Abstract
A transposon-based approach for the construction of sequencing libraries is an effcient way of preparing samples for processing on both Illumina and Ion Torrent platforms. However, PCR-mediated incorporation of adaptors in tagged DNA fragments leaves behind self-complementary regions fanking the DNA fragment. These regions are capable of forming hairpin structures and, together with adaptors, create conditions for the potential formation of template hetero-duplexes. These negatively affect the sequencing process on the Ion Torrent platform and can lead to a more than 3-fold decline in output data compared with sequencing of conventional libraries. To address this problem, we have developed MuPlus, a transposon-based protocol for barcoded library preparation for Ion Torrent, in which one adaptor is integrated by PCR and the second is integrated by ligation as a single-stranded oligonucleotide after enzymatic cleavage of a complementary part on one strand of the tag. The resulting library does not contain self-complementary, hairpin-forming regions, is free of hetero-duplexes, and can be analyzed on the Ion Torrent platform with the same effciency as a library created with a ligation-based protocol.
Keywords: library construction, next-generation sequencing, Ion Torrent, Ion Proton, transposon
Preparation of sequencing libraries is an essential step in the next-generation sequencing (NGS) workfow. Both Illumina and Ion Torrent, the most common NGS platforms, require attachment of adaptors (i.e., oligonucleotides of a particular sequence) at the ends of the DNA fragments to be sequenced. Conventional methods for library preparation use ligase activity to anneal adaptors to the fragmented and end-repaired DNA. Although straightforward, these methods have several disadvantages, including the need to fragment DNA, the requirement for relatively large amounts of starting material, and the high risk of creating artifcial chimeric sequences due to ligation of DNA fragments to each other. The latter results in chimeric fragments, mimicking genome rearrangements, thereby precluding reliable identifcation of such ultra-low abundant somatic structural variants. The alternative to the ligation-based protocol is the transposon-based approach for library construction (1). This method uses transposase activity to simultaneously fragment DNA and incorporate tags of known sequence at the ends of the fragments, allowing addition of sequencing adaptors by PCR. Thus, transposon-based library preparation requires less DNA because there is no need for a separate DNA fragmentation step, and it does not suffer from artifcially created chimeric DNA sequences.
There are commercially available transposon-based library preparation kits for both the Illumina (Illumina, Inc., San Diego, CA) and Ion Torrent (Life Technologies Corporation, Carlsbad, CA) platforms: the Nextera DNA Sample Preparation Kit (Illumina) and the MuSeek Library Preparation Kit (Thermo Fisher Scientifc Inc., Waltham, MA), respectively. However, although Illumina libraries prepared with the Nextera kit do not pose any problems during sequencing (data not shown), the barcoded MuSeek libraries produce ∼3-fold less output data when analyzed on the Ion Proton compared with sequencing of conventional libraries. This output reduction is due to an unexpectedly high level of polyclonal particles and a high fraction of low-quality reads (Table 1).
Table 1.
Sequencing metrics and data output on the Ion Proton NGS platform of libraries constructed with three different approaches.
| Metrics | Library preparation protocol | ||
|---|---|---|---|
| Ligation-based | MuSeek | MuPlus | |
| ISP density (%) | 84 | 82 | 78 |
| Polyclonal (%) | 31 | 51 | 23 |
| Low quality (%) | 9 | 55 | 6 |
| Usable reads (%) | 63 | 22 | 72 |
| Reads (n) | 77,160,045 | 26,265,788 | 81,970,145 |
Representative results out of 24 MuSeek-based and 15 MuPlus-based sequencing runs (with 6–9 libraries multiplexed per run) are shown.
We reasoned that there could be two potential causes of this problem (Figure 1A). The first stems from the presence of complementary regions fanking each DNA fragment in the sequencing library prepared with the MuSeek kit. These 16-bp fanking regions are inherited from tags introduced during transposase-mediated tagmentation. They are capable of forming stable hairpin structures that hinder progression of the polymerization reaction during the sequencing process. This leads to production of low-quality reads, presumably due to de-synchronization of the addition of the next nucleotide in the synthesized strand between clonal fragments occupying one bead. In the worst case scenario, the fraction of fragments failing to incorporate an appropriate nucleotide at one or several fows leads to phasing of that part of the cluster, which will then be identifed as polyclonal. The other, less likely, potential cause could be formation of template heteroduplexes during PCR amplifcation of tagged DNA fragments. Formation of heteroduplexes is characteristic of PCR amplifcation of mixed templates using universal primers (2). This problem becomes even more severe when amplifying sequencing libraries because the shared sequences (i.e., adaptors) at the ends of the DNA fragments are long and comparable in size to the DNA inserts. In the case of barcoded Ion Torrent libraries constructed with the transposon-based MuSeek kit, the size of shared sequences amounts to more than 40% of the total barcoded library length. When such a heteroduplex serves as a template during amplifcation onto a bead, the bead becomes populated by two different clones and will later be recognized as polyclonal.
