Abstract
Telomerase replicates chromosome ends, a function necessary for maintaining genome integrity. We have identified the gene that encodes the catalytic reverse transcriptase (RT) component of this enzyme in the malaria parasite Plasmodium falciparum (PfTERT) as well as the orthologous genes from two rodent and one simian malaria species. PfTERT is predicted to encode a basic protein that contains the major sequence motifs previously identified in known telomerase RTs (TERTs). At ∼2500 amino acids, PfTERT is three times larger than other characterized TERTs. We observed remarkable sequence diversity between TERT proteins of different Plasmodial species, with conserved domains alternating with hypervariable regions. Immunofluorescence analysis revealed that PfTERT is expressed in asexual blood stage parasites that have begun DNA synthesis. Surprisingly, rather than at telomere clusters, PfTERT typically localizes into a discrete nuclear compartment. We further demonstrate that this compartment is associated with the nucleolus, hereby defined for the first time in P.falciparum.
INTRODUCTION
Telomeres are nucleoprotein complexes that protect chromosome ends from degradation and fusion. In most eukaryotes, telomeric DNA consists of tandem arrays of short G-rich repeats that are maintained by telomerase, an RNA-dependent DNA polymerase. Loss of telomerase activity leads to a gradual shortening of telomere length, resulting in growth arrest and eventually senescence (1–3). The core components of telomerase enzyme consist of a catalytic protein component, called telomerase reverse transcriptase (TERT), and an RNA molecule that serves as an internal template from which repeats are copied to the 3′ end of the telomere G-rich strand. Other proteins have been identified that associate with telomerase and, some at least, are required for its action in vivo (4,5).
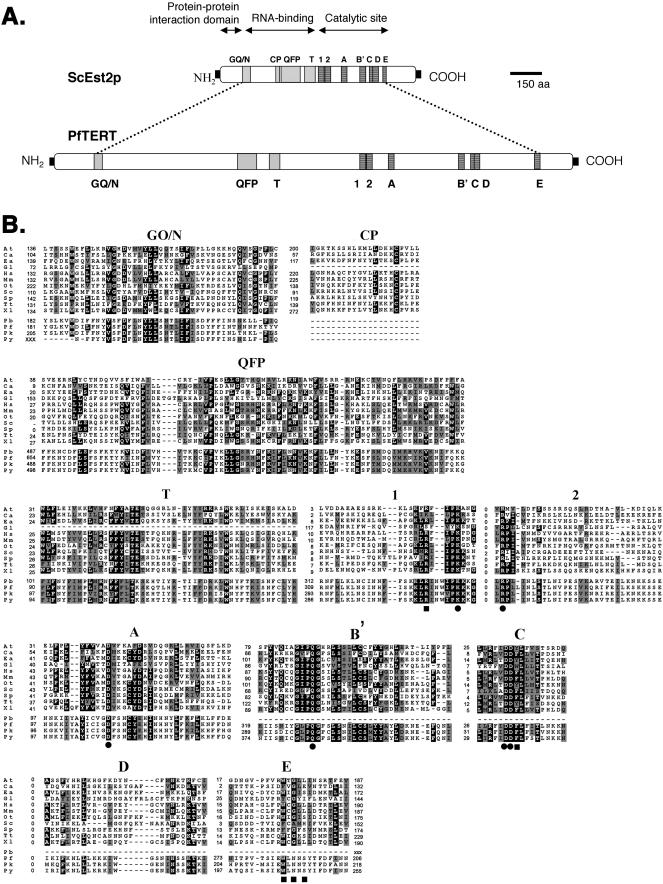
TERT was first purified from the ciliate Euplotes aediculatus (1). By sequence homology, other TERTs were subsequently identified, such as ciliates (6,7) and diplomonads (8), several yeasts (9–11), plants (12) and animals, including humans (10,13) and mouse (14). All TERTs contain telomerase-specific motifs within the N-terminal half of the protein (GQ/N, CP, QFP and T) and RT-specific motifs in the C-terminal half (1, 2 and A–E; Figure 1) (1,8,15,16). The RT domain of TERT is essential for catalytic activity, whereas the N-terminal half is required for efficient binding of the RNA template, defining the 5′ RNA template boundary, multimerization and interactions with associated proteins [reviewed in (17)].
Figure 1.
Plasmodium TERT proteins contain telomerase conserved motifs. (A) In-scale diagram showing the difference in size of PfTERT and the S.cerevisiae homologue, Est2p. Grey boxes represent telomerase-specific motifs: GQ/N, CP, QFP and T. Black boxes represent RT motifs. (B) Multiple sequence alignment of the conserved motifs of all TERT proteins described to date and the four Plasmodium TERT proteins identified in this work: PbTERT (P.berghei), PfTERT (P.falciparum), PkTERT (P.knowlesi) and PyTERT (P.yoelii). For a given position within the alignment, identical or similar amino acids to the consensus sequence are shaded in black or grey, respectively. Note: The putative C.elegans TERT gene was excluded because it lacks all TERT-specific motifs. Squares mark telomerase-specific amino acids that are not present in viral RTs; circles indicate amino acids essential for RT activity. The numbers accompanying each protein aligned in (B) represent the number of amino acids that exist between two motifs, or between a terminal motif and the protein end. When an N- or C-terminus is unknown, the amino acid distance is shown as ‘xxx’. Motifs CP and T have not been found in G.lamblia and accordingly are absent from these alignments. At, Arabidopsis thaliana; Ca, Candida albicans; Ea, Euplotes aediculatus; Gl, Giardia lamblia; Hs, Homo sapiens; Mm, Mus musculus; Ot, Oxytricha trifallax; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Tt, Tetrahymena thermophila; Xl, Xaenopus laevis; Pb, P.berghei; Pf, P.falciparum; Pk, P.knowlesi; and Py, P.yoelii.
Telomerase is developmentally regulated in higher eukaryotes (18). In a human being, telomerase activity is undetectable in most somatic tissues. Interestingly, telomerase activity is detectable in the germline and in highly proliferative cells of renewal tissues, such as bone marrow progenitor cells and intestinal mucosa (19). Telomerase is necessary for the unlimited proliferation of human somatic cells in vitro (20). These observations along with the prevalence of telomerase expression in human cancers, makes telomerase an attractive candidate for cancer therapy [reviewed in (21)].
Human malaria is re-emerging as one of the world's most lethal infections. In tropical regions of the globe, 300 million people are infected and of those 1–2 million are killed annually (WHO, 1998, Malaria, http://www.who.int:inj-fs/en/fact094.html). It is caused by intracellular protozoa of the genus Plasmodium, of which Plasmodium falciparum is the most virulent species. P.falciparum is transmitted from one person to another by the Anopheles mosquito. After a short hepatic stage, parasites are released into the bloodstream of the host, where they infect erythrocytes. Massive proliferation of the blood stage parasites causes the clinical symptoms [reviewed in (22)]. Although several anti-malarial drugs exist, resistance is becoming a major problem, and there is an urgent need for new therapeutics (23).
The haploid nuclear genome of P.falciparum is ∼30 Mb in size and is organized in 14 linear chromosomes [reviewed in (24)]. The ends of the chromosomes consist of a tandem array of a degenerate G-rich heptamer, most frequently GGGTT(T/C)A (25). The mean telomere length is ∼1.2 kb and is maintained at a constant size during blood-stage proliferation (26). Analysis of nuclear architecture reveals that chromosome ends form clusters of 4–7 telomeres that localize around the nuclear periphery (27). This arrangement was proposed to facilitate gene conversion events between the sub-telomeric regions of heterologous chromosomes. As a result, there is a continuous sequence diversification among gene families coding for surface antigens, which promotes the generation of new antigenic and adhesive phenotypes.
