Abstract
Evidence is accumulating that small, noncoding RNAs are important regulatory molecules. Computational and experimental searches have led to the identification of ∼60 small RNA genes in Escherichia coli. However, most of these studies focused on the intergenic regions and assumed that small RNAs were >50 nt. Thus, the previous screens missed small RNAs encoded on the antisense strand of protein-coding genes and small RNAs of <50 nt. To identify additional small RNAs, we carried out a cloning-based screen focused on RNAs of 30–65 nt. In this screen, we identified RNA species corresponding to fragments of rRNAs, tRNAs and known small RNAs. Several of the small RNAs also corresponded to 5′- and 3′-untranslated regions (UTRs) and internal fragments of mRNAs. Four of the 3′-UTR-derived RNAs were highly abundant and two showed expression patterns that differed from the corresponding mRNAs, suggesting independent functions for the 3′-UTR-derived small RNAs. We also detected three previously unidentified RNAs encoded in intergenic regions and RNAs from the long direct repeat and hok/sok elements. In addition, we identified a few small RNAs that are expressed opposite protein-coding genes and could base pair with 5′ or 3′ ends of the mRNAs with perfect complementarity.
INTRODUCTION
An increasing number of regulatory RNAs are being characterized in organisms from all three domains of life. In Escherichia coli, these regulatory RNAs, also termed noncoding RNAs or small RNAs, have been found to have a wide variety of biological functions including the repression and activation of translation and the protection and degradation of mRNAs via base pairing with the target transcripts [reviewed in (1,2)]. Another group of small RNAs found in E.coli binds to and modifies the activities of specific proteins in part by mimicking the structures of other nucleic acids [reviewed in (3)].
Despite their wide range of functions, until recently, the regulatory RNAs were largely ignored because they were hard to detect. RNA genes have not been annotated during genome sequence analysis due to their lack of defined sequence features. RNA genes are also poor targets for mutation screens due to their small size and because they are resistant to frameshift and nonsense mutations since they do not encode proteins. In addition, RNAs are often missed in biochemical assays.
However, in the past three years, several systematic searches have led to the identification of more than 60 small RNA genes in E.coli [reviewed in (4)]. Four studies employed predominantly computational approaches to predict small RNA genes (5–8). These screens were primarily based on searches for sequence conservation among closely related bacteria and/or searches for promoter and terminator sequences in intergenic regions. The expression of many of the predicted small RNAs was confirmed by northern analysis of total RNA isolated from a set number of growth conditions. Other studies were based on direct detection of E.coli small RNAs. In one approach, signals on high-density oligonucleotide probe arrays that did not correspond to mRNAs were classified as small RNAs (9). In another study, a shotgun cloning approach (RNomics) was used to generate cDNA libraries of RNAs between 50 and 500 nt (10). Finally, immunoprecipitation with the RNA chaperone protein Hfq and direct detection of the bound RNAs on genomic microarrays were used to identify candidate small RNAs (11).
While the computational and direct detection-based approaches have led to the identification of many new small RNAs, it is certain that not all small RNAs have been detected. The computational approaches all focused on the intergenic regions of the genome, and several of the studies assumed that small RNAs were >50 nt. Thus, the screens likely missed small RNAs expressed from the noncoding strand of known genes and small RNAs of <50 nt.
To circumvent some of the limitations of the previous screens and to identify additional small RNAs in E.coli, we carried out a cloning-based screen of RNAs <65 nt. In this screen, we identified several 5′- and 3′-UTR (untranslated region)-derived small RNAs as well as RNAs encoded in cis to the 5′ or 3′ ends of mRNAs. Preliminary characterization of these transcripts is described.
MATERIALS AND METHODS
Oligonucleotides
The sequences of all DNA oligonucleotides used in this study are provided in Supplementary Table S1.
RNA isolation
For the samples of total RNA used in the cloning, the wild-type MG1655 strain was grown in Luria–Bertani broth (LB) at 37°C to A600 ≈ 0.47 (fractions I and II) and A600 ≈ 1.7 (4 h after 1:100 dilution from an overnight culture) (fractions III and IV). For the northern analysis, MG1655 and the corresponding hfq-1::Ω (cmr) mutant strain (GSO67) were grown in LB or M9 medium with 0.2% glycerol at 37°C to the exponential (A600 ≈ 0.3) and stationary phase (18 and 24 h after 1:100 dilution from an overnight culture for the cells grown in LB and M9-glycerol, respectively). In all the cases, RNA was isolated by acid hot-phenol extraction (12).
RNA cloning
The protocol used to clone the small RNAs in this study was based on the approach used to clone Drosophila microRNAs (miRNAs) (13,14). Total RNA (≈500 μg) was fractionated on a denaturing 8% polyacrylamide gel. The regions of the gel corresponding to RNAs in the size range of 30–50 nt (fractions I and III) and 50–65 nt (fractions II and IV) were excised, and the RNA was eluted by an overnight incubation at 4°C in 2 ml of 0.3 M NaCl in siliconized tubes. The eluate was extracted with chloroform to remove residual gel fragments, and the RNA was recovered by ethanol precipitation with 40 μg of glycogen. The isolated RNA was dissolved in 30 μl of diethylpyrocarbonate (DEPC)-treated H2O. The small RNA samples were dephosphorylated (30 μl reaction, 50°C, 60 min, 20 U alkaline phosphatase; Roche Applied Science, Indianapolis, IN), extracted with phenol/chloroform, precipitated with ethanol and dissolved in 25 μl of DEPC-treated H2O. Subsequently, the 3′ adapter oligonucleotide phosphorylated at the 5′ end (5′-PuuuAACCGCGAATTCCAG idT-3′; uppercase = DNA, lowercase = RNA, idT = inverted deoxythymidine 3′ modification; Dharmacon RNA Technologies, Lafayette, CO) was ligated to the dephosphorylated small RNA (30 μl reaction, 15°C, overnight, 10 μM 3′ adapter, 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 1 mM ATP, 10 mM DTT, 0.1 mg/ml acetylated BSA and 40 U T4 RNA ligase; Amersham Biosciences Inc., Piscataway, NJ). The ligation reaction was stopped by the addition of an equal volume of gel loading buffer II (Ambion Inc., Austin, TX). The RNA fragments were purified by size selection on a denaturing 8% acrylamide gel as described above. The ligation products were phosphorylated at their 5′ ends (30 μl reaction, 37°C, 1 h, 2 mM ATP and 5 U T4 polynucleotide kinase; New England Biolabs Inc., Beverly MA). The phosphorylation reaction was stopped by phenol/chloroform extraction. The RNA then was recovered by ethanol precipitation and dissolved in 25 μl of DEPC-treated H2O. The 5′ adapter oligonucleotide (5′-ACGGAATTCCTCACTaaa-3′; uppercase = DNA, lowercase = RNA; Dharmacon RNA Technologies) was ligated to the phosphorylated ligation product as described above. Again, the new ligation products were gel purified and eluted from the gel slice. Reverse transcription reactions (20 μl reaction, 50°C, 1 h, 200 U Superscript III; Invitrogen Corp., Carlsbad, CA) were followed by PCR using 5′ forward primer and 3′ reverse transcription primers. The PCR products then were purified by phenol/chloroform extraction, ethanol precipitated, dissolved in 30 μl of TE and digested with EcoRI. Digested products were concatamerized using T4 DNA ligase (New England Biolabs), and concatamers in the size range of 200–650 bp were recovered from a 1.5% agarose gel using a gel extraction kit (Qiagen, Valencia, CA). The extracted DNA was phenol extracted, ethanol precipitated and dissolved in 42 μl of ultra pure H2O. The single-stranded ends were filled in by incubation with Taq polymerase under standard conditions at 72°C for 15 min. The DNA products were ethanol precipitated, dissolved in 10 μl of TE and ligated directly into the pCRII-TOPO vector (Invitrogen Corp.). The TOP10F′ strain was transformed and single colonies were screened for inserts by PCR using M13 Reverse and M13 Forward sequencing primers. PCR products corresponding to inserts were sequenced using BigDye Terminator (Applied Biosystems, Foster City, CA) and an M13 Reverse primer.
