Abstract
DEK is an abundant chromatin protein in metazoans reaching copy numbers of several millions/nucleus. Previous work has shown that human DEK, a protein of 375 amino acids, has two functional DNA-binding domains, of which one resides in a central part of the molecule and contains sequences corresponding to the scaffold attachment factor-box (SAF-box) domain as found in a growing number of nuclear proteins. Isolated SAF-box peptides (amino acids 137–187) bind weakly to DNA in solution, but when many SAF-box peptides are brought into close proximity on the surface of Sephadex beads, cooperative effects lead to a high affinity to DNA. Furthermore, a peptide (amino acids 87–187) that includes a sequence on the N-terminal side of the SAF-box binds efficiently to DNA. This peptide prefers four-way junction DNA over straight DNA and induces supercoils in relaxed circular DNA just like the full-length DEK. Interestingly, however, the 87–187 amino acid peptide introduces negative supercoils in contrast to the full-length DEK, which is known to introduce positive supercoils. We found that two adjacent regions (amino acids 68–87 and 187–250) are necessary for the formation of positive supercoils. Our data contribute to the ongoing characterization of the abundant and ubiquitous DEK chromatin protein.
INTRODUCTION
The human DEK protein was discovered as a fusion with a nuclear pore protein in a subset of patients with acute myeloid leukemia (1). It was also identified as an autoantigen in a relatively high percentage of patients with autoimmune diseases (2–5). In addition, DEK mRNA levels are higher in transcriptionally active and proliferating cells than in resting cells, and elevated mRNA levels are found in several transformed and cancer cells (6,7).
Human DEK is an abundant nuclear protein of 375 amino acids that occurs in copy numbers of more than a million/nucleus. Most nuclear DEK protein is bound to chromatin throughout the cell cycle (8). Its interactions with positive and negative transcription factors (9,10) suggest that DEK may have a role in the transcription complex formation at promoters and enhancers [reviewed in (11)].
Work with the isolated and the recombinant human protein has shown that DEK has an intrinsic DNA-binding activity preferring four-way junction and superhelical over straight DNA and introducing positive supercoils into relaxed circular DNA (12,13). More detailed analysis revealed that DEK has two DNA-binding domains. A first domain is located in a central peptide that includes a conserved sequence element, the SAF (scaffold attachment factor) or SAP (after SAF-A/B; acinus; Pias) box. A second DNA-binding domain is in the C-terminal region of DEK and overlaps with most of the identified phosphorylation sites (Figure 1A). In fact, the DNA-binding properties of DEK are clearly influenced by phosphorylation as phosphorylated DEK binds weaker to DNA than unmodified DEK and induces the formation of DEK multimers (14,15).
Figure 1.
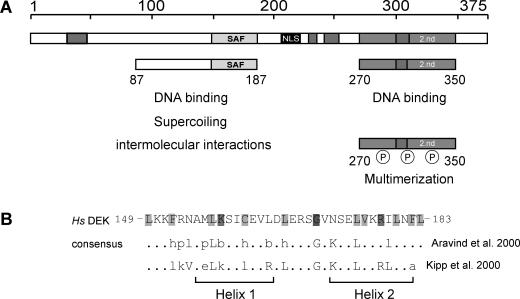
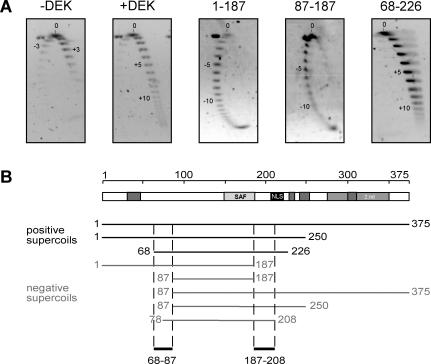
The human DEK protein—an overview. (A) Schematic representation of DEK with the known functional domains. The central SAF or SAP box (149–183) is shown in light gray, the nuclear localization sequence (NLS) in black, the four highly acidic regions in dark gray and the second DNA-binding domain, located at the C-terminal region, is represented by bright gray (2.nd). The 87 to 187 peptide mediates DNA binding, induces supercoiling in relaxed circular DNA and promotes the formation of large nucleoprotein complexes (‘intermolecular interactions’) (14). The region from residues 270 to 350 harbors most of the mapped phosphorylation sites. When phosphorylated, the overall DEK–DNA interaction is reduced and DEK–DEK interactions are promoted (‘multimerization’) (11,14,15). (B) Primary structure of the SAF or SAP box of human DEK in comparison with consensus sequences derived by Kipp et al. (17) and Aravind and Koonin (16) and a schematic representation of the predicted secondary structure (two amphipathic helices separated by a region with an invariant glycine). Light gray, small and hydrophobic residues (A, V, L, I, M, F, W); bright gray, charged residues (D, E, R, K); dark gray, glycin (modified from: http://www.sanger.ac.uk/Software/Pfam, search term: SAP). Abbreviations used: h (hydrophobic) and l (aliphatic): Y, F, W, L, I, V, M, A; p (polar): S, T, Q, N, E, D, R, K, H; and b (bulky): K, R, E, Q, W, F, Y, L, M, I; Hs, Homo sapiens. Amino acids indicated in capitals contribute to the consensus sequence in more then 90% of all investigated SAP box proteins (16).