P.falciparum telomeres are partially organized in a non-nucleosomal structure (26), suggesting that specific proteins may constitute the telomeric chromatin. In yeast and humans, numerous proteins have been shown to interact directly or indirectly with telomeric DNA [for a review see (28)]. In P.falciparum, several putative homologues of known telomeric-specific proteins have been identified and are currently being characterized (29). Telomerase activity has been detected in semi-purified nuclear extracts of P.falciparum blood stages (30). Furthermore, it was shown that P.falciparum telomerase can be used as a substrate oligonucleotide that mimic not only telomeric sequences, but also chromosome breaks, suggesting that this enzyme contributes to telomere maintenance and to the de novo telomere formation necessary to repair broken chromosomes.
Plasmodial telomerase is likely to be necessary to maintain telomeres at a constant length during the highly replicative stages of the parasite's life cycle, such as the bloodstream stages. We hypothesize that drugs that block telomerase activity would induce telomere shortening, chromosome loss and eventually death of the parasite. In the present work, we report the identification and characterization of a putative TERT from the human malaria parasite P.falciparum (PfTERT). We also identified TERT candidate genes from two murine and one simian Plasmodial species. Using immunolocalization techniques, PfTERT and telomeres were shown to localize in different regions of the nucleus. After identifying the nucleolus as a sub-nuclear compartment in P.falciparum for the first time, we were able to show that PfTERT generally localizes within the nucleolar region. The implications of such spatial segregation are discussed.
MATERIALS AND METHODS
Plasmodium falciparum cultures
Plasmodium falciparum blood stage parasites from the FCR3 strain were cultured as described previously (31). Synchronization of cultures for time-course immunofluorescence (IF) analysis consisted of two consecutive Plasmagel treatments, at a 48 h interval, followed by Sorbitol treatment. Parasites were collected at the beginning of the next cycle (∼40 h later). We estimated that the parasites were synchronized within a window of ∼6 h.
Genome searches and sequence analysis
Genes coding for putative TERTs in Plasmodium species were obtained using TBLASTN from both NCBI Custom Blast in Malaria Genetics & Genomics (http://www.ncbi.nlm.nih.gov/Malaria/Plasmodiumblcus.html) and PlasmoDB (www.plasmodb.org). To identify the first Plasmodial TERT gene (PfTERT), motif T and other RT motifs were used as queries. The PfTERT putative gene has been annotated with the accession no. chr13_400065.gen2 in PlasmoDB database and as GenBank accession no. AX112155. TERT genes from Plasmodium yoelii (GenBank accession no. AABL01000132) and Plasmodium knowlesi (PlasmoDB accession no. Pk_812g03q1c) were found by blasting the entire PfTERT gene against the relevant database. For Plasmodium berghei, the best hit was an expressed sequence tag clone (GenBank accession no. AZ525829). Using oligonucleotides that match PfTERT, we amplified a larger fragment containing the putative PbTERT gene. The 3942 bp fragment was cloned and sequenced. Although it lacks the 3′ end of the open reading frame (ORF), it spans motifs GQ/N, QFP, T, 1 and 2.
The number of significant large tandem repeats in a gene was computed with the mreps software [(32), http://www.loria.fr/mreps/]. This program identifies all maximal repeats in a given DNA sequence, compares them with the repeats obtained in simulations of DNA sequences of the same length and composition and eventually outputs the ones that are statistically relevant.
Construction of motif alignment and phylogenetic tree
Known alignments of the motifs in other characterized species were used to identify the motifs in the Plasmodium species by independently using Clustal W and HMMer (33,34). Both analyses provided similar results. Once identified and aligned, motifs were manually checked, in order to remove the positions showing a very large variability (Figure 1).
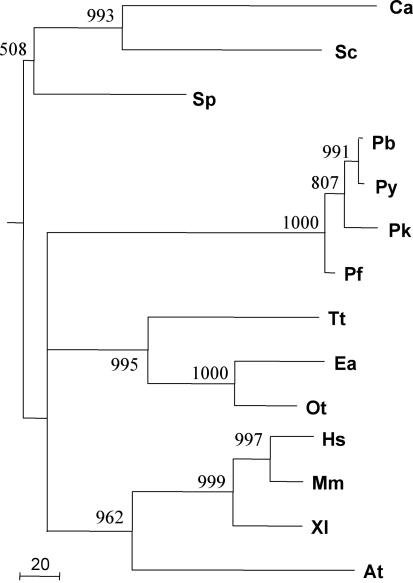
For each species (except Giardia lamblia and Caenorhabditis elegans), the amino acid sequences of all motifs were concatenated in order to produce a single continuous sequence. In P.falciparum, for example, this resulted in a concatenated sequence with 343 amino acids. Phylogenetic trees were constructed using PROML from the PHYLIP package (35), using the JTT model and the Gamma correction with eight discrete classes. The 1000 bootstraps values were computed using SEQBOOT and CONSENSE, also from the PHYLIP package (Figure 3).
Figure 3.
The phylogenetic tree based on sequences of concatenated telomerase motifs is fully compatible with the known phylogeny for these species. The tree was built as described in Materials and Methods. A total of 1000 bootstraps experiments were performed, and bootstraps >50% are shown. Abbreviations of the species are as in Figure 1. The unit of branch length is the expected fraction of changed amino acids.
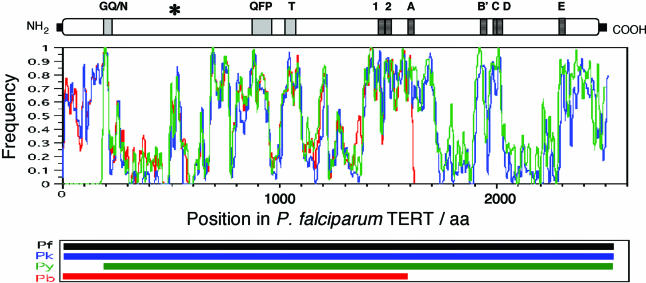
To compute the average similarity of P.falciparum TERT with the other Plasmodium TERTs, a multiple alignment was constructed using the entire sequences of the four Plasmodium species. We then used a 20 amino acid sliding window along the multiple alignments to score for alignment quality (Figure 2).
Figure 2.
Plasmodium TERTs are highly divergent outside telomerase and RT motifs. Average similarity of PfTERT to PkTERT (blue), Py TERT (green) and PbTERT (red). Similarity was computed in sliding windows of 20 amino acids over the multiple alignment. A score of 1 means that the 20 amino acids of the window are identical, whereas 0 indicates that the sequences do not share a single identical amino acid for a given position within the alignment (either because of substitutions or insertions/deletions). Above the graphics, the diagram indicates the in-scale position of all motifs in PfTERT protein. The asterisk points to an example of a region of PfTERT that is highly conserved among Plasmodium TERTs, but where no motif has yet been assigned. The lower panel shows the contribution of each Plasmodium TERT to the multiple alignments. Note that PyTERT and PbTERT sequences are not complete.