The sequences, for which strand specificity could be determined by the ligated primer sequences, were mapped to the E.coli genomic sequence using BLAST (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi), and classified as rRNA sequences, tRNA sequences, sequences within, outside of or on the antisense strand of an ORF, repetitive sequences or unknown sequences. cDNAs, which corresponded to mRNA sequences outside of an ORF region, were initially grouped into three categories. Those for which only one end of the RNA was within the ORF were categorized as ‘border UTRs’ (10 cDNA species), those for which one end of the RNA was within 30 nt of the ORF were categorized as ‘UTRs’ (16 cDNA species) and those for which neither end of the RNA was within 30 nt of the ORF were categorized as ‘intergenic regions’ (13 cDNA species). Upon closer inspection of some of the intergenic regions and perusal of the literature, 12 of the intergenic regions were reclassified as UTR sequences. In addition, one of the border UTR cDNA species was reclassified as intergenic, since northern analysis and end mapping revealed that the corresponding RNA spanned nearly the entire intergenic region. All of the sequences and their map positions are given in Supplementary Tables S2–S7.
Northern analysis
Total RNA (5 μg per lane) was separated on denaturing 8% polyacrylamide gels, alongside radiolabeled RNA markers comprising a mixture of the Decade (Ambion Inc.) and Perfect RNA Markers (Novagen, Madison, WI). The RNA was electrophoretically transferred to Zeta-Probe GT membranes (Bio-Rad Laboratories, Hercules, CA). Membranes were ultraviolet-crosslinked, probed with 32P-labeled oligonucleotides in ULTRAhyb-Oligo buffer (Ambion Inc.) at 45°C and washed as described in the manual for Zeta-Probe GT membranes.
The 5′ and 3′ rapid amplification of cDNA ends (RACEs)
The 5′ and 3′ RACEs were carried out essentially as described previously (5). The results of these analyses are given in Supplementary Tables S8 and S9.
RESULTS
Cloning strategy
To identify more small RNAs in E.coli, we used a direct cloning strategy. Our approach was based on the method used to clone 21–22 nt miRNAs from Drosophila melanogaster embryo and HeLa cell lysates (13,14). Briefly, total RNA isolated from the exponential (A600 ≈ 0.47) and early stationary phase (A600 ≈ 1.7) cultures was size-fractionated on polyacrylamide gels. RNA of 30–50 nt and 50–65 nt was isolated, and specialized 5′ and 3′ adapter molecules were ligated to the ends of the transcripts. cDNAs were synthesized by reverse transcription, and the cDNA products were amplified by PCR. Finally, the amplified products were concatamerized, cloned, sequenced and mapped to the genome. The existence of RNA species corresponding to some of the cloned sequences was confirmed by northern analysis.
A total of 641 cDNAs were sequenced in this screen, ∼300 cDNAs corresponding to each growth condition (Table 1). The most abundant class of cDNA clones was derived from rRNAs and tRNAs (38.8%). The second most abundant class of cDNAs corresponded to antisense RNAs encoded by the repetitive sequences (33.1%) known as long direct repeats (LDRs) (12) and hok/sok toxin–antitoxin modules (15). We also identified cDNAs corresponding to known small RNAs (1.9%), the coding regions (ORFs) of mRNAs (7.2%) and the UTRs of mRNAs (8.0%). Other cDNAs corresponded to RNAs expressed from the intergenic regions (Ig) (0.3%) and the opposite strands of ORFs (cis-encoded antisense RNA) (1.2%). Some cloned cDNAs (9.5%), which might be the result of spurious ligation of RNA fragments, could not be mapped to the genome because the cloned cDNA sequences were only partially homologous to the genomic sequence.
Table 1.
Summary of cDNAs corresponding to E.coli RNAs of 30–65 nt
| Category | Total | Log | Early stationary | Species | % |
|---|---|---|---|---|---|
| rRNA | 193 | 75 | 118 | 30.1 | |
| tRNA | 56 | 28 | 28 | 8.7 | |
| ORF | 46 | 21 | 25 | 44 | 7.2 |
| UTR | 51 | 25 | 26 | 37 | 8.0 |
| Intergenic (RyfC and RyfD) | 2 | 2 | 0 | 2 | 0.3 |
| Known sRNA | 12 | 4 | 8 | 8 | 1.9 |
| Sok RNA | 90 | 68 | 22 | 4 | 14.1 |
| Rdl RNA | 122 | 57 | 65 | 4 | 19.0 |
| cis-antisense | 8 | 1 | 7 | 7 | 1.2 |
| Unknown | 61 | 42 | 19 | 9.5 | |
| Total | 641 | 323 | 318 | 100 |
Fragments of rRNAs and tRNAs
Approximately 200 cDNAs corresponded to fragments of rRNAs (Supplementary Table S2). Clones corresponding to the leader, trailer and spacer regions of rRNA precursors were more abundant in the exponential phase (45 of 75), consistent with higher rates of rRNA synthesis and ribosome assembly during rapid growth. On the other hand, cDNAs corresponding to fragments of the mature rRNAs were predominant in the early stationary phase (113 of 118), consistent with increased degradation of rRNAs upon entry into the stationary phase (16). We also obtained 56 clones derived from fragments of tRNA species (Supplementary Table S2). The fragments were cloned at equal levels from the exponential and early stationary phase samples (Table 1). Some cDNAs corresponded to stem–loop structural regions of the rRNAs and tRNAs. Presumably, the ends of the RNA fragments are paired and cut by the endoribonuclease RNase III, which cleaves at double-stranded RNA structures (17). The cloned fragments are likely to provide clues to the intermediates in rRNA and tRNA processing and degradation. Whether any of these RNA fragments have independent biological functions remains to be elucidated.
