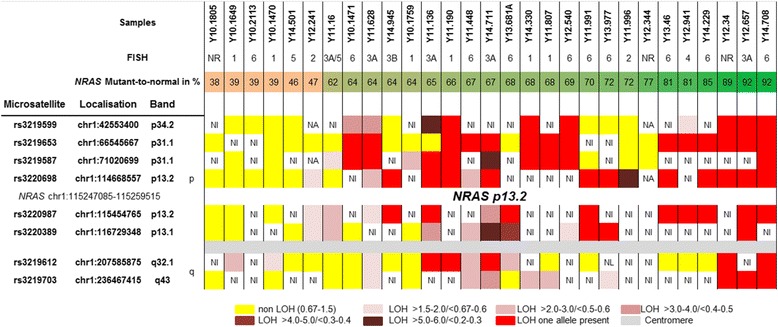
Chromosome 1 microsatellite analyses in NRAS mutated melanomas. Summary of AFLP analysis of eight chromosome 1 microsatellite markers performed in 29 NRAS Q61 mutated melanomas. Each row contains following information for one patient: identification n°, NRAS/chromosome 1 FISH result, NRAS mutant allele percentage, results of microsatellite markers LOH. NRAS gene and centromere localization are indicated. NA – non analysable, NI – non informative, NR – not realized. FISH: 1 - disomy, 2 - disomy with rares polysomic cells, 3 polysomy (A:3–4 copy, B: >4 copy), 4 - amplification, 5- monosomy, 6- high intra-tumor copy number variations of NRAS/chromosome 1
FISH analysis
FISH analysis was performed on tissue microarray (TMA) containing 94 melanomas and on tissue sections from 7 melanoma patients. FISH probe preparation and FISH technique was performed as already described [20, 25]. All samples were analyzed with RP11-245I3 and RP11-269F19 probes covering NRAS and a region of chromosome 1 telomeric to NRAS gene (chr1:45.142.760–45.303.288, (2009 GRCh37/hg19)), respectively. Chromosome 1/NRAS disomie was concluded if two FISH signals for each probe were observed and polysomy if three or more FISH signals were detected in the majority of cell. Chromosome 1/NRAS signal ratios 2:1 and 1:2 were described as NRAS monosomy and NRAS gain, respectively. Innumerable NRAS FISH signals were interpreted as amplification. Intra-tumor heterogeneity was defined as a presence of cell populations with different chromosome 1/NRAS status; some cells with increase, some with normal and some with loss of NRAS allele.
ATGC data analysis
Sequencing data about NRAS mutation of 479 cutaneous skin melanomas were extracted from cBioPortal platform as describe by Gao and colleagues [26]. The NRAS Q61 mutant allele percentage in 85 melanomas was compared to the data of our series.
Statistical analysis
Overall survival (OS) was defined as the period between the date of the primary melanoma diagnosis to the date of death (all causes) or last follow-up evaluation. Survival was censored at the last follow-up evaluation. Distant metastasis free survival (DMFS) was defined as the period between the date of the primary melanoma diagnosis to the date of onset of stage IV melanoma or death. The date of onset of stage IV melanoma was defined as the date of the clinical examination or imaging procedure that provided an unequivocal diagnosis of distant visceral metastasis. Progression-free survival was defined as the period between the date of the primary melanoma diagnosis (with or without lymph node sentinel biopsy procedure) to the date of the first regional (node or cutaneous) recurrence (stage IIIB minimum). Progression-free survival, distant metastasis-free survival and overall survival curves were estimated using the Kaplan-Meier method, and differences between PFS, DMFS, and OS curves were assessed using the log-rank test.
Survival and histoprognostic markers (age, gender, Breslow index, ulceration and mitotic activity) of NRAS Q61 mutated primary melanomas were compared between two groups: one group of 48 melanomas with <60% of M%NRAS and another group of 24 tumors with ≥60% of M%NRAS.
Student tests were performed for quantitative values, and Chi2 tests for qualitative values. The results were considered significant when P < 0.05.
