Abstract
Despite the loss of sequence-specific DNA binding, mutant p53 (mutp53) proteins can induce or repress transcription of mutp53-specific target genes. To date, the molecular basis for transcriptional modulation by mutp53 is not understood, but increasing evidence points to the possibility that specific interactions of mutp53 with DNA play an important role. So far, the lack of a common denominator for mutp53 DNA binding, i.e. the existence of common sequence elements, has hampered further characterization of mutp53 DNA binding. Emanating from our previous discovery that DNA structure is an important determinant of wild-type p53 (wtp53) DNA binding, we analyzed the binding of various mutp53 proteins to oligonucleotides mimicking non-B DNA structures. Using various DNA-binding assays we show that mutp53 proteins bind selectively and with high affinity to non-B DNA. In contrast to sequence-specific and DNA structure-dependent binding of wtp53, mutp53 DNA binding to non-B DNA is solely dependent on the stereo-specific configuration of the DNA, and not on DNA sequence. We propose that DNA structure-selective binding of mutp53 proteins is the basis for the well-documented interaction of mutp53 with MAR elements and for transcriptional activities mediates by mutp53.
INTRODUCTION
Sequence-specific transactivation underlies the growth suppressing and apoptotic functions of wild-type p53 (wtp53) (1,2). Substitution of a single amino acid residue within the central DNA-binding domain (DBD) (the characteristic feature of the p53 mutational spectrum) affects sequence-specific DNA binding (SSDB). Loss of p53-SSDB and impaired ability to elicit the same transcriptional response as wtp53 are the hallmarks of mutant p53 (mutp53) proteins, which are generally considered to be transcriptionally inactive. However, the identification of genes that can be induced specifically by mutp53, but not by wtp53 (3), indicates that mutp53 proteins may not necessarily be disqualified as transcriptional activators. The functional spectrum of the so far known mutp53-responsive genes strongly suggests that some mutp53 proteins target a set of genes that is different from those controlled by wtp53. Furthermore, transactivation by mutp53 not only appears to be target gene-specific, but also mutp53-specific to date, as promoters regulated by a specific mutp53 protein may not be responsive to other p53 mutants (4). In addition, the cellular context seems to be important for mutp53 target gene specificity. So far, neither the mechanism of mutp53 transactivation nor how the specificity of mutp53 transactivation is achieved is known. Although modulation of transcription by mutp53 proteins, similar to wtp53, can occur via protein–protein interactions (4–9), the ability of mutp53 proteins to regulate transcription by directly binding to DNA is a possibility (10–12). The assumption is consistent with the requirement of the central DNA binding and of the regulatory C-terminal domains for transcriptional activity of mutp53 (13,14) in addition to N-terminal transactivation domains (15).
The identification of parameters important for mutp53 interaction with DNA is of paramount importance in light of the possibility that the ability to activate transcription by direct binding to mutp53-regulated promoters may underlie the cancer-promoting effects of mutp53 proteins (16–18). DNA binding of mutp53 proteins can be supported either by partially retained SSDB (19,20) or/and by the acquisition of novel DNA-binding activities that appear specific yet distinct from SSDB (10,12,21). However, the lack of a common denominator, such as the presence of a specific sequence-motif in the DNA that is recognized by mutp53 proteins, poses a major difficulty in delineating the parameters that determine specificity of mutp53 DNA binding. Several cognate motifs identified as putative binding sites for different mutp53 proteins exhibit no sequence similarity, suggesting that sequence-specific DNA recognition may not be the mode determining DNA binding of mutp53.
An important consideration in delineating the parameters of mutp53 DNA binding is that this activity most probably derives from DNA-binding activities inherent to wtp53. Wtp53 binds DNA by various modes that can be formally divided into SSDB and non-SSDB. Non-SSDB of wtp53 includes high-affinity binding to double-stranded DNA (22) and single-stranded DNA ends (23), secondary DNA structures such as Holliday junctions (24), t-loop junctions in telomeric DNA (25), cruciforms (26), and to aberrant sites in DNA such as mismatched bases and DNA bulges (27,28). It has been proposed that the various modes of DNA binding are associated with different activities of wtp53, with SSDB-mediating p53 transcriptional control, mismatch/bulged DNA binding being associated with p53 activities in DNA repair or recombination, and binding to single-stranded DNA ends implicated as an initial step in DNA strand transfer (29). However, it appears that the various types of DNA binding are not only specific for certain activities of wtp53, but also contribute to its major activity—SSDB and transcriptional activation [reviewed in (30)]. Wtp53-SSDB can occur in different modes depending on the conformation of p53-binding sites, either sequence-specific to linear (B-) DNA, or sequence- and structure-specific to non-B DNA (31–36). In contrast to SSDB to linear DNA, which most probably is mediated solely by the p53 core DBD (37,38), sequence-specific and DNA structure-dependent SSDB to non-linear DNA and non-SSDB modes of DNA interaction involve both the DBD and the CTD (C-terminal domain) (33,34,39,40). The p53 CTD, which has been previously implicated exclusively in ‘unspecific’ DNA binding, appears to be an important constituent of SSDB as it stabilizes sequence-specific binding of the p53 DBD to wtp53-cognate motifs when they adopt a non-B DNA conformation (33). Supporting the notion, the potentiating effects of the CTD were revealed when wtp53-SSDB and transactivation was examined under conditions more closely resembling p53 interaction with chromatin (40). The complex picture emerging from these findings reveals that not only the various DNA-binding activities of the p53 core domain, but also the DNA-binding activity of the p53 CTD has strong impact on wtp53-SSDB.
The changing picture of wtp53-SSDB also puts the issue of mutp53-specific DNA binding in a new perspective. Our laboratory has previously analyzed DNA binding of mutp53 proteins in some detail (41–44). The complex interactions of mutp53 with DNA were shown to require both the mutated p53 DBD and the intact p53 CTD (43). As mutp53 proteins have not only lost the wtp53-SSDB activity, but are also impaired for high-affinity binding to unspecific linear DNA, an activity which in wtp53 is mediated by the DBD and the CTD [reviewed in (45)]; unspecific binding to linear DNA is unlikely to underlie the interactions of mutp53 with DNA. The conclusion is compatible with our previous findings that the DNA sequences bound by mutp53 in vitro and in vivo predominantly contain repetitive sequences (44) with a high propensity to undergo structural transitions.
As DNA conformation plays an important role in the DNA binding of wtp53 and mutp53, and as DNA structure-dependent binding of wtp53 and mutp53 requires the CTD, which is intact in most of the frequently encountered mutp53 proteins, we investigated whether DNA binding of mutp53 proteins may be determined by the recognition of DNA structure. We analyzed DNA binding of different mutp53 proteins to conformationally distinct forms of DNA by using electromobility shift assay (EMSA), confocal fluorescence lifetime analysis (cFLA) and a novel p53 protein array-based DNA-binding assay. Our results demonstrate that ‘hot spot’ mutp53 proteins, while having lost sequence-specific DNA recognition, have retained the potential to bind non-linear DNA in a DNA structure-dependent manner. DNA structure-dependent binding of mutp53 does not require the presence of specific sequence motifs. Nevertheless, DNA binding of mutp53 is not unspecific, as it is highly selective for secondary DNA structures that fulfill specific structural criteria. Therefore, we termed the mode of mutp53 DNA binding as ‘DNA structure-selective binding’ (DSSB) and propose that it has important implications for the activities elicited by mutp53 in cancer cells.
MATERIALS AND METHODS
Protein purification and EMSA
Recombinant p53 proteins expressed in insect cells were isolated as described previously (33) and purified by ion-exchange chromatography (FPLC; Amersham Bioscience). DNA-binding experiments were performed using 50 ng or the indicated amounts of recombinant p53 proteins in a reaction mixture containing 5 ng of poly(dI:dC) (Amersham Bioscience) and 2 μg of BSA in 50 mM Tris–HCl (pH 7.5), 0.1 mM EDTA, 1 mM dithiothreitol (DTT) and 50 mM NaCl. After a 20 min preincubation at room temperature, 2 ng of the radioactively labeled DNA probe (20–30 000 c.p.m.) was added, and the incubation was continued for another 25 min. Samples were loaded onto a 4% native polyacrylamide gel and separated by electrophoresis in 10 mM Tris–HCl (pH 7.8), 0.2 mM EDTA, 1.25 mM NaOAc and 8 mM acetic acid at 200 V for 2.5 h at room temperature. After electrophoresis, the gels were dried and subjected to autoradiography.