The SAF or SAP box is a conserved 35 amino acid motif that contains a number of conserved hydrophobic and charged amino acids, which form two amphipathic α-helices that resemble helix 1 and helix 2 of the homeodomain (16,17) (Figure 1B). The SAF box occurs in a number of other chromatin proteins including the matrix or scaffold attachment factors SAF-A and SAF-B, the transcriptional repressor PIAS as well as DNA repair proteins, such as Ku70, the poly(ADP-ribose) polymerase and the fission yeast cruciform-cutting endonuclease (SpCCE) (16). It has been experimentally shown for a few cases that the SAF box can serve as a DNA-binding module.
Here, we investigate DEK-derived peptides that contain the SAF-box motif. We show that the SAF box alone (amino acids 137–187) (Figure 1B) appears not to interact with DNA in solution. However, when many SAF boxes are brought into close proximity, cooperative effects lead to high affinity for DNA. A peptide with amino acids 87–187 (including a sequence of ∼50 amino acids on the N-terminal side of the SAF box) binds to DNA much like the intact DEK preferring four-way junction DNA over straight DNA. This peptide forms large aggregates in the presence of DNA and is also able to introduce supercoils into relaxed circular DNA. Interestingly, however, the 87–187 amino acid peptide induces negative rather than positive supercoils (as the full-length DEK does). Relatively short stretches of adjacent amino acid sequences are required for the introduction of positive supercoils.
Our experiments contribute to current efforts to better understand the ubiquitous and abundant, yet still enigmatic, chromatin protein DEK.
MATERIALS AND METHODS
Cloning and expression of recombinant DEK fragments
Cloning and expression of his-tagged DEK full-length as well as peptides 1–250, 1–187, 87–187, 187–375 and Δ87–187 were performed as described previously (15). The fragments 78–208 and 68–226 were cloned and expressed in Escherichia coli. The fragments 87–250, 137–187 and 87–138 with 5′ EcoRI and 3′ XhoI cutting sites were generated by PCR from DEK cDNA cloned in pRSET-A (15). A stop codon was introduced before the XhoI site. The PCR products were subcloned by topo cloning into the pCR Blunt II TOPO vector according to the manufacturer's protocol (Invitrogen). The cDNA of the DEK fragments were digested with EcoRI and XhoI and then ligated with the linearized pRSET-A vector.
For protein expression, the vectors were transformed in E.coli BL2A (DE3) Lys S. The purification of the his-tagged DEK fragments was performed by Ni-NTA Agarose according to the manufacturer's protocol (Qiagen) and as described previously (15).
Topology assay and 2D gel electrophoresis
Topology assays with the different DEK fragments were performed exactly as described previously (15). Recombinant proteins produced in the baculovirus system had to be dephosphorylated prior to use (14,15). Bacterially expressed recombinant proteins were treated in the same way.
The reaction products were analyzed by 2D gel electrophoresis. The first dimension was on a 0.8% agarose gel in 0.5× TBE at 2 V/cm for 16 h. After electrophoresis, the gel was soaked for 2 h in 0.25 μg/μl chloroquine diluted in 0.5× TBE and rotated by 90° for a run in the second dimension in 0.5× TBE + 0.25 μg/μl chloroquine at 4 V/cm for 3 h. The gels were stained with SybrGold (MobiTec).
Duplex and four-way junction DNA
The preparation of duplex and four-way junction DNA has been described previously (13).
The annealed oligonucleotides were radiolabeled with [γ-32P]ATP using the T4 polynucleotide kinase. Labeled oligonucleotides were incubated with DEK or its derivates and separated on 8% native polyacrylamid gels in 0.5× TBE and visualized by autoradiography.
Electrophoretic mobility shift assay (EMSA) with SV40 DNA
Purified DEK fragments were treated as described for the topology assay. Aliquots containing 0.014 pmol of linearized SV40 DNA were incubated with increasing amounts of DEK as indicated in the Figure 2 legend. The reactions were performed in a total volume of 50 μl and loaded directly onto a 0.6% agarose gel in 0.5× TBE (50 mM Tris-borate, 1 mM EDTA, adjusted to pH 7.8 with boric acid) and run at 2 V/cm overnight. The nucleoprotein complexes were analyzed by SybrGold staining (MobiTec).
Figure 2.
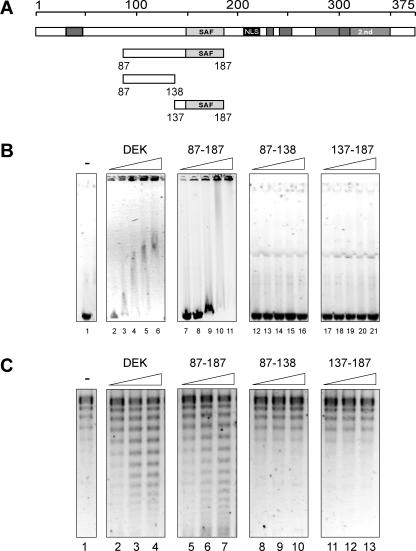
DNA binding of recombinant his-tagged DEK and DEK-derived peptides. (A) Schematic overview of the investigated fragments (for explanation, see Figure 1) (B) Electrophoretic mobility shift assay. Aliquots containing 175 ng (0.05 pmol) of linearized SV40 DNA were incubated with 2.5, 7.5, 15, 22.5 and 30 pmol of dephosphorylated full-length DEK (lanes 2–6), dephosphorylated 87–187 (lanes 7–11), 87–138 (lanes 12–16), 137–187 (lanes 17–21) or without protein (lane 1) for 1 h at 37°C. The nucleoprotein complexes were separated on 0.6% agarose gels for 16 h at 2 V/cm and visualized by ethidium bromide staining. (C) Topology assay with fragments as in (A). An aliquot of 10 ng (0.003 pmol) of circular SV40 DNA was incubated in the absence (lane 1) or presence of 1.2; 2.4 and 5 pmol of dephosphorylated full-length DEK (lanes 2–4), dephosphorylated 87–187 (lanes 5–7), 87–138 (lanes 8–10) or 137–187 (lanes 11–13) and in the presence of 1 U topoisomerase I. After 1 h at 37°C, the samples were deproteinized with Proteinase K, the DNA was purified and analyzed by agarose gel electrophoresis (0.8%, 2 V/cm) and analysis by SybrGold staining. Note that the SV40 DNA was not fully relaxed and DNA topoisomers with low superhelical densities appeared below the fully relaxed DNA (lane 1) [see also Figure 5, panel −DEK and (12)].
