Abstract
The ribonuclease A (RNase A) superfamily has been the subject of extensive studies in the areas of protein evolution, structure and biochemistry and are exciting molecules in that they appear to be responding to unique selection pressures, generating proteins capable of multiple and diverse activities. The RNase 4 and RNase 5/ang 1 shared locus breaks a pattern that is otherwise canonical among the members of the RNase A gene superfamily. Conserved among humans, mice and rats, the locus includes two non-coding exons followed by two distinct exons encoding RNase 4 and RNase 5/ang 1. Transcription from this locus is controlled by differential splicing and tissue-specific expression from promoters located 5′ to each of the non-coding exons. Promoter 1, 5′ to exon I, is universally active, while Promoter 2, 5′ to exon II, is active only in hepatic cells in promoter assays in vitro. Transcription from Promoter 2 is dependent on an intact HNF-1 consensus binding site which binds the transcription factor HNF-1α. In summary, RNase 4 and RNase 5/ang 1 are unique among the RNase A ribonuclease genes in that they maintain a complex gene locus that is conserved across species with transcription initiated from tissue-specific dual promoters followed by differential exon splicing.
INTRODUCTION
The ribonuclease A (RNase A) superfamily has been the subject of extensive studies in the areas of protein evolution, structure and biochemistry. RNase A (bovine pancreatic ribonuclease), the prototype of this gene superfamily, was one of the first proteins to have its crystal structure solved and to be chemically synthesized de novo (1,2). The molecular evolution of the RNase A vertebrate-specific enzyme family is startlingly complex, as the various gene lineages (13 in humans and, 9 in mice and rats) appear to be responding to unique selection pressures, generating proteins capable of reducing the infectivity of viruses, killing systemic pathogens and inducing the growth of blood vessels while maintaining conserved signature motifs (3–7). Until recently, the signature motifs of this family included 6–8 cysteines which form the disulfide bonds necessary to achieve the required 3D structure and a catalytic triad composed of two histidines and a lysine within a CKXXNTF motif. With the discovery of RNases -9, -10, -11, -12 and -13 (3,8,9), these requirements have been somewhat relaxed as these genes are clearly the derivatives of the RNase A lineage based on sequence similarity and the presence of the characteristic disulfide bridges, but they lack the active site signature motif and, as such, are unlikely to have ribonucleolytic activity.
With the elucidation of the human genome, Lander et al. (10) noted the unique nature of the RNase A ribonucleases as the only gene family with enzymatic activity clearly limited to vertebrate species. The genes encoding RNase A ribonucleases cluster on human chromosome 14; all with a presumed simple genomic structure consisting of a small non-coding exon I followed by an uninterrupted coding sequence on exon II (11–14). Of Interest, the transcripts encoding each of these ribonucleases are initiated from single promoters located 5′ to the non-coding exons and rely on the presence of enhancers in the introns to achieve maximum levels of transcription.
Notwithstanding the rich research history of the RNase A superfamily, relatively little is known specifically about the RNase 4 lineage. Human RNase 4 was first described by Shapiro et al. (15) after isolating the protein from conditioned medium from an adenocarcinoma cell line. Two groups isolated the cDNA encoding human RNase 4 (16,17). Rosenberg and Dyer (17) found human RNase 4 to be a single copy gene expressed in all tissues examined by northern analysis with the exception of brain and placenta, with the highest expression found in liver.
Conversely, more is known about the RNase 5/angiogenin lineage. The angiogenins are structurally distinct from other members of the RNase A superfamily in that these proteins have only six of the eight standard cysteines. Angiogenin (ang) was originally identified as a factor stimulating blood vessel growth [reviewed in (5)]. Recently, mouse angiogenin (ang) 1 and ang 4, and human angiogenin have been identified as microbicidal agents (6). Evolutionary analysis of the angiogenin lineage from non-mammalian species suggests that this group may be the most ancient form of the superfamily (3,5). While examining the expressed sequence tag (EST) database, Strydom (5) noted that both human angiogenin and mouse angiogenin 1 (ang 1) shared 5′-untranslated regions (5-UTRs) with RNase 4 and suggested that these genes might exist in close proximity along the chromosome and that the different mRNAs could be the products of differential splicing. Alternatively, but less likely, the shared 5′-UTRs could have resulted from very recent duplications of the non-coding exons, as duplication events are certainly very prominent in the evolutionary history of the RNase A gene family (18,19). In this work, we characterize RNase 4 and RNase 5/ang 1 genes demonstrating that the two genes share a common locus consisting of four exons and three intervening introns in the mouse, rat and human genome. We further demonstrate that transcription from the RNase 4–RNase 5/ang 1 locus is controlled by two promoters that operate in a tissue-specific manner and that the proximal promoter is absolutely dependent on an intact HNF-1α consensus binding site.
MATERIALS AND METHODS
Cell culture
The following cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA) and were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, l-glutamine and penicillin/streptomycin at 37°C with 5% CO2 in a humidified incubator: MMSV (CCL-163.2) and K-Balb (CCL-163.3) which are derivatives of the mouse 3T3 fibroblast cell line; RAW264.7 (TIB-71), a mouse macrophage cell line; Hepa 1-6 (CRL-1830), a mouse hepatoma cell line; Hep-G2 (HB-8065), a human liver cell line and H-4-II-E (CRL-1548), a rat hepatoma cell line.
Rapid amplification of cDNA ends (5′ and 3′ RACE)
Complementary DNA was synthesized from 1 μg RNA from mouse liver, kidney and bone marrow with MMLV reverse transcriptase as per the manufacturer's instructions (SMART RACE cDNA Amplification kit, Clontech, Palo Alto, CA) and then amplified using mouse RNase 4 specific oligonucleotides. Sequences used are as follows:
5′ RACE, 5′-CTGAGGGTCCACATGCTGTCGAAG-3′
3′ RACE, 5′-ATGATGCAGAGACGGAAGATGACT-3′
The PCR conditions were as follows: 5 cycles of 94°C for 5 s followed by 72°C for 3 min; then 5 cycles of 94°C for 5 s followed by 70°C for 10 s followed by 72°C for 3 min; and finally 25 cycles of 94°C for 5 s followed by 68°C for 10 s followed by 72°C for 3 min. The cDNAs were gel purified (BIO 101, Alta Vista, CA) and subcloned into the pCR2.1 (Invitrogen, San Diego, CA); multiple colonies were sequenced in both directions. The sequences were assembled using Sequencher 4.1 (GeneCodes, Ann Arbor, MI) to obtain the full-length cDNA sequence. The GenBank accession number for the full length liver transcript is AY762362. The RNase 4 and RNase 5/ang 1 locus maps to mouse chromosome 14 (NT_039599, nt 6 429 305–6 445 565).