Preparation of p53-binding substrates in linear or non-linear DNA conformation
DNA probes used in EMSA were prepared from synthetic oligonucleotides listed in Table 1. p53-binding sites presented in linear or non-linear DNA conformation were prepared as described previously (33). Briefly, short radiolabeled oligonucleotide T3-1 lacking the p53-specific sequence was annealed with long unlabeled oligonucleotides p21stem–loop or SCRstem–loop to obtain specific or unspecific stem–loop structures, respectively. Resulting stem–loop structures were separated from non-annealed single-stranded oligonucleotides by electrophoresis in 8% native polyacrylamide gels and purified after elution. Linear DNA was prepared by conventional annealing of complementary oligonucleotides p21lin/s and p21lin/as of which one was radioactively labeled. Double-stranded linear DNA was eluted and purified after separation in 8% native polyacrylamide gels. A four-way junction DNA was prepared as described in the original paper by Duckett et al. (46) by hybridizing four oligonucleotides (b-, h-, r- and x-strand) of which one was radioactively labeled at the 5′ end. The resulting four-way junction structure was purified using Sephadex G-25 columns.
Table 1.
Oligonucleotides used in the study
| Oligonucleotides | Sequence | DNA substrate | Figure |
|---|---|---|---|
| T3-1 | 5′-ccgcggtaccattacctaaggcgtc-3′ | Linear DNAunspec | 1 and 4 |
| T3-2 | 5′-gacgccttaggtaatggtaccgcgg-3′ | ||
| b-strand | 5′-tccgtcctagcaagccgctgctaccggaag-3′ | Four-way junctiona | |
| h-strand | 5′-cttccggtagcagcgagagcggtggttgaa-3′ | 1 and 2 | |
| r-strand | 5′-ttcaaccaccgctcttctcaactgcagtct-3′ | ||
| x-strand | 5′-agactgcagttgagagcttgctaggacgga-3′ | ||
| T3-1 | 5′-ccgcggtaccat–tacctaaggcgtc-3′ | Stem–loop DNAunspec | 2–4 |
| SCRstem–loop | 5′-gacgccttaggta–cctgccctcgctcgctcacc–gtgcgctggctggatggtaccgcgg-3′ | ||
| T3-1 | 5′-ccgcggtaccat–tacctaaggcgtc-3′ | Stem–loop DNAunspec2 | 3 |
| SCRstem–loop2 | 5′-gacgccttaggta–ccagcctccgcacgctcacc–gagcgctggcagg–atggtaccgcgg-3′ | ||
| T3-1 | 5′-ccgcggtaccat–tacctaaggcgtc-3′ | Stem–loop DNAspecb | 3–5 |
| p21stem–loop | 5′-gacgccttaggta–cctgccGAACATGTCCCAACATGTTGggcctg–atggtaccgcgg-3′ | ||
| T3-1 | 5′-ccgcggtaccat–tacctaaggcgtc-3′ | Stem–loop DNACONb | 3C |
| CON-loop | 5′-gacgccttaggta–cctggcctgcctGGACTTGCCTggcctgcctgg–atggtaccgcgg-3′ | ||
| T3-1 | 5′-ccgcggtaccat–tacctaaggcgtc-3′ | Stem–loop DNACON-2b | 3C |
| CON-2 | 5′-gacgccttaggta–cctggcctgccaGGACTTGCCTggcaggccagg–atggtaccgcgg-3′ | ||
| p21lin/s | 5′-gctctgccGAACATGTCCCAACATGTTGccgctctg-3′ | Linear DNAspec | 5 |
| p21lin/as | 5′-cagagcggCAACATGTTGGGACATGTTCggcagagc-3′ |
aSequences and nomenclature of oligonucleotides used for the preparation of four-way junction DNA were adopted from the original study by Duckett et al. (46).
bSequence corresponding to the p53 binding site from the p21 promoter or to the p53 consensus are shown in upper case letters. Underlined sequences correspond to complementary regions. Regions forming the stem of stem–loop structures are shown by ‘–’.
DNA binding using Mut p53 express array
Mutant p53 protein arrays containing 49 recombinant p53 proteins (200 pg of each protein spotted in duplicate) were purchased from SenseProteomics (UK). DNA-binding experiments were performed according to the manufacturer's recommendations. Briefly, arrays were washed three times with 2 ml of DNA-binding buffer (25 mM HEPES, pH 7.4, 50 mM KCl, 1 mg/ml BSA, 20% glycerol, 0.1% Triton X-100 and 1 mM DTT) for 5 min. After washing, membranes were incubated with purified radiolabeled DNA in 2 ml of the assay buffer for 30 min at room temperature. Unbound DNA was removed by three washing steps (5 min each) with 2 ml of DNA-binding buffer. In competition assays, the binding step was followed by 30 min incubation at room temperature with unlabeled DNAspec that contains p53-cognate motif in linear conformation at a molar ratio of 1:1 and 1:5 of labeled DNA probe/unlabeled competitor DNA. In experiments addressing the influence of p53 antibodies on DNA binding, the arrays were first incubated with 4 μg of purified PAb421 antibody for 30 min at room temperature, followed by the addition of DNA. In all the experiments p53-bound DNA was detected by autoradiography.
DNA binding by cFLA
All measurements were performed with a FCS+plus research reader (Evotec Technologies, Hamburg). The working principal of cFLA and the mathematics underlying the quantification of read-outs has been described elsewhere (47,48). In brief, the sample is optically excited by a fast pulsed laser system (Lynx-VAN-532; Time-Bandwidth, Switzerland) running at 80 MHz repetition rate and an average power of ∼5 mW. The pulse width is <350 ps and the pulse-to-pulse distance is <20 ns. The laser beam passes through an objective lens and is focused onto the sample. The fluorescence light is collected by the same objective lens and separated from the excitation light by suitable optical filters. A photon counting device, i.e. an avalanche photodiode (APD) (SPCM-AQ-131; EG&G Optoelectronics, Vaudreuil, Quebec, Canada), records the emitted fluorescence light. The electronic data processing is performed by a high time resolution acquisition electronics based on Time-to-Digital-Converters (TDCs). The TDCs thereby measure and digitize the temporal distances between the arrival times of detected fluorescence photons and the last excitational laser pulse.
In vitro ubiquitination assay
Insect cells expressing recombinant GST-Mdm2 protein (49) were suspended in PBS and briefly sonicated. The supernatant was centrifuged at 15 000 g for 30 min and applied to the glutathione–Sepharose 4B (Pharmacia) column. GST-Mdm2 was eluted with 10 mM reduced glutathione and 50 mM Tris–HCl (pH 8.0) and analyzed using western blot with anti-MDM2 antibody Ab2A10. An aliquot of 50 ng of recombinant p53 protein was incubated with increasing amounts of DNA in 50 mM Tris–HCl (pH 7.5), 0.1 mM EDTA, 1 mM DTT, 50 mM NaCl, 2 μg BSA and 5 ng poly(dI−dC) for 20 min. After preincubation, 150 ng GST-Mdm2, 250 ng E1 (Calbiochem), 500 ng UbcH5 (Calbiochem) and 15 μg ubiquitin (Sigma) were added. Ubiquitination was performed in a reaction mixture (40 μl) containing 50 mM Tris–HCl (pH 7.4), 5 mM MgCl2, 2 mM ATP and 2 mM DTT for 90 min at 25°C. Samples were resolved by 10% SDS–PAGE and p53 was detected by immunoblotting using anti-p53 antibody DO1.