Pull down DNA-binding assays
Aliquots containing 10 μl of settled Protein-A Sepharose beads were incubated with 6 μg of monospecific polyclonal DEK antibodies for 1 h at 4°C in nE450 buffer (20 mM HEPES, pH 7.6, 450 mM NaCl, 10 mM sodium bisulfite and 1 mM EDTA) in the presence of 1 μg/μl BSA (New England Biolabs) + 0.5% NP40. The beads were washed twice with 1 ml nE450. The fragments 87–138 and 137–187 were dialyzed against nE300 buffer (20 mM HEPES, pH 7.6, 300 mM NaCl, 10 mM sodium bisulfite, 1 mM EDTA and 1 μg/μl BSA) overnight and incubated at 200 ng polypeptide/200 μl nE450 for 1 h at 4°C with the coupled Protein-A Sepharose beads. The beads were then washed three times with nE100 buffer (20 mM HEPES, pH 7.6, 100 mM NaCl, 10 mM sodium bisulfite and 1 mM EDTA) and 30 ng of radiolabeled and double-digested pMII plasmid (17,18) were added. Competition was carried out with unlabeled, MspI-digested pBlueBacHis2A-vector DNA (fragments ranging between 50 and 1500 bp). After 1 h incubation at 4°C, the beads were washed three times with nE100 and then eluted three times with 100 μl 2% SDS. The eluates were pooled and precipitated by the Wessel–Flügge method (19). The first supernatant was precipitated with 3 M sodium acetate for DNA analysis. The DNA was separated on a 0.6% agarose gel in 0.5× TBE at 2 V/cm and analyzed by autoradiography. Proteins were loaded on an 18% SDS–PAGE and analyzed by immunoblotting.
For the investigation of the full-length DEK, a DNA precipitation assay was used. An aliquot of 600 ng of recombinant DEK (14,15) was incubated in a total volume of 30 μl buffer P (10 mM Tris–HCl, pH 8, 80 mM NaCl and 1 mM MgCl2) in the presence of radiolabeled DNA and competitor DNA for 15 min at room temperature. The resulting protein–DNA aggregates were centrifuged for 15 min at 14 000 r.p.m. and the supernatant was collected. The pellets were resolved in 2% SDS and both fractions were analyzed on an agarose gel as described above. For the investigation of a preferential binding of fragments 1–187 and 68–226 to different topological DNA forms, we used the DNA precipitation assay as just described. DNA substrate was partially relaxed SV40 DNA. The assay was performed in a total volume of 30 μl buffer P with 20 ng of DNA and increasing amounts (12.5, 25, 50, 100 and 200 ng) of dialyzed (against nE300) DEK-fragments. After incubation for 15 min at room temperature, the samples were centrifuged for 15 min at 14 000 r.p.m. The pellets were dissolved in 1% SDS and deproteinized with Proteinase K for 30 min at 55°C. The DNA was precipitated by ethanol, dried and finally resolved in loading buffer. Final analysis was carried out by agarose gel electrophoresis (0.8%, 17 h, 0.5× TBE, 2 V/cm) and SybrGold staining (MobiTec).
RESULTS
The isolated SAF box binds to DNA
To investigate the DNA-binding properties of the SAF box in DEK, we expressed a peptide comprising amino acids 137–187, the SAF-box peptide (Figure 1B), and compared it with the larger central peptide from amino acids 87 to 187 that we have studied previously (14) and with the full-length recombinant DEK (12,13,15) (Figure 2). We first used EMSAs to determine the interaction of these peptides with linearized SV40 DNA. We found that both full-length DEK and the central peptide (amino acids 87–187) induced the formation of large DNA–protein complexes that cannot migrate into the gel (Figure 2B) (14). A minor portion of the protein–DNA complexes carried submaximal amounts of protein and, therefore, migrated into the gel (Figure 2B, DEK lanes 3–6; 87–187 lanes 9–11). In contrast, the SAF-box peptide was unable to bind to DNA under these conditions (Figure 2B, lanes 17–21). An explanation would be that the amino acid sequence which precedes the SAF box and which is present in the central peptide, but absent in the SAF-box peptide, may be responsible for DNA binding. But this could be excluded because a peptide from residues 87 to 138 failed to bind to DNA (Figure 2B, lanes 12–16).
A topology assay is shown in Figure 2C. Both, the full-length DEK and the central peptide could change the topology of closed circular DNA, while the SAF-box peptide was unable to do so (as expected from Figure 2B) (14).