To determine the precise transcriptional start site(s) of the human and rat RNase 4 gene, 5′ RACE was performed using liver and kidney mRNA as described above. Sequences of the primers are as follows:
Generation of the pGL3 enhancer reporter constructs
Fragments 500 bp 5′ of exon 1 and exon II were generated by PCR from genomic DNA and ligated into the BglII and HindIII sites of the pGL3 Basic reporter vector (Promega, Madison, WI). The resulting constructs are named according to the gene segment contained: pGL3 Basic has no promoter, pGL3-Pr1 contains the promoter region of 5′ to exon I and pGL3-Pr2 contains the promoter region 5′ to exon II. The following primers were used to generate the indicated fragments:
5′ Pr1 5′-ctctctcctc AGATCT TCGTTGGTCTAGGGGTATGATTCTCGCTTT-3′
3′ Pr1 5′-ctcctcctc AAGCTT TGGGTGAGGTCGGCGTTGCCTGGCCACGCA-3′
5′ Pr2 5′-ctctctcctc AGATCT TGAGTAGCCATAAATATATATTCTATTTTC-3′
3′ Pr2 5′-ctcctcctc AAGCTT CTAGAGGCTCCTCACAGGCTGGTAATATACA-3′
The underlined nucleotides encode the BglII restriction enzyme site on the 5′ primer and HindIII on the 3′ primers and the lower case letters indicate a sequence added to aid in the restriction digestion of the PCR generated fragments. All constructs were confirmed by automated sequencing using the dRhodamine sequencing kit (Applied Biosystems, Foster City, CA) on the ABI377 Automated Sequencer followed by analysis using Sequencher 3.1.1 software (Gene Codes Corporation, Ann Arbor, MI).
Identification of potential transcription factor binding sites
Analysis of the potential transcription binding sites was done using Transcription element search system (TESS) available at http://www.cbil.upenn.edu/tess.
Mutagenesis of the HNF-1 transcription factor binding site in the mouse RNase 4 Pr2 construct
The putative transcription factor binding site HNF-1, identified within the sequence of Pr2 (defined above), was mutated from GTTAATATTTGAC to ATGAAGATTTGAC (underlined residues highlight mutations) by site directed mutagenesis (QuickChange Kit, Stratagene, La Jolla, CA) according to the manufacturer's instructions. Presence of the mutation (pGL3 Pr2mHNF-1) was confirmed by DNA sequencing and the effect of the mutation was assessed in the dual luciferase assay as described below. The residues selected for mutation were those shown by Ryffel et al. (21) to be necessary for the liver-specific transcription.
Transient expression and the promoter reporter assay
The cell lines were plated at 300 000 cells per well in a six-well plate 24 h prior to transfection. About 5–10 μl of lipofectin (Life Technologies, Rockville, MD) was added according to the manufacturer's recommendation to transfect 0.5 μg of the pGL3-derived constructs (pGL3 Basic, vector only; pGL3 Pr1, Pr1; pGL3 Pr2, Pr2; and pGL3 Pr2mHNF-11) and 0.1 μg pRL CMV reporter plasmid into the cell line. The dual luciferase assay (Promega, Madison, WI) was used to determine transcriptional activity of the reporter constructs according to manufacturer's suggestions. The firefly luciferase (experimental promoter construct) activities of each of the experimental constructs were normalized to the Renilla luciferase (co-transfectant control promoter construct) activity. All experiments were performed in triplicate and statistical significance was determined using the two-tailed t-test assuming unequal variance (Microsoft Excel). All data are reported as mean ± standard error of the mean (SEM).
Biotin labeling of 3′-OH ends of EMSA probes
Five picomoles of 3′-OH ends of oligonucleotides were end-labeled with biotin using terminal deoxynucleotidyl transferase (TdT, Pierce, Rockford, IL). Briefly, 5 pmol of the complementary oligonucleotides was labeled separately in 37°C incubation, with 1× TdT reaction buffer, 0.5 μM Biotin-N4-CTP and 10 U TdT in a 50 μl reaction. After 30 min, 2.5 μl of 0.2 M EDTA was added to terminate the reaction. The biotin-labeled oligonucleotides were extracted in the aqueous phase of CHCl3:isoamyl alcohol (24:1). A double stranded electromobility shift assay (EMSA) probe was made by annealing a 1:1 mix of the biotin-labeled complementary oligonucleotides by heating to 65°C for 10 min followed by slowly cooling to room temperature. The biotin-labeled EMSA probe was stored at −20°C until needed. Unlabeled Self and mutated Self (mSelf) complementary oligonucleotides were also annealed and stored at −20°C.
The oligonucleotide sequences are as follows:
Pr2HNF-1(Self) 5′-TGGTCCACCAGGTGGTTAATATTTGACCCAG-3′
Pr2mHNF-1(mSelf) 5′-TGGTCCACCAGGTGATGAAGATTTGACCCAG-3′
The underlined residues are those mutated in the pGL3 Pr2mHNF-1 promoter construct.
EMSA
About 6 μg of the rat liver nuclear extract (3 μg/μl, Active Motif) was pre-incubated at 4°C for 20 min in the presence or absence of 2 μl antibody. Competing unlabeled oligonucleotide (100-fold molar excess, Self or mSelf) was added prior to the addition of 2 μl of the biotin-labeled probe. After a 20 min incubation at 4°C, 1 μl of high-density sample buffer (Novex, San Diego, CA) was added and the samples were evaluated by electrophoresis in a 6% polyacrylamide DNA retardation gel (Invitrogen, San Diego, CA) with 0.25× TBE running buffer at a constant 100 V at 4°C for 1.5 h. The gel was transferred to a positively charged nylon membrane (Pierce, Rockford, IL) in 0.5× TBE at a constant 30 V at 4°C for 1 h. The DNA was fixed to the membrane by UV cross-linking (UV Stratalinker 1800, Stratagene). The membrane was developed using the Chemiluminescent Nucleic Acid Detection Module (Pierce). Briefly, the membrane was blocked for 15 min at room temperature followed by a 15 min incubation in streptavidin-horseradish peroxidase conjugate. The membrane was then washed extensively followed by a 5 min incubation in substrate equilibration buffer. Finally, the membrane was developed in a 1:1 mixture of luminol enhancer solution and peroxide for 5 min. The membrane was exposed to X-ray film for 15–60 s and developed. The following rabbit polyclonal antibodies were used: anti-HNF-1α either from Active Motif (Nushift HNF1α kit) and/or from Santa Cruz (sc10791X, 2 μg/μl); anti-HNF-1β (sc-22840X, 2 μg/μl, Santa Cruz) and anti-GATA4 (sc-1237X, 2 μg/μl, Santa Cruz).