RESULTS
Mutp53 proteins bind preferentially to non-linear DNA
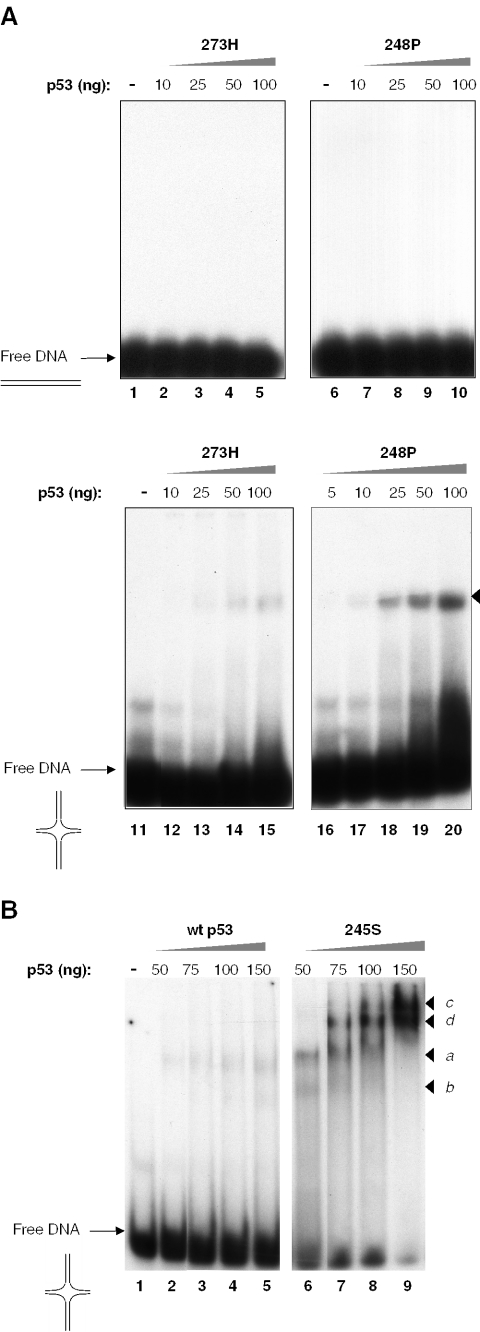
To test whether DNA topology may be a relevant parameter for mutp53 DNA binding, we used EMSA to examine the binding of mutp53 proteins to DNA substrates, comprising four-way junction DNA or stem–loop structures, which are the two types of secondary DNA structures to which wtp53 can bind with high affinity (24,33). Figure 1A shows that mutp53 proteins, 273H, 248P and 245S, while being unable to bind linear double-stranded DNA (shown for mutp53 proteins 273H and 248P, lanes 1–10), did bind to a DNA structure resembling a four-way junction in Holliday structures (Figure 1A and B, lanes 11–20 and lanes 6–9, respectively). Wtp53 also bound four-way junction DNA albeit rather weakly (Figure 1B, lanes 1–5). The binding of wtp53 and different mutp53 proteins to four-way junction DNA exhibited marked quantitative differences. The apparent binding affinity was higher for 248P and 245S mutants compared with 273H mutant [compare lanes 16–20 in Figure 1A and 6–9 in Figure 1B with lanes 11–15 in Figure 1A, respectively], which bound weakly, with an efficiency comparable with that of wtp53 (compare lanes 11–15 in Figure 1A with lanes 1–5 in Figure 1B). Furthermore, mutp53 proteins also exhibited different patterns of binding. Mutp53 273H and mutp53 248P formed one major complex with four-way junction DNA, which increased correspondingly with protein input (Figure 1A, lanes 11–20). A different pattern was observed with mutp53 245S: two major complexes a and b were formed at low concentrations of the 245S protein (Figure 1B, lane 6). Further increasing the protein input led to the appearance of slower migrating complexes c and d (lanes 7 and 8, complexes indicated by arrowheads), which paralleled the concomitant decrease of complexes a and b that eventually disappeared at higher 245S concentrations (lanes 7–9 in Figure 1B). Complexes a, b, c and d could be further supershifted by the p53-specific antibody DO-1 confirming that they were formed by mutp53 245S p53 (data not shown). The binding pattern exhibited by mutp53 245S at low concentrations qualitatively resembled that of wtp53, which also formed two complexes with four-way junction DNA at high concentrations, albeit very weakly (Figure 1B, lane 5), in line with the much lower capacity of wtp53 to bind four-way junction DNA compared with mutp53 245S. Although the precise composition of complexes a, b, c and d is unclear, the pattern suggests a highly co-operative mode of binding with the slowly migrating complexes representing higher order oligomeric forms of G245S. Interestingly, mutp53 proteins 273H and 248P formed only one complex and did not form higher order oligomeric forms with four-way junction DNA even at high concentrations (Figure 1A, lower panel) indicating that different p53 mutants bind four-way junction DNA by different modes.
Figure 1.
Analysis of mutp53 DNA binding by EMSA. (A) Hot-spot p53 mutp53 proteins 273H, 245S and 248P were incubated with linear DNAunspec lacking a p53-specific cognate motif (upper panel). Binding of mutp53 proteins 273H and 248P to a four-way junction structure (lower panel). Four-way junction structure was prepared as described in Materials and Methods by annealing four intercomplementary oligonucleotides b, h, r and x (the resulting structure is shown schematically), of which one was radioactively labeled. Arrowheads indicate p53 complexes formed with different types of DNA probes, as shown by the corresponding symbols in this and in other figures. (B) Binding of wtp53 and 245P proteins to a four-way junction structure.
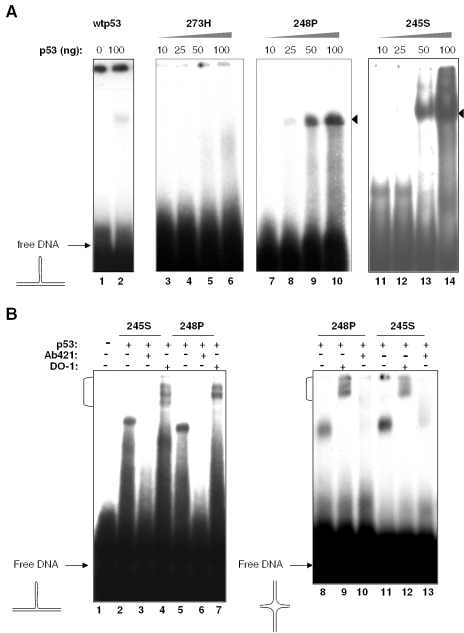
We next examined mutp53 binding to stem–loop DNA, which represent another type of secondary DNA structures bound by wtp53 (33). Confirming our previous findings, wtp53 bound weakly to stem–loop structures formed by unspecific (i.e. lacking the p53 consensus) stem–loop DNAunspec (Figure 2A). In contrast, mutp53 proteins 248P and 245S bound strongly to stem–loop DNAunspec (Figure 2A, compare lanes 7–10 and 11–14, respectively). Of note, the complex formed by mutp53 with stem–loop migrated with a mobility similar to that of the complex formed by wtp53 (Figure 2A, compare the complex in lane 2 with the complex in lanes 9, 10, 13 and 14). Since wtp53 binds stem–loop as a tetramer (33), we conclude from this observation that mutp53 also binds stem–loop structure as tetramer. Interestingly, only very little higher order protein/DNA complexes were formed between mutp53 245S and stem–loop DNA, underscoring the notion that this protein binds stem–loop and four-way junction DNA in different modes. In contrast to 245S and 248P proteins, mutant 273H did not form stable complexes with stem–loop DNAunspec even at high concentrations (Figure 2A, lanes 3–6).
Figure 2.
(A) Binding of mutp53 proteins 273H, 245S and 248P to a stem–loop structure formed by unspecific DNA (stem–loop DNAunspec). (B) Effects of p53-specific antibodies PAb421 and DO-1 on mutp53 interaction with non-linear DNA structures. An aliquot of 50 ng of p53 proteins were incubated with DNA in the presence or absence of 200 ng of the purified antibody.
SSDB of wtp53 to non-linear DNA requires the C-terminal domain and is strongly inhibited by the C-terminal antibody PAb421 (31–34). Examination of the effects of p53 specific antibodies revealed that PAb421, but not the N-terminal antibody DO-1, strongly inhibited the binding to stem–loop DNA (Figure 2B, lanes 1–7) as well as to four-way junction DNA (lanes 8–13). The inhibitory effect of PAb421 indicates that the C-terminus is involved in non-linear DNA binding of mutp53. Significantly, incubation of all mutp53 proteins with stem–loop DNAunspec led to the appearance of a smeary tail (lanes 6, 10 and 14 in Figure 2A and lanes 2 and 5 in Figure 2B), which may be due to the dissociation of unstable protein/DNA complex during electrophoresis or due to the DNA structure disturbing effects caused by p53 binding. Interestingly, we repeatedly observed that the addition of DO-1 led to the appearance of several supershifted bands (indicated by parentheses in Figure 2B). One explanation of such effects of DO-1 may be that not all p53 molecules may be equally accessible for antibody binding in the mutp53 tetramer bound to non-linear DNA. Alternatively, it is possible that DO-1 may have promoted the formation of high-order p53 oligomers on non-linear DNA.