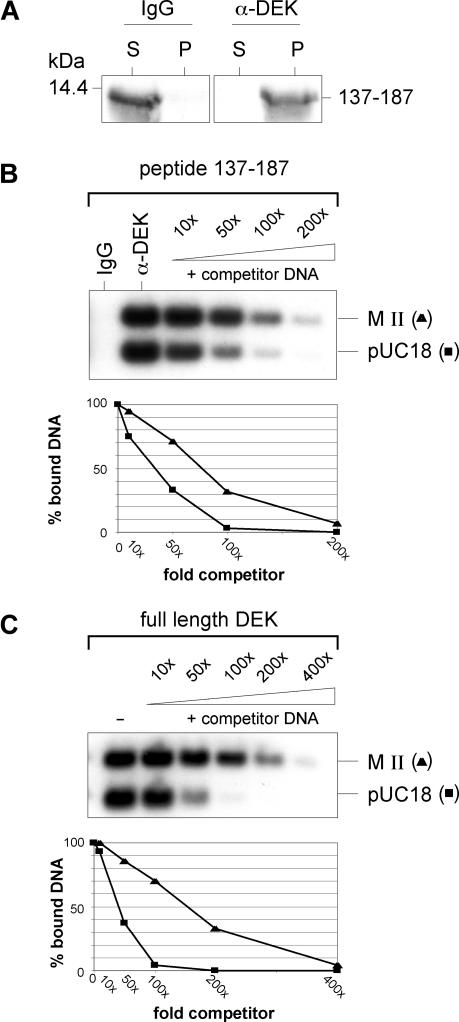
The inability of the SAF-box peptide to bind to DNA was surprising, as the SAF-box motif is assumed to be a major DNA-binding domain. However, we note that Kipp et al. (17) studying an isolated SAF-box peptide of the SAF-A obtained similar results. These authors observed efficient and specific DNA binding when many SAF-box elements were immobilized at closely adjacent sites (17), a phenomenon described as a ‘mass binding mechanism’ that allows many weak DNA-binding proteins to gain efficiency and specificity when they together associate with DNA (20). To determine whether this could also be the case for DEK's SAF-box peptide, we first loaded Sephadex-protein A beads with DEK-specific antibodies and then added SAF-box peptides that were effectively captured (Figure 3A) and thereby placed in close proximity to each other. The beads were then incubated with labeled DNA fragments derived from a plasmid with a cloned human matrix attachment region (MAR) (17,18). We determined that the immobilized SAF-box peptides efficiently retained the labeled DNA. Competition with unlabeled restricted plasmid DNA revealed a slight, but distinct preference for the MAR fragment MII over the pUC18 plasmid fragment (Figure 3B). Since a preference of DEK for an AT-rich MAR-sequence has not been determined earlier, we repeated the experiment using the full-length DEK instead of the SAF-box peptide. Competition experiments revealed again a preferred binding to the MAR-DNA fragment (Figure 3C). We note though that this preference is less pronounced than that of the SAF box in SAF-A as described by Kipp et al. (17), and it is quite possible that DEK recognizes not so much the sequence, but the unusual DNA structures that are known to occur in DNA with long sequences of AT base pairs (21,22).
Figure 3.
DNA pull-down assay. (A) Sephadex protein A beads were coupled with 6 μg of either unspecific rabbit IgGs or rabbit monospecific DEK antibodies for 1 h at 4°C, followed by the addition of 200 ng of the recombinant SAF-box fragment 137–183. After immobilization, supernatants (S) and the SDS-extracted beads (P) were analyzed by SDS–PAGE (18%) and immunoblotting with DEK specific antibodies. A molecular weight marker is indicated in kDa. (B) Binding of immobilized SAF-box peptides to DNA. Sephadex beads coated with the SAF-box peptide 137–183 prepared exactly as in (A) were incubated with a radiolabeled equimolar mixture of MAR (MII) and non-MAR DNA (pUC18) in the absence (IgG; α-DEK) or presence of increasing amounts of unspecific competitor DNA (+competitor DNA). Bound DNA was eluted from the beads and analyzed on 0.6% agarose gels followed by autoradiography. The relative intensities of both DNA fragments were determined by a densitometry program (NIH-Imager) and are shown in the lower panel of the graph. (C) An aliquot of 600 ng of full-length his-DEK was incubated with a radiolabeled equimolar mixture of MAR (MII) and non-MAR DNA (pUC18) in the absence or presence of increasing amounts of unspecific competitor DNA. The samples were centrifuged and the pellets were analyzed by agarose gel electrophoresis followed by autoradiography. The relative intensities of the both DNA forms were analyzed like in (B) and shown the lower panel.
In conclusion, we show that the isolated SAF-box peptide of the DEK protein possesses an intrinsic DNA-binding activity that could, however, not be detected in solution, but became apparent after the peptides were immobilized and densely packed on a Sephadex-bead matrix. A central peptide (amino acids 87–187) that includes the SAF box plus ∼50 N-terminal residues binds as efficiently as the full-length DEK to DNA. Therefore, the sequence preceding the SAF box appears to induce an interaction between single peptides creating a platform that favors a ‘mass binding mode’. Indeed, when incubated with DNA, the central peptide formed large complexes that remained in the slots of the agarose gel exactly as the full-length DEK does (Figure 2B). We investigated whether the central peptide shares other properties with intact DEK.