Results
NRAS mutation frequency and allele quantification
The frequencies of BRAF V600 and NRAS Q61 mutations were evaluated in 267 FFPE melanoma patients of the series (flow chart in Additional file 2: Figure S2) diagnosed between March 2013 and May 2015. NRAS mutation was detected in 48 melanomas, corresponding to 18% (48/267) of all melanomas and 33.8% (48/142) of BRAF V600 wild type cases. Additionally, 63 NRAS Q61 mutated melanomas, diagnosed before this period, were included into the molecular analyses. In total, 111 NRAS Q61 mutated tumors were collected with 60–95% tumor cells.
Characterization of M%NRAS were performed in a larger group of 104 NRAS Q61 mutated melanomas containing ≥80% tumor cells. The corresponding mutations were c.182A > G p.Q61R in 58.6% (61/104), c.181C > A p.Q61K in 23.1% (24/104), c.182A > T p.Q61L in 13.5% (14/104), c.183A > T p.Q61H in 2.9% (3/104) and c.183A > C p.Q61H in 1.9% (2/104) of samples. M%NRAS was highly heterogeneous, ranging from 15.5 to 94% (Fig. 2). The majority of cases (60.6%, 63/104) had ≥30 to 60% M%NRAS and was thus considered as heterozygous (HET). The remaining 41 cases were considered as having non-heterozygous M%NRAS: 11.5% (12/104) had <30% of M%NRAS (Low non-HET) and 27.9% (29/104) had ≥60% of M%NRAS (High non-HET).
Fig. 2.
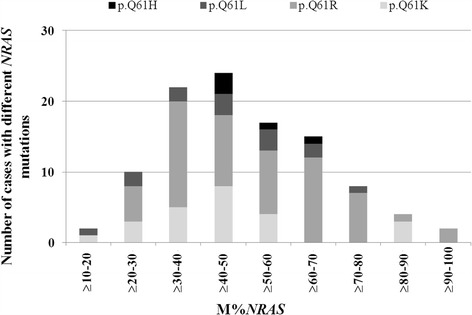
NRAS Q61 mutant allele burden in melanomas. Histogram representation of NRAS Q61 mutant allele quantity (in percentage) in 104 NRAS mutated melanomas. The X and Y axis correspond to the percentage of NRAS mutant and to the number of cases, respectively
We then compared our results with database of the Cancer Genome Atlas (TCGA) analyzed on the CBioPortal platform. Among the 85 NRAS mutated cases that were available only 50.6% (43/85) had a heterozygous status of NRAS mutation, while 34.1% (29/85) was High non-HET and 15.3% (13/85) was Low non-HET (Additional file 3: Figure S3).
FISH analysis
Among 101 samples (41 NRAS WT and 60 NRAS Q61) analyzed by FISH with NRAS locus and chromosome 1 specific fluorescent probes, different types of alternations were observed: no alteration of NRAS/chromosome 1 (disomy), disomy but rare cells with polysomy, polysomy and monosomy, which were detected in 23.1% (24/101), 19.2% (20/101), 9.6% (10/101) and 3.8% (4/101) of cases, respectively. NRAS amplification was a rare alteration and observed in 6.7% of melanoma samples (7/101). In 23.1% of samples (24/101), we observed intra-tumor cellular heterogeneity in NRAS/chromosome 1 copy numbers. Finally, FISH analysis of NRAS gene was non informative in 14.4% of cases (15/101).
To better understand the chromosomal mechanisms leading to M%NRAS increase, we compared M%NRAS and NRAS/chromosome 1 copy number status between 32 BRAF/NRAS WT and 57 NRAS Q61 mutated melanomas (Fig. 3). Disomy of NRAS/chromosome 1 (with or without polysomy in few cells) were detected in 59.4% (19/32) of BRAF/NRAS WT, 50% (15/30) of HET, but in only 28.6% (6/21) of High non-HET melanomas (P = 0.08). Polysomy of NRAS/chromosome 1 was detected in 13.3% (4/30) of HET, and 28.6% (6/21) of High non-HET samples, but was absent in BRAF/NRAS WT melanomas (P < 0.05). Amplification of NRAS was detected in 6.3% (2/32) and 9.5% (2/21) of BRAF/NRAS WT and High non-HET melanomas, respectively. Additionally, in 3.3% (1/30) of HET and 9.5% (2/21) of High non-HET cases, a gain of NRAS gene was observed. Deletion of NRAS was also a rare event and was observed in 9.4% (3/32) of BRAF/NRAS WT and 3.3% (1/30) of HET melanomas. A high intra-tumor copy number variation of NRAS/chromosome 1 was observed in 25% (8/32) of BRAF/NRAS WT, 30% (9/30) of HET and 23.8% (5/21) of High non-HET. FISH results of Low non-HET group were excluded from the comparative analysis because of insufficient numbers of samples (only 6 samples).