Fragments of known small RNAs
Among the ∼60 previously reported small RNAs, fragments of eight (RybD, RydB, SsrS/6S, RyeA/SraC, SsrA/tmRNA, RygD/QUAD1d, Spf/Spot42 and RyjB) were cloned in this screen (Supplementary Table S3). All of the cDNAs corresponded to truncated versions of the full-length transcripts; two were derived from the 5′ end (SsrS and Spf), three from the 3′ end (RydB, SsrA and RyjB) and three were internal fragments (RybD, RyeA and RygD).
Fragments of mRNAs
Cloned cDNAs derived from mRNAs, for which both ends are within the protein-coding sequence, were categorized as ORFs in this screen (Supplementary Table S4). A total of 7.2% of the cloned cDNAs fell into this category. Approximately equal numbers of cDNAs were derived from the exponential phase (21 cDNAs) and early stationary phase (25 cDNAs) samples. We isolated three cDNAs corresponding to the aidB gene encoding a putative acyl coenzyme A dehydrogenase. Two of the three cDNAs contained overlapping sequence encompassing a possible inverted repeat, while the third cDNA mapped to a separate location. These cDNAs are likely to correspond to intermediates in the degradation of the aidB mRNA; however, we were not able to detect distinct fragments by northern analysis (data not shown). All other cDNAs corresponding to mRNA fragments were cloned only once in this screen. We found 9 of the 46 ORF cDNAs had an additional one to five A residues at the 3′ end, indicating that these RNA fragments were polyadenylated. This result shows that 3′ ends of the cleaved transcripts as well as full-length transcripts are polyadenylated. Polyadenylation has been demonstrated to facilitate degradation by 3′ to 5′ exoribonucleases, such as PNPase and RNase II [reviewed in (18)]. The small RNA fragments that we cloned are probably degradation intermediates and may give clues to the steps in mRNA degradation; however, we cannot rule out the possibility that some of the mRNA fragments have biological functions.
Fragments of UTRs
A total of 37 sequences were classified as being derived from UTR sequences (Supplementary Table S5). Of the 37 UTR RNAs, 17 appear to be derived from 5′-UTR sequences and 20 appear to be derived from 3′-UTR sequences. To determine whether specific RNA species could be detected for all of these regions, northern analysis was carried out using total RNA isolated from the exponential and stationary phase in rich (LB) and minimal media (M9-glycerol). Given that the RNA chaperone Hfq is thought to be required for the functions of all of the known E.coli small RNAs that act by base pairing and impacts the stability of many of these RNAs, we also carried out northern analysis on RNA isolated from hfq-1 mutant cells. In all the cases, one or more labeled oligonucleotides complementary to the sequence of the cloned RNA were used to probe for these transcripts. We did not detect a distinct signal for 10 of the UTR regions. The cDNA transcripts corresponding to these regions were all only cloned once in our screen. We did detect transcripts for the other 27 UTR regions, but noted that the RNAs were longer than the fragments cloned for these regions. The northern analysis of the 5′-UTR sequences showed some distinct bands together with other faint and somewhat diffuse bands. We predict that most of these transcripts are the byproducts of transcriptional or post-transcriptional regulation, such as attenuation. Five examples of small RNAs with the strongest signals are described below. The northern analysis of several 3′-UTR sequences reveals very strong specific signals raising the possibility that these RNAs have separate functions in the cell. Again, examples of RNAs with strong signals are described below.
5′-UTRs
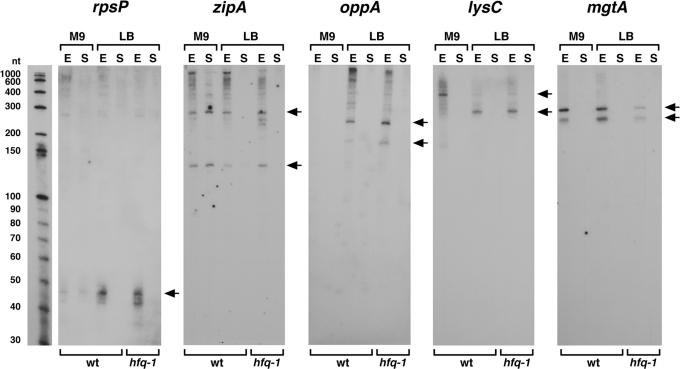
The 5′-UTR sequence cloned most frequently (five times) corresponded to the leader of the rpsP gene encoding the ribosomal protein S16 (Supplementary Table S5). Thirty-one nucleotides are identical among the five clones, which range in size from 34 to 51 nt. The region encompassing this cloned sequence was reported to make a stable structure in vitro (19). In addition, two-thirds of the rpsP transcripts synthesized in vitro were found to prematurely terminate resulting in an attenuated transcript of 51 nt (19), the size of the RNA detected in our northern analysis (Figure 1).
Figure 1.
Northern analysis of RNAs derived from 5′-UTR sequences. Samples were isolated from MG1655 wild type (wt) and hfq-1 mutant cells from exponential (E) and stationary (S) phase cells. Arrows indicate the predominant bands. RNA molecular weight markers were run with each set of samples for direct estimation of RNA transcript length. The RNA marker lane for one of the panels is shown, but only represents the approximate sizes for the other panels because there was slight variation in the running of the gels.
A 5′-UTR for which we obtained one clone corresponds to the sequence upstream of zipA, an essential gene that encodes a protein thought to be a structural component of the septal ring. The fact that the sequence of the 162 nt zipA leader is highly conserved among E.coli and Salmonella species suggests that it has a function. Possibly the long 5′ leader of zipA plays a role in pausing translation to allow protein trafficking machinery to associate with the mRNA as has been proposed for other transcripts (20). In northern analysis, we detected distinct ∼135 nt and ∼290 nt bands (Figure 1), which could be products of the predicted translational pausing.