Northern and Southern analysis
Multiple-tissue and cell line Northern membranes from Clontech (Palo Alto, CA) were prehybridized and hybridized following the manufacturer's instructions. The hybridization was performed with either a radiolabeled or a biotin labeled (BrightStar Psoralen-biotin, Ambion) open reading frame probe of mouse RNase 4 (GenBank accession no. AY762362, nt 93–537). The membranes were washed and exposed to the film overnight. The actin gene was used as a control for RNA loading and integrity. For the Southern blot, 10 μg of 129SV mouse genomic DNA provided by Jiliang Gao (National Institutes of Health, Bethesda, MD) was digested with the indicated restriction enzymes overnight at 37°C. The digested DNA was electrophoresed on a 0.8% agarose gel and then transferred to a positively charged membrane. Following prehybridization, the blot was hybridized with the biotin labeled probe.
Generation of the mouse RNase 4 expression construct and production of recombinant protein
The portion of the open reading frame encoding the predicted mature RNase 4 peptide (GenBank accession no. AY762362, nt 178–534) was amplified from liver cDNA using the following primers:
Forward: 5′-ctcctcctcAAGCTTCTCAGGATCGAATGTACCAAC-3′
Reverse: 5′-ctcctcctcGAATTCTCTGTCAAAGTGCACTGGGAC-3′
(underlined residues encode engineered restriction enzyme sites). The PCR product was subcloned into pCR2.1 for sequencing and then the RNase 4 insert was excised using HindIII and EcoRI and then ligated in frame into the prepared pFlag CTS bacterial expression vector (International Biotechnologies, Inc., New Haven, CT). The fidelity of the expression construct was confirmed by sequencing. Production and isolation of recombinant mouse RNase 4 from bacterial transfectants was performed as described previously (22). The concentration of recombinant protein was determined by the comparison with serial dilutions of known concentrations of a FLAG-conjugated protein standard on western blots. The western blot was incubated with monoclonal anti-FLAG antibody (Sigma) followed by HRP conjugated sheep anti-mouse IgG and developed with reagents from Amersham.
Determination of ribonuclease activity against tRNA substrate
The assay used was adapted from the procedure of Slifman et al. (23) as described previously (22). Calculations included the following approximations: the average molecular weight (Mr) of tRNA as 28 100 (75–90 ribonucleotides/tRNA molecules × Mr 341/ribonucleotide), with A260 of 1.0 corresponding to 40 μg/ml RNA. All time points represent averages of the triplicate samples.
Evolutionary analysis of DNA sequences
RNase 4 mature protein sequence from Mus musculus—house mouse, Q9JJH1; Rattus norvegicus—Norway rat, O55004; Homo sapiens—human, P34096; Pan troglodytes—chimpanzee, Q8HZQ0; Bos taurus—cow, P15467 and Sus scrofa—pig, P15468 were aligned using ClustalW. A neighbor-joining phylogenetic tree was reconstructed with p-distances and bootstrap percentages derived from 2000 replications were used to evaluate the reliability of the tree (24).
RESULTS
Identification and chromosomal localization of the mouse RNase 4 gene
A genomic sequence encoding mouse (M.musculus) RNase 4 was identified by homology to the human RNase 4 coding region (D37931) in the Celera Mouse Genome Database. This sequence was used to identify the mouse and rat sequences in the public domain genome sequences and for oligonucleotide design in order to amplify mouse full-length cDNA from reverse transcribed liver RNA. Additionally, RNase 4 from R.norvegicus (Norway rat), H.sapiens (human), P.troglodytes (chimpanzee), B.taurus (cow) and S.scrofa (pig) were collected from the GenBank on the basis of their homology to the mouse RNase 4 utilizing the BLAST algorithm.
Northern analysis and RACE
Transcripts encoding mouse RNase 4 (∼1.4 kb) were detected in the prostate, thyroid, salivary glands, uterus, heart, lung, liver, and kidney of mice (Figure 1A). No hybridization signals were detected in the large intestine, stomach, thymus, brain, spleen, skeletal muscle or testis. Mouse RNase 4 has a pattern of tissue expression similar to that of human RNase 4, which was readily detected by northern analysis in heart, lung liver, skeletal muscle, kidney and pancreas (17). Transcripts encoding mouse RNase 4 were also detected in several cell lines with highest levels of expression in the RAW 264.7 mouse macrophage and the Hepa 1-6 hepatoma cell lines (Figure 1B). Since liver expressed such abundant amounts of RNase 4 message, liver RNA was used to determine the full transcript size. In addition to the experimentally derived cDNAs, several liver ESTs were found by homology search (AI787202, AI96225, AI530414, AI663624 and AW412859). The total size of the liver RNase 4 transcript is 1409 nt, which is in agreement with the northern analysis.
Figure 1.