The central peptide binds to four-way junction DNA
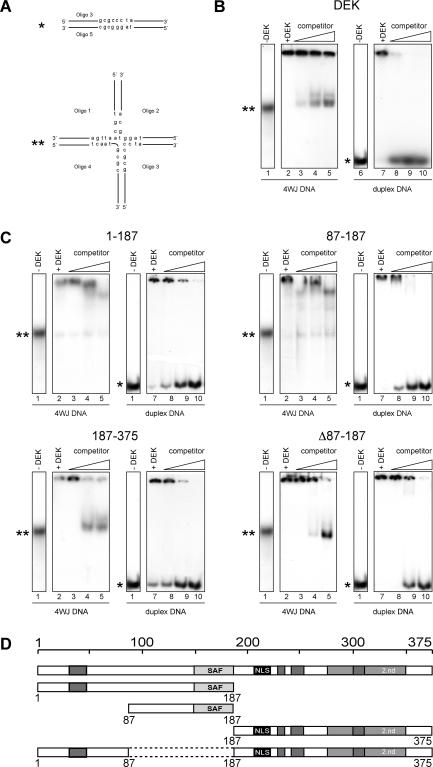
We have recently shown that the full-length DEK preferentially binds to four-way junction DNA (13), and we investigate here whether the central peptide and other selected DEK-derived peptides share this DNA-binding mode. The DNA substrates were a set of labeled oligonucleotides that could be annealed to form either linear or four-way junction DNA (Figure 4A). The full-length DEK as well as the DEK-derived protein fragments bound readily to both DNA forms as detected by the mobility shifts of the resulting protein–DNA complexes (Figure 4B and C, lanes 2 and 7). Their preference for four-way junction DNA was assayed in competition experiments using unlabeled linear DNA as competitor. As expected, the binding to labeled linear DNA could be easily suppressed by competing with unlabeled linear DNA (Figure 4B and C, lanes 8–10), but the binding to four-way junction DNA remained largely resistant to competing straight DNA in cases when the full-length DEK (Figure 4B, lanes 3–5) and peptides including the sequence between residues 87 and 187 were used as binding partners (Figure 4C, 1–187, 87–187, lanes 3–5, respectively). Peptides that lacked the central DNA-binding domain but contained the C-terminal DNA-binding domain (Figure 1) bound efficiently to DNA (Figure 4C, 187–375 and Δ87–187), but showed no preference for four-way junction DNA, since competing linear DNA suppressed the binding to both labeled linear and labeled four-way junction DNA (Figure 4C, 187–375 and Δ87–187).
Figure 4.
Binding of DEK and DEK-derived peptides to four-way junction (4WJ) and straight DNA. (A) Duplex (asterisk) and four-way junction (4WJ) DNA (double asterisk), for molecular sizes see (13). Oligonucleotides 1–4 are partially complementary to each other and assemble to the indicated four-way junction DNA upon annealing. (B) Full-length his-tagged DEK in bandshift assays. Oligonucleotides were annealed, purified on native polyacrylamid gels and then labeled. Labeled 4WJ DNA (lanes 1–5) and labeled duplex DNA (lanes 6–10) was incubated without (lanes 1 and 6) or with recombinant his-DEK (at molar ratio of DEK/DNA 50 for 4WJ and 100 for duplex DNA) without (lane +DEK) or in the presence of increasing amounts of unspecific, unlabeled duplex competitor DNA (at molar ratios: 10, 100 and 500) for 1 h at 37°C. After separation on native 8% polyacrylamid gels, the bands were visualized by autoradiography. (C) Preferential binding of SAF box carrying fragments to 4WJ DNA. Bandshifts were performed as in (B) (molar ratios for all investigated peptides DEK/DNA: 50 for 4WJ DNA, lanes 2–5; 100 for duplex DNA, lanes 7–10 or 0; lane 1, −DEK). Unspecific duplex competitor DNA was present in molar ratios 10, 100 and 500 or absent (lane +DEK). The used DEK derived fragments are indicated above the figures (1–187, 87–187, 187–375 and Δ87–187) and are schematically represented in (D).
We thus conclude that it is the central DNA-binding domain, including the SAF box, which is responsible for the specificity of DEK for non-linear four-way junction DNA.
The SAF box containing fragment induces negative supercoils
The full-length DEK protein changes the topology of closed circular DNA by introducing positive supercoils (12). As shown previously (14) and above in Figure 2C, the central fragment is also able to induce topological changes in closed circular DNA. However, the directionality of the supercoils introduced by this fragment has not been determined. To investigate this point, we first incubated DEK-derived peptides with closed circular SV40 DNA in the presence of type I DNA topoisomerase. The DNA samples were analyzed after deproteinization by 2D gel electrophoresis using chloroquine in the second dimension.
We confirmed that the full-length DEK induced positive superhelical turns, but were surprised to find that the central peptide (amino acids 87–187) induced negative supercoils (Figure 5A). We investigated a number of other DEK-derived peptides and found that sequences on both, the N-terminal side and on the C-terminal side of the central peptide, are necessary for the formation of positive supercoils. The smallest unit that we have found to be capable of inducing positive supercoils includes the amino acids 68–226. Deletions of 18–20 amino acids on either the C-terminal or the N-terminal side of this unit produced peptides that changed the topology of DNA by introducing negative turns.
Figure 5.
Changing the topology of relaxed circular DNA. (A) The 2D gel electrophoresis. A standard topology assay was performed without protein (−DEK) or with recombinant full-length his-DEK (+DEK), 1–178, 87–187 or 68–226 at molar ratios of 150 (DEK/DNA). The purified DNA was separated by standard agarose gel electrophoresis for the first dimension (16 h, 2 V/cm) (from top to bottom). The gel was rotated by 90°, incubated in 0.25 μg/μl chloroquin (in 0.5× TBE) followed by a run for the second dimension (3 h, 4 V/cm) (from left to right). DNA was visualized by SybrGold staining. Direction (−; +) and number of introduces supercoils are indicated in the figures. (B) Schematic overview of all DEK-derived peptides tested in 2D gel electrophoresis. The direction of introduced supercoils is indicated for each peptide. Sequences 68–87 and 187–208 are necessary for the introduction of positive supercoils.