The 5′-UTR of the oppA gene encoding an oligopeptide permease was cloned once. The intergenic region between ychE and oppA is extremely long (737 nt) but is not highly conserved between E.coli and other closely related bacteria. It has been reported that the expression of oppA is lower in LB media compared with minimum medium [reviewed in (21)] and might be regulated by base pairing with the GcvB RNA (22). We detected ∼200 and 240 nt transcripts derived from the oppA 5′-UTR in cells grown to the exponential phase in LB medium (Figure 1), raising the possibility that these fragments accumulate in response to GcvB RNA repression of oppA.
One cDNA contained the 5′ terminal end of lysC mRNA, encoding aspartokinase III. Recently, it has been shown that the long 5′-UTR, including L-box element of the lysC gene, serves as a lysine-responsive riboswitch (23–25). We detected two different 5′-UTR species in the exponential phase (Figure 1). The predominant transcript in rich medium is ∼220 nt, while the predominant transcript in minimal medium is ∼310 nt. Since the ∼220 nt transcript covers the 180 nt L-box element, we suggest that the transcript is generated by the attenuation of lysC transcription under lysine-rich conditions when the L-box element binds lysine. In Bacillus subtilis, a 270 nt RNA species was detected during growth with saturating lysine (26). Under lysine-poor conditions, the L-box element has a different conformation that allows transcription read-through and synthesis of the full-length mRNA. We propose that the ∼310 nt band we observe for cells grown in minimal medium corresponds to an intermediate in the degradation of the full-length transcript. Interestingly, two other E.coli riboswitch elements, the thiamine-responsive THI box of thiB and the flavin-responsive RFN box of ribB, were cloned by Vogel et al. (10) in their screen for small RNAs.
Another cDNA mapped to the 378 bp intergenic region between treR and mgtA, 38 nt upstream of mgtA initiation codon. The expression of mgtA, encoding a magnesium importer, is controlled by two promoters (mgtAP1 and mgtAP2) located 263 and 345 nt upstream of the initiation codon, respectively. The downstream mgtAP1 promoter is activated under Mg(II)-limiting conditions (27). Northern analysis showed two distinct transcripts (∼240 nt and ∼290 nt) that were expressed in the exponential phase (Figure 1), most likely corresponding to 5′-UTR fragments of the mRNA transcribed from the constitutive mgtAP2 promoter.
3′-UTRs
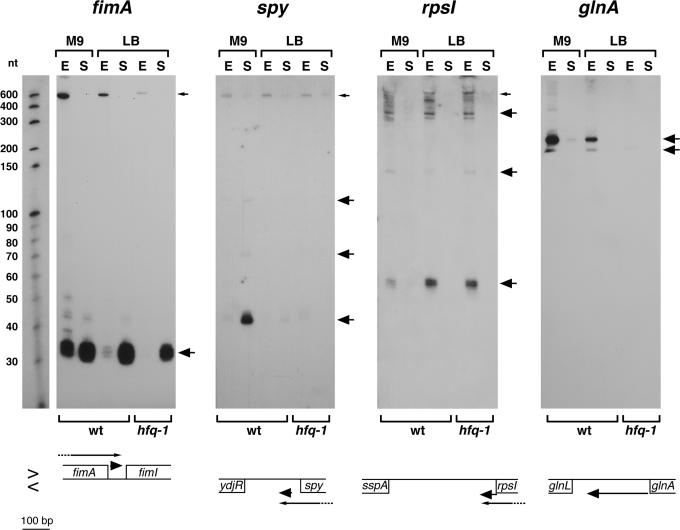
Four independent cDNAs corresponding to the 3′-UTR of fimA, which encodes a component of type 1 fimbriae, were obtained. All of the cDNAs contain an identical 32 nt (Supplementary Table S5). In the middle of this sequence, there is an inverted repeat sequence of 5 nt. Northern analysis revealed a ∼700 nt signal in the exponential phase and a strong signal of ∼35 nt in the stationary phase (Figure 2). Because the fimA gene is 549 nt in length, the ∼700 nt signal probably corresponds to the fimA mRNA. The short fragment is likely to be a processed form of the fimA mRNA. Based on 5′ and 3′ RACE experiments (Supplementary Tables S8 and S9), the 5′ end of the small RNA is somewhat heterogeneous ends map to within the fimA stop codon as well as 10 and 11 nt downstream of the fimA stop codon (4541230, 4541241 and 4541242). The longest 3′ end maps to 19 nt upstream of the fimI initiation codon (4541277), in good correspondence with the cDNAs that were isolated. Interestingly, the expression of the 35 nt 3′-UTR RNA and the mRNA is opposing; the levels of the ∼700 nt fimA mRNA are highest in exponential phase cells, while the levels of the 35 nt RNA are highest in stationary phase cells.
Figure 2.
Northern analysis of RNAs derived from 3′-UTR sequences. The samples were the same as in Figure 1, and the RNA marker lane similarly corresponds to one of the panels. Small arrows indicate the full-length mRNA and large arrows indicate processed transcripts. The arrangement of the adjacent genes is shown below each panel. Genes present on the clockwise or Watson strand are denoted above the central line (>) and the genes on the counterclockwise or Crick strand are denoted below the central line (<).
cDNAs corresponding to the 3′-UTR of the spy mRNA, which encodes a periplasmic protein, were also obtained four times, once from the exponential phase sample and three times from the early stationary phase sample. The cloned cDNAs share a core of 48 nt. In northern analysis, a strong signal of ∼45 nt and weaker signals of 70 and 120 nt were detected for cells grown to stationary phase in minimal media (Figure 2). We were not able to detect the full-length spy mRNA with the oligonucleotide probe for the 3′-UTR, though a ∼600 nt transcript corresponding to the mRNA was detected using an oligonucleotide complementary to the central portion of the spy ORF. 5′ and 3′ RACE experiments revealed that the 5′ end of the ∼45 nt RNA is 33 nt downstream of the spy stop codon (1823131) and the longest 3′ end is 123 nt upstream of ydjR (1823084), exactly matching the ends of the cDNA clones. The expression of the 3′-UTR RNA and the spy mRNA is opposing similar to what is seen for the fimA transcripts.