Fig. 3.
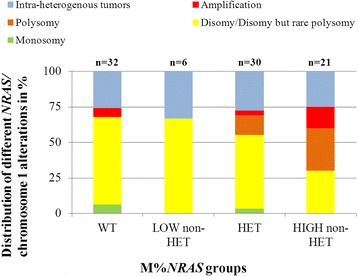
NRAS/chromosome 1 aberrations in NRAS mutated (n = 57) and NRAS WT (n = 32) melanomas. Histogram representation of prevalence of NRAS/chromosome 1 abnormalities evaluated by FISH in 104 NRAS mutated melanomas depending on the amounts of NRAS Q61 mutations. WT – wild-type, HET – heterozygous
Chromosome 1 microsatellite analysis
Eight polymorphic microsatellite markers were analyzed in 6 HET and 23 High non-HET tumors melanoma samples. In all HET melanomas, non LOH of NRAS locus surrounding microsatellites was detected but one for which microsatellites presented a low LOH, near the upper LOH detection threshold. In the majority of samples of High non-HET group, the microsatellite analysis revealed the presence of only one marker in the regions surrounding NRAS gene (Fig. 1). The probability of one NRAS allele loss was evaluated as very high in 30% (7/23) and mean in 26.1% (6/23) of tumors. In 26.1% (6/23) of tumors, the LOH results were non informative. In 17.4% (4/23), LOH with the presence of two alleles rather than one allele loss was detected.
Correlation of M%NRAS and clinical data
Information on primary tumor was available for 72 patients. The main histological subtypes were SSM and nodular melanomas, in 44.4% (32/72) and 38.9% (28/72) of cases, respectively. Median Breslow was 2.84 mm [0.4–10]. An ulceration was present in 47.2% (34/72) of melanomas, and 58.3% (42/72) had a mitotic activity (> 1 mitosis / mm2). Among these cases, we found NRAS Q61R mutation in 63.9% (46/72) of melanomas, NRAS Q61K in 25% (18/72), NRAS Q61L in 6.9% (5/72) and NRAS Q61H in 4.2% (3/72). M%NRAS was quantified by analysis of primary melanoma in 18.1% (13/72) of cases and of metastasis (lymph node, cutaneous or visceral) in 81.9% (59/72). Patients were divided into two groups depending on % of NRAS mutant allele. M%NRAS was ≥60% in the first group (n = 24, 33.3%) and <60% in the second cohort (n = 48, 66.7%). The second group contained 41 HET M%NRAS and 7 low not-HET M%NRAS cases. Clinical and pathological features of primary NRAS mutated melanomas were compared between these two groups and they are summarized in Table 2. No statistically significant differences were observed between these two groups with baseline criteria (P > 0.05). The median follow-up of the patients was 40 months (range [1–445]). PFS, DMFS and OS in both groups were not different (Fig. 4).
Table 2.