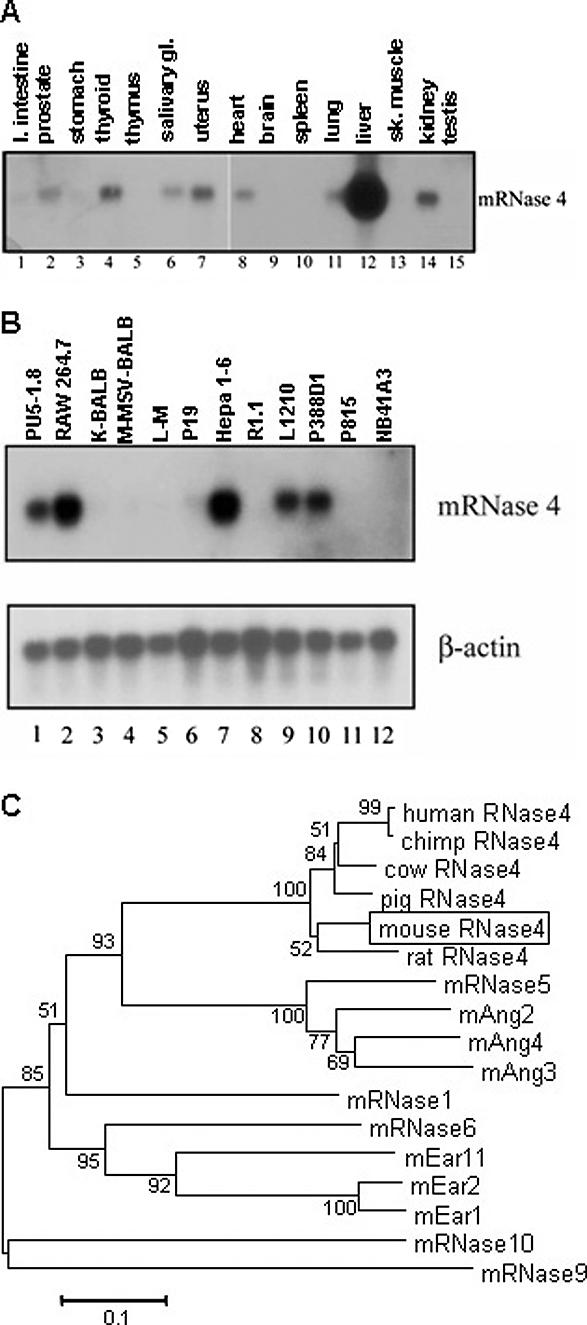
(A) Northern analysis of RNase 4 expression in mouse tissues. A 460 nt probe inclusive of the open reading frame of mouse RNase 4 was used to hybridize mouse multi-tissue northern blots. (B) Northern analysis of RNase 4 expression in mouse cell lines. A 460-nucleotide probe inclusive of the open reading frame of mouse RNase 4 was used to hybridize a mouse cell line northern blot. Lane 1, lymphoid; lane 2, leukemia; lanes 3 and 4, fibroblast; lane 5, connective; lane 6, carcinoma; lane 7, hepatoma; lanes 8 and 10, lymphoma; lane 9, lymphocytic leukemia; lane 11, mast cell; lane 12, neuroblastoma. (C) Neighbor-joining phylogenetic tree of RNase A superfamily members. Swiss protein database accession numbers for amino acid sequences are as follows: mouse RNase 4 (boxed), Q9JJH1 (box); rat RNase 4, O55004 (29–147); human RNase 4, P34096 (29–147); chimpanzee RNase 4, Q8HZQ0 (29–147); cow RNase 4, P15467 (1–119); pig RNase 4, P15468 (29–147); mRNase 5/Ang 1, P21570 (25–145); mAng2/Angrp, Q64438 (25–145); mAng4, Q9JJH1 (30–148); mAng3/Anglp, P97802 (24–144); mRNase 1, P00683 (26–149); mEar11, (predicted from AF512015); mEar2, P97425 (28–156); and mEar1, P97426 (28–155); mRNase 6, AY545655; mRNase 9, AY226989; mRNase 10, AY226990. Only three of the 10–15 mEar genes are shown due to space constraints. Unrooted phylogenetic tree and bootstrap percentages (2000 replications, at branch points) were obtained using MEGA2 (24).
Southern analysis reveals that mouse RNase 4 is a single copy gene (data not shown). RNase 4 is localized to mouse chromosome 14 between RNase 5/ang 1 and RNase 6 (25,26) in both the mouse and human genome and on chromosome 15 (NW_047453, Build 2) in rat indicating a syntenic relationship as would be expected of orthologous genes. Phylogenetic analysis demonstrates that mouse RNase 4 is more closely related to RNase 4 of non-mouse species than it is to other mouse RNase A gene lineages (Figure 1C). Sequence analysis reveals that the RNase 4 predicted protein sequences are 83–99% identical to each other over the length of the mature protein. Taken together, these data suggest strong orthologous relationships among these ribonuclease sequences.
Ribonuclease activity of mouse RNase 4
The RNases 4 from each species contain the eight cysteines necessary for folding, the histidines (amino acid position 40 and 144) and the lysine (position 68) that form the catalytic site, as well as the CKXXNTF motif found in all members of the RNase A superfamily (Figure 2A). Recombinant mouse RNase 4 was isolated from the periplasm of bacteria using the pFLAG CTS expression system (Figure 2B) and 2.4 pmol recombinant protein was used to generate a double reciprocal plot (Figure 2C). The catalytic constants determined using the yeast tRNA substrate are kcat = 0.17 s−1, Km = 0.43 μM and the catalytic efficiency of kcat/Km = 3.8 × 105 M−1s−1, which are similar to those of human and pig RNase 4 against the UpA substrate [kcat/Km = 2.46 × 105 M−1s−1 (15) and 2.5 × 105 M−1s−1 (27), respectively]. The catalytic efficiency of mouse RNase 4 against yeast tRNA is within the range of other RNase A enzymes against this substrate [human RNase 2, kcat/Km = 1.3 × 106 M−1s−1 (17); mEar 6, kcat/Km = 8.7 × 105 M−1s−1 (28); mRNase 6, kcat/Km = 4 × 105 M−1s−1 (25)] and establishes mouse RNase 4 as an active enzyme.
Figure 2.
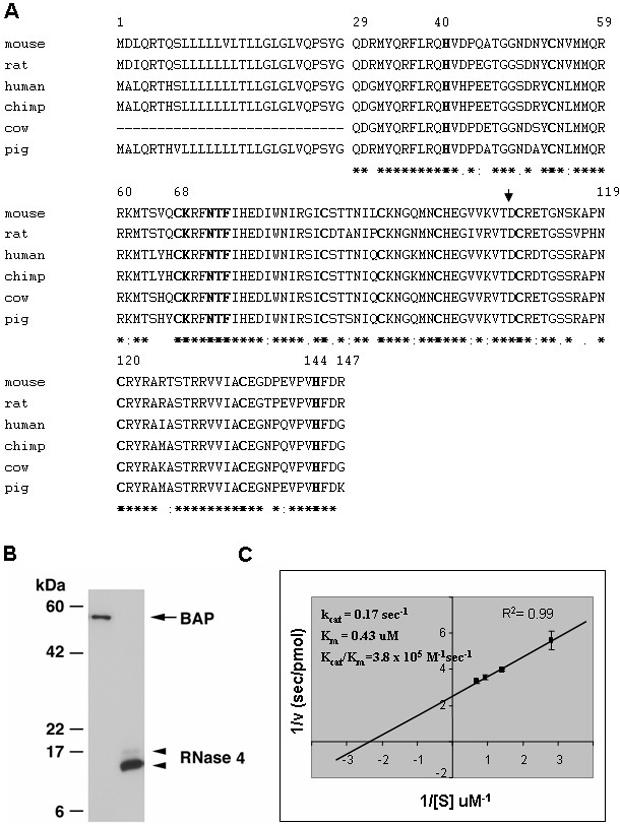
(A) An alignment of RNase 4 amino acid sequences. Amino acids 1–28 are components of the signal sequence and amino acids 29–147 form the mature protein. Residues 40, 68 and 144 form the catalytic triad and the eight cysteines (amino acids 53, 67, 85, 92, 99, 109, 120 and 135) form the disulfide bonds. The residues in bold are those shown to be conserved in all RNase A family members. The aspartic acid (D) at position 108 (arrow) has been shown to be involved in substrate specificity (41). Consensus line highlights identical (*) and conserved (. or :) amino acids. (B) Western analysis of recombinant mouse RNase 4 protein. The arrow highlights bacterial alkaline phosphatase (BAP) used as a positive control and the arrow heads indicates mouse RNase 4 with the lower, darker band corresponding to the fully processed, active enzyme. (C) Enzymatic activity of recombinant mouse RNase 4 against yeast tRNA substrate. Double reciprocal plot used to generate enzymatic constants for mouse RNase 4.