Antibodies
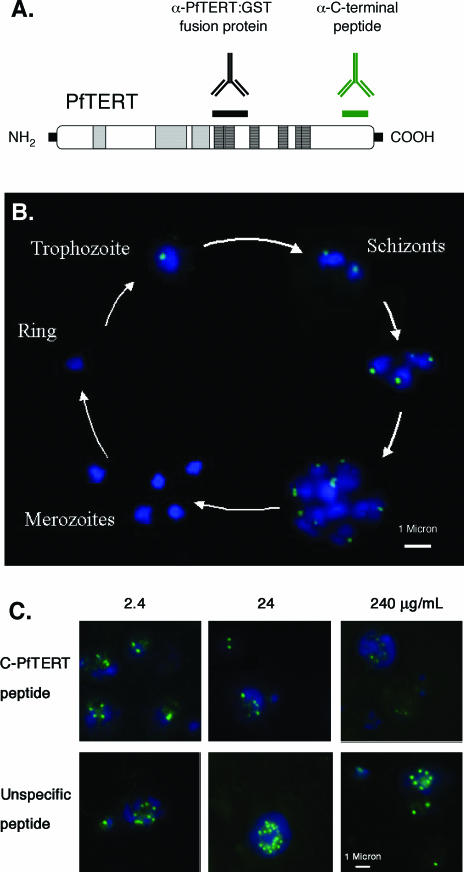
Two different sera were produced against distinct regions of the PfTERT protein. A rabbit serum was obtained by immunizing rabbits with C-PfTERT peptide: a 15 amino acid peptide from the PfTERT C-terminus coupled with keyhole limpet hemocyanin (KLH) carrier protein (Sigma–Aldrich), N-CKIKKRLINKYKIGH-C. This peptide corresponds to a region of PfTERT unique to Plasmodium species. A mouse serum was prepared from the immunization of C129 mice with a glutathione S-transferase (GST)-fusion protein corresponding to the 220 amino acids that span motifs 1 and 2.
Human autoimmune serum (S63) directed against fibrillarin (Nop1 is the Saccharomyces cerevisiae orthologue) was identified in the extracts of fibrillarin–green fluorescent protein human cell line using western–blot analysis. These sera were found to cross-react with fibrillarin in many species, including yeast Nop1 (36). A rabbit anti-PfNop1 serum was obtained by immunizing rabbits with two synthetic peptides coupled with KLH, such as N-GRGNKDRKSFKKDNK-C and N-DLTNMSKKRSNIVPI-C.
Immunofluorescence microscopy
Parasitized erythrocytes from a fresh culture of 8–12% parasitemia were centrifuged and washed once in RPMI 1640 (Gibco-BRL) and once in phosphate-buffered saline (PBS) (0.15 M NaCl and 10 mM sodium phosphate, pH 7.2). Cell pellets were re-suspended in 5 vol of PBS and a monolayer was set onto microscope slides as described previously (37). Cells were air-dried for 30 min and fixed for 10 min at room temperature in freshly prepared 2% paraformaldehyde in PBS. Slides were blocked for 10 min in PBS 0.1% BSA. Rabbit anti-PfTERT and human anti-Nop1 sera were diluted 1:200 in PBS 0.1% BSA and incubated with cells for 1 h. After washing in PBS 0.1% BSA, cells were incubated for 45 min in a 1:200 dilution of anti-rabbit or anti-human IgG-, fluorescein isothiocyanate- or rhodamine-conjugated antibodies (Sigma). Slides were washed thoroughly in PBS and mounted in VECTASHIELD anti-fading with DAPI (4′,6-diamidino-2-phenylindole) (Vector Labs). Images were captured using a Nikon E800 optical microscope.
Peptide competition assays were performed to test the specificity of the anti-PfTERT antibodies. Increasing concentrations of a specific (C-PfTERT) and an unspecific peptide were added to the serum diluted 1:3 in PBS. The mixture was incubated overnight in a rotating wheel at 4°C. The following day, antibody/peptide complexes were diluted 1:70 in PBS, in order to obtain a final dilution of the serum of around 1:200. IF assay was carried out as described above. Images were captured with constant acquisition parameters.
Immunofluorescence combined with in situ hybridization
Cells were prepared as for standard IF except following fixation cells were permeabilized in 0.1% NP-40 in PBS for 5 min. After incubation with primary and secondary antibodies, cells were post-fixed in 2% paraformaldehyde in PBS for 5–10 min. Fluorescence in situ hybridization (FISH) was performed as described previously (37). Hybridization was performed overnight with 0.5–1 ng/μl of probe in 50% formamide (Roche-Boehringer), 10% dextran sulfate (Quantum), 2× SSPE and 250 μg/ml herring sperm DNA. The probe used to visualize the sub-telomeric Rep20 repetitive DNA consists of a 1.4 kb fragment containing ∼70 Rep20 units, in which biotin was incorporated using the Biotin High-Prime kit (Roche). To detect biotinylated probes (Rep20), slides were incubated for 30 min at room temperature with 50 ng/ml of Rhodamine-conjugated Avidin (Roche).
Ultrastructure analysis
Cells were fixed using 0.1 M glutaraldehyde in phosphate buffer containing 0.05 M sucrose for 1 h at 4°C. After centrifugation at 8000 g, cells were washed for 10 min in phosphate buffer and twice for 10 min in 0.1 M cacodylate buffer. Post-fixation was performed in 1% osmium tetroxide in cacodylate buffer for 1 h, followed by a 30 min washing step in cacodylate buffer. Cells were washed in 30% methanol for 10 min and stained in 2% uranyl acetate in 30% methanol for 1 h at room temperature. After rinsing in 30% methanol, cells were dehydrated in a series of increasing concentrations of methanol (30–100%). Dehydrated cells were washed twice in propyl oxide and embedded in Epon 812 (Polysciences). Ultra-thin sections were cut with a Leica UltraCut UCT. Cuts were post-stained with uranyl acetate and Reynold's lead citrate. Observations were made on a Jeol 1200 EX II at 80 kV accelerating voltage electron microscope.
Immuno-electron microscopy
Cells were fixed for 1 h at 4°C in 4% paraformaldehyde in phosphate buffer. After washing in phosphate buffer twice for 5 min, cells were incubated for 30 min at room temperature in 0.25% NH4Cl in phosphate buffer in order to inactivate residual aldehyde groups. Cells were washed twice in phosphate buffer and finally embedded in 10% gelatin buffer on ice. Specimens were next infused in 1.7 M sucrose + 15% polyvinylpirrolidone for at least 4 h at 4°C. Small blocks (1 mm3) were cryofixed by immersion in liquid nitrogen at −196°C and by adding substitution medium in an automate freeze substitution (Leica). Substitution was performed for 16 h at −90°C in anhydrous methanol containing 0.5% uranyl acetate, followed by temperature increments of 5°C/h, until 4°C were reached. After incubation in LR White resin at 20°C, samples were cut into ultra-thin sections for immunolabelling. They were then incubated with anti-PfTERT rabbit serum at two different dilutions: 1:50 or 1:25 in TBG (0.1% gelatin and 0.1% BSA in 0.1 M Tris buffer) for 1 h at room temperature. After six 5 min washes in PBG, sections were incubated for 1 h at room temperature with protein A conjugated with 10 nm colloidal gold (Auroprobe TM/EM protein A.G10 RPN438; Batch 158170) diluted 1:20 in PBG. Controls with either no primary antibody or with pre-immune serum were performed. Ultra-thin sections were washed six times for 5 min in TBG and TBS. After fixation to grids with 1% glutaraldehyde, thin sections were washed in distilled water and post-stained for 10 min in uranyl acetate and 3 min in diluted lead citrate (1:2). Observations were made as described for ultrastructure analysis.