The local structure of non-linear DNA, not the presence of specific sequence motifs determines the selectivity of mutp53 DNA binding
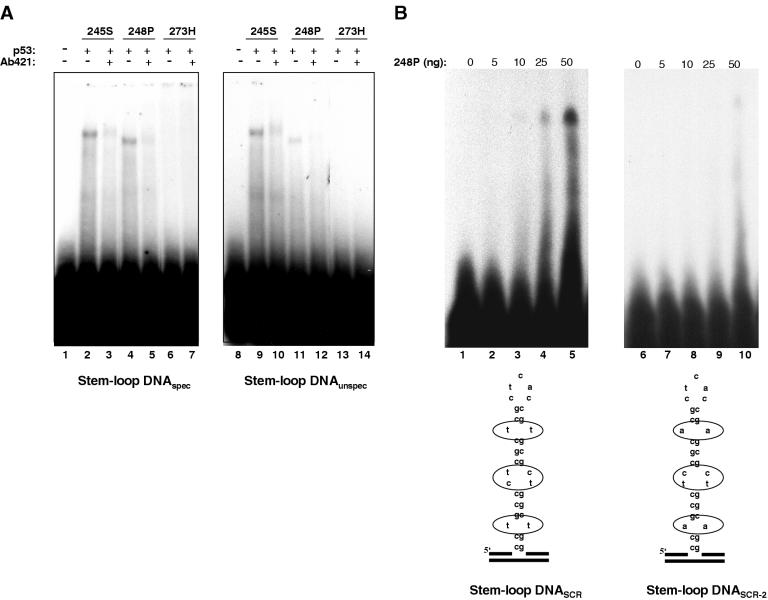
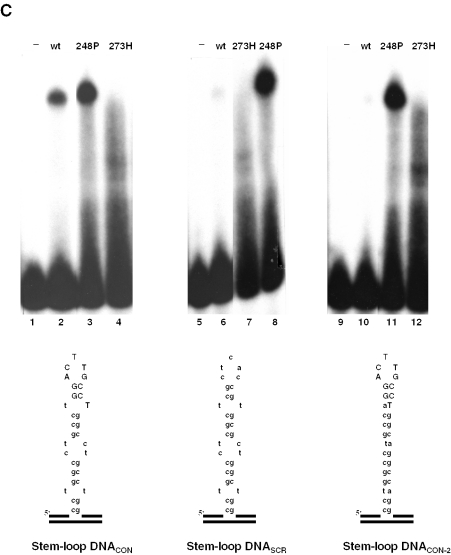
High-affinity SSDB of wtp53 is dependent on two crucial parameters, the presence of the p53-specific sequence and the appropriate conformation of the DNA (33,34). In contrast, mutp53 proteins bind to non-B DNA also in the absence of the consensus sequence (Figure 2A). Indeed, as shown in Figure 3A, mutp53 proteins 245S and 248P bound stem–loop DNAspec containing p53-specific sequence (lanes 1–7) and stem–loop DNAunspec (lanes 8 and 9) with comparable efficiencies, indicating that mutp53 DNA binding is not determined by a specific sequence. Whereas the presence or absence of a p53 consensus sequence had no significant impact on mutp53 DNA binding, the base composition of structural elements in the stem–loop DNA, such as mismatches or bulges, strongly influenced the efficiency of binding (Figure 3B, shown for mutp53 248P). We reckon that the most probably explanation for the strong influence of individual bases on mutp53 binding to non-linear DNA is a different 3D configuration of the secondary DNA structure, which greatly depends on the base composition. Therefore, we conclude that non-linearity of DNA as such is not sufficient to qualify for mutp53 DNA binding and that stereo-specific criteria have to be fulfilled to promote mutp53 binding to secondary DNA structures. Intriguingly, stem–loop structures that either lack or contain mismatched bases were bound with comparable efficiency by mutp53 proteins 248P (Figure 3C, lanes 3 and 11) and 245S (data not shown). The ability of mutp53 to bind stem–loop structures lacking mismatched bases is in striking contrast to the requirement of mismatched bases for efficient stem–loop binding by wtp53 (Figure 3C, compare lanes 2 and 10), as reported previously (33). These results indicate that the potential of mutp53 to bind non-linear DNA not only is retained in mutp53 proteins, but also can be altered by mutations in the central DBD. As a consequence, a different spectrum of DNA structures can be bound by mutp53 proteins compared with that of wtp53.
Figure 3.
Impact of the p53-specific sequence-motif and of mismatched bases on DNA binding of mutp53. (A) Stem–loop structures either containing (stem–loop DNAspec) or lacking (stem–loop DNAunspec) a p53-binding site from the p21 promoter were incubated with mutp53 proteins in the presence or the absence of PAb421. (B) Binding of 248P mutant to stem–loop structures lacking a p53-specific cognate motif and differing in the composition of non-matching bases within the stem (encircled bases). (C) Mutp53 and wtp53 bind differently to stem–loop structure lacking mismatched bases in the stem.
Quantitative analysis of mutp53 binding to non-linear DNA
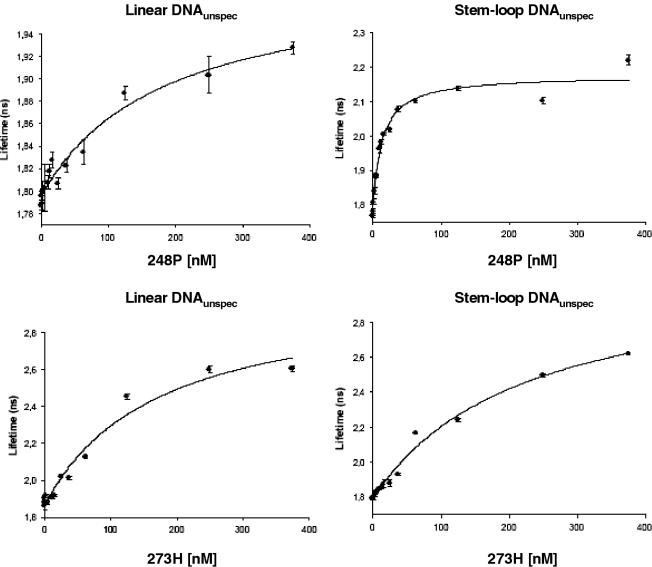
Our EMSA experiments suggested that mutp53 proteins 245S and 248P bind non-linear DNA with high affinity, whereas mutp53 273H appeared to be a weak binder. We evaluated the binding affinities of mutp53 proteins 273H, 245S and 248P with different types of DNA, using cFLA. Fluorescence lifetime represents an intrinsic molecular property of the fluorophore and is able to detect minute changes in the fluorophore's direct environment, like binding processes. In brief, cFLA is explained as follows: using an objective lens, the pulsed laser light emitting at a wavelength of 532 nm is focused onto a sample, which illuminates a volume of ∼1 fl (with a diameter of ∼1 μm). The fluorescence light is collected by the same objective lens and separated from the excitation light by suitable optical filters. A photon counting device, i.e. an APD, records the emitted fluorescence light. The electronic data processing is performed by a high time resolution acquisition electronics based on TDC. The TDCs thereby measure and digitize the temporal distances between the arrival times of detected fluorescence photons and the last excitational laser pulse. From the recorded data, the lifetime from the bound and unbound state of the used samples can be determined and quantified. The binding curves for mutp53 248P and 273H are shown in Figure 4 and visualize the differences in binding affinities between mutp53 248P and mutp53 273H in binding to stem–loop DNAunspec, and in binding to linear DNAunspec. The read-outs of the cFLA experiments are summarized in Table 2 and show that mutp53 proteins 248P and 245S bind stem–loop DNAunspec with high affinity (11.2 ± 5.0 and 40.2 ± 6.0 nM, respectively), which is comparable with that determined for SSDB of wtp53 to stem–loop DNAspec (35.9 ± 5.1 nM). In contrast, binding of mutp53 273H was significantly weaker (248 ± 70.3 nM). Furthermore, confirming the results of our EMSA experiments, all mutp53 proteins exhibited a low affinity toward linear DNA (Table 2), which is in contrast to wtp53 that binds linear DNA with high affinity (22). Thus, the results of the cFLA studies fully support the conclusions drawn from our EMSA experiments, and show that mutp53 proteins 245S and 248P bind to non-linear DNA with high affinity in a DNA structure-selective mode.
Figure 4.
Graphic representation showing typical results of mut p53 DNA-binding analyses using cFLA (shown for 273H and 248P). The results of all the cFLA experiments are summarized in Table 2.
Table 2.