Thus, a central peptide including the SAF box can change the topology of closed circular DNA much like the full-length DEK does, but the direction of supercoiling is negative rather than positive as in the case of intact DEK. The reason seems to be that the central peptide lacks adjacent sequences which are somehow involved in changing the sense of supercoiling from negative to positive.
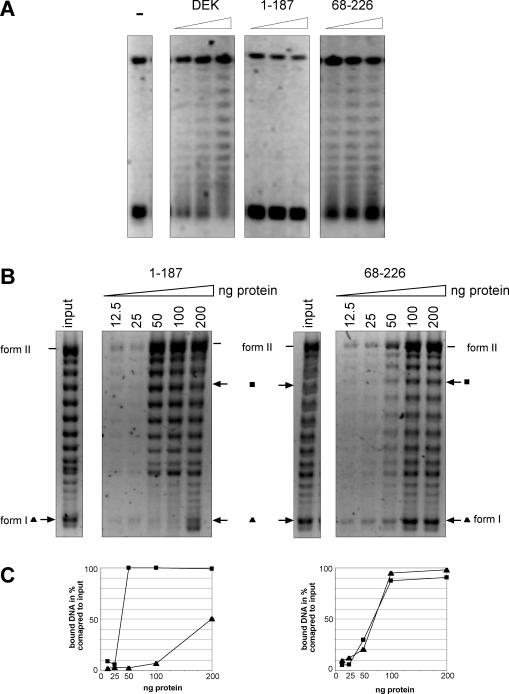
The data of Figure 5 lead to the prediction that the 68–226 fragment, but not the 1–187 fragment, may reduce the negative superhelicity of minichromosomal SV40 DNA with its 25–28 constrained supercoils. This was indeed the case, and the data of Figure 6A show that the 68–226 fragment [like the full-length DEK, Figure 6A; DEK and see (12)] changes the topology of superhelical SV40 DNA, whereas the 1–187 fragment did not.
Figure 6.
The binding of DEK-derived polypeptides to superhelical DNA. (A) Topology assay using SV40 minichromosomes. Left panel, control DNA. Full-length DEK, the 1–187 fragment and the 68–226 fragment were incubated at molar ratios of 50, 100 and 150 protein/DNA for 1 h at 37°C in the presence of topoisomerase I as described previously (12). The DNA was purified and analyzed by agarose gel electrophoresis and SybrGold staining. (B) DNA precipitation assay. Partially relaxed SV40 DNA (lane input, 20 ng) was incubated with increasing amounts of the polypeptides as indicated. After incubation for 15 min at room temperature, the protein–DNA complexes were precipitated. The DNA was purified and analyzed by agarose gel electrophoresis and SybrGold staining. (C) Densitometric analysis. The intensities of a selected DNA topoisomer with low superhelical density (filled square) and the intensities of highly supercoiled form I DNA (filled triangle) were determined by the densitometric program AIDA (Raytest). The results were plotted as relative amounts of precipitated DNA versus amounts of protein compared with input DNA.
The data could be explained by a different binding preference of the fragments tested. To investigate this point, we used the DNA precipitation assay with partially relaxed SV40 DNA as a binding substrate and increasing amounts of the 1–187 and the 68–226 fragments.
We detected that the 1–187 fragment bound preferentially to DNA topoisomers with reduced negative superhelicity, whereas the 68–226 fragment also bound to highly supercoiled DNA [like the full-length DEK, Figure 6B and C; see (13)]. Thus, the ability of the 68–226 fragment to associate with highly supercoiled DNA may at least partially explain why it reduces the negative superhelicity of SV40 DNA.
DISCUSSION
The SAF or SAP box is an amino acid motif that is detected in an increasing number of nuclear proteins that are involved in various activities, such as nuclear architecture, transcription, RNA processing and DNA repair (16). Most of these activities require an interaction of the protein with DNA. However, experimental evidence that the SAF-box motif directly participates in DNA binding has so far been obtained only for SAF-A (17), PIAS (23), Ku70 (24) and SpCCE1 (25).
In the SAF-A protein, the SAF box appears to be the only DNA-binding motif. It directs the protein to the scaffold/matrix attachment regions (S/MAR) which are AT-rich stretches of DNA that associate with nuclear matrix preparations (26). In addition, SAF-A has a C-terminal RGG motif as an additional module, which is most likely involved in mRNA processing (27–30).
Interestingly, in other proteins, the SAF-box is not the only DNA-binding domain; it increases the specificity and stability of the interactions. Protein Ku70 together with Ku80 forms a protein ring that encircles DNA at their ends as an important step in the non-homologous end-joining pathway. The SAF domain is located in an extreme C-terminal region of Ku70, outside the major DNA contacts, but could function as a barrier to inward movement of Ku70 bound to DNA ends (24).
The SAF box of SpCCE clearly stabilizes the protein's interaction with four-way junction DNA, but is not its major DNA-binding determinant. However, SAF box is involved in DNA binding as demonstrated by specific amino acid exchanges that reduce the half-life of contacts between SpCCE and DNA. The crucial residues are two basic amino acids close to the second helix in the helix–loop–helix domain that constitute the SAF box (25) (Figure 1).