RESULTS
Identification of P.falciparum TERT gene
With the perspective of using telomerase as a target for antimalarials, we have characterized the protein component of telomerase in P.falciparum. Two previous observations indicated the existence of such an enzyme in P.falciparum. First, telomeric DNA is composed of canonical G-rich repeats (25), suggesting that telomeres are maintained by telomerase. Second, telomerase enzymatic activity has previously been detected in semi-purified nuclear extracts (30). Attempts to PCR amplify a PfTERT gene based on consensus motifs generated from other genomes were not successful, and only after advances in the sequencing project of malaria genomes we were able to identify a candidate TERT gene. The P.falciparum genome database was searched with the conserved motifs described previously: the T motif and the seven RT motifs (10). Two overlapping contigs displayed a high score. Their alignment resulted in a sequence of ∼10 kb, containing a large ORF of 7554 bp, which was predicted to encode a protein of 2518 amino acids. We have named this gene PfTERT and its corresponding protein PfTERT.
Features of P.falciparum TERT protein
PfTERT contains all of the canonical motifs of RTs (motifs 1, 2 and A–E; Figure 1A), as well as those conserved amino acids known to be critical for the RT activity (1,38) (marked with circles in Figure 1B). In the N-terminal half of PfTERT, we found three previously described motifs specific to TERTs: GQ/N, QFP and T (16), but we failed to identify the CP motif. Amino acids known to be specific for telomerases were found in PfTERT (marked with squares in Figure 1B): Arg in motif 1, an aromatic residue (Phe or Tyr) following the two critical Asp residues in motif C and a motif similar to the Trp-X-Gly-X-Ser/Leu in motif E.
The PfTERT predicted molecular weight is ∼280 kDa, which is almost three times the size of other TERTs described previously (e.g. S.cerevisiae Est2p is 103 kDa). Among most of the TERTs, slight differences in length are chiefly due to size variations in the N-terminal half of the protein and between motifs A and B′ (6). In contrast, the difference in size of the Plasmodial TERT results from increased distance between nearly all motifs. The in-scale diagram of Figure 1A shows that the spacing between the motifs is much greater in PfTERT than in ScEst2p. Only two blocks of motifs show relatively conserved spacing: motifs 1+2 and motifs B′+ C+D (Figure 1B). As observed in other P.falciparum proteins, PfTERT contains insertions of basic amino acids that are absent from its orthologues in other genomes (39). For example, one of these segments (between motifs T and 1) consists of a stretch of 30 asparagines, interspaced by a few lysines, isoleucines and tyrosines.
Since Plasmodium proteins are systematically larger than their orthologues in yeast or humans (40), we tested whether that difference is the same for PfTERT and other DNA and RNA polymerases (Table 1). As expected, most polymerases genes are larger in P.falciparum (Pf/Yeast ratio between 1.00 and 1.75), but surprisingly the difference between TERT sequences is much bigger (Pf/Yeast ratio = 2.85). To test whether this is related to the number of stretches of basic amino acids easily detectable in the PfTERT protein sequence, we computed the number of significant large tandem repeats in these genes (Table 1). We observed a larger number of repeats in PfTERT, 28, than in other Plasmodium polymerases of similar size. This difference is mainly due to the higher number of nucleotide repeats, 11, such as tandem A or T nucleotides. This is consistent with the higher A+T content of this gene (80.8%) relative to the others. In conclusion, we assume that the large size of PfTERT gene is due partially to a general trend of Plasmodium genes being larger than the yeast orthologues, and also due to the presence of more A/T repeats.
Table 1.
Comparative analysis of the DNA coding sequence of some polymerases in P.falciparum and S.cerevisiae
| Gene | Function | Length of Pf gene (nt) | Pf/Yeast ratio | % (A+T) of Pf gene | No. of tandem repeats in Pf gene (no. single nt repeats) |
|---|---|---|---|---|---|
| PFE0465c | RNA pol I (LSU) | 8745 | 1.75 | 76.6 | 20 (2) |
| PF11_0264 | DNA-directed RNA pol, mitochondrial | 4596 | 1.13 | 76.3 | 16 (7) |
| PF13_0150 | DNA-directed RNA pol 3 (LSU) | 7071 | 1.61 | 76.4 | 27 (4) |
| PFD0590c | DNA pol α | 5739 | 1.30 | 77.7 | 19 (4) |
| PFC0805w | DNA-directed RNA pol II | 7374 | 1.42 | 72.3 | 22 (0) |
| PFC0340w | DNA pol δ (SSU) | 1497 | 1.02 | 76.3 | 10 (1) |
| PF10_0165 | DNA pol δ | 3285 | 1.00 | 74.8 | 2 (0) |
| PfTERT | TERT | 7554 | 2.85 | 80.8 | 28 (11) |
Pf/Yeast ratio is the length of the P.falciparum gene divided by the length of the yeast orthologue (as defined in PlasmoDB). The far right column indicates the number of tandem repeats in each Pf gene (‘mreps’ software). The number of nucleotide repeats is shown in parentheses.
Abbreviations: LSU, larger subunit; and SSU, smaller subunit.
Identification of other Plasmodial TERT candidate genes
For future tests of anti-telomerase drugs in experimental malaria models (mice and monkeys), it will be useful to know the TERT sequences of the Plasmodium species that cause malaria in these animals. To search for TERT homologues in non-human malaria species, we searched the genome databases of rodent and simian Plasmodium species with the PfTERT protein sequence. We found three candidate TERT proteins: PkTERT, in the simian P.knowlesi species and PyTERT and PbTERT, in the rodent species P.yoelii and P.berghei, respectively. PkTERT shares 42% amino acid identity with PfTERT and contains all motifs present in PfTERT. PyTERT also contains all PfTERT motifs, but it appears to lack a complete N-terminus, probably as a consequence of the ongoing status of this genome project. Nevertheless, PyTERT and PfTERT share 43% identical amino acids. PbTERT gene, although incomplete, spans motifs GQ/N, QFP, T, 1 and 2. The amino acid identity between PfTERT and this truncated PbTERT candidate is 54%. The two species that infect rodents, PyTERT and PbTERT are highly homologous (76% identity), suggesting closer ancestry.
The overall percentage of amino acid identity between PfTERT and the other Plasmodial TERTs is surprisingly low for a protein that has a conserved function in eukaryotes (e.g. hTERT and msTERT share 63% amino acid identity). We carried out a multiple alignment of the four Plasmodial TERT proteins and plotted the frequency of identical amino acids along the alignment (Figure 2). As expected, the regions containing the telomerase motifs are most highly conserved. Notably, we can also see regions of high similarity, where motifs have not yet been described (asterisk in Figure 2). Nevertheless, large domains share very little similarity between Plasmodium candidate TERT proteins. These typically correspond to the regions showing repetitive sequences and characterized by many small insertions and deletions, as well as an unusual sequence composition, as discussed above.