DNA-binding affinities determined by cFLA
| Protein | KD (nM) | DNA | Number of experiments |
|---|---|---|---|
| 273H | 155 ± 15 | Linear DNAunspec | 3 |
| 248P | 159 ± 32 | — | 3 |
| 245S | 464 ± 104 | — | 2 |
| 273H | 248 ± 70 | Stem–loop DNAunspec | 3 |
| 248P | 11.2 ± 5.0 | — | 4 |
| 245S | 40 ± 6.0 | — | 2 |
Structure-selective DNA binding is a common feature of many mutp53 proteins
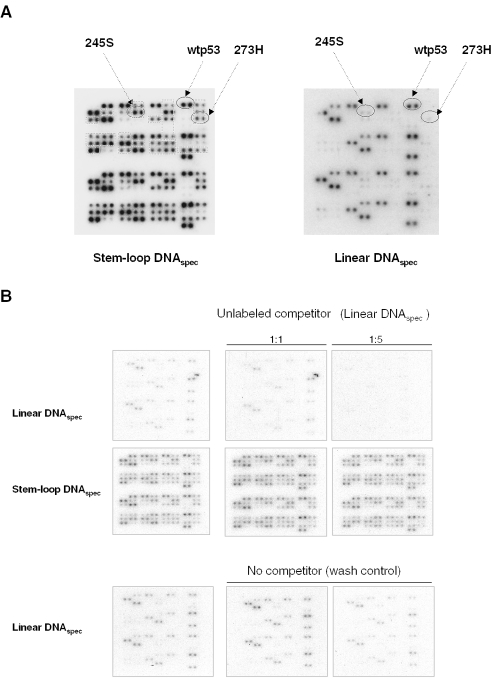
We next investigated whether non-linear DNA binding may be a peculiar feature of the G245S and R248P mutp53 proteins, or whether it may be a more general feature of mutp53 proteins. Since only a limited number of recombinant mutp53 proteins were at our disposal, we took advantage of a mutp53 protein array that contains 49 different mutp53 proteins and allows simultaneous on-array monitoring of DNA binding (graphic representation of the mutp53 protein array and on-array immunodetection of p53 proteins with PAb240 and PAb421 are shown in Supplementary Figures 1A and B, respectively). We examined DNA binding of mutp53 proteins to radioactively labeled DNA in stem–loop or in linear conformation. Since linear as well as non-linear DNA binding of wtp53 is dependent on the presence of a p53 consensus sequence (33,34), we used DNAspec containing a p53 consensus sequence, allowing us to compare non-linear DNA binding of mutp53 with that of wtp53, which is also present on the array. The arrays were incubated with radioactively labeled DNAspec in stem–loop or linear DNA conformation, and bound DNA was detected after repeated cycles of washing as described in Materials and Methods. While very few non-hot spot mutp53 proteins with mutations outside the DBD bound both stem–loop DNAspec and linear DNAspec, none of the hot-spot mutp53 proteins present on the array appreciably bound linear DNA (Figure 5A). In contrast, most mutp53 proteins, including all hot-spot mutants, did bind strongly to stem–loop DNA (data summarized in Table 3), indicating that non-linear DNA is a preferred binding substrate for the majority of mutp53 proteins. In this assay, also mutp53 273H bound to stem–loop DNA, which is in apparent contrast to our EMSA and cFLA data. Although we cannot rule out the possibility that the difference is due to the set-up of the assay, we consider it likely that it is due to the different (PAb240-positive) conformation of the bacterially expressed mutp53 273H protein as compared with the PAb240-non-reactive form, which is the predominant conformation of 273H expressed in insect cells. As PAb240-positivity reflects the accessibility of an epitope in the p53 DBD, the ability of 273H in a PAb240-reactive form to bind non-linear DNA further supports the notion that the p53 DBD is involved in non-linear DNA binding. The efficiency of non-linear DNA binding by mutp53 proteins correlated directly with their reactivity to PAb240, suggesting that an ‘open’ DBD in mutp53 proteins may promote binding to non-linear DNA. Supporting our cFLA results, competition experiments demonstrated that non-linear DNA binding of mutp53 proteins is a high affinity interaction, as an excess of unlabeled linear double-stranded DNA, while effectively displacing radioactive linear DNA from the complex with mutp53 (arrays shown in the upper panel of Figure 5B), had only a marginal effect on stem–loop DNA binding (Figure 5B, middle panel). The specificity of binding was further confirmed by the finding that PAb421 significantly inhibited binding of stem–loop DNAspec, as was the case in our EMSA experiments (Supplementary Figure 1C). Altogether, the results demonstrate that DNA structure-selective binding is a property inherent to many mutp53 proteins, which bind preferentially and with high affinity to non-linear DNA.
Figure 5.
Analyses of DNA binding by using mutp53 protein arrays. (A) Autoradiograph of radioactively labeled DNAspec bound to p53 proteins. Mutp53 arrays were incubated with DNAspec present either in linear or in stem–loop conformation, washed and exposed to X-ray film for autoradiographic detection, amplified with the aid of an intensifying screen at −70°C. Positions of wtp53 and mutp53 proteins 273H and 245S that were analyzed also by other assays are indicated by circles. Dotted lines indicate exclusive binding of stem–loop DNAspec by those mutants that failed to bind linear DNAspec. The results of binding experiments are summarized in Table 3, in which different mutp53 proteins have been grouped according to their ability to bind non-linear DNA. The apparent binding affinity was evaluated by densitometry whereby the intensity of the signal produced by p53-306X mutant lacking the oligomerization and the C-terminal domains was considered as background (designated as ‘−’ in Table 3). To score the binding to linear DNA, intensities that were equal to or consisted at least 50% of the value corresponding to wtp53 binding were designated as ‘++’. For stem–loop DNA binding, intensities higher than at least 10% of the value obtained for the same protein with linear DNA were designated as ‘+++’. (B) DNA competition assay. Arrays were first incubated with linear DNAspec (upper panel) or with stem–loop DNAspec (lower panel), and bound DNA was detected using autoradiography. The much weaker signal compared with images shown in (A) is due to more gentle conditions of autoradiography (shorter exposure time at room temperature under moist conditions) that were used for the sake of preserving protein–DNA complex. After the documentation of DNA binding, the p53–DNA complex were challenged by two sequential rounds of competition with a 1:1 and 1:5 molar excess of unlabeled linear DNAspec. The lower panel shows a control array that was treated under identical conditions except that the competitor DNA was omitted.
Table 3.
DNA-binding patterns of wt and 49 mutp53 proteins
| Protein | Linear DNAspec | Stem–loop DNAspec |
|---|---|---|
| wtp53 | ++ | +++ |
| 273H; 245S; 273C; 245C; 245D; 248W; 248Q; 219S; 220C; 233D; 235D; 241F; 252P; 256I; 257Q; 265P; 266A; 272L; 278L; 280K; 133T; 152L; 141Y; 151S;154V; 175H; 180K; 193R;82L | − | +++ |
| 181H; 82L; 23G; 23A; 72P; 181C; 227T; 306P; 392A | ++ | +++ |
| Δ196; Δ209; Δ213; Δ306; 235S; 251M; 258K; 344P; 337C | − | − |
DISCUSSION
We demonstrate in this study that mutp53 proteins bind specifically and selectively to DNA in a non-linear DNA conformation, and that binding is determined by recognition of DNA topology. Therefore, we termed this mode of DNA binding as DSSB. In contrast to the high-affinity binding of wtp53 to non-linear DNA, which requires the presence of a p53-specific sequence motif (33,34), high-affinity DSSB of mutp53 is determined solely by a favorable stereo-specific DNA conformation (this study). In addition to having lost the ability to bind DNA sequence specifically, mutp53 proteins are also impaired for high-affinity interaction with linear DNA (22) (Figure 1A). High-affinity DNA binding of mutp53-DSSB, thus is restricted to non-linear DNA. In contrast to the previous view that the mutp53 proteins are either DNA binding inactive or bind to DNA in an unspecific manner, the striking selectivity toward non-linear DNA, and the requirement for a stereo-specific DNA conformation provide mutp53 proteins with features of DNA structure-specific DNA-binding proteins.