Differential splicing and exon sharing between RNase 4 and RNase 5/ang 1
We determined the gene structure of mouse RNase 4 (Figure 3A) by mapping our experimentally-determined RACE products from the liver and kidney as well as EST sequences onto the mouse genomic sequence. Furthermore, RNA from various tissues was screened by RT–PCR for the presence of the non-coding exons together with the coding exons of RNase 5/ang 1 and RNase 4. As can be seen in Figure 3B, the transcript containing exon II is found primarily in the liver (lane 5) while exon 1 can be detected in all tissues. This differential non-coding exon usage is observed for both mouse RNase 4 and RNase 5/ang 1. Note that in the kidney (lane 7), RNase 4 is seen in conjunction with both exon I and exon II. The four exon gene structure and 5′-UTR sharing between RNase 4 and RNase 5/ang 1 were determined to be a feature of the mouse, human and rat genome by analysis of the 5′ RACE products from at least two RNA sources from each species followed by mapping of the RACE products and database ESTs onto the appropriate genome. ESTs were identified by BLAST analysis of the protein coding region against mouse, human and rat EST databases. EST fragments extending the sequence in the 5′ direction were used in this analysis to confirm experimental RACE data shown in Figure 4. In the human, mouse and rat transcriptomes, exon I (Figure 4B) is seen in transcripts from liver, kidney and placenta while exon II (Figure 4C) has a more restricted expression being detected primarily in the liver and occasionally in the kidney.
Figure 3.
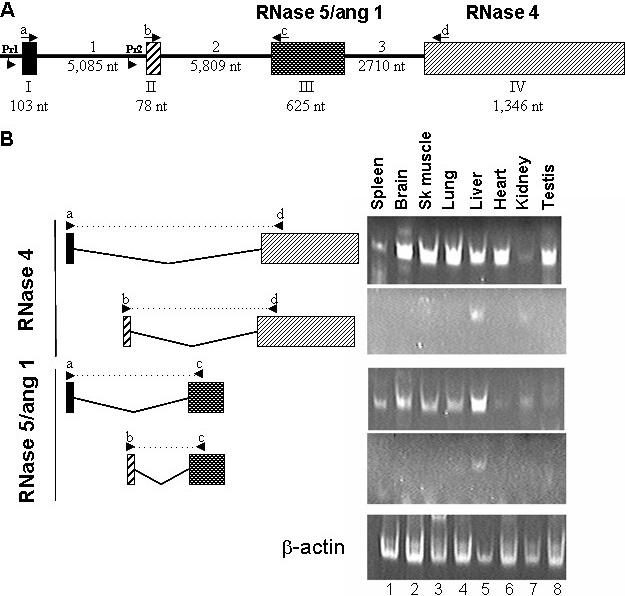
(A) Gene structure of the mouse RNase 4 and RNase 5/ang 1 locus. Exons are represented as boxes and introns are the intervening lines. The numbering denotes the size of each segment in the mouse genome. The RNase 5/ang 1 coding sequence is on exon III and the RNase 4 coding sequence is on exon IV. Lowercase letters and arrows indicate location and direction of RT–PCR primers used to generate data featured in (B). Pr1 and Pr2 refer to promoters examined in Figure 5. (B) Tissue-specific detection of exon I/exon II variants. RT–PCR was performed using primers complementary to sequences in either exon I (primer a, upper panel of each pair) or exon II (primer b, lower panel of each pair) in conjunction with primers complementary to the sequence of the coding region of RNase 5/ang 1 (primer c, exon III) or RNase 4 (primer d, exon IV) to identify tissue-specific expression of individual variants. Splicing pattern and relative position of primers are shown in the left hand side of Figure while RT–PCR data is shown in the right. Actin primers were used as a positive control.
Figure 4.
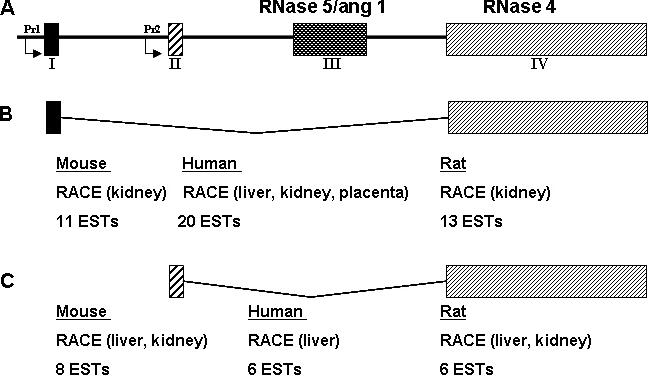
(A) Schematic of mouse, rat and human RNase 4 and RNase 5 locus. Support for the existence of each transcript was generated by 5′ RACE from the tissue indicated and by the detection of ESTs in the mouse, human and other EST databases. Pr1 and Pr2 refer to the promoters examined in Figure 5. (B) RNase 4—exon I-containing transcripts found in most tissues (C) RNase 4—exon II-containing transcripts found primarily in the liver.