Like SpCCE and Ku70, DEK has two DNA-binding domains, namely a central domain including the SAF-box and a C-terminal domain of as yet undetermined structure (Figure 1A). However, it appears that the SAF-box domain is the main DNA-binding activity because a central fragment that includes the SAF box, but not the C-terminal binding domain, has DNA-binding properties that are similar to those of the entire DEK molecule. More precisely, DEK and the central domain prefer four-way junction over straight B-form DNA and induce supercoils into relaxed circular DNA. Peptides including the C-terminal domain bind well to DNA, but do not have the specificities of the full-length DEK or the central peptide. We assume that the second DNA-binding domain stabilizes the interaction of DEK with DNA and modulates its interaction in vivo and in vitro as it overlaps with major phosphorylation sites. Indeed, previous work has shown that a phosphorylation of DEK weakens DEK–DNA contacts (15).
Like SAF-A, DEK's isolated SAF box has a very low affinity to DNA that cannot be determined by standard DNA-binding assays in solution (Figure 2B). Rather, it is necessary to fix many SAF boxes on a Sephadex matrix forming a DNA-binding platform that firmly associates with DNA. The combined binding activities of immobilized SAF boxes show a slight preference for AT-rich S/MAR sequences (Figure 3B). The full-length DEK protein has a similar preference (Figure 3C), but it is in any case much less pronounced than the affinity of SAF-A for S/MAR (17). It is therefore not clear whether DEK's preference for S/MAR is physiologically significant. In addition, AT-rich sequences are known to assume unusual DNA structures (21,22) and it is possible that DEK recognizes these structures and not the sequence. We note though that one-tenth or so of all nuclear DEK fractionates with standard nuclear matrix preparations (31,32) (data not shown). However, the majority of nuclear DEK can be released from chromatin by mild digestion with micrococcal nuclease (8). It will be an interesting problem for future research to determine how DEK is distributed on chromatin, and how it is directed to its binding sites on chromatin. A possibility is that DEK associates with transcription factors at promoters or enhancers changing the path of the DNA to which it is bound, much like the better known HMGA or HMGB architectural proteins (33–36). Like DEK, these proteins are known to preferentially bind to four-way junction and superhelical DNA (36–38).
Another similarity is that HMGA and HMGB proteins, such as DEK, induce supercoils into relaxed circular DNA. Whether this property has physiological consequences in vivo is not known. The topological changes are probably caused by the extensive twisting of DNA that occurs in the large DEK–DNA complexes which form in vitro (13). In any case, it is interesting to note that the central peptide (amino acids 87–187) introduces negative supercoils, whereas a slightly larger peptide (amino acids 68–226) induces positive supercoils (like the full-length DEK). This change in directionality cannot simply be explained by the known properties of DEK. Clearly, a determination of the 3D structure would help to successfully address this issue.
In conclusion, we have shown here that the SAF box is the major DNA-binding domain of DEK because a central fragment of ∼100 amino acids that includes the SAF box has many of the DNA properties of the full-length DEK, while the C-terminal second DNA-binding domain appears to influence the overall protein–DNA contacts.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft. Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.von Lindern M., Poustka A., Lerach H., Grosveld G. The (6;9) chromosome translocation, associated with a specific subtype of acute nonlymphocytic leukemia, leads to aberrant transcription of a target gene on 9q34. Mol. Cell. Biol. 1990;10:4016–4026. doi: 10.1128/mcb.10.8.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong X., Michelis M.A., Wang J., Bose R., DeLange T., Reeves W.H. Autoantibodies to DEK oncoprotein in a patient with systemic lupus erythematosus and sarcoidosis. Arthritis Rheum. 1998;41:1505–1510. doi: 10.1002/1529-0131(199808)41:8<1505::AID-ART23>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 3.Dong X., Wang J., Kabir F.N., Shaw M., Reed A.M., Stein L., Andrade L.E., Trevisani V.F., Miller M.L., Fujii T., et al. Autoantibodies to DEK oncoprotein in human inflammatory disease. Arthritis Rheum. 2000;43:85–93. doi: 10.1002/1529-0131(200001)43:1<85::AID-ANR11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 4.Sierakowska H., Williams K.R., Szer I.S., Szer W. The putative oncoprotein DEK, part of a chimera protein associated with acute myeloid leukaemia, is an autoantigen in juvenile rheumatoid arthritis [Erratum (1994) Clin. Exp. Immunol., 96, 177.] Clin. Exp. Immunol. 1993;94:435–439. doi: 10.1111/j.1365-2249.1993.tb08214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wichmann I., Respaldiza N., Garcia-Lozano J.R., Montes M., Sanchez-Roman J., Nunez-Roldan A. Autoantibodies to DEK oncoprotein in systemic lupus erythematosus (SLE) Clin. Exp. Immunol. 2000;119:530–532. doi: 10.1046/j.1365-2249.2000.01154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kondoh N., Wakatsuki T., Ryo A., Hada A., Aihara T., Horiuchi S., Goseki N., Matsubara O., Takenaka K., Shichita M., Tanaka K., Shuda M., Yamamoto M. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59:4990–4996. [PubMed] [Google Scholar]
- 7.Savli H., Aalto Y., Nagy B., Knuutila S., Pakkala S. Gene expression analysis of 1,25(OH)2D3-dependent differentiation of HL-60 cells: a cDNA array study. Br. J. Haematol. 2002;118:1065–1070. doi: 10.1046/j.1365-2141.2002.03734.x. [DOI] [PubMed] [Google Scholar]