Despite the distinct modes of DNA recognition, some mechanistic aspects are similar in mutp53-DSSB and in wtp53-SSDB. Both, mutp53-DSSB and wtp53-SSDB require the p53 DBD and the CTD for high-affinity binding. The CTD is important for mediating stable complex formation of p53 with non-linear DNA in mutp53-DSSB (this study) and in wtp53-SSDB (33,34,40). However, the possibility that the C-terminus might impact non-linear DNA binding of mutp53 by supporting the appropriate quarternary structure of mutp53 proteins should also be considered. In fact, our on-array binding analyses showing that none of the oligomerization-deficient mutp53 proteins was capable of binding to DNA is in accordance with the idea. While the CTD is a major regulator of mutp53-DSSB, the core DBD appears to play an important role in the specificity of mutp53-DSSB. Underscoring the importance of non-linear DNA binding as a major mode of interaction of mutp53 with DNA, we found that mutants with an ‘open’, i.e. PAb40-positive DBD also bind better to non-linear DNA. The requirement of an open DBD for non-linear DNA binding would explain why DNA contact mutants such as 273H bound less efficiently than conformational p53 mutants to the DNA substrates analyzed here. Another possibility is that the differing affinity of mutp53 proteins may be due to the fact that the ‘designed’ DNA structures used here, while better suited for binding of some (245S and 248P) mutp53 proteins, may be poorly compatible with binding of other mutants such as 273H. Again, such mutation-dependent selectivity toward different DNA structures would not be surprising considering that individual mutations affect the conformation of the p53 core domain differently (50). Further supporting the idea, our results show that the spectrum of secondary DNA structures bound by mutp53 proteins is different and probably much broader from that of wtp53. The identification of genomic sequences bound by individual mutp53 proteins in chromatin might be helpful for delineating the optimal structural binding sites for mutp53 proteins such as 273H, which while being potent DNA-binding proteins in vivo may behave as weak binders with rationally designed structures in vitro. We are currently analyzing a library of genomic sequences obtained by a ChIP-based approach (44) that represents sequences bound by mutp53 273H in vivo. Preliminary analyses reveal that sequences bound by mutp53 273H in chromatin exhibit repetitiveness, which is a characteristic feature of structurally flexible DNA, and to an above-average percentage contain variations of the DNA unwinding motif ‘AATATATTT’ (M. Brazdova, G. Tolstonog and W. Deppert, manuscript in preparation), which had been previously shown to be recognized by mutp53 273H in vitro (42). Repetitiveness and flexibility thus seem to be common denominators of DNA bound by mutp53 proteins. Such parameters are characteristic for MAR/SAR elements, which were the initially identified DNA sequences bound by various mutp53 proteins (42,43). Secondary DNA structures formed by MAR/SAR sequences are extremely flexible due to their high content of repetitive sequences (often AT-rich) and difficult to reconstitute in vitro with short DNA. In chromatin, however, MAR/SAR elements are very stable and can form facultative secondary structures due to their large size (kilobase range). Taken together, all data available support the idea that DNA-structure-selective binding of mutp53 proteins as described here is the base for MAR/SAR binding of mutp53 proteins described originally by our laboratory (42,43) and confirmed by other groups (51).
The finding that mutp53 can bind with high affinity to non-canonic DNA structures formed by p53-specific sequences seems discrepant to the fact that mutp53 cannot activate transcription from wtp53 responsive elements in vivo. One explanation may be that mutp53 proteins may in fact bind to wtp53-response elements when they adopt a non-linear conformation, but the outcome of such binding could be different from wtp53-SSDB. Transcriptional activation requires the assembly of a stereo-specific nucleoprotein complex (pre-initiation complex) between the activator and the components of the basal transcriptional machinery at the specific promoters. Wtp53 and mutp53 proteins interact differently with general transcription co-activators, such as TAFII31 (52), TAFII40 and TAFII60 (53). Therefore, the complex of mutp53 bound to non-linear DNA may not be favorable for the assembly of an active pre-initiation complex. Alternatively, the stability of secondary structures formed within p53-response elements may be the limiting factor. Considering that self-complementarity within the p53-cognate motifs is not continuous but, as a rule, is interrupted by individual non-matching bases, stem–loop structures formed by p53-response elements are unlikely to be stable. Therefore, it is possible that specific conditions may be required to support the formation of a non-linear conformation within wtp53-response elements. The possibility that such conditions may not be fulfilled in cells with mutp53 is intriguing. In such a scenario, one would have to assume that some activity inducing alterations in the local DNA topology within or nearby p53-binding sites should be present in cells with wtp53, but not in cells with mutp53. In fact, such a hypothetic activity may be inherent to the wtp53 protein itself. In this regard, it is worth considering that binding to linear DNA, which is severely impaired in mutp53 proteins [(22); this study] appears to be an important intermediate step and may be even a pre-requisite for wtp53-SSDB. According to the recently proposed model of ‘p53 linear diffusion’, unspecific binding to linear DNA may be of crucial importance for wtp53-SSDB, as it may allow wtp53 to ‘slide’ along the duplex until it comes across a specific site where it then forms a stable sequence-specific complex (36). In contrast to wtp53, mutp53 proteins are impaired in their ability to bind linear DNA. This may explain why mutp53 will not bind to non-canonic DNA structures formed by wtp53-response elements in vivo: they cannot find such structures because the potential to slide along linear DNA duplex is diminished in mutp53 proteins. An important implication would be that mutp53-binding sites may be limited to DNA structures that are constitutively present in a conformation that favors binding of mutp53, e.g. MAR/SAR DNA elements.
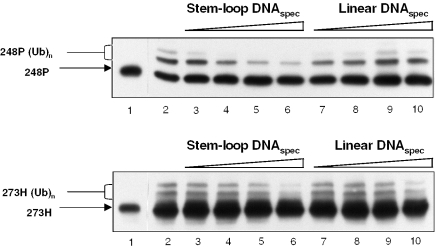
Constitutive binding to non-B DNA structures might also contribute to the strongly increased metabolic stability of mutp53 proteins in vivo. We have observed that in vitro ubiquitination of mutp53 proteins by Mdm2 is effectively inhibited in the presence of stem–loop DNA (Figure 6, lanes 3–6) whereas the impact of linear DNA is minimal (lanes 7–10). The DNA-dependent protection from ubiquitination reflected the potential of different mutp53 proteins to bind non-linear DNA. Indeed, stem–loop DNA had virtually no effect on the ubiquitination of mutp53 protein R273H, which binds weakly (lanes 3–6 in the lower panel), whereas the strongly binding mutp53 protein R248P was efficiently protected under the same conditions (lanes 3–6 in the upper panel). Whether such mechanism operates in cells remains unknown. However, considering that the basal expression of Mdm2 is sustained by wtp53-independent mechanisms either transcriptionally, via the p53-independent P1 promoter (54–57), or post-transcriptionally (58), the constitutive binding to secondary DNA structures may be relevant for protecting mutp53 from degradation by basal levels of Mdm2.
Figure 6.
Non-linear DNA binding protects mutp53 from Mdm2-mediated ubiquitination. In vitro ubiquitination of recombinant p53 proteins in the absence (lane 2) or in the presence of DNAspec in stem–loop or in linear conformation as indicated. Lane 1 show control samples that were treated under identical conditions as samples in lane 2 except that ubiquitin was omitted from the reaction mixture.
The binding of mutp53 proteins to non-B DNA structures might also be the basis for the proposed augmentation of recombination by mutp53 (59,60). Unusual secondary DNA structures are intrinsically recombinogenic as they can be recognized as high-affinity substrates for DNA topology-dependent recombinogenic factors, such as topoisomerases, ligases and DNA structure-dependent binding proteins. The constitutive interaction of mutp53 proteins with topoisomerases I and II correlates with higher rates of gene amplification (60) and raises the intriguing possibility that mutp53 proteins bound to secondary structures in DNA may attract recombinogenic factors and thereby promote genomic instability.
In conclusion, our findings reveal for the first time that mutant forms of p53 are DNA-binding active proteins, which specifically bind DNA in a DNA structure-selective mode that is different from sequence-specific DNA interaction of wtp53. The striking selectivity of mutp53 proteins toward non-linear DNA is accompanied by the loss of linear DNA binding, which is an important component of wtp53-SSDB. We propose that the loss of sequence-specific and unspecific binding to linear DNA combined with enhanced binding to non-linear DNA is an important parameter underlying the oncogenic activities, increased stability and nuclear accumulation and the gain-of-function phenomenon associated with mutp53 proteins.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank Korden Walter for helping with the preparations of the p53 protein and Martina Hintz-Malchow for assistance during the preparation of the manuscript. This research was supported by the Deutsche Forschungsgemeinschaft (De 212/19-4), and by EC FP6 funding. This publication reflects the author's views and not necessarily those of the EC. The Community is not liable for any use that may be made of the information contained herein. The Heinrich-Pette-Institut is financially supported by Freie und Hansestadt Hamburg and by Bundesministerium für Gesundheit und Soziale Sicherung. Funding to pay the Open Access publication charges for this article was provided by the Heinrich-Pette-Institut, Hamburg.