Phylogenetic analysis of TERT proteins
To test the extent of conservation of the four Plasmodium TERTs among eukaryotes, we undertook a phylogenetic analysis of all eukaryotic TERTs described to date. By concatenating all the conserved motifs within each TERT, we built a phylogenetic tree using the maximum-likelihood algorithm. The statistical significance of branches was tested by bootstrap analysis (35). We eliminated G.lamblia and C.elegans from the analysis, as both lack several of the motifs present in the other TERTs. Besides, the low conservation of the G.lamblia and C.elegans TERT has been pointed out previously (8), and evidence has not yet been obtained of telomerase activity in these species. The final tree (Figure 3), despite displaying a few branches with low bootstrap values, is compatible with the phylogeny based on the small subunit ribosomal RNA for these species. Moreover, it lends support to the hypothesis that telomerase was present in early eukaryotes.
Sub-nuclear localization of PfTERT in blood stages
Given the role of telomerase in DNA replication, a nuclear localization is expected for this enzyme. It is known that the nuclear localization signals (NLS) are conserved among eukaryotes. Based on an algorithm that predicts sorting signals and localization sites encoded in protein sequences (PSORTII) (http://psort.ims.u-tokyo.ac.jp/; Kenta Nakai, Human Genome Center, Tokyo, Japan), we found that PfTERT contains several NLS-like motifs. In fact, the k-nearest neighbours classifier (k-NN) algorithm predicts that PfTERT primary sequence refers to a protein that has 78.3% probability to be nuclear (for Tetrahymena TERT, for example, the probability is 52.2%).
In order to test this prediction, we performed IF assays on blood stage parasites. The blood stage cycle takes ∼48 h to be completed. It begins with the invasion of non-infected erythrocytes by merozoites present in the bloodstream. Once inside the erythrocyte, the parasite will undergo three stages: ring (∼0–18 h), trophozoite (∼18–38 h) and schizont (∼38–48 h) stages. S phase begins in the trophozoite stage, ∼28–31 h after merozoite invasion. Nuclear division occurs throughout schizogony, leading to the production of up to 32 individual merozoites. At the end of the 48 h cycle, erythrocytes burst releasing merozoites and the cycle starts anew. Previous studies have shown that telomerase activity is detectable by TRAP only in trophozoite and schizont stages (41). To follow the expression and localization of PfTERT during the blood stage cycle by IF, we collected infected erythrocytes from a synchronized culture at different time points: 0, 12, 24, 36, 40 and 48 h. Studies using rabbit α-PfTERT-peptide (Figure 4), revealed no staining in the first 12 h (ring stages). When the parasite matures into trophozoite stage, however, a clear pattern was observed: a single dot within a region of the nucleus. During schizogony, the number of PfTERT foci increased proportionally to the number of nuclei. When schizonts become fully mature (48 h) and merozoites are released into the blood stream, PfTERT was no longer detectable.
Figure 4.
PfTERT localizes in a discrete nuclear region in certain blood stream stages. (A) Diagram indicating the source of two different antibodies used to characterize PfTERT: a mouse serum raised against a GST–PfTERT fusion protein and a rabbit serum raised against a 15 amino acid peptide at the PfTERT C-terminus (C-PfTERT peptide). (B) The 48 h time course in blood stages, during which parasites develop inside an erythrocyte. PfTERT (green) was detected by IF using the rabbit anti-peptide antibody; nuclear DNA was stained with DAPI (blue). (C) Peptide competition assay. The specificity of PfTERT serum was determined by IF after pre-incubating the antibodies with increasing concentrations of a specific (C-PfTERT) or an unspecific peptide. Detection of anti-PfTERT antibodies and staining of DNA was performed as in (B).
Two lines of evidence confirm the specificity of the PfTERT signal. First, when the above IF assay was undertaken in the presence of competing epitopes, the PfTERT signal disappeared with increasing concentrations of the C-terminal PfTERT peptide (C-PfTERT); in contrast, when an unspecifc peptide was used as competitor, the PfTERT signal intensity remained unchanged (Figure 4C). Second, the mouse sera produced against a recombinant PfTERT protein generated a similar localization pattern (data not shown).
Thus, our results show that PfTERT is expressed at detectable levels in trophozoites and schizonts, the same stages that were previously shown to have telomerase activity and where DNA replication occurs. Moreover, our studies also revealed that PfTERT is not detectable throughout the entire nucleoplasm, but rather in a discrete sub-nuclear compartment.
PfTERT: localization distinct from telomere clusters
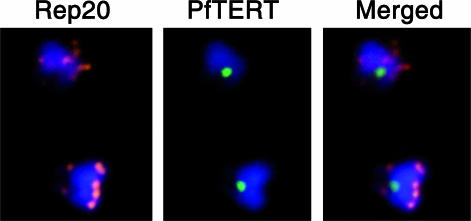
The sub-nuclear localization of PfTERT is surprising considering that telomeres, the substrate, are organized in 4–7 clusters around the nuclear periphery (27). We hypothesized that the plasmodial telomerase complex may localize to a single cluster and then ‘hop’ to another. To test this hypothesis, we combined IF with FISH, using a probe (Rep20) that recognizes a sub-telomeric repeat (telomeric DNA and Rep20 FISH signals co-localize; Scherf et al., unpublished data) and examined if PfTERT co-localizes with a telomere cluster (Figure 5). Optimization of the IF/FISH technique allowed to have >50% of the cells double-labelled. Strikingly, we concluded that in the majority of the cells, telomeres and PfTERT did not co-localize. We observed occasional colocalization of PfTERT with telomere clusters, which may be due to errors prone to the 2D analysis of the images. We conclude that, within the detection limits of IF, PfTERT is located in a sub-nuclear compartment that is distinct from telomeric clusters.
Figure 5.
PfTERT is not detectable at telomeres. Indirect-IF of PfTERT (green) combined with FISH of Rep20 (red), a sub-telomeric repetitive motif. DNA was stained with DAPI (blue). Both cells are blood stage trophozoites.
PfTERT: localization in a sub-compartment of the nucleolus
The largest sub-nuclear compartment of eukaryotic cells is the nucleolus, the main function of which is the transcription of rRNA genes and the assembly of ribosomes. Some ribonucleoproteins are also assembled in this compartment. In recent studies, human and yeast TERTs were shown to be enriched at the nucleolus [reviewed in (4)]. Given PfTERT's localization to a discrete domain of the nucleus away from the telomeres, we hypothesized that it localizes to the nucleolus.
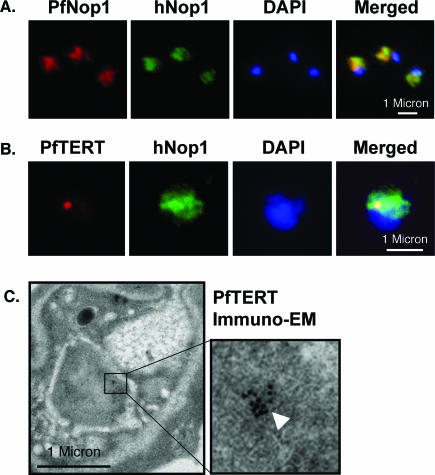
In P.falciparum, a nucleolus has never been identified and its presence has even been questioned given the low number of ribosomal genes in the genome (40). Thus, in order to test if PfTERT localizes in the nucleolus, we searched for nucleolar markers. By IF, we tested different sera against highly conserved nucleolar proteins. Strikingly, human anti-hNop1 serum revealed a hat-like structure polarized towards one side of the nucleus (Figure 6A), very similar to that described for S.cerevisiae. Rabbit antibodies raised against a recombinant PfNop1 protein showed an identical staining pattern (Figure 6A). Immuno-electron microscopy (EM) observations further support that the anti-hNop1 serum is a nucleolar marker, as it preferentially stains a sub-nuclear region (data not shown). Taken together, our data provide evidence for the existence of a nucleolus in P.falciparum.