Figure 1. The MuPlus method overcomes limitations of the original transposon-based MuSeek protocol.
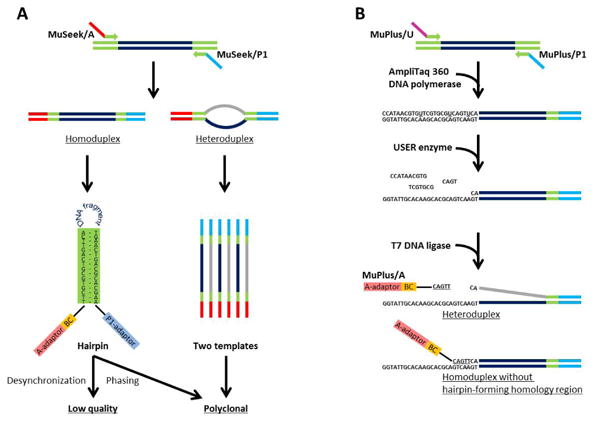
(A) The presence of stable hairpins and possible formation of template heteroduplexes are two potential causes of low data output with MuSeek libraries. (B) Schematic for MuPlus library preparation protocol workflow. Enzymatic cleavage of the complementary part on one end of the library prevents formation of hairpins and ensures addition of the second adaptor only to homoduplexes.
To overcome these limitations, we have developed a modifed protocol for barcoded library preparation, called MuPlus, which retains all the benefts of the transposon-based approach but provides normal data output on the Ion Proton sequencing platform (Figure 1B). The frst step, transposase-mediated fragmentation and tagmentation of genomic DNA, is performed using components of the MuSeek Library Preparation Kit according to the manufacturer's protocol. The second step, addition of the first sequencing adaptor, is performed by PCR amplifcation of the tagged DNA fragments using MuPlus/P1 (5′-/5SpC3/CCACTAC-GCCTCCGCTTTCCTCTCTATGGGCAGTC-GGTGATTTCGTGCGTCAGTTCA-3′) and MuPlus/U (5′-CCATAACGTGUTCGTGC-GUCAGTUCA-3′) oligonucleotides as primers. The MuPlus/P1 primer contains the P1 sequencing adaptor and is essentially identical to the MuSeek/P1 primer provided in the MuSeek kit, except for the addition of a C3 spacer phosphora-midite at the 5′-end to prevent unwanted ligation in subsequent steps. Because the MuPlus/U primer contains several deoxyuridines, PCR is carried out with AmpliTaq 360 DNA Polymerase (Life Technologies). After completion of PCR, the reaction mixture is treated with USER enzyme (New England BioLabs Inc., Ipswich, MA). The USER enzyme is a cocktail of uracil DNA glycosylase, which forms an abasic site in place of uracil (3), and the DNA glycosylaselyase Endonuclease VIII, which catalyzes breakage of the phosphodiester backbone at the 3′ and 5′ sides of the abasic site (4). As a result, the incorporated MuPlus/U primer, sharing a 16-nucleotide identical region with MuPlus/P1, is cleaved into small oligomers, which dissociate from the amplicons and expose the 3′ single-stranded overhang on each fragment containing MuPlus/U. In the fnal step, the addition of the second sequencing adaptor, a single-stranded oligonucleotide (MuPlus/A) containing an A-adaptor and a barcode, is ligated to the DNA fragments. The effciency of the ligation is ensured by the presence of a CAGTT sequence on the 3′-end of the oligonucleotide. This pentaplet is complementary to the 5′-end of the exposed overhang and ensures ligation of the fragment to the oligonucleotide. The ligation reaction is carried out by T7 DNA ligase (New England BioLabs) to avoid blunt end ligation. The resulting library lacks complementary regions capable of forming hairpins; moreover, whereas only homodupexes have DNA strands equipped with both adaptors and are suitable for sequencing, heteroduplexes lack the second adaptor and do not participate in clonal amplifcation (Figure 1B).