REFERENCES
- 1.Vousden K.H., Lu X. Live or let die: the cell's response to p53. Nature Rev. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura Y. Isolation of p53-target genes and their functional analysis. Cancer Sci. 2004;95:7–11. doi: 10.1111/j.1349-7006.2004.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigal A., Rotter V. Oncogenic mutations of the p53 tumor suppressor: the demons of the guardian of the genome. Cancer Res. 2000;60:6788–6793. [PubMed] [Google Scholar]
- 4.Sampath J., Sun D., Kidd V.J., Grenet J., Gandhi A., Shapiro L.H., Wang Q., Zambetti G.P., Schuetz J.D. Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J. Biol. Chem. 2001;276:39359–39367. doi: 10.1074/jbc.M103429200. [DOI] [PubMed] [Google Scholar]
- 5.Kim E., Gunther W., Yoshizato K., Meissner H., Zapf S., Nusing R.M., Yamamoto H., Van Meir E.G., Deppert W., Giese A. Tumor suppressor p53 inhibits transcriptional activation of invasion gene thromboxane synthase mediated by the proto-oncogenic factor ets-1. Oncogene. 2003;22:7716–7727. doi: 10.1038/sj.onc.1207155. [DOI] [PubMed] [Google Scholar]
- 6.Scian M.J., Stagliano K.E., Deb D., Ellis M.A., Carchman E.H., Das A., Valerie K., Deb S.P., Deb S. Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene. 2004;23:4430–4443. doi: 10.1038/sj.onc.1207553. [DOI] [PubMed] [Google Scholar]
- 7.Gualberto A., Baldwin A.S., Jr p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J. Biol. Chem. 1995;270:19680–19683. doi: 10.1074/jbc.270.34.19680. [DOI] [PubMed] [Google Scholar]
- 8.Bargonetti J., Chicas A., White D., Prives C. p53 represses Sp1 DNA binding and HIV-LTR directed transcription. Cell. Mol. Biol. 1997;43:935–949. [PubMed] [Google Scholar]
- 9.Lee Y.I., Lee S., Das G.C., Park U.S., Park S.M. Activation of the insulin-like growth factor II transcription by aflatoxin B1 induced p53 mutant 249 is caused by activation of transcription complexes; implications for a gain-of-function during the formation of hepatocellular carcinoma. Oncogene. 2000;19:3717–3726. doi: 10.1038/sj.onc.1203694. [DOI] [PubMed] [Google Scholar]
- 10.Zalcenstein A., Stambolsky P., Weisz L., Muller M., Wallach D., Goncharov T.M., Krammer P.H., Rotter V., Oren M. Mutant p53 gain of function: repression of CD95(Fas/APO-1) gene expression by tumor-associated p53 mutants. Oncogene. 2003;22:5667–5676. doi: 10.1038/sj.onc.1206724. [DOI] [PubMed] [Google Scholar]
- 11.Chicas A., Molina P., Bargonetti J. Mutant p53 forms a complex with Sp1 on HIV-LTR DNA. Biochem. Biophys. Res. Commun. 2000;279:383–390. doi: 10.1006/bbrc.2000.3965. [DOI] [PubMed] [Google Scholar]
- 12.Gualberto A., Hixon M.L., Finco T.S., Perkins N.D., Nabel G.J., Baldwin A.S., Jr A proliferative p53-responsive element mediates tumor necrosis factor alpha induction of the human immunodeficiency virus type 1 long terminal repeat. Mol. Cell. Biol. 1995;15:3450–3459. doi: 10.1128/mcb.15.6.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanyi A., Deb D., Seymour R.C., Ludes-Meyers J.H., Subler M.A., Deb S. ‘Gain of function’ phenotype of tumor-derived mutant p53 requires the oligomerization/nonsequence-specific nucleic acid-binding domain. Oncogene. 1998;16:3169–3176. doi: 10.1038/sj.onc.1201857. [DOI] [PubMed] [Google Scholar]
- 14.Frazier M.W., He X., Wang J., Gu Z., Cleveland J.L., Zambetti G.P. Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol. Cell. Biol. 1998;18:3735–3743. doi: 10.1128/mcb.18.7.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin J., Teresky A.K., Levine A.J. Two critical hydrophobic amino acids in the N-terminal domain of the p53 protein are required for the gain of function phenotypes of human p53 mutants. Oncogene. 1995;10:2387–2390. [PubMed] [Google Scholar]
- 16.Sigal A., Matas D., Almog N., Goldfinger N., Rotter V. The C-terminus of mutant p53 is necessary for its ability to interfere with growth arrest or apoptosis. Oncogene. 2001;20:4891–4898. doi: 10.1038/sj.onc.1204724. [DOI] [PubMed] [Google Scholar]
- 17.Deppert W. The nuclear matrix as a target for viral and cellular oncogenes. Crit. Rev. Eukaryot. Gene Expr. 2000;10:45–61. [PubMed] [Google Scholar]
- 18.Deppert W., Gohler T., Koga H., Kim E. Mutant p53: “Gain of function” through perturbation of nuclear structure and function? J. Cell. Biochem. 2000;(Suppl.):115–122. [PubMed] [Google Scholar]
- 19.Resnick M.A., Inga A. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proc. Natl Acad. Sci. USA. 2003;100:9934–9939. doi: 10.1073/pnas.1633803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato S., Han S.Y., Liu W., Otsuka K., Shibata H., Kanamaru R., Ishioka C. Understanding the function–structure and function–mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc. Natl Acad. Sci. USA. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gagnebin J., Kovar H., Kajava A.V., Estreicher A., Jug G., Monnier P., Iggo R. Use of transcription reporters with novel p53 binding sites to target tumour cells expressing endogenous or virally transduced p53 mutants with altered sequence-specificity. Oncogene. 1998;16:685–690. doi: 10.1038/sj.onc.1201568. [DOI] [PubMed] [Google Scholar]
- 22.Wolcke J., Reimann M., Klumpp M., Gohler T., Kim E., Deppert W. Analysis of p53 “latency” and “activation” by fluorescence correlation spectroscopy: evidence for different modes of high affinity DNA binding. J. Biol. Chem. 2003;278:32587–32595. doi: 10.1074/jbc.M303615200. [DOI] [PubMed] [Google Scholar]
- 23.Bakalkin G., Selivanova G., Yakovleva T., Kiseleva E., Kashuba E., Magnusson K.P., Szekely L., Klein G., Terenius L., Wiman K.G. p53 binds single-stranded DNA ends through the C-terminal domain and internal DNA segments via the middle domain. Nucleic Acids Res. 1995;23:362–369. doi: 10.1093/nar/23.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S., Cavallo L., Griffith J. Human p53 binds Holliday junctions strongly and facilitates their cleavage. J. Biol. Chem. 1997;272:7532–7539. doi: 10.1074/jbc.272.11.7532. [DOI] [PubMed] [Google Scholar]
- 25.Stansel R.M., Subramanian D., Griffith J.D. p53 binds telomeric single strand overhangs and t-loop junctions in vitro. J. Biol. Chem. 2002;277:11625–11628. doi: 10.1074/jbc.C100764200. [DOI] [PubMed] [Google Scholar]
- 26.Jett S.D., Cherny D.I., Subramaniam V., Jovin T.M. Scanning force microscopy of the complexes of p53 core domain with supercoiled DNA. J. Mol. Biol. 2000;299:585–592. doi: 10.1006/jmbi.2000.3759. [DOI] [PubMed] [Google Scholar]
- 27.Lee S., Elenbaas B., Levine A., Griffith J. p53 and its 14 kDa C-terminal domain recognize primary DNA damage in the form of insertion/deletion mismatches. Cell. 1995;81:1013–1020. doi: 10.1016/s0092-8674(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 28.Degtyareva N., Subramanian D., Griffith J.D. Analysis of the binding of p53 to DNAs containing mismatched and bulged bases. J. Biol. Chem. 2001;276:8778–8784. doi: 10.1074/jbc.M006795200. [DOI] [PubMed] [Google Scholar]
- 29.Bakalkin G., Yakovleva T., Selivanova G., Magnusson K.P., Szekely L., Kiseleva E., Klein G., Terenius L., Wiman K.G. p53 binds single-stranded DNA ends and catalyzes DNA renaturation and strand transfer. Proc. Natl Acad. Sci. USA. 1994;91:413–417. doi: 10.1073/pnas.91.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim E., Deppert W. The complex interactions of p53 with target DNA: we learn as we go. Biochem. Cell Biol. 2003;81:141–150. doi: 10.1139/o03-046. [DOI] [PubMed] [Google Scholar]
