Abstract
Objective:
Osteosarcoma (OS) is the most common malignant bone tumor in children and young adults. Many studies have shown that microRNAs play a critical role in proliferation and metastasis with this tumour type. However, whether aberrant expression might contribute to a metabolism switch in osteosarcoma cases is not clearly understood. In this study, we explored expression and function of miR-150 in osteosarcoma cells.
Materials and methods:
Expression of miR-150 was assessed by real-time PCR in cell lines and human patient tissues. Scramble siRNA, miR-150 inhibitor, and miR-150 mimics were transfected into osteosarcoma cells to determine their effects on proliferation rate, glucose uptake and lactate secretion. Finally, the relationship between Glut1 and the miR-150 level was explored by luciferase reporter assay and western blotting.
Result:
miR-150 was consistently decreased in cell lines and osteosarcoma tissues as compared to osteoblast cells and normal bone. Ectopic overexpression of miR-150 inhibited osteosarcoma cell proliferation and suppressed glucose uptake and lactate secretion. Loss of function of miR-150, on the other hand, enhanced osteosarcoma cell proliferation and increased glucose uptake and lactate secretion. Western blot and luciferase reporter assays showed that miR-150 may function by regulating Glut1 expression.
Conclusion:
These data suggest that miR-150 is involved in regulation of glycolysis in osteosarcoma cells by influencing Glut1 expression.
Keywords: miR-150, glycolysis, Glut1, osteosarcoma
Introduction
Osteosarcoma (OS) is the most common bone tumor in children, representing 6% of all childhood cancers (Ferguson and Goorin, 2001). Although great efforts have been made to understand the underlying mechanisms of osteosarcoma carcinogenesis, the 5-year survival and the prognosis of advanced osteosarcoma remains largely poor (Luetke et al., 2014; Wu et al., 2016). Therefore, the comprehensive understanding of the mechanisms for effective therapeutic strategies and novel diagnostic biomarkers for osteosarcoma are needed.
A major source of cellular energy and new cell mas is glucose (Cairns et al., 2011). Glucose consumption in cancer cells is markedly different from that in normal tissues. Normal cells rely primarily on mitochondrial oxidative phosphorylation to generate the energy needed for cellular processes, which is a more efficient energy production process than glycolysis. However, most cancer cells instead rely on aerobic glycolysis, a phenomenon is known as “the Warburg effect” (Hsu and Sabatini, 2008; Kroemer and Pouyssegur, 2008). Aerobic glycolysis confers cancer cells a selective advantage that leads to the rapid growth. There is increasing evidence that altered metabolism in cancer is a potential therapeutic target.
MicroRNAs (miRNAs) are a class of small non-coding RNAs, which regulate gene function by targeting mRNA for translational repression or degradation (Treiber et al., 2012). miRNAs have been implicated in diverse biological events including cell proliferation, apoptosis and metabolism (Mitra et al., 2015). Overexpression of oncogenic miRNAs or knockdown of tumor suppressor miRNAs promote tumor development. A growing evidence have reported an association between aberrant expression of miRNAs and osteosarcoma (Geller and Gorlick, 2010; Luetke et al., 2014). Recently, many microRNAs have been identified as key regulators of metabolism. For example, miR-181a mediates a metabolic switch in colon cancer cells by regulating the PTEN/AKT pathway (Wei et al., 2014), and miR-424 regulates metabolic reprogramming in cancer-associated fibroblasts by decreasing the expression of IDH3a (Zhang et al., 2015). miR-33a and miR-33b play critical roles in regulating cholesterol homeostasis and fatty acid degradation (Davalos et al., 2011), microRNA-495 regulates metabolic shift in glioma cells by targeting Glut1 (Nie et al., 2015), and knockdown of miR-182 is found to increase glucose uptake and glycolysis (Hu et al., 2014).
Aberrant expression of miR-150 has been reported in various types of cancers and appears to be significantly associated with the clinical outcome of human tumor patients. For example, elevated miR-150 targets FOXO4 expression and therefore regulates multiple genes expression, resulting in cervical cancer cell growth and survival (Li et al., 2015a); increased miR-150 expression promoted prostate cancer cell development by suppressing p27Kip1 (Liu et al., 2015); downregulation of miR-150 promotes pertuzumab resistance in ovarian cancer cells via AKT activation (Wuerkenbieke et al., 2015). Li X et.al reported that miR-150 expression was significantly decreased in osteosarcoma cell lines and miR-150 inhibits cell proliferation, invasion by regulating the expression of Sp1 (Li et al., 2015b), however the effect of miR-150 on glycolysis in osteosarcoma remains unknown.