Clinicopathologic characteristics of studied subjects. Comparison of clinical and pathological features of NRAS mutated primary melanomas according to the NRAS mutant allelic burden (<60% or ≥60%)
| ≥60% οf Μ% ΝRΑS n = 24/72 (33.3%) | <60% οf Μ% ΝRΑS n = 48/72 (66.7%) | P-value (statistic test) | |
|---|---|---|---|
| Origin of pyrosequencing sample | |||
| From primary melanoma | 5 | 8 | 0.91 (Chi2with Yates’ correction) |
| From metastasis | 19 | 40 | |
| Age | |||
| Mean, years (SD) | 63.1 ± 17.4 | 65.2 ± 14.7 | 0.61 (Student) |
| Median, year [range] | 60 [37–93] | 66 [32–97] | |
| Gender | |||
| Ratio M/F | 13/11 | 30/18 | 0.49 (Chi2) |
| Breslow index | |||
| Mean, mm (SD) | 3.0 ± 2.1 | 3.3 ± 2.2 | 0.57 (Student) |
| Median, mm [range] | 3.00 [0.40–10.00] | 2.61 [0.50–9.00] | |
| Ulceration | 14 | 20 | 0.14 (Chi2) |
| Mitotic activity | 11 | 31 | 0.45 (Chi2) |
| Initial AJCC stage | |||
| I | 5 | 10 | 0.7 (Chi2) |
| IA | 4 | 2 | |
| IB | 1 | 8 | |
| II | 11 | 26 | |
| IIA | 4 | 10 | |
| IIB | 6 | 5 | |
| IIC | 1 | 11 | |
| III | 7 | 9 | |
| IIIA | 0 | 2 | |
| IIIB | 4 | 2 | |
| IIIC | 3 | 6 | |
| IV | 1 | 0 | |
| NA | 0 | 2 | |
| Histologic subtype | |||
| Nodular melanoma | 9 | 19 | 0.63 (Chi2) |
| SSM | 10 | 22 | |
| Acral melanoma | 1 | 0 | |
| Lentigo maligna melanoma | 0 | 1 | |
| Mucosal melanoma | 0 | 1 | |
| On congenital naevus | 0 | 1 | |
| Inclassable | 1 | 2 | |
| NA | 3 | 2 | |
SMM superficial spreading melanoma
NA data not available
SD standard deviation
M/F male/female
Fig. 4.
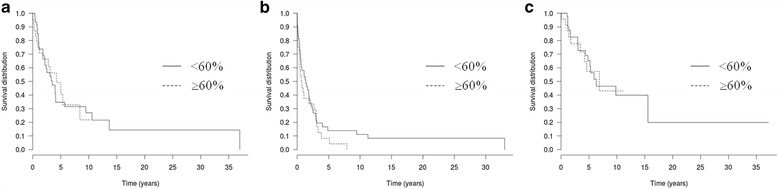
Survival curves. Kaplan-Meir survival curves of DMFS (a), PFS (b) and OS (c) of two distinct groups of NRAS mutated melanoma patients, according to the NRAS mutant allelic burden (<60% or ≥60%)
Discussion
In this study, we reported the prevalence of NRAS Q61 mutation and, for the first time, the variations of NRAS mutant alleles (M%NRAS), in a large series of human melanoma samples. We have demonstrated that M%NRAS was highly heterogeneous; indeed, only 61% of NRAS mutated melanomas were heterozygous, while 30% of cases had a significantly increased M%NRAS (≥60%). Our results were confirmed by analysis of the cases of the TCGA database.
NRAS pyrosequencing assays used in this study were developed to identify all hot-spot mutations in the codon 61 of NRAS gene. The specificity of these assays for different mutations was confirmed by Sanger sequencing. Additionally, the genotyping accuracy of 40 NRAS mutated melanomas, 27 of which were p.Q61R was confirmed by immunohistochemistry with an antibody against Q61R [27]. In a recent study, we have demonstrated that pyrosequencing was a robust molecular technique for oncogenic mutant allele quantification, by comparing it with quantitative real time PCR and picodroplet digital PCR [20]. This previous study was focused on BRAF mutations, and similar M%BRAF heterogeneity was demonstrated in melanomas, with 19% of cases having an increased M%BRAF. Altogether, we estimate from both series that 36.2% of melanomas with BRAF/NRAS mutations have a non-heterozygous oncogenic allele.
Few studies have investigated M%NRAS in melanomas. Recently, we reported two cases with an increase of M%NRAS during metastatic melanoma progression; suggesting that M%NRAS may enhance metastatic capacities of melanomas [28]. Additionally, a large screening study of 833 cells lines from the database of Cancer Genome Project, Sanger Institute, focused on frequently mutated genes (six suppressor gene and five oncogenes), has identified NRAS homozygous mutation in 10% of cell lines [29]. However, the zygosity status was only determinate by manual examination of sequencing electropherograms.
Interestingly, in vitro studies of mutant RAS family members had demonstrated a high oncogenic potential of increased mutant allele frequency. The oncogenic potential of NRAS G12D/G12D was highly increased as compared to heterozygous or hemizygous NRAS cells in NRAS-driven hematopoietic transformation [30]. Additionally, progenitors of hematopoietic cells expressing the highest levels of NRAS G12D demonstrated cytokine-independent CFU-GM colony growth and exhibited an increased level of pAkt, pErk and pS6 proteins. Endogenous expression of HRAS G12V promotes papilloma and angiosarcoma development and these neoplasm initiations have been strongly associated with HRAS G12V allelic and gene copy number imbalances [31, 32].