- 31.Kim E., Albrechtsen N., Deppert W. DNA-conformation is an important determinant of sequence-specific DNA binding by tumor suppressor p53. Oncogene. 1997;15:857–869. doi: 10.1038/sj.onc.1201412. [DOI] [PubMed] [Google Scholar]
- 32.Kim E., Rohaly G., Heinrichs S., Gimnopoulos D., Meissner H., Deppert W. Influence of promoter DNA topology on sequence-specific DNA binding and transactivation by tumor suppressor p53. Oncogene. 1999;18:7310–7318. doi: 10.1038/sj.onc.1203139. [DOI] [PubMed] [Google Scholar]
- 33.Gohler T., Reimann M., Cherny D., Walter K., Warnecke G., Kim E., Deppert W. Specific interaction of p53 with target binding sites is determined by DNA conformation and is regulated by the C-terminal domain. J. Biol. Chem. 2002;277:41192–41203. doi: 10.1074/jbc.M202344200. [DOI] [PubMed] [Google Scholar]
- 34.McKinney K., Prives C. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol. Cell. Biol. 2002;22:6797–6808. doi: 10.1128/MCB.22.19.6797-6808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagaich A.K., Appella E., Harrington R.E. DNA bending is essential for the site-specific recognition of DNA response elements by the DNA binding domain of the tumor suppressor protein p53. J. Biol. Chem. 1997;272:14842–14849. doi: 10.1074/jbc.272.23.14842. [DOI] [PubMed] [Google Scholar]
- 36.McKinney K., Mattia M., Gottifredi V., Prives C. p53 linear diffusion along DNA requires its C terminus. Mol. Cell. 2004;16:413–424. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y., Reed M., Wang P., Stenger J.E., Mayr G., Anderson M.E., Schwedes J.F., Tegtmeyer P. p53 domain: identification and characterization of two autonomous DNA-binding regions. Genes Dev. 1993;7:2575–2586. doi: 10.1101/gad.7.12b.2575. [DOI] [PubMed] [Google Scholar]
- 38.Anderson M.E., Woelker B., Reed M., Wang P., Tegtmeyer P. Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53: implications for regulation. Mol. Cell. Biol. 1997;17:6255–6264. doi: 10.1128/mcb.17.11.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakovleva T., Pramanik A., Terenius L., Ekstrom T.J., Bakalkin G. p53 latency—out of the blind alley. Trends Biochem. Sci. 2002;27:612–618. doi: 10.1016/s0968-0004(02)02209-0. [DOI] [PubMed] [Google Scholar]
- 40.Espinosa J.M., Emerson B.M. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell. 2001;8:57–69. doi: 10.1016/s1097-2765(01)00283-0. [DOI] [PubMed] [Google Scholar]
- 41.Weissker S.N., Muller B.F., Homfeld A., Deppert W. Specific and complex interactions of murine p53 with DNA. Oncogene. 1992;7:1921–1932. [PubMed] [Google Scholar]
- 42.Will K., Warnecke G., Wiesmuller L., Deppert W. Specific interaction of mutant p53 with regions of matrix attachment region DNA elements (MARs) with a high potential for base-unpairing. Proc. Natl Acad. Sci. USA. 1998;95:13681–13686. doi: 10.1073/pnas.95.23.13681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller B.F., Paulsen D., Deppert W. Specific binding of MAR/SAR DNA-elements by mutant p53. Oncogene. 1996;12:1941–1952. [PubMed] [Google Scholar]
- 44.Koga H., Deppert W. Identification of genomic DNA sequences bound by mutant p53 protein (Gly245→Ser) in vivo. Oncogene. 2000;19:4178–4183. doi: 10.1038/sj.onc.1203745. [DOI] [PubMed] [Google Scholar]
- 45.Yakovleva T., Pramanik A., Kawasaki T., Tan-No K., Gileva I., Lindegren H., Langel U., Ekstrom T.J., Rigler R., Terenius L., et al. p53 latency. C-terminal domain prevents binding of p53 core to target but not to nonspecific DNA sequences. J. Biol. Chem. 2001;276:15650–15658. doi: 10.1074/jbc.M100482200. [DOI] [PubMed] [Google Scholar]
- 46.Duckett D.R., Murchie A.I., Diekmann S., von Kitzing E., Kemper B., Lilley D.M. The structure of the Holliday junction, and its resolution. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 47.Turconi S., Shea K., Ashman S., Fantom K., Earnshaw D.L., Bingham R., Haupts U., Brown M.J., Pope A. Real experiences of uHTS: a prototypic 1536-well fluorescence anisotropy-based uHTS screen and application of well-level quality control procedures. J. Biomol. Screen. 2001;6:275–290. doi: 10.1177/108705710100600502. [DOI] [PubMed] [Google Scholar]
- 48.Eggeling C., Brand L., Ullmann D., Jäger S. Highly sensitive fluorescence detection technology currently available for HTS. Drug Discov. Today. 2003;8:632–641. doi: 10.1016/s1359-6446(03)02752-1. [DOI] [PubMed] [Google Scholar]
- 49.Honda R., Yasuda H. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Friedler A., Veprintsev D.B., Hansson L.O., Fersht A.R. Kinetic instability of p53 core domain mutants: implications for rescue by small molecules. J. Biol. Chem. 2003;278:24108–24112. doi: 10.1074/jbc.M302458200. [DOI] [PubMed] [Google Scholar]
- 51.Jiang M., Axe T., Holgate R., Rubbi C.P., Okorokov A.L., Mee T., Milner J. p53 binds the nuclear matrix in normal cells: binding involves the proline-rich domain of p53 and increases following genotoxic stress. Oncogene. 2001;20:5449–5458. doi: 10.1038/sj.onc.1204705. [DOI] [PubMed] [Google Scholar]
- 52.Lu H., Levine A.J. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc. Natl Acad. Sci. USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thut C.J., Chen J.L., Klemm R., Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 54.Mendrysa S.M., Perry M.E. The p53 tumor suppressor protein does not regulate expression of its own inhibitor, MDM2, except under conditions of stress. Mol. Cell. Biol. 2000;20:2023–2030. doi: 10.1128/mcb.20.6.2023-2030.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berberich S.J., Litteral V., Mayo L.D., Tabesh D., Morris D. mdm-2 gene amplification in 3T3-L1 preadipocytes. Differentiation. 1999;64:205–212. doi: 10.1046/j.1432-0436.1999.6440205.x. [DOI] [PubMed] [Google Scholar]
- 56.Phelps M., Darley M., Primrose J.N., Blaydes J.P. p53-independent activation of the hdm2-P2 promoter through multiple transcription factor response elements results in elevated hdm2 expression in estrogen receptor alpha-positive breast cancer cells. Cancer Res. 2003;63:2616–2623. [PubMed] [Google Scholar]
- 57.Chang C.J., Freeman D.J., Wu H. PTEN regulates Mdm2 expression through the P1 promoter. J. Biol. Chem. 2004;279:29841–29848. doi: 10.1074/jbc.M401488200. [DOI] [PubMed] [Google Scholar]
- 58.Trotta R., Vignudelli T., Candini O., Intine R.V., Pecorari L., Guerzoni C., Santilli G., Byrom M.W., Goldoni S., Ford L.P., et al. BCR/ABL activates mdm2 mRNA translation via the La antigen. Cancer Cell. 2003;3:145–160. doi: 10.1016/s1535-6108(03)00020-5. [DOI] [PubMed] [Google Scholar]
- 59.Gobert C., Skladanowski A., Larsen A.K. The interaction between p53 and DNA topoisomerase I is regulated differently in cells with wild-type and mutant p53. Proc. Natl Acad. Sci. USA. 1999;96:10355–10360. doi: 10.1073/pnas.96.18.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.El-Hizawi S., Lagowski J.P., Kulesz-Martin M., Albor A. Induction of gene amplification as a gain-of-function phenotype of mutant p53 proteins. Cancer Res. 2002;62:3264–3270. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.