In this study, we explored the expression of miR-150 in osteosarcoma tissues and cell lines. The reduced expression of miR-150 promotes glycolysis as measured by increases in glucose uptake and lactate secretion through regulating Glut1 expression.
Materials and Methods
Cell culture, reagents and clinical samples
HOS, G-292, U2OS, MG-63, Saos2, hFOB 1.19 and 293T cells were maintained in Dulbeco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 4.5 g/L glucose (FBS, Invitrogen, California) and 100 units/ml penicillin/streptomycin in a humidified atmosphere of 5% CO2 at 37°C. Primary antibodies against actin and Glut1 (catalog no.12939), HK2 (catalog no.2867) and PFKL (catalog no.13029) were purchased from Cell Signaling Technology (Boston, MA, USA). A total of four osteosarcoma tissues and paired normal adjacent tissues at least 4 cm away from the tumor sites were acquired from each patient by biopsy prior to any treatment.
Transfections
miR-150 inhibitor, miR-150 mimics and scramble controls were ordered from GenePharma (Shanghai, China). For the transient transfection, the cells were plated at 80% confluence and transfected with the indicated RNA molecules at a final concentration of 20 nM with the Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Quantitative real-time PCR
Total RNA of cells was extracted with TRIzol reagent as described in the manufacturer’s instructions. Reverse transcription of microRNA and mRNA were synthesized using the TaqMan qPCR miRNA assay kit according to the manufacturer’s protocol. RT-qPCR detection of miR-150 was done with SYBR Green qPCR Supermix. RNU48 was used as an endogenous internal loading control. The primers used for miR-150 and RNU48 were as follows: miR-150 forward 5’-GTCGTATCCAGTGCAGGGTCC-3’, miR-150 reverse 5’-CCCTTGUACCAGTG-3’; RNU48 forward 5’-TGATGATGACCCCAGGTAACTC-3’ and reverse 5’-GAGCGCTGCGGTGATG-3’.
Western blot analysis
Cells were lysed by RIPA buffer. The 30 µg total protein of was separated with 10% SDS-PAGE and transferred into the PVDF membrane. After blocking with 5% nonfat dried milk in TBST for 1h, the membrane was incubated with the primary antibody at 4°C overnight. After rinsing three times for 10 min, the secondary antibody was incubated with the membranes for 1hour at room temperature. After washing with TBST three times, the membranes were then detected with an enhanced chemiluminescent kit.
Dual-luciferase reporter assay
Dual-luciferase reporter assay was used to confirm whether miR-150 down-regulates Glut1 via binding to the 3’UTR region. Thus, HEK 293T cells were plated in a 48-well plate at 80-90% confluence. After co-transfection with either 20 nM of miR-150 mimics, scramble controls, together with 40 ng of pGL-Glut1-3’UTR-wt or pGL-Glut1-3’UTR-mut, cells were collected to measure the firefly activity with Dual Luciferase Assay (Promega, Madison, WI). Experiments of luciferase activity assay was performed in triplicate independently.
Cell proliferation assay
For the cell proliferation assay, cells were plated at 5000 cells/well at 96-well plates and transfected with the indicated small RNA molecules. After maintaining in serum-free medium for 6h, the cells recovered to the culture medium for 48h. 48 hours later, the cells were treated with the Cell Counting kit-8 assay kit (CCK8) (DOJINDO. Tabaru, Japan) for 1 h at 37°C. Cell viability was measured at an absorbance of 450 nm.
Measurement of glucose uptake and extracellular lactate secretion
The miR-150 inhibitor or mimics were transiently transfected into osteosarcoma cells. The medium was collected after transfection for 48 h. Lactate production was measured using the lactate assay kit (Sigma-Aldrich). Glucose uptake was measured using a colorimetric assay according to the manufacturer’s instructions (Glucose Uptake Colorimetric Assay Kit, BioVision).