Mutation in one allele of an oncogene is sufficient for activation of its targets and M%NRAS is expected to be around 50% in diploid cells. However, in tumours with high chromosome instability, chromosome number is rarely disomic and M%NRAS could widely exceeded 50%. To better understand the chromosome mechanisms leading to NRAS mutant allele increase in the proportion of NRAS mutated melanoma, we firstly performed FISH analyses with 2 BAC probes covering NRAS region and another region of chromosome 1, telomeric to this gene, in a large series of 104 melanomas. Different types of NRAS/chromosome 1 status were observed. Polysomy was mainly observed in NRAS mutated tumours and disomic and/or disomic but rare polysomic cells were less frequent in High non-HET M%NRAS than in M%NRAS WT tumours. Amplification and deletion of NRAS gene were rarely observed and were seen in both NRAS WT and NRAS mutated melanomas. Genomic analysis of human cutaneous melanoma genomes have been described in several studies. However, in most of them, only melanoma cell lines were studied. The analysis of 60 melanoma cell lines by Gast [33] have revealed targeted focal amplifications of NRAS genes in 11% of them (n = 7/60) and amplification were detected in both NRAS mutated and NRAS WT melanomas. This frequency is higher that the frequency of NRAS amplification detected in the present series and in other reports. In a subset of cutaneous melanocytic lesions, NRAS amplification was found to be restricted to a few cases with NRAS mutations [34]. Additionally, Stark and colleagues reported rare instances of focal amplification including NRAS gene in two cell lines with NRAS mutation; however, a poor correlation between copy number increase and concomitant mutation in this oncogene was described [35]. Polysomy of chromosome 1 and intra-tumour NRAS/chromosome 1 heterozygosity were frequently found in our series and was preferentially observed in NRAS mutated cancers. Correlation between mutant burden and gene copy gains have already been described for KRAS [36] and BRAF gene [20]. To our knowledge, NRAS/chromosome 1 copy number variations have never been described in melanomas with regard to the NRAS mutant allele burden.
Secondly, we analyzed chromosome 1 microsatellite polymorphism in normal and tumor DNA in a group of 29 NRAS mutated cancers by ALFP method. As expected, LOH with WT allele loss was mostly restricted to the High non-HET M%NRAS group. However, unlike in haematological malignancy [37, 38], acquired copy-neutral LOH was not a predominant mechanism of mutant allele imbalance in NRAS Q61 mutated melanomas; indeed this aberration was detected in only 23% of our samples. Other mechanisms of High non-HET M%NRAS were amplification and gain of NRAS gene (14%) and polysomy of chromosome 1 (23.8%). In 9 melanomas (38.1%), an intra-tumor copy number variation of NRAS/chromosome 1 was detected. As most melanomas have copy number variations of whole chromosomes and of chromosome segment, NRAS mutant allele increase could be a consequence of chromosome instability and clonality in these tumors.
Correlation of M%NRAS with clinical data revealed no association with age, sex, histological melanoma subtypes, nor with histoprognostic markers of the patients with NRAS Q61 mutated melanomas. Moreover, no differences in patient survival outcomes were observed between patients with <60% and ≥60% of M%NRAS. However, this cohort is a retrospective monocentric study, and the analyses were limited by small number of patients. Furthermore, the value of M%NRAS has to be investigated for prediction of response to targeted therapy, as done for M%NRAS with promising results. We hypothesize that High non-HET M%NRAS could have an oncogenic addiction effect, which could improve the sensitivity of targeted therapy in this subgroup of NRAS Q61 mutated melanoma.
Conclusion
We report herein for the first time that 30% of cutaneous NRAS mutant melanomas have a high M%NRAS. Chromosome instability, (chromosome 1 polysomy, intra-tumor copy number variation of chromosome1/NRAS) rather than the acquired copy neutral LOH seems to be responsible for most of the cases with high M%NRAS. Histoprognostic markers and survivals were not different when comparing patients with <60% and ≥60% of M%NRAS; however this should be checked in a larger and multicentric series.