Figure 6.
PfTERT localizes in a sub-compartment of the nucleolus. (A) IF with an anti P.falciparum Nop1 serum (red) shows the nucleolus as a hat-like structure in the nucleus of a trophozoite stage. An anti-human Nop1 serum (green) recognizes exactly the same sub-nuclear region. (B) Double-labelling reveals that PfTERT (red) co-localizes with part of the nucleolus (green); yellow spot in the merge image. (C) Immuno-EM shows that PfTERT (white arrow head) localizes to a peripheral region of the nucleus. In (A and B), DNA was stained with DAPI (blue).
In order to test whether PfTERT localizes to the nucleolus, we performed IF in which cells were double-labelled using the rabbit anti-PfTERT and the human anti-hNop1 sera. Strikingly, PfTERT co-localized with the Nop1-defined region, indicating it localizes to the nucleolus. Remarkably, PfTERT is not found all over the nucleolus, but is restricted to a sub-compartment of this organelle (Figure 6B). This contrasts with the situation in other eukaryotes (42–44), in which ectopically expressed TERT accumulates uniformly throughout the nucleolus. Taken together, these data strongly suggest that PfTERT localizes in a sub-compartment of the nucleolus. This finding is further supported by immuno-EM studies. The anti-PfTERT serum revealed a single cluster of gold beads close to the nuclear membrane (Figure 6C).
DISCUSSION
In the present work, we identified and characterized a gene in P.falciparum that has the characteristics of the protein component of telomerase, named here PfTERT. It contains structural features that are common with the telomerases known in other species: RT motifs, telomerase-specific motifs and pI ≥ 10. We also identified homologous genes in other Plasmodial species: the simian P.knowlesi, PkTERT, and two murine species, P.yoelii and P.berghei (PyTERT and PbTERT genes).
A comparison of PfTERT and other eukaryote TERTs shows very different degrees of conservation along the sequence. This is in agreement with experimental data in S.cerevisiae, where hypermutable segments separate conserved and vital regions of the molecule (15). Within the conserved regions, we were able to identify almost all of the TERT-specific and all of the RT-specific motifs. The CP motif, which was not found in any Plasmodia, is the only exception. This motif is also absent from G.lamblia TERT, and given its very low-sequence conservation, doubts have been put forward for its significance outside the ciliates group (6,15). It is thus unclear if this motif has diverged beyond recognition or if it has appeared after the separation between Plasmodia and Giardia and the other eukaryotes analysed.
PfTERT contains several stretches of 10–20 basic amino acids, such as asparagines, which are encoded by A-rich codons. Interestingly, among P.falciparum polymerases, the TERT gene is the one that contains more A+T nucleotides, more repeats and a higher increase in size relative to the yeast orthologue (Table 1), suggesting that PfTERT sequence is less functionally constrained than that of the other polymerases. These repeats are therefore more likely to reflect the propensity of the P.falciparum genome by polymerase slippage events due to the presence of stretches of A and/or T (45,46), instead of selection process that leads to increased gene length. If this is so, the extreme A+T richness of Plasmodia genomes (80% A+T for P.falciparum) may favour the increase in gene size and led to the accumulation of the repetitive regions in non-essential gene areas.
Given the common biological role of telomerase in eukaryotes, we expected its catalytic component to be much conserved among species of the same genus. Surprisingly, we found very little sequence conservation between the TERT sequences of the different Plasmodium species: PfTERT shares 42% identical amino acids with PkTERT, 43% with PyTERT and 54% with PbTERT. The regions between the conserved motifs show lower amino acid identity (Figure 2). The existence of such hypervariable regions is indicative of a continuous process of genome evolution of malaria species, as it was previously shown by a phylogenetic analysis based on comparison of the small-subunit ribosomal RNA gene sequences (47). These differences among Plasmodial TERT proteins may play a role in the specific mean telomere length observed for the different species (2.0 kb for P.yoelii, 1.2 kb for P.falciparum and 6.7 kb for P.vivax) (26).
A phylogenetic tree built from sequences consisting of a concatenation of the conserved motifs, ignoring the hypervariable stretches, respects the known phylogenetic relationships, both among Plasmodia species and among eukaryotes in general. Our results confirm previous analysis made with a smaller dataset (6), and reinforce the hypothesis that telomerase was present in early eukaryotes. The phylogenetic analysis is complicated by a few branches lacking robustness (low-bootstrap values) and by the lack of motifs in the sequences of the putative TERT of C.elegans and G.lamblia. Addition of newly sequenced eukaryotic genomes will allow to further resolve these questions.
It was previously shown that, in P.falciparum cell extracts, telomerase can be efficiently inhibited by RT type of drugs, such as nucleoside analogues (30). These same drugs are currently being tested on in vitro cultures and preliminary data show killing of P.falciparum parasites after 3–5 blood stage cycles at micromolar concentrations (data not shown). Moreover, we were unable to generate a knock-out of the PfTERT gene, supporting the idea that telomerase activity is needed for blood stage parasite proliferation. Thus, a prophylactic therapy based on plasmodial telomerase inhibitors might be possible. In this work, the comparison of the TERT sequences from human, rodent and simian Plasmodium species provides valuable information for the design of specific anti-telomerase drugs. Obviously, drugs should be targeted to conserved regions and functional domains, in order to avoid rapid emergence of drug resistance.
The cell-cycle progression in P.falciparum is not yet completely understood (48). DNA synthesis begins in the relatively small trophozoites (intra-erythrocytic form), but nuclear subdivision, which leads to the formation of multinucleate cells, occurs only during schizogony. Whether or not any gap phases (G) exist between each round of DNA synthesis and mitosis is unknown. Eventually, a schizont composed of 8–32 nuclei undergoes segmentation, which culminates with the formation of individual merozoites that burst from the erythrocyte into the blood stream. Our observations indicate that PfTERT is only detectable in trophozoites and schizonts, the stages where multiple rounds of DNA synthesis occur and where telomerase activity can be detected (41). Thus, our data reveal a correlation between the expression of PfTERT protein and the pattern of telomerase activity in the parasite. As is known to occur in human cells, we speculate that the regulation of PfTERT expression may be one of the mechanisms that P.falciparum uses to control telomerase activity (49).
Studies in S.cerevisiae and human cells have shown that ectopically expressed TERTs are partially enriched in the nucleolus (42–44). In humans, the telomerase RNA component also seems to be enriched at the nucleolus (50,51) or Cajal bodies (52,53). These observations led to the speculation that the nucleolus may play a dual role: providing an environment for the biogenesis of telomerase [reviewed in (4)] and regulating the release of telomerase to its telomeric substrates (54).
In this work, we described the localization of the endogenous telomerase catalytic component. We observed that PfTERT is not evenly distributed in the nucleoplasm, but accumulates in a discrete peripheral site of the nucleus, which we identified as a sub-compartment of the nucleolus. In the P.falciparum related apicomplexan parasite Toxoplasma, the nucleolus appears as a dot in the middle of the nucleus (as detected by anti-human fibrillarin or a YFP-tagged nucleolar protein) (55). In P.falciparum, we observe a significantly larger structure with a hat-like pattern mostly at the periphery of the nucleus. This pattern was observed with human serum against hNop1 and confirmed with antibodies that we raised against P.falciparum Nop1. Thus, our experimental data strongly suggest that the nucleolus of P.falciparum has a more expanded structure than in Toxoplasma.