Tissue-specific promoters
One way to regulate the expression of RNase 4 and invoke differential expression of exons I and II would be by the use of two different promoters. We characterized the promoter activity of 500 nt 5′ to exon I (Promoter 1, Pr1, NT_039599, nt 6 429 595–6 430 094) and 500 nt 5′ to exon II (Promoter 2, Pr2, NT_039599, nt 6 434 259–6 434 758) in several cell lines (Figure 5A). The activity from the Promoter 1-containing construct was detected at 4- to 8-fold over background levels and was more or less equivalent in all cell lines examined. This is consistent with the expression of exon I-containing transcripts in multiple tissues. Interestingly, the Promoter 2-containing construct has minimal transcriptional activity in all but the Hepa 1-6 liver cell line (2-fold over background compared with 9-fold in the Hepa 1-6 cells). Promoter 2 contains a sequence similar to the consensus binding site for the hepatocyte nuclear factor (HNF)-1 family of transcription factors (29). When mutations are introduced into the putative HNF-1 site, the mutated promoter construct (Pr2mHNF-1) lost significant transcriptional activity in mouse, rat and human hepatic cell lines (Figure 5B–D, respectively). EMSA analysis revealed specific binding of the transcription factor HNF-1α to this site in rat liver nuclear extract (Figure 5E). The supershift seen with two distinct anti-HNF-1α antibodies (lanes 5 and 6) was not replicated with an antibody directed toward the related HNF-1β protein which was used here as an isotype control (lane 7).
Figure 5.
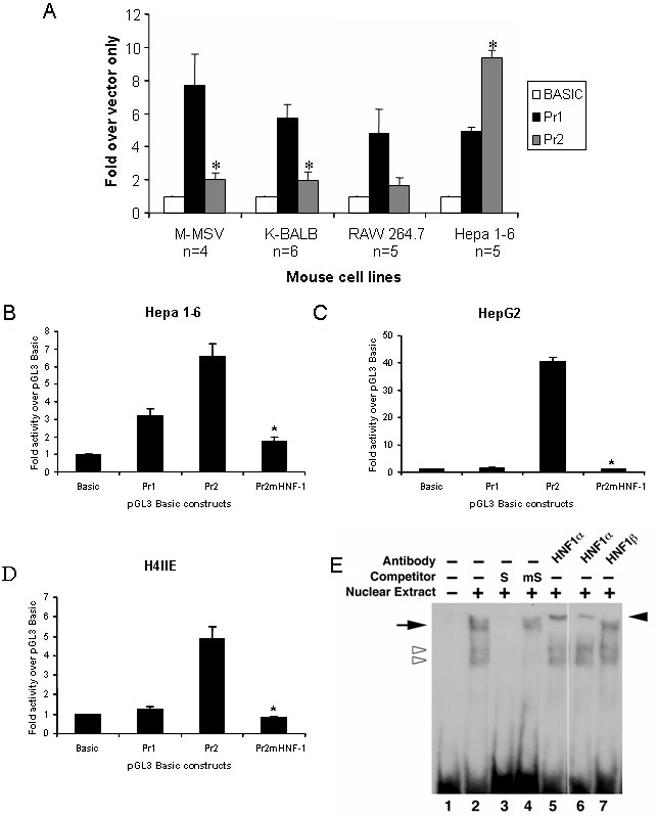
(A) Differential activity of Promoter 1 (Pr1, 500 nt 5′ of exon I) and Promoter 2 (Pr1, 500 nt 5′ of exon II) in various mouse cell lines. Each experiment was performed in triplicate; the number of experiments performed is indicated (n). Values are normalized to pGL3 Basic which lacks a promoter and data are reported as mean ± SEM; *P < 0.01 between Pr2 and Pr1. Transcription from Pr2 in response to mutagenesis of the putative HNF-1 site in the (B) mouse Hepa 1-6 cell line; (C) human HepG2 cell line; (D) rat H4IIE cell line. *P < 0.001 between Pr2 and Pr2mHNF-1, n = 3. (E) EMSA analysis. Biotin-labeled probe corresponding to nt 6 434 577–6 434 607 of GenBank accession no. NT_039599 in the mouse liver Pr 2 sequence was incubated with rat liver extract in the absence (lane 2) or presence of excess unlabeled probe (lane 3) or probe in which the HNF-1site had been mutated (lane 4). Pre-incubation with two different anti-HNF1α antibodies (lanes 5 and 6); isotype control (HNF1β-lane 7). The open arrowheads denote non-specific DNA–protein interactions; the arrow (left) denotes specific DNA–protein interaction while the filled arrow head on right highlights the DNA–protein–antibody complex or ‘supershift’.
DISCUSSION
In this work, we explore the RNase 4 and RNase 5/ang 1 gene locus in three species and demonstrate an unusual gene structure consisting of four exons and three introns (Figure 3A). We demonstrate that the shared 5′-UTRs of RNase 4 and RNase 5/ang 1 arise from differential splicing of two shared non-coding exons confirming the results of Strydom (5). We also demonstrate the potential for tissue-specific expression regulated by dual promoters and the presence of tissue-specific expression mediated by at least one unique consensus binding site in the proximal 5′ promoter. Transcription from multiple promoters is not uncommon in the mammalian genome and provides a means for differential production of gene transcripts in response to specific environmental cues. Among some examples, Li et al. (30) described the dual promoters regulating the expression of eosinophil granule major basic protein (MBP) during different stages of eosinophil development. Similarly, CD94, a C-type lectin expressed on NK cells and T-cell subsets, is controlled by dual promoters that differ in their responsiveness to cytokines IL-2 and IL-15 (31) and determine expression in T-cell subsets (32). Interestingly, we have known for quite some time that at least one of the RNase A ribonucleases, RNase 3/eosinophil cationic protein, is expressed uniquely in eosinophils (33) and yet the mechanism underlying this tissue-specific expression has been elusive. Not only is this work the first demonstration of dual promoters among the RNase A ribonucleases, it is likewise the first demonstration of a mechanism underlying tissue-specific expression.
Taken together, our promoter analysis and the RACE/EST data indicate that the distal promoter, Pr1, directs the transcription of RNase 4 and angiogenin utilizing the first non-coding exon (Figure 3B). In this situation, when transcripts for RNase 4 are produced, exon II and exon III (encoding RNase 5/ang 1) will be spliced out or not recognized as exons. The proximal promoter, Pr2 directs liver-specific expression resulting in the splicing of the second of the two non-coding exons with the appropriate coding exon. Again, when RNase 4 transcripts are produced, exon III encoding RNase 5/ang 1 is spliced out or not recognized as an exon.