Additional files
Identification of NRAS WT and different NRAS mutations. Sequence of NRAS wild type allele (A) and of different NRAS mutant alleles and the corresponding pyrosequencing profiles. Pyrosequencing profiles by assays 1 (A, B, D, E) and by assay 2 (C and F) are present (TIFF 178 kb)
Flow chart. Flow chart for melanoma samples regarding the NRAS gene status and the type of molecular analysis carried out (TIFF 170 kb)
NRAS Q61 mutant allele burden in ATGC melanomas. Histogram representation of NRAS Q61 mutant allele quantity (in percentage) in 85 ATGC NRAS mutated melanomas. The X and Y axis correspond to the percentage of NRAS mutant and to the number of cases, respectively (TIFF 102 kb)
Acknowledgements
The authors thank Mariama Bakari, Dominique Pechaud, Yolaine Pothin and Catherine Le Gall for technique contribution and/or data collection.
Funding
This work was supported partly by grants from the Association Vaincre le Mélanome, Ligue Contre le Cancer (Comité 92 WB2013–232), and Association pour la Recherche et l’Enseignement en Pathologie (AREP).
Availability of data materials
Please contact the corresponding author for all data requests.
Abbreviations
- ALFP
Amplified fragment length polymorphism
- DMFS
Distant metastasis free survival
- FISH
Fluorescence in situ hybridization
- HET
Heterozygous
- LOH
Loss of heterozygosity
- M%NRAS
NRAS mutant allele frequency
- OS
Overall survival
- PFS
Progression-free survival
Authors’ contributions
Conception and design: ZHR, EFB, JFE; Development of the methodology, Acquisition of data: ZHR, EFB; NT, CM, UZ; Analysis and interpretation of data: ZHR, EFB, AB, NT, UZ, CM, PS, JFE; Writing and revision of the manuscript: ZHR, EFB, JFE; Study supervision: JEF, PS. All authors have read and approved the final manuscript.
Ethical approval and consent to participate
The research was performed in compliance with the ethical principles of the Helsinki Declaration (1964) and with the French ethics laws. Patients were informed and approved the use of their samples for research purpose. Tumor samples collection was declared to the French Ministry of Research (DC 2009–933). Melan-Cohort study was approved by CPP IDF 8 Ethics committee (030209) and registered with https://www.clinicaltrials.gov/ct2/search (NCT00839410).
Consent for publication
Not applicable
Competing interests
JF.E. received honoraria from Roche and Glaxo Smith Kline for counseling on patients with melanomas on the diagnosis and/or treatment with BRAF inhibitors. P.S. received honoraria for counseling on diagnosis and/or treatment of patients with melanomas from Roche and Glaxo Smith Kline.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12895-017-0061-x) contains supplementary material, which is available to authorized users.
Contributor Information
Zofia Hélias-Rodzewicz, Phone: +33-149095725, Email: zofia.helias-ext@aphp.fr.
Elisa Funck-Brentano, Email: elisa.funckbrentano@gmail.com.
Nathalie Terrones, Email: nathalie.terrones@yahoo.fr.
Alain Beauchet, Email: alain.beauchet@aphp.fr.
Ute Zimmermann, Email: ute.zimmermann@aphp.fr.
Cristi Marin, Email: cristi.marin@aphp.fr.
Philippe Saiag, Email: philippe.saiag@uvsq.fr.
Jean-François Emile, Email: jean-francois.emile@uvsq.fr.