One of the 3′-UTR cDNAs extended from the last 7 nt of rpsI, which encodes the ribosomal protein S9, into the rpsI 3′-UTR. Using oligonucleotide probes to this region, we only detected transcripts in exponential phase (Figure 2). The longest ∼900 nt band is likely to correspond to the mRNA coding rpsM and rpsI, while the 60, 150 and 390 nt bands are truncated fragments of the mRNA. The 5′ and 3′ ends of the RNA fragments in this region were found to be heterogeneous; the shortest 5′ end maps to 21 nt upstream of the rpsI stop codon (3375473) and the longest 3′ end is located downstream of a Rho-independent terminator (3375389). In this case, the expression of the 3′-UTR transcripts and the rpsI transcripts is elevated under the same conditions.
Another cDNA mapped to the 285 bp intergenic region between glnL/ntrB and glnA, which encode a regulator of the E.coli response to nitrogen starvation and a glutamine synthetase, respectively. The nitrogen-regulated glnALG genes consist of an operon whose expression is regulated by GlnG/NtrC at three promoters; two located upstream of glnA (glnAP1 and glnAP2) and a third located upstream of glnL (glnLP) [reviewed in (28)]. The intergenic region between glnA and glnL contains a 37 bp repetitive extragenic palindromic (REP) (29) sequence and a Rho-independent terminator downstream of glnA. Our northern analysis showed that two short transcripts (∼190 nt and ∼220 nt) were expressed from this region in the exponential phase (Figure 2). The two short transcripts were absent in the hfq-1 mutant strain indicating that Hfq is required to produce or maintain these transcripts. The 5′ end of the longest transcript mapped 4 nt downstream of the glnA stop codon (4054201) and the longest 3′ end mapped downstream of a Rho-independent terminator, 88 nt upstream of the glnL initiation codon (4054007). The expression of the glnA 3′-UTR fragment is not dependent on the REP sequence, since RNAs of ∼155–170 nt were expressed from the glnA-glnL region in Salmonella typhimurium, which does not contain the REP sequence (data not shown).
Intergenic regions
We also obtained clones that correspond to independent RNAs expressed from two intergenic regions (Ig1583 and Ig1603).
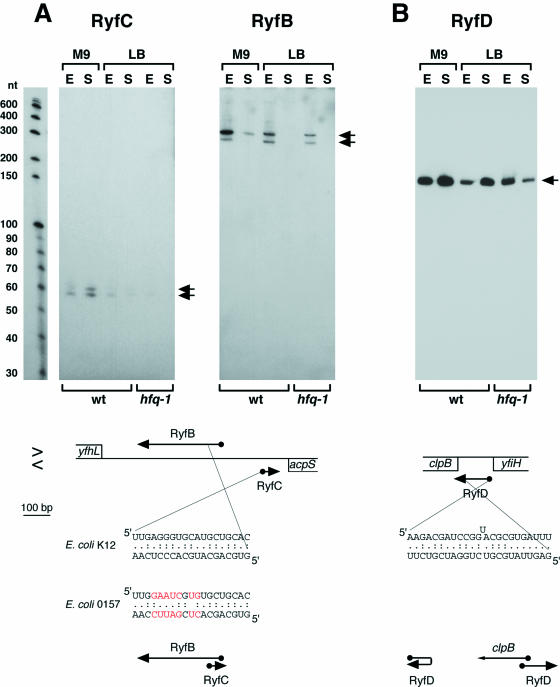
ryfB and ryfC
One clone was mapped to a long intergenic region (694 bp) between yfhL and acpS (Ig1583). Potential σ70-dependent promoter and Rho-independent terminator sequences are found upstream and downstream of the cloned cDNA, respectively. Northern analysis showed that distinct RNAs were expressed from this region [denoted RyfC using the same convention as in (8) and (11)]. Although we cloned a 32 nt RNA, ∼60 nt and ∼63 nt transcripts were detected under all conditions tested, and were highly expressed in cells grown to stationary phase in minimal medium (Figure 3A). The results of the 5′ and 3′ RACE experiments were consistent with the RyfC RNA initiating (2698540) and terminating (2698616) at the predicted promoter and terminator sequences. The 3′ end of the RyfC RNA potentially overlaps the 3′-UTR of the acpS mRNA encoded on the opposite strand.
Figure 3.
Northern analysis of the (A) RyfB, RyfC and (B) RyfD RNAs derived from intergenic sequences. The samples were the same as in Figure 1, and the RNA marker lane similarly corresponds to one of the panels. Arrows indicate predominant bands. The arrangement of adjacent genes is shown below each panel as in Figure 2. Sequences capable of base pairing and diagrams of possible pairing arrangements are given below the gene arrangements. Bases that differ between E.coli 0157 and E.coli K12 are indicated in red. The 5′ ends of the RyfB, RyfC and RyfD RNAs (indicated by balls) all correspond to the actual transcription start.
In BLAST searches, a 19 nt sequence identical to a portion of the RyfC RNA was detected in the same yfhL-acpS intergenic region. A signal detected in transcriptome analyses using high-density oligonucleotide probe arrays indicated that an RNA was expressed in this second region on the strand opposite the RyfC RNA (9). Northern analysis confirmed that a second RNA of ∼280 nt and ∼320 nt (denoted RyfB) was expressed from this region in the exponential phase. A potential σ70-dependent promoter was centered ∼120 nt upstream of the 5′ end of the RyfC RNA, and 5′ RACE analysis showed that the longest RyfB RNAs initiated at this site (2698397). The 3′ ends mapped in RACE experiments correspond to RNAs of the size detected by northern analysis (2698079). Interestingly, both the 5′ and 3′ ends of the RyfB RNA and the 3′ end of the RyfC RNA were heterogeneous, and several of the ends map adjacent to the 19 nt sequence that is complementary between RyfB and RyfC. We propose that the two RNAs base pair at the region of complementarity. This hypothesis is strengthened by the observation that, although the 19 nt sequence is different in E.coli O157 and CFT073, the complementarity is maintained (Figure 3A). We also noted that the RyfB RNA encodes a putative ORF of 26 amino acids and the RyfC RNA encodes a putative ORF of 9 amino acids, and in both the cases, the ORFs are preceded by putative Shine–Dalgarno sequences; however, additional experiments need to be carried out to determine the functions of these two RNAs.