As a proof of principle, we used our modifed protocol to prepare barcoded libraries from mouse genomic DNA. After size selection using a Pippin Prep instrument (Sage Science, Beverly, MA) and quantifcation with the KAPA Library Quantification Kit (KapaBiosystems, Woburn, MA), the completed library was sequenced on the Ion Proton using the 200 kit v.3 (Life Technologies). Sequencing results demonstrated a signifcant decline in both the fraction of polyclonal particles and low-quality reads compared with the original MuSeek protocol. The data output was similar to that observed with the ligation-based library (Table 1). We also compared the frequencies of chimeric sequences in the libraries prepared by the different approaches using our analytical tool for quantitative assessment of somatic structural variants (Maslov et al., unpublished data). In short, the chimeric sequences are identifed as soft-clipped reads (5) in which the aligned segment has a mapping quality ≥30, and the clipped part can be independently realigned with the same or higher quality. The results of this analysis demonstrated that the frequency of chimeric reads in the library prepared by our approach is similar to that in the MuSeek library and ∼224-fold less than in the library constructed using the ligation-based approach (Figure 2).
Figure 2. Frequency of chimeric sequences present in libraries constructed with the three different approaches.
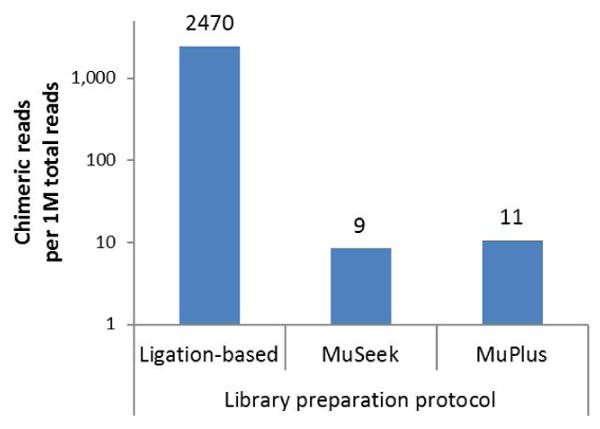
Ligation-based library was prepared using the NEBNext Fast DNA Fragmentation & Library Prep Set for Ion Torrent (New England BioLabs); the MuSeek Library Preparation Kit (Thermo Fisher) and the MuPlus method were used to prepare transposon-based libraries. Numbers above bars indicate frequency of chimeric reads per 1,000,000 total reads. Chimeric reads are defined as described in the text.
Thus, our MuPlus protocol for transposon-based library construction for the Ion Torrent sequencing platform has all the benefts of the commercial MuSeek protocol but provides signifcantly higher data output.
Method Summary.
Here we present a transposon-based library preparation method for the Ion Torrent sequencing platform providing a significantly higher data output than the commercial MuSeek protocol.
Acknowledgments
This research was supported by National Institutes of Health grant AG 17242 (J.V.), Ministry of Education and Science of the Russian Federation grant 14.574.21.0027-RFMEFI57414X0027 (V.N.P.), Albert Einstein College of Medicine Human Genome Program Pilot project grant (A.Y.M.), and Einstein-Nathan Shock Center of Excellence Pilot and feasibility grant 5P30AG038072–05 (A.Y.M.). This paper is subject to the NIH Public Access Policy.
Footnotes
Author contributions: A.Y.M., J.V., and V.N.P. conceived the idea for the study and analyzed data. T.G. and W.Q. performed the experiments. A.Y.M., J.V., and T.G. wrote the manuscript.
Competing interests: The authors declare no competing interests.
Supplementary material for this article is available at www.BioTechniques.com/article/114277.
References
- 1.Adey A, Morrison HG, Asan X, Xun JO, Kitzman EH, Turner B, Stackhouse AP, MacKenzie, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11:R119. doi: 10.1186/gb-2010-11-12-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson JR, Marcelino LA, Polz MF. Heteroduplexes in mixed-template amplifcations: formation, consequence and elimination by ‘reconditioning PCR’. Nucleic Acids Res. 2002;30:2083–2088. doi: 10.1093/nar/30.9.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindahl T. DNA repair enzymes. Annu Rev Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 4.Melamede RJ, Hatahet Z, Kow YW, Ide H, Wallace SS. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33:1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki S, Yasuda T, Shiraishi Y, Miyano S, Nagasaki M. ClipCrop: a tool for detecting structural variations with single-base resolution using soft-clipping information. BMC Bioinformatics. 2011;12(Suppl 14):S7. doi: 10.1186/1471-2105-12-S14-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]