References
- 1.Albino AP, Nanus DM, Mentle IR, Cordon-Cardo C, McNutt NS, Bressler J, et al. Analysis of ras oncogenes in malignant melanoma and precursor lesions: correlation of point mutations with differentiation phenotype. Oncogene. 1989;4:1363–1374. [PubMed] [Google Scholar]
- 2.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 3.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 4.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev. 2012;26:1131–1155. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. 2015. Cell 2015, 161:1681–1696. [DOI] [PMC free article] [PubMed]
- 6.Long GV, Menzies AM, Nagrial AM, Haydu LE, Hamilton AL, Mann GJ, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29:1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 7.Greaves WO, Verma S, Patel KP, Davies MA, Barkoh BA, Galbincea JM, et al. Frequency and spectrum of BRAF mutations in a retrospective, single-institution study of 1112 cases of melanoma. J Mol Diagn. 2013;15:220–226. doi: 10.1016/j.jmoldx.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Omholt K, Platz A, Kanter L, Ringborg U, Hansson J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clin Cancer Res. 2003;9:6483–6488. [PubMed] [Google Scholar]
- 9.Akslen LA, Angelini S, Straume O, Bachmann IM, Molven A, Hemminki K, et al. BRAF and NRAS mutations are frequent in nodular melanoma but are not associated with tumor cell proliferation or patient survival. J Invest Dermatol. 2005;125:312–317. doi: 10.1111/j.0022-202X.2005.23788.x. [DOI] [PubMed] [Google Scholar]
- 10.Thumar J, Shahbazian D, Aziz SA, Jilaveanu LB, Kluger HM. MEK targeting in N-RAS mutated metastatic melanoma. Mol Cancer. 2014;13:45. doi: 10.1186/1476-4598-13-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devitt B, Liu W, Salemi R, Wolfe R, Kelly J, Tzen CY, et al. Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. Pigment Cell Melanoma Res. 2011;24:666–672. doi: 10.1111/j.1755-148X.2011.00873.x. [DOI] [PubMed] [Google Scholar]
- 12.Ellerhorst JA, Greene VR, Ekmekcioglu S, Warneke CL, Johnson MM, Cooke CP, et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin Cancer Res. 2011;17:229–235. doi: 10.1158/1078-0432.CCR-10-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jakob JA, Bassett RL, Jr, Ng CS, Curry JL, Joseph RW, Alvarado GC, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118:4014–4023. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. BRIM-3 study group. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 16.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371:1877–1888. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 17.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet. 2015;386:444–451. doi: 10.1016/S0140-6736(15)60898-4. [DOI] [PubMed] [Google Scholar]
- 18.Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 19.Dummer R, Schadendorf D, Ascierto PA, Arance Fernández AM, Dutriaux C, Maio M,et al. Results of NEMO: A phase III trial of binimetinib (BINI) vs dacarbazine (DTIC) in NRAS-mutant cutaneous melanoma. J Clin Oncol 2016, ASCO Meeting Abstracts 34:9500.
- 20.Hélias-Rodzewicz Z, Funck-Brentano E, Baudoux L, Jung CK, Zimmermann U, Marin C, et al. Variations of BRAF mutant allele percentage in melanomas. BMC Cancer. 2015;15:497. doi: 10.1186/s12885-015-1515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebbé C, How-Kit A, Battistella M, Sadoux A, Podgorniak MP, Sidina I, et al. BRAF(V600) mutation levels predict response to vemurafenib in metastatic melanoma. Melanoma Res. 2014;24:415–418. doi: 10.1097/CMR.0000000000000088. [DOI] [PubMed] [Google Scholar]
- 22.Colomba E, Hélias-Rodzewicz Z, Von Deimling A, Marin C, Terrones N, Pechaud D, et al. Detection of BRAF p.V600E mutations in melanomas: comparison of four methods argues for sequential use of immunohistochemistry and pyrosequencing. J Mol Diagn. 2013;15:94–100. doi: 10.1016/j.jmoldx.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Moreau S, Saiag P, Aegerter P, Bosset D, Longvert C, Hélias-Rodzewicz Z, et al. Prognostic value of BRAFV600 mutations inmelanoma patients after resection of metastatic lymph nodes. Ann SurgOncol. 2012;2012(19):4314–4321. doi: 10.1245/s10434-012-2457-5. [DOI] [PubMed] [Google Scholar]