It is possible that, as in S.cerevisiae, the sub-nucleolar PfTERT-containing structure is functionally equivalent to the mammalian Cajal bodies (56). The RNA component of P.falciparum telomerase has not yet been identified and thus we cannot test where the assembled telomerase complex localizes. We speculate that the presence of PfTERT in the nucleolus may be necessary for the assembly of the telomerase complex, given that in some eukaryotes this sub-nuclear compartment is involved in the assembly of ribonucleoproteins (57).
Given the role of telomerase at telomeres, one might predict a release of the telomerase activity from the nucleolus to the nucleoplasm during the replication of telomeric DNA. Surprisingly, a combination of immunolabelling with anti-PfTERT sera and FISH, did not reveal a co-localization of PfTERT with the telomeric clusters. The general view point in the telomerase field is that only a few telomerase molecules are necessary to lengthen telomeres. This may explain the perceptible absence of telomerase at telomeres using immunolocalization assays. By chromatin immunoprecipitation, however, it was shown that anti-telomerase antibodies were capable of precipitating telomere repeats, providing evidence that telomerase can be associated with telomeres in vivo (58).
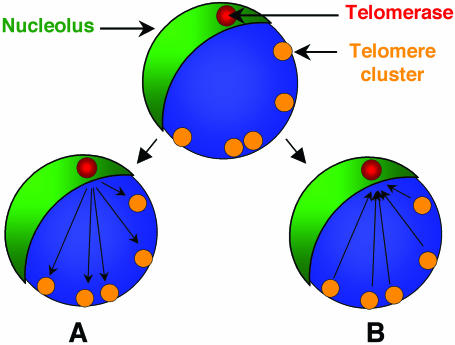
We propose two models that could explain our observations (Figure 7). In the first model, we predict that a fraction of the activated telomerase is released from the nucleolus to the nucleoplasm, to allow complete telomeric DNA replication, as observed with ectopically expressed hTERT (54). However, in P.falciparum, levels of telomerase released to the nucleoplasm may be insufficient to be detected by IF. After the addition of telomeric repeats onto the chromosome ends, telomerase is either degraded or shuttled back to the nucleolus. The second model speculates that telomerase is not released to the nucleoplasm. Instead, it remains in a sub-nucleolar compartment where telomeres are elongated. In this scenario, chromosome ends would move to the nucleolus (or its proximity) when telomere replication takes place. In support of this second model, preliminary studies in our laboratory show a bouquet-like structure of telomeres proximal to the nucleolus during DNA replication (data not shown). In both models, the telomerase sub-nucleolar compartment may fulfil a role in the assembly of the telomerase complex and/or a role in the regulation of telomerase activity by determining when telomerase should have access to its substrate. Further studies are required to test these hypotheses.
Figure 7.
Model for the mechanism of action of telomerase in P.falciparum. We speculate that a sub-compartment of the nucleolus (green) is the location where telomerase complex (red) is assembled and stored. When telomeres (yellow) replicate, two scenarios can be envisioned. A, A subfraction (undetectable by IF) of the assembled/active telomerase is released from the sub-nucleolar compartment and goes to the telomeres, where replication takes place. B, Telomeres form a bouquet-like structure during mitosis and move to the proximity of the nucleolus. Telomerase replicates the chromosome ends, while remaining anchored in the nucleolus.
Acknowledgments
We thank Christine Scheidig Benatar for technical assistance, Joanna Lowell for the revision of the manuscript and Stuart Ralph for help with electron microscopy. This work was supported by a grant from the European Community (Network of excellence BioMalPar). Funding to pay the open access publication charges for this article was provided by the same grant. L.M.F and L.M.S. were supported by fellowships (Ref. PRAXIS-XXI/16020/98 and SFRH/BD/11756/2003, respectively) from the Fundação para a Ciência e Tecnologia, Portugal.
REFERENCES
- 1.Lingner J., Hughes T.R., Shevchenko A., Mann M., Lundblad V., Cech T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura T.M., Cooper J.P., Cech T.R. Two modes of survival of fission yeast without telomerase. Science. 1998;282:493–496. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- 3.Blasco M.A., Lee H.W., Hande M.P., Samper E., Lansdorp P.M., DePinho R.A., Greider C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 4.Vega L.R., Mateyak M.K., Zakian V.A. Getting to the end: telomerase access in yeast and humans. Nature Rev. Mol. Cell. Biol. 2003;4:948–959. doi: 10.1038/nrm1256. [DOI] [PubMed] [Google Scholar]
- 5.Evans S.K., Lundblad V. Positive and negative regulation of telomerase access to the telomere. J. Cell. Sci. 2000;113:3357–3364. doi: 10.1242/jcs.113.19.3357. [DOI] [PubMed] [Google Scholar]
- 6.Bryan T.M., Sperger J.M., Chapman K.B., Cech T.R. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl Acad. Sci. USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins K., Gandhi L. The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl Acad. Sci. USA. 1998;95:8485–8490. doi: 10.1073/pnas.95.15.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malik H.S., Burke W.D., Eickbush T.H. Putative telomerase catalytic subunits from Giardia lamblia and Caenorhabditis elegans. Gene. 2000;251:101–108. doi: 10.1016/s0378-1119(00)00207-9. [DOI] [PubMed] [Google Scholar]
- 9.Counter C.M., Meyerson M., Eaton E.N., Weinberg R.A. The catalytic subunit of yeast telomerase. Proc. Natl Acad. Sci. USA. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura T.M., Morin G.B., Chapman K.B., Weinrich S.L., Andrews W.H., Lingner J., Harley C.B., Cech T.R. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- 11.Metz A.M., Love R.A., Strobel G.A., Long D.M. Two telomerase reverse transcriptases (TERTs) expressed in Candida albicans. Biotechnol. Appl. Biochem. 2001;34:47–54. doi: 10.1042/ba20010021. [DOI] [PubMed] [Google Scholar]
- 12.Oguchi K., Liu H., Tamura K., Takahashi H. Molecular cloning and characterization of AtTERT, a telomerase reverse transcriptase homolog in Arabidopsis thaliana. FEBS Lett. 1999;457:465–469. doi: 10.1016/s0014-5793(99)01083-2. [DOI] [PubMed] [Google Scholar]
- 13.Meyerson M., Counter C.M., Eaton E.N., Ellisen L.W., Steiner P., Caddle S.D., Ziaugra L., Beijersbergen R.L., Davidoff M.J., Liu Q., et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg R.A., Allsopp R.C., Chin L., Morin G.B., DePinho R.A. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 15.Friedman K.L., Cech T.R. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 1999;13:2863–2874. doi: 10.1101/gad.13.21.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia J., Peng Y., Mian I.S., Lue N.F. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 2000;20:5196–5207. doi: 10.1128/mcb.20.14.5196-5207.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J.-P. Molecular mechanisms regulating telomerase activity. In: Mattson M.P., Pandita T.K., editors. Telomerase, Aging and Disease. The Netherlands: Elsevier Science; 2002. pp. 33–59. [Google Scholar]
- 18.Shay J.W., Wright W.E. Telomerase activity in human cancer. Curr. Opin. Oncol. 1996;8:66–71. doi: 10.1097/00001622-199601000-00012. [DOI] [PubMed] [Google Scholar]