Pr2 contains a consensus binding site for the transcription factor, HNF-1α, and we demonstrate that point mutations in the consensus sequence reduce promoter activity to baseline levels in luciferase assays performed in vitro in relevant liver cell lines. HNF-1α is a dimerizing homeodomain protein that, together with other transcription factors, coordinates the expression of transcripts encoding hepatocyte proteins (29,34). Originally perceived to be hepatocyte-specific, HNF-1α expression itself is not limited to hepatocytes (35,36). Moreover, specific mutations in HNF-1α result in the genetic syndrome known as maturity onset diabetes of the young (MODY) (29,37), and the targeted HNF-1α gene-deletion likewise has elucidated clear alterations in gene-expression in mouse pancreatic tissue (38–40). The RNase 4 and RNase 5/ang 1 proximal promoter Pr2 also has consensus binding sites for the transcription factors HNF-3 and the CCAAT-enhancer binding protein (C/EBP) family which may coordinate with HNF-1α in promoting gene expression. Clearly, the predominant expression of RNase 4 and RNase 5/ang 1 in the liver tissue may be due to the action of Pr2 in vivo in humans, mice and rats as suggested by the RACE, EST and promoter data shown here.
While other members of the RNase A superfamily have intriguing evolutionary patterns, unusual expression patterns and/or unique protein structure and function, there has been only limited interest in the highly conserved RNase 4 lineage. However, in this work, we demonstrate the rather unusual nature of its regulatory mechanism and demonstrate the first molecular mechanism underlying tissue-specific expression in this gene family. These findings certainly provide greater impetus to study RNase 4 and its specific function vis à vis hepatocytes and liver metabolism. Furthermore, given that RNase 4 and RNase 5/ang 1 are transcribed from the same promoters and can be expressed in concert, equally intriguing is the possibility of coordinate activity of the two gene products (5).
Acknowledgments
The authors would like to thank the members of the Eosinophil Biology Section of LAD/NIAID for their review of this work with a special thanks to Dr Takeaki Nitto for his critical reading of the manuscript. Funding to pay the Open Access publication charges for this article was provided by Division of Intramural Research, NIAID, NIH.
REFERENCES
- 1.Beintema J.J. Introduction: the ribonuclease A superfamily. Cell. Mol. Life Sci. 1998;54:763–765. doi: 10.1007/s000180050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D'Alessio G., Riordan J.F. Ribonucleases: Structure and Functions. San Diego, CA: Academic Press; 1997. [Google Scholar]
- 3.Cho S., Beintema J.J., Zhang J. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories by comparative genomics. Genomics. 2005;85:208–220. doi: 10.1016/j.ygeno.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Beintema J.J., Kleineidam R.G. The ribonuclease A superfamily: general discussion. Cell. Mol. Life Sci. 1998;54:825–832. doi: 10.1007/s000180050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Strydom D.J. The angiogenins. Cell. Mol. Life Sci. 1998;54:811–824. doi: 10.1007/s000180050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hooper L.V., Stappenbeck T.S., Hong C.V., Gordon J.I. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nature Immunol. 2003;4:269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg H.F., Domachowske J.B. Eosinophils, eosinophil ribonucleases, and their role in host defense against respiratory virus pathogens. J. Leukoc. Biol. 2001;70:691–698. [PubMed] [Google Scholar]
- 8.Castella S., Fouchecourt S., Teixeira-Gomes A.P., Vinh J., Belghazi M., Dacheux F., Dacheux J.L. Identification of a member of a new RNase A family specifically secreted by epididymal caput epithelium. Biol. Reprod. 2004;70:319–328. doi: 10.1095/biolreprod.103.022459. [DOI] [PubMed] [Google Scholar]
- 9.Penttinen J., Pujianto D.A., Sipila P., Huhtaniemi I., Poutanen M. Discovery in silico and characterization in vitro of novel genes exclusively expressed in the mouse epididymis. Mol. Endocrinol. 2003;17:2138–2151. doi: 10.1210/me.2003-0008. [DOI] [PubMed] [Google Scholar]
- 10.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 11.Dyer K.D., Nitto T., Moreau J.M., McDevitt A.L., Rosenberg H.F. Identification of a purine-rich intronic enhancer element in the mouse eosinophil-associated ribonuclease 2 (mEar 2) gene. Mamm. Genome. 2004;15:126–134. doi: 10.1007/s00335-003-2304-x. [DOI] [PubMed] [Google Scholar]
- 12.Handen J.S., Rosenberg H.F. Intronic enhancer activity of the eosinophil-derived neurotoxin (RNS2) and eosinophil cationic protein (RNS3) genes is mediated by an NFAT-1 consensus binding sequence. J. Biol. Chem. 1997;272:1665–1669. doi: 10.1074/jbc.272.3.1665. [DOI] [PubMed] [Google Scholar]
- 13.van Dijk T.B., Caldenhoven E., Raaijmakers J.A., Lammers J.W., Koenderman L., de Groot R.P. The role of transcription factor pu.1 in the activity of the intronic enhancer of the eosinophil-derived neurotoxin (RNS2) gene. Blood. 1998;91:2126–2132. [PubMed] [Google Scholar]
- 14.Tiffany H.L., Handen J.S., Rosenberg H.F. Enhanced expression of the eosinophil-derived neurotoxin ribonuclease (RNS2) gene requires interaction between the promoter and intron. J. Biol. Chem. 1996;271:12387–12393. doi: 10.1074/jbc.271.21.12387. [DOI] [PubMed] [Google Scholar]
- 15.Shapiro R., Fett J.W., Strydom D.J., Vallee B.L. Isolation and characterization of a human colon carcinoma-secreted enzyme with pancreatic ribonuclease-like activity. Biochemistry. 1986;25:7255–7264. doi: 10.1021/bi00371a002. [DOI] [PubMed] [Google Scholar]
- 16.Seno M., Futami J., Tsushima Y., Akutagawa K., Kosaka M., Tada H., Yamada H. Molecular cloning and expression of human ribonuclease 4 cDNA. Biochim. Biophys. Acta. 1995;1261:424–426. doi: 10.1016/0167-4781(95)00040-n. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg H.F., Dyer K.D. Human ribonuclease 4 (RNase 4): coding sequence, chromosomal localization and identification of two distinct transcripts in human somatic tissues. Nucleic Acids Res. 1995;23:4290–4295. doi: 10.1093/nar/23.21.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J., Dyer K.D., Rosenberg H.F. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc. Natl Acad. Sci. USA. 2000;97:4701–4706. doi: 10.1073/pnas.080071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J., Zhang Y.P., Rosenberg H.F. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nature Genet. 2002;30:411–415. doi: 10.1038/ng852. [DOI] [PubMed] [Google Scholar]