ryfD
One cloned cDNA overlapped the first seven codons of clpB as well as the clpB ribosome-binding site (Ig1603). Using a probe to this region, we detected a highly abundant ∼140 nt transcript expressed under all conditions tested (Figure 3B). The 5′ RACE experiments revealed that the 5′ end of this transcript mapped to 8 nt downstream of the yfiH stop codon. A near consensus σ70 promoter corresponding to this start site is present within the yfiH gene. The 3′ end of the transcript determined by 3′ RACE corresponds to the 3′ end of our cloned cDNA. These results suggest that the ∼140 nt transcript (denoted RyfD) is expressed independent of the heat-shock σ32 promoter located directly upstream of clpB (30,31). Secondary structure predictions of the ∼140 nt transcript indicate that ∼20 nt sequences of 5′ and 3′ termini of the ∼140 nt transcript could base pair with each other (Figure 3B). The 5′ end of ryfD could also hybridize to the clpB mRNA and possibly act as an antisense RNA. Another possibility is that transcription of ∼140 nt transcript itself prevents the binding of σ32 RNA polymerase complex to the promoter region as has been found for the ∼550 nt SRG-1 transcript expressed upstream of the Saccharomyces cerevisiae SER3 gene (32).
cis-encoded antisense RNAs
Multiple examples of antisense regulation have been described in bacterial systems. Thus far, however, most of the chromosomally encoded antisense RNAs that have been studied in E.coli are trans-encoded; the antisense RNA is encoded at a location separate from the target gene [reviewed in (2)]. In contrast, most of the plasmid, phage and transposon-encoded antisense RNAs have been found to be cis-encoded; the antisense RNA is encoded on the strand opposite the target gene [reviewed in (33)]. Aside from the RyeA/RyeB RNAs (8) and the GadY RNA described below, the only distinct cis-encoded antisense RNAs found to be expressed from the E.coli chromosome thus far have been associated with repetitive sequences (12,15). In our screen, we obtained clones corresponding to the cis-encoded antisense RNAs from several repetitive sequences; all of the LDR regions and four out of the six hok/sok repeats. A limited number of clones also corresponded to RNAs that are encoded opposite the 5′ or 3′ ends of protein-coding genes.
Repetitive sequence RNAs
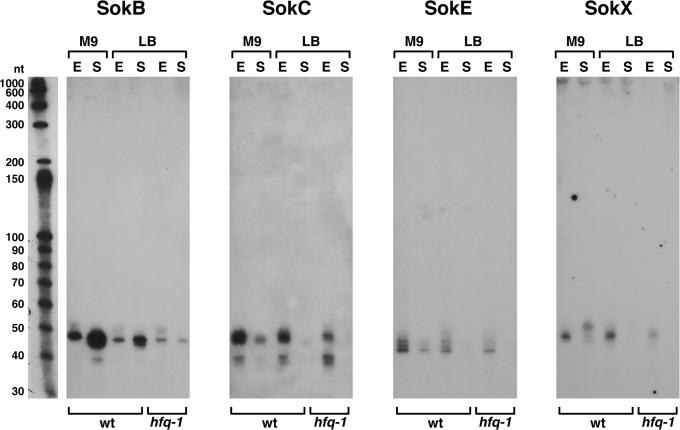
Aside from the rRNA- and tRNA-derived cDNAs, the clones obtained most frequently were derived from LDR and hok/sok repetitive sequences. Most of these elements contain genes that encode a toxic peptide (ldr and hok) and an antisense RNA (rdl and sok) that regulates the expression of the toxic peptide at a post-transcriptional level (12,15). The physiological functions of the sequences are not clear in E.coli, but the hok/sok locus found on the R1 plasmid contributes to the maintenance of the R1 plasmid in host cells [reviewed in (34)].
There are four LDR sequences in E.coli (A, B, C and D). We obtained cDNAs corresponding to each of the rdl-encoded RNAs (RdlA, RdlB, RdlC and RdlD), though few RdlD clones were isolated possibly reflecting the low expression of this RNA (Supplementary Table S6). The chromosome of E.coli K-12 codes for six regions (A, B, C, D, E and X) that show homology to the plasmid hok/sok loci. We isolated cDNAs that corresponded to antisense RNAs for four of these loci (SokB, SokC, SokE and SokX) (Supplementary Table S6). The SokB and SokC expression was also shown by Pedersen and Gerdes (15), but the SokE and SokX expression was not reported previously. Northern analysis (Figure 4) revealed that the four Sok RNAs showed differences in the expression between each other and between the exponential and stationary phase; SokB was most abundant in the stationary phase, while SokC, SokE and SokX were predominant in the exponential phase. In addition, the predominant SokX transcripts from the exponential and stationary phase cells differed slightly in size. We did not isolate any clones corresponding to the hokA and hokD loci consistent with the fact that these regions contain deletions that affect the sok sequences. It is not clear whether all of the Sok RNAs act as antisense RNAs. Given that the sokE and sokX regions are predicted not to express hok transcripts (15), it is possible that the SokE and SokX RNAs have functions independent of acting as cis-encoded antisense RNAs or are simply remnants of previously functional hok/sok systems.
Figure 4.
Northern analysis of RNAs derived from hok/sok repetitive sequences. The samples were the same as in Figure 1, and the RNA marker lane similarly corresponds to one of the panels.
5′ antisense RNAs
We also cloned one RNA encoded on the strand opposite the 5′ end of the yjiW gene, which is flanked by the mcrB and hsdS restriction genes. The function of the yjiW gene product is not known, but the yjiW expression has been shown to be induced during the SOS response (35). A strong signal for the yjiW antisense RNA, denoted RyjC, was detected by northern analysis under all growth conditions as well as after the SOS induction (Figure 5A) (data not shown). Thus, the yjiW mRNA and the corresponding 5′ antisense RNA are expressed simultaneously under some conditions. Primer extension and 3′ RACE analysis revealed that the RyjC RNA is 77 nt in length (from 4577858 to 4577934) and overlaps 6 nt of the yjiW ORF. The σ70 promoter for the ryjC gene is found within yjiW gene, and the antisense RNA is completely complementary to the 5′-UTR and part of the yjiW ORF.
Figure 5.