- 8.Kappes F., Burger K., Baack M., Fackelmayer F.O., Gruss C. Subcellular localization of the human proto-oncogene protein DEK. J. Biol. Chem. 2001;276:26317–26323. doi: 10.1074/jbc.M100162200. [DOI] [PubMed] [Google Scholar]
- 9.Hollenbach A.D., McPherson C.J., Mientjes E.J., Iyengar R., Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein DEK. J. Cell. Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 10.Campillos M., Garcia M.A., Valdivieso F., Vazquez J. Transcriptional activation by AP-2alpha is modulated by the oncogene DEK. Nucleic Acids Res. 2003;31:1571–1575. doi: 10.1093/nar/gkg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldmann T., Scholten I., Kappes F., Hu H.G., Knippers R. The DEK protein—an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 12.Waldmann T., Eckerich C., Baack M., Gruss C. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J. Biol. Chem. 2002;277:24988–24994. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- 13.Waldmann T., Baack M., Richter N., Gruss C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–7010. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kappes F., Scholten I., Richter N., Gruss C., Waldmann T. Functional domains of the ubiquitous chromatin protein DEK. Mol. Cell. Biol. 2004;24:6000–6010. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kappes F., Damoc C., Knippers R., Przybylski M., Pinna L.A., Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol. Cell. Biol. 2004;24:6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aravind L., Koonin E.V. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 17.Kipp M., Gohring F., Ostendorp T., van Drunen C.M., van Driel R., Przybylski M., Fackelmayer F.O. SAF-box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 2000;20:7480–7489. doi: 10.1128/mcb.20.20.7480-7489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romig H., Ruff J., Fackelmayer F.O., Patil M.S., Richter A. Characterisation of two intronic nuclear-matrix-attachment regions in the human DNA topoisomerase I gene. Eur. J. Biochem. 1994;221:411–419. doi: 10.1111/j.1432-1033.1994.tb18753.x. [DOI] [PubMed] [Google Scholar]
- 19.Wessel D., Flügge U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984;138:141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 20.Zuckerkandl E., Villet R. Generation of high specificity of effect through low-specificity binding of proteins to DNA. FEBS Lett. 1988;231:291–298. doi: 10.1016/0014-5793(88)80836-6. [DOI] [PubMed] [Google Scholar]
- 21.Lobov I.B., Tsutsui K., Mitchell A.R., Podgornaya O.I. Specificity of SAF-A and lamin B binding in vitro correlates with the satellite DNA bending state. J. Cell. Biochem. 2001;83:218–229. doi: 10.1002/jcb.1220. [DOI] [PubMed] [Google Scholar]
- 22.Pommier Y., Cockerill P.N., Kohn K.W., Garrard W.T. Identification within the simian virus 40 genome of a chromosomal loop attachment site that contains topoisomerase II cleavage sites. J. Virol. 1990;64:419–423. doi: 10.1128/jvi.64.1.419-423.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okubo S., Hara F., Tsuchida Y., Shimotakahara S., Suzuki S., Hatanaka H., Yokoyama S., Tanaka H., Yasuda H., Shindo H. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J. Biol. Chem. 2004;279:31455–31461. doi: 10.1074/jbc.M403561200. [DOI] [PubMed] [Google Scholar]
- 24.Walker J.R., Corpina R.A., Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 25.Ahn J.S., Whitby M.C. The role of the SAP motif in promoting Holliday junction binding and resolution by SpCCE1. J. Biol. Chem. 2003;278:29121–29129. doi: 10.1074/jbc.M302314200. [DOI] [PubMed] [Google Scholar]
- 26.Boulikas T. Chromatin domains and prediction of MAR sequences. Int. Rev. Cytol. 1995;162A:279–388. doi: 10.1016/s0074-7696(08)61234-6. [DOI] [PubMed] [Google Scholar]
- 27.Romig H., Fackelmayer F.O., Renz A., Ramsperger U., Richter A. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 1992;11:3431–3440. doi: 10.1002/j.1460-2075.1992.tb05422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fackelmayer F.O., Dahm K., Renz A., Ramsperger U., Richter A. Nucleic-acid-binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. Eur. J. Biochem. 1994;221:749–757. doi: 10.1111/j.1432-1033.1994.tb18788.x. [DOI] [PubMed] [Google Scholar]
- 29.Fackelmayer F.O., Richter A. Purification of two isoforms of hnRNP-U and characterization of their nucleic acid binding activity. Biochemistry. 1994;33:10416–10422. doi: 10.1021/bi00200a024. [DOI] [PubMed] [Google Scholar]
- 30.Gohring F., Fackelmayer F.O. The scaffold/matrix attachment region binding protein hnRNP-U (SAF-A) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry. 1997;36:8276–8283. doi: 10.1021/bi970480f. [DOI] [PubMed] [Google Scholar]
- 31.Mirkovitch J., Mirault M.E., Laemmli U.K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 32.de Graaf A., Meijne A.M., van Renswoude A.J., Humbel B.M., van Bergen en Henegouwen P.M., de Jong L., van Driel R., Verkleij A.J. Heat shock-induced redistribution of a 160-kDa nuclear matrix protein. Exp. Cell. Res. 1992;202:243–251. doi: 10.1016/0014-4827(92)90071-f. [DOI] [PubMed] [Google Scholar]
- 33.Agresti A., Bianchi M.E. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 34.Bustin M., Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 35.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 36.Thomas J.O. HMG1 and 2: architectural DNA-binding proteins. Biochem. Soc. Trans. 2001;29:395–401. doi: 10.1042/bst0290395. [DOI] [PubMed] [Google Scholar]
- 37.Thomas J.O., Travers A.A. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 38.Zlatanova J., van Holde K. Binding to four-way junction DNA: a common property of architectural proteins? FASEB J. 1998;12:421–431. [PubMed] [Google Scholar]