- 20.Zhao W., Kote-Jarai Z., van Santen Y., Hofsteenge J., Beintema J.J. Ribonucleases from rat and bovine liver: purification, specificity and structural characterization. Biochim. Biophys. Acta. 1998;1384:55–65. doi: 10.1016/s0167-4838(97)00213-6. [DOI] [PubMed] [Google Scholar]
- 21.Ryffel G.U., Kugler W., Wagner U., Kaling M. Liver cell specific gene transcription in vitro: the promoter elements HP1 and TATA box are necessary and sufficient to generate a liver-specific promoter. Nucleic Acids Res. 1989;17:939–953. doi: 10.1093/nar/17.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenberg H.F., Dyer K.D. Eosinophil cationic protein and eosinophil-derived neurotoxin. Evolution of novel function in a primate ribonuclease gene family. J. Biol. Chem. 1995;270:30234. [PubMed] [Google Scholar]
- 23.Slifman N.R., Loegering D.A., McKean D.J., Gleich G.J. Ribonuclease activity associated with human eosinophil-derived neurotoxin and eosinophil cationic protein. J. Immunol. 1986;137:2913–2917. [PubMed] [Google Scholar]
- 24.Kumar S., Tamura K., Jakobsen I.B., Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 25.Dyer K.D., Rosenberg H.F., Zhang J. Isolation, characterization, and evolutionary divergence of mouse RNase6: evidence for unusual evolution in rodents. J. Mol. Evol. 2004 doi: 10.1007/s00239-004-2657-0. in press. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., Dyer K.D., Rosenberg H.F. RNase 8, a novel RNase A superfamily ribonuclease expressed uniquely in placenta. Nucleic Acids Res. 2002;30:1169–1175. doi: 10.1093/nar/30.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vicentini A.M., Hemmings B.A., Hofsteenge J. Residues 36–42 of liver RNase pl3 contribute to its uridine-preferring substrate specificity. Cloning of the cDNA and site-directed mutagenesis studies. Protein Sci. 1994;3:459–466. doi: 10.1002/pro.5560030311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nitto T., Dyer K.D., Mejia R.A., Bystrom J., Wynn T.A., Rosenberg H.F. Characterization of the divergent eosinophil ribonuclease, mEar 6, and its expression in response to Schistosoma mansoni infection in vivo. Genes and Immunity. 2004 doi: 10.1038/sj.gene.6364143. in press. [DOI] [PubMed] [Google Scholar]
- 29.Ryffel G.U. Mutations in the human genes encoding the transcription factors of the hepatocyte nuclear factor (HNF)1 and HNF4 families: functional and pathological consequences. J. Mol. Endocrinol. 2001;27:11–29. doi: 10.1677/jme.0.0270011. [DOI] [PubMed] [Google Scholar]
- 30.Li M.S., Sun L., Satoh T., Fisher L.M., Spry C.J. Human eosinophil major basic protein, a mediator of allergic inflammation, is expressed by alternative splicing from two promoters. Biochem. J. 1995;305:921–927. doi: 10.1042/bj3050921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieto L.D., Borrego F., You C.H., Coligan J.E. Human CD94 gene expression: dual promoters differing in responsiveness to IL-2 or IL-15. J. Immunol. 2003;171:5277–5286. doi: 10.4049/jimmunol.171.10.5277. [DOI] [PubMed] [Google Scholar]
- 32.Wilhelm B.T., Landry J.R., Takei F., Mager D.L. Transcriptional control of murine CD94 gene: differential usage of dual promoters by lymphoid cell types. J. Immunol. 2003;171:4219–4226. doi: 10.4049/jimmunol.171.8.4219. [DOI] [PubMed] [Google Scholar]
- 33.Rosenberg H.F., Ackerman S.J., Tenen D.G. Human eosinophil cationic protein. Molecular cloning of a cytotoxin and helminthotoxin with ribonuclease activity. J. Exp. Med. 1989;170:163–176. doi: 10.1084/jem.170.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendel D.B., Crabtree G.R. HNF-1, a member of a novel class of dimerizing homeodomain proteins. J. Biol. Chem. 1991;266:677–680. [PubMed] [Google Scholar]
- 35.Baumhueter S., Mendel D.B., Conley P.B., Kuo C.J., Turk C., Graves M.K., Edwards C.A., Courtois G., Crabtree G.R. HNF-1 shares three sequence motifs with the POU domain proteins and is identical to LF-B1 and APF. Genes Dev. 1990;4:372–379. doi: 10.1101/gad.4.3.372. [DOI] [PubMed] [Google Scholar]
- 36.Kuo C.J., Conley P.B., Hsieh C.L., Francke U., Crabtree G.R. Molecular cloning, functional expression, and chromosomal localization of mouse hepatocyte nuclear factor 1. Proc. Natl Acad. Sci. USA. 1990;87:9838–9842. doi: 10.1073/pnas.87.24.9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellard S. Hepatocyte nuclear factor 1 alpha (HNF-1 alpha) mutations in maturity-onset diabetes of the young. Hum. Mutat. 2000;16:377–385. doi: 10.1002/1098-1004(200011)16:5<377::AID-HUMU1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 38.Pontoglio M., Sreenan S., Roe M., Pugh W., Ostrega D., Doyen A., Pick A.J., Baldwin A., Velho G., Froguel P., et al. Defective insulin secretion in hepatocyte nuclear factor 1 alpha-deficient mice. J. Clin. Invest. 1998;101:2215–2222. doi: 10.1172/JCI2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y.H., Sauer B., Gonzalez F.J. Laron dwarfism and non-insulin-dependent diabetes mellitus in the HNF-1 alpha knockout mouse. Mol. Cell. Biol. 1998;18:3059–3068. doi: 10.1128/mcb.18.5.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shih D.Q., Screenan S., Munoz K.N., Philipson L., Pontoglio M., Yaniv M., Polonsky K.S., Stoffel M. Loss of HNF-1alpha function in mice leads to abnormal expression of genes involved in pancreatic islet development and metabolism. Diabetes. 2001;50:2472–2480. doi: 10.2337/diabetes.50.11.2472. [DOI] [PubMed] [Google Scholar]
- 41.Hofsteenge J., Moldow C., Vicentini A.M., Zelenko O., Jarai-Kote Z., Neumann U. A single amino acid substitution changes ribonuclease 4 from a uridine-specific to a cytidine-specific enzyme. Biochemistry. 1998;37:9250–9257. doi: 10.1021/bi9803832. [DOI] [PubMed] [Google Scholar]