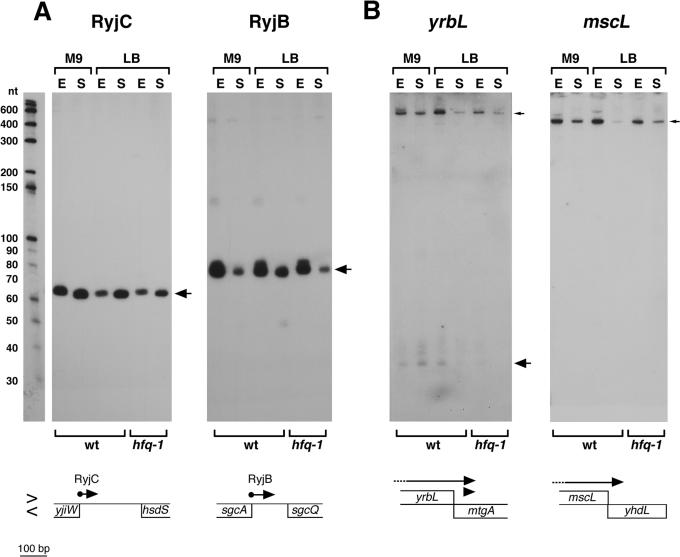
Northern analysis of antisense RNAs expressed near the (A) 5′ end or (B) 3′ end of the mRNA encoded on the opposing strand. The samples were the same as in Figure 1, and the RNA marker lane similarly corresponds to one of the panels. Small arrows indicate the full-length mRNA and large arrows indicate processed transcripts. The arrangement of adjacent genes is shown below each panel as in Figure 2. The 5′ ends of the RyjB and RyjC (indicated by balls) correspond to the actual transcription start.
Sequence inspection and 5′ and 3′ RACE analysis revealed that RyjB, a previously described small RNA (11), which was also cloned in our screen and gave a strong signal of ∼80 nt in northern analysis (Figure 5A), similarly overlaps the sgcA ORF by 4 nt. Again, a σ70 promoter for the antisense RNA is predicted to be within the sgcA gene, which encodes a putative phosphotransferase system transport protein (36).
3′ antisense RNAs
cDNA species were derived from the opposite strands of the 3′ end of five annotated protein-coding genes (mtgA, yhdL, yhdZ, yhbQ and rtcR) (Supplementary Table S7). In addition, one cloned cDNA corresponded to an RNA encoded opposite the 3′ end of the GadY (IS183) small RNA (37). The 3′ RACE analysis confirmed that the 3′-UTR of the gadX mRNA overlaps the GadY small RNA and subsequent experiments showed that this region of overlap is required for GadY RNA stabilization of the gadX mRNA (37). Among the other cDNAs, we only detected the strong expression for the antisense strands of the mtgA and yhdL genes (Figure 5B).
Two cDNAs were found to correspond to sequences within, but on the opposite strand of the mtgA gene, which encodes a monofunctional transglycolase. The two clones are overlapping and are centered 48 bp downstream of the stop codon of yrbL, the adjacent gene of unknown function. By northern analysis using an oligonucleotide probe to the antisense strand of mtgA, we detected a faint ladder of bands of ∼35–40 nt as well as a distinct ∼700 nt transcript likely corresponding to the yrbL mRNA (Figure 5B), and RACE experiments confirmed that the 3′ ends of these RNAs map within the mtgA coding sequence (3346826).
Another cDNA was derived from a transcript opposite yhdL, a gene of unknown function. The clone begins 13 bp downstream of the stop codon of the adjacent mscL gene. A band of ∼450 nt was clearly seen in both the exponential and stationary phase by northern analysis using an oligonucleotide probe corresponding to this region (Figure 5B), and the 3′ end mapped to within the yhdL coding sequence (3436136). The transcript we detect is likely the full-length mscL mRNA given that the mscL-encoded membrane channel protein is 136 amino acids. These results show that the yrbL and mscL mRNAs overlap the genes encoded on the opposite strands. We did not detect the expression for mtgA and yhdL mRNAs under our growth conditions, but it is conceivable that there are circumstances under which transcripts are expressed from both strands.
DISCUSSION
By employing the approach used to clone miRNAs in eukaryotic cells, we have been able to identify a number of previously unknown small RNAs in E.coli. Several of these RNAs were not detected in the other screens due to their small size (such as the 35 nt fimA-derived RNA), short 129 nt intergenic region (such as RyfD), lack of conservation (such as RyfC) and proximity to protein-coding genes (such as the RyjC encoded opposite yjiW). We suggest that direct cloning of RNAs of a specific size can easily be applied to the identification of small RNAs in other bacteria grown under a variety of conditions. The only limitation of the approach is the need to evaluate large numbers of sequences.
Including the 5′- and 3′-UTR-derived small RNAs but excluding the 3′ antisense RNAs, a total of 79 small RNAs have now been documented by northern analysis in E.coli (http://dir2.nichd.nih.gov/nichd/cbmb/segr/segr.html). The total number of small RNA genes is not yet known for any organism. However, given that many of the small RNAs have now been identified in multiple screens, it is likely that only a limited number of small RNAs remain to be identified in E.coli. We also isolated RNA of 10–30 nt but with the exception of one mRNA fragment, only obtained clones that were partially homologous to the genomic sequence (data not shown). These initial cloning results suggest that E.coli cells may not express noncoding RNAs that are the size of eukaryotic miRNAs.
A large number of the clones that were isolated in our screen corresponded to fragments of rRNA, tRNA and mRNA species. We believe many of these fragments are intermediates in processing and degradation pathways and may provide insights into these pathways. rRNA fragments were the most abundant species consistent with the high abundance of this class of RNAs. However, there was not necessarily a correspondence between mRNA abundance and the number of cDNAs that were obtained for a particular mRNA. Certain intermediates are likely to be particularly abundant or amenable to linker ligation. It should be noted that we were not able to detect some of the intermediates by northern analysis. In addition, the length of cloned cDNAs was not always consistent with transcript sizes observed by northern analysis. Thus, there are populations of RNA species that are sufficiently abundant for linker ligation yet are not sufficiently abundant to be detected by northern analysis.
We only obtained a limited number of clones corresponding to cis-encoded antisense RNAs. One study of whole genome expression using oligonucleotide microarrays reported that transcripts could be detected from the antisense strand of between 3000 and 4000 predicted ORFs (38). In addition, various algorithms predict promoter sequences at high frequency in the E.coli genome (39). However, given that we observed distinct transcripts for only a minority of the antisense regions, we examined in this study as well as in a plasmid-based screen for promoter activity within ORF sequences (R. G. Martin, M. Kawano, G. Storz, B. S. Rao and J. L. Rosner, manuscript in preparation), we suggest that most of the antisense transcripts do not persist and thus may not be significant for the cell.