- 24.Balschun K, Haag J, Wenke AK, von Schönfels W, Schwarz NT, Röcken C. KRAS, NRAS, PIK3CA exon 20, and BRAF genotypes in synchronous and metachronous primary colorectal cancers diagnostic and therapeutic implications. J Mol Diagn. 2011;2011(13):436–445. doi: 10.1016/j.jmoldx.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lourenço N, Hélias-Rodzewicz Z, Bachet JB, Brahimi-Adouane S, Jardin F. Tran van Nhieu J, Peschaud F, Martin E, Beauchet A, Chibon F. Emile JF Copy-neutral loss of heterozygosity and chromosome gains and losses are frequent in gastrointestinal stromal tumors Mol Cancer. 2014;13:246. doi: 10.1186/1476-4598-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO,et al Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013, 6:pl1. [DOI] [PMC free article] [PubMed]
- 27.Ilie M, Long-Mira E, Funck-Brentano E, Lassalle S, Butori C, Lespinet-Fabre V, et al. Immunohistochemistry as a potential tool for routine detection of the NRAS Q61R mutation in patients with metastatic melanoma. J Am Acad Dermatol. 2015;72:786–793. doi: 10.1016/j.jaad.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Funck-Brentano E, Hélias-Rodzewicz Z, Longvert C, Mokhtari K, Saiag P, Emile JF. Increase in NRAS mutant allele percentage during metastatic melanoma progression. Exp Dermatol. 2016;25:472–474. doi: 10.1111/exd.13001. [DOI] [PubMed] [Google Scholar]
- 29.Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Haigis KM, Firestone AJ, McNerney ME, Li Q, Davis E, et al. Dominant role of oncogene dosage and absence of tumor suppressor activity in Nras-driven hematopoietic transformation. Cancer Discov. 2013;3:993–1001. doi: 10.1158/2159-8290.CD-13-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Mitsutake N, LaPerle K, Akeno N, Zanzonico P, Longo VA, et al. Endogenous expression of Hras(G12V) induces developmental defects and neoplasms with copy number imbalances of the oncogene. Proc Natl Acad Sci U S A. 2009;106:7979–7984. doi: 10.1073/pnas.0900343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Makarewicz JM, Knauf JA, Johnson LK, Fagin JA. Transformation by Hras(G12V) is consistently associated with mutant allele copy gains and is reversed by farnesyl transferase inhibition. Oncogene. 2014;33:5442–5449. doi: 10.1038/onc.2013.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gast A, Scherer D, Chen B, Bloethner S, Melchert S, Sucker A, et al. Somatic alterations in the melanoma genome: a high-resolution array-based comparative genomic hybridization study. Genes Chromosomes Cancer. 2010;49:733–745. doi: 10.1002/gcc.20785. [DOI] [PubMed] [Google Scholar]
- 34.Dubruc E, Balme B, Dijoud F, Disant F, Thomas L, Wang Q, et al. Mutated and amplified NRAS in a subset of cutaneous melanocytic lesions with dermal spitzoid morphology: report of two pediatric cases located on the ear. J Cutan Pathol. 2014;2014(41):866–872. doi: 10.1111/cup.12389. [DOI] [PubMed] [Google Scholar]
- 35.Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67:2632–2642. doi: 10.1158/0008-5472.CAN-06-4152. [DOI] [PubMed] [Google Scholar]
- 36.Modrek B, Ge L, Pandita A, Lin E, Mohan S, Yue P, et al. Oncogenic activating mutations are associated with local copy gain. Mol Cancer Res. 2009;2009(7):1244–1252. doi: 10.1158/1541-7786.MCR-08-0532. [DOI] [PubMed] [Google Scholar]
- 37.Rumi E, Pietra D, Guglielmelli P, Bordoni R, Casetti I, Milanesi C, et al. Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative. Acquired copy-neutral loss of heterozygosity of chromosome 1p as a molecular event associated with marrow fibrosis in MPL-mutated myeloproliferative neoplasms. Blood. 2013;121:4388–4395. doi: 10.1182/blood-2013-02-486050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunbar AJ, Gondek LP, O'Keefe CL, Makishima H, Rataul MS, Szpurka H, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;2008(68):10349–10357. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Identification of NRAS WT and different NRAS mutations. Sequence of NRAS wild type allele (A) and of different NRAS mutant alleles and the corresponding pyrosequencing profiles. Pyrosequencing profiles by assays 1 (A, B, D, E) and by assay 2 (C and F) are present (TIFF 178 kb)
Flow chart. Flow chart for melanoma samples regarding the NRAS gene status and the type of molecular analysis carried out (TIFF 170 kb)
NRAS Q61 mutant allele burden in ATGC melanomas. Histogram representation of NRAS Q61 mutant allele quantity (in percentage) in 85 ATGC NRAS mutated melanomas. The X and Y axis correspond to the percentage of NRAS mutant and to the number of cases, respectively (TIFF 102 kb)