- 19.Marciniak R., Guarente L. Human genetics. Testing telomerase. Nature. 2001;413:370–371. doi: 10.1038/35096663. 373. [DOI] [PubMed] [Google Scholar]
- 20.Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., Wright W.E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 21.Mergny J.L., Riou J.F., Mailliet P., Teulade-Fichou M.P., Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knell A.J. Malaria. Oxford: Oxford University Press; 1991. Trustees of the Welcome Trust. [Google Scholar]
- 23.Ridley R.G. Medical need, scientific opportunity and the drive for antimalarial drugs. Nature. 2002;415:686–693. doi: 10.1038/415686a. [DOI] [PubMed] [Google Scholar]
- 24.Hoffman S.L., Subramanian G.M., Collins F.H., Venter J.C. Plasmodium, human and Anopheles genomics and malaria. Nature. 2002;415:702–709. doi: 10.1038/415702a. [DOI] [PubMed] [Google Scholar]
- 25.Vernick K.D., McCutchan T.F. Sequence and structure of a Plasmodium falciparum telomere. Mol. Biochem. Parasitol. 1988;28:85–94. doi: 10.1016/0166-6851(88)90055-2. [DOI] [PubMed] [Google Scholar]
- 26.Figueiredo L.M., Pirrit L.A., Scherf A. Genomic organisation and chromatin structure of Plasmodium falciparum chromosome ends. Mol. Biochem. Parasitol. 2000;106:169–174. doi: 10.1016/s0166-6851(99)00199-1. [DOI] [PubMed] [Google Scholar]
- 27.Freitas-Junior L.H., Bottius E., Pirrit L.A., Deitsch K.W., Scheidig C., Guinet F., Nehrbass U., Wellems T.E., Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 28.McEachern M.J., Krauskopf A., Blackburn E.H. Telomeres and their control. Annu. Rev. Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 29.Scherf A., Figueiredo L.M., Freitas-Junior L.H. Plasmodium telomeres: a pathogen's perspective. Curr. Opin. Microbiol. 2001;4:409–414. doi: 10.1016/s1369-5274(00)00227-7. [DOI] [PubMed] [Google Scholar]
- 30.Bottius E., Bakhsis N., Scherf A. Plasmodium falciparum telomerase: de novo telomere addition to telomeric and nontelomeric sequences and role in chromosome healing. Mol. Cell. Biol. 1998;18:919–925. doi: 10.1128/mcb.18.2.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scherf A., Hernandez-Rivas R., Buffet P., Bottius E., Benatar C., Pouvelle B., Gysin J., Lanzer M. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolpakov R., Bana G., Kucherov G. mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res. 2003;31:3672–3678. doi: 10.1093/nar/gkg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eddy S.R. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 34.Thomson J.D., Higgins D.G., Gibson T.J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felsenstein J. 1993. PHYLIP (Phylogeny Inference Package) version 3.6a. Department of Genetics, University of Washington, Seattle, WA.
- 36.Gautier T., Fomproix N., Masson C., Azum-Gelade M.C., Gas N., Hernandez-Verdun D. Fate of specific nucleolar perichromosomal proteins during mitosis: cellular distribution and association with U3 snoRNA. Biol. Cell. 1994;82:81–93. doi: 10.1016/s0248-4900(94)80010-3. [DOI] [PubMed] [Google Scholar]
- 37.Freitas-Junior L.H., Scherf A. Methods in Malaria Research. Waldorf, MD: ATCCC Books; 2000. [Google Scholar]
- 38.Weinrich S.L., Pruzan R., Ma L., Ouellette M., Tesmer V.M., Holt S.E., Bodnar A.G., Lichtsteiner S., Kim N.W., Trager J.B., et al. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet. 1997;17:498–502. doi: 10.1038/ng1297-498. [DOI] [PubMed] [Google Scholar]
- 39.Aravind L., Iyer L.M., Wellems T.E., Miller L.H. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 40.Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sriwilaijareon N., Petmitr S., Mutirangura A., Ponglikitmongkol M., Wilairat P. Stage specificity of Plasmodium falciparum telomerase and its inhibition by berberine. Parasitol. Int. 2002;51:99–103. doi: 10.1016/s1383-5769(01)00092-7. [DOI] [PubMed] [Google Scholar]
- 42.Etheridge K.T., Banik S.S., Armbruster B.N., Zhu Y., Terns R.M., Terns M.P., Counter C.M. The nucleolar localization domain of the catalytic subunit of human telomerase. J. Biol. Chem. 2002;277:24764–24770. doi: 10.1074/jbc.M201227200. [DOI] [PubMed] [Google Scholar]
- 43.Yang Y., Chen Y., Zhang C., Huang H., Weissman S.M. Nucleolar localization of hTERT protein is associated with telomerase function. Exp. Cell. Res. 2002;277:201–209. doi: 10.1006/excr.2002.5541. [DOI] [PubMed] [Google Scholar]
- 44.Teixeira M.T., Forstemann K., Gasser S.M., Lingner J. Intracellular trafficking of yeast telomerase components. EMBO Rep. 2002;3:652–659. doi: 10.1093/embo-reports/kvf133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Achaz G., Netter P., Coissac E. Study of intrachromosomal duplications among the eukaryote genomes. Mol. Biol. Evol. 2001;18:2280–2288. doi: 10.1093/oxfordjournals.molbev.a003774. [DOI] [PubMed] [Google Scholar]
- 46.Levinson G., Gutman G.A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 47.Waters A.P., Higgins D.G., McCutchan T.F. Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proc. Natl Acad. Sci. USA. 1991;88:3140–3144. doi: 10.1073/pnas.88.8.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnot D.E., Gull K. The Plasmodium cell-cycle: facts and questions. Ann. Trop. Med. Parasitol. 1998;92:361–365. doi: 10.1080/00034989859357. [DOI] [PubMed] [Google Scholar]
- 49.Cong Y.S., Wright W.E., Shay J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. Table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mitchell J.R., Cheng J., Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol. Cell. Biol. 1999;19:567–576. doi: 10.1128/mcb.19.1.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lukowiak A.A., Narayanan A., Li Z.H., Terns R.M., Terns M.P. The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. RNA. 2001;7:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Y., Tomlinson R.L., Lukowiak A.A., Terns R.M., Terns M.P. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol. Biol. Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jady B.E., Bertrand E., Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J. Cell. Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong J.M., Kusdra L., Collins K. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nature Cell. Biol. 2002;4:731–736. doi: 10.1038/ncb846. [DOI] [PubMed] [Google Scholar]
- 55.Gubbels M.J., Wieffer M., Striepen B. Fluorescent protein tagging in Toxoplasma gondii: identification of a novel inner membrane complex component conserved among Apicomplexa. Mol. Biochem. Parasitol. 2004;137:99–110. doi: 10.1016/j.molbiopara.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 56.Verheggen C., Lafontaine D.L., Samarsky D., Mouaikel J., Blanchard J.M., Bordonne R., Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 2002;21:2736–2745. doi: 10.1093/emboj/21.11.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taggart A.K., Teng S.C., Zakian V.A. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science. 2002;297:1023–1026. doi: 10.1126/science.1074968. [DOI] [PubMed] [Google Scholar]