Several of the small RNAs identified in our cloning-based screen and that of Vogel et al. (10) represent classes different from those identified in the computational screens. First, we detected several small RNAs derived from the 5′- or 3′-UTRs of mRNAs. Many of the 5′-UTR-derived transcripts, such as the fragments corresponding to the L-box, THI box and RFN box elements, overlap regions known to be required for the attenuation or regulation of translation. Whether these RNA fragments have independent functions, such as the binding and storage of lysine, thiamine or other small molecules is an intriguing question that remains to be addressed. Given that a number of the 3′-UTR-derived small RNAs are very abundant and show different expression patterns from the mRNAs from which they are likely to be derived, we suggest that the 3′-UTR-derived RNAs have independent functions. The RyfD RNA identified in our screen is unique in that it is an independent transcript that overlaps but does not encompass the downstream mRNA. We also identified one novel cis-encoded antisense small RNA. The next challenge will be to elucidate the functions of these new classes of small RNA molecules.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
We thank S. Gottesman, B. Martin, L. Rosner and members of our laboratory for comments on the manuscript. M.K. was supported by a research fellowship from the Japan Society for the Promotion of Science. Funding to pay the Open Access publication charges for this article was provided by the intramural program of the National Institute of Child Health and Human Development.
REFERENCES
- 1.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 2.Storz G., Opdyke J.A., Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Storz G., Altuvia S., Wassarman K.M. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74 doi: 10.1146/annurev.biochem.74.082803.133136. in press. [DOI] [PubMed] [Google Scholar]
- 4.Hershberg R., Altuvia S., Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G.H., Margalit H., Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 6.Chen S., Lesnik E.A., Hall A.T., Sampath R., Griffey R.H., Ecker D.J., Blyn L.B. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. BioSystems. 2002;65:157–177. doi: 10.1016/s0303-2647(02)00013-8. [DOI] [PubMed] [Google Scholar]
- 7.Rivas E., Klein R.J., Jones T.A., Eddy S.R. Computational identification of noncoding RNAs in E.coli by comparative genomics. Curr. Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 8.Wassarman K.M., Repoila F., Rosenow C., Storz G., Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tjaden B., Saxena R.M., Stolyar S., Haynor D.R., Kolker E., Rosenow C. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 2002;30:3732–3738. doi: 10.1093/nar/gkf505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vogel J., Bartels V., Tang T.H., Churakov G., Slagter-Jäger J.G., Hüttenhofer A., Wagner E.G.H. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang A., Wassarman K.M., Rosenow C., Tjaden B.C., Storz G., Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 12.Kawano M., Oshima T., Kasai H., Mori H. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-aminoacid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 2002;45:333–349. doi: 10.1046/j.1365-2958.2002.03042.x. [DOI] [PubMed] [Google Scholar]
- 13.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 15.Pedersen K., Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 1999;32:1090–1102. doi: 10.1046/j.1365-2958.1999.01431.x. [DOI] [PubMed] [Google Scholar]
- 16.Deutscher M.P. Degradation of stable RNA in bacteria. J. Biol. Chem. 2003;278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- 17.Nicholson A.W. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 18.Grunberg-Manago M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- 19.Byström A.S., von Gabain A., Bjork G.R. Differentially expressed trmD ribosomal protein operon of Escherichia coli is transcribed as a single polycistronic. J. Mol. Biol. 1989;208:575–586. doi: 10.1016/0022-2836(89)90149-6. [DOI] [PubMed] [Google Scholar]
- 20.Gottesman S., Storz G., Rosenow C., Majdalani N., Repoila F., Wassarman K.M. Small RNA regulators of translation: mechanisms of action and approaches for identifying new small RNAs. Cold Spring Harb. Symp. Quant. Biol. 2001;66:353–362. doi: 10.1101/sqb.2001.66.353. [DOI] [PubMed] [Google Scholar]
- 21.Monnet V. Bacterial oligopeptide-binding proteins. Cell. Mol. Life Sci. 2003;60:2100–2114. doi: 10.1007/s00018-003-3054-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urbanowski M.L., Stauffer L.T., Stauffer G.V. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 23.Grundy F.J., Lehman S.C., Henkin T.M. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl Acad. Sci. USA. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodionov D.A., Vitreschak A.G., Mironov A.A., Gelfand M.S. Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch? Nucleic Acids Res. 2003;31:6748–6757. doi: 10.1093/nar/gkg900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudarsan N., Wickiser J.K., Nakamura S., Ebert M.S., Breaker R.R. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kochhar S., Paulus H. Lysine-induced premature transcription termination in the lysC operon of Bacillus subtilis. Microbiology. 1996;142:1635–1639. doi: 10.1099/13500872-142-7-1635. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto K., Ogasawara H., Fujita N., Utsumi R., Ishihama A. Novel mode of transcription regulation of divergently overlapping promoters by PhoP, the regulator of two-component system sensing external magnesium availability. Mol. Microbiol. 2002;45:423–438. doi: 10.1046/j.1365-2958.2002.03017.x. [DOI] [PubMed] [Google Scholar]
- 28.Magasanik B. Regulation of transcription of the glnALG operon of Escherichia coli by protein phosphorylation. Biochimie. 1989;71:1005–1012. doi: 10.1016/0300-9084(89)90104-1. [DOI] [PubMed] [Google Scholar]
- 29.Stern M.J., Ames G.F., Smith N.H., Robinson E.C., Higgins C.F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984;37:1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- 30.Kitagawa M., Wada C., Yoshioka S., Yura T. Expression of ClpB, an analog of the ATP-dependent protease regulatory subunit in Escherichia coli, is controlled by a heat shock sigma factor (sigma 32) J. Bacteriol. 1991;173:4247–4253. doi: 10.1128/jb.173.14.4247-4253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Squires C.L., Pedersen S., Ross B.M., Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martens J.A., Laprade L., Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 33.Wagner E.G., Altuvia S., Romby P. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 34.Gerdes K., Gultyaev A.P., Franch T., Pedersen K., Mikkelsen N.D. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 1997;31:1–31. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Fernandez De Henestrosa A.R., Ogi T., Aoyagi S., Chafin D., Hayes J.J., Ohmori H., Woodgate R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 36.Reizer J., Charbit A., Reizer A., Saier M.H., Jr Novel phosphotransferase system genes revealed by bacterial genome analysis: operons encoding homologues of sugar-specific permease domains of the phosphotransferase system and pentose catabolic enzymes. Genome Sci. Technol. 1996;1:53–75. [Google Scholar]
- 37.Opdyke J.A., Kang J.-G., Storz G. GadY, a small RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selinger D.W., Cheung K.J., Mei R., Johanson E.M., Richmond C., Blattner F.R., Lockhart D.J., Church G.M. RNA expression analysis using a 30 base pair resolution Eschericia coli genome array. Nat. Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 39.Huerta A.M., Collado-Vides J. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.