Statistical analysis
Statistical analyses were performed using SPSS 17.0 statistical software. Data are presented as the mean ± SD. Results were considered to have statistically significant differences if p<0.05.
Results
miR-150 is downregulated in osteosarcoma tissues and cell lines
To explore the expression of miR-150 in osteosarcoma carcinogenesis, we downloaded the microRNA expression profile from a published datasheet (Gene expression omnibus accession: 39040). The miR-150 expression in this microRNA assay was lower in human osteosarcoma than in the human normal tissues. To further verify the decreased expression of miR-150 in osteosarcoma tissues, we examined 4 pairs of osteosarcoma and matched normal tissues using RT-PCR analysis. Relative to matched normal tissues, all four osteosarcoma tissues exhibited down-regulated expression of miR-150. In addition, we tested the expression of miR-150 in human osteosarcoma cell lines. As expected, miR-150 expression level was obviously decreased in G-292, Saos2, MG-63, HOS and U2OS cells compared with normal osteoblastic cell line hFOB19.
miR-150 suppressed osteosarcoma cell proliferation
The reduced expression of miR-150 in both human osteosarcoma tissues and cell lines prompted us to examine the function in osteosarcoma carcinogenesis. We transfected HOS cells with miR-150 mimics, miR-150 inhibitor or scramble controls at a final concentration of 20 nM and performed the RT-PCR to test the miR-150 level. The data showed that miR-150 expression was dramatically increased in the mimic-transfected cells and decreased in the inhibitor-transfected cells. Cell proliferation was measured using CCK-8 assays. Ectopic overexpression of miR-150 led to a decrease proliferation rate in osteosarcoma cells. As expected, knockdown of miR-150 expression enhanced the cell growth rate. In addition, the colony formation assay also showed that loss-of-function of the miR-150 enhances osteosarcoma cell colony formation, and gain-of function of miR-150 reduces cell colony formation. Taken together, these results indicate that miR-150 functions as tumor suppressor in osteosarcoma cell proliferation.
miR-150 induces glycolysis in HOS and U2OS cells
To further explore the biological function of miR-150 on glycolysis in the osteosarcoma cells, we examine the effect of miR-150 on the glucose uptake rate and lactate secretion. The result demonstrated that ectopic overexpression of miR-150 in HOS and U2OS cells transient transfected with miR-150 mimics suppressed the lactate secretion and glucose uptake (Figure 3. A, B). As expected, knockdown of miR-150 with the miR-150 inhibitors in HOS and U2OS cells enhanced the lactate secretion and glucose uptake (Figure 3. A, B). To further validate the effect of miR-150 on glycolysis, we performed the RT-PCR and western blot to detect Glut1, HK2 and PFKL expression in mRNA level and protein level. The data showed that ectopic expression of miR-150 with the mimics reduced the expression of Glut1, HK2 and PFKL both in mRNA and protein level.
Figure 1.
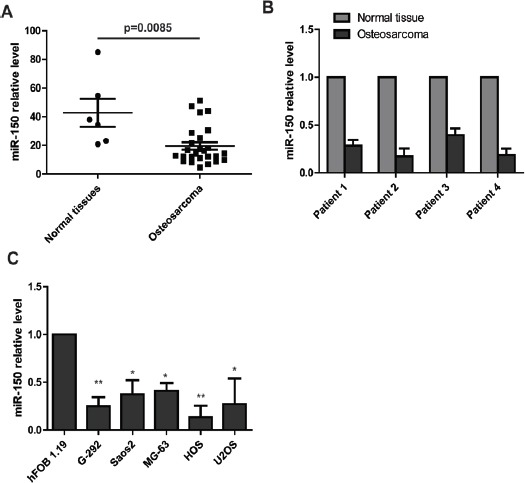
Mir-150 Expression is Decreased in Osteosarcoma. (A) The microRNA expression assay indicates that miR-150 level is obviously lower in osteosarcoma tissue than in normal tissue. (B) Real-time PCR analysis shows that miR-150 is markedly down-regulated in osteosarcoma tissues compared with normal tissues. (C) Real-time PCR analysis shows that miR-150 expression is significantly decreased in osteosarcoma cell lines compared to the normal cell line hFOB 1.19
Figure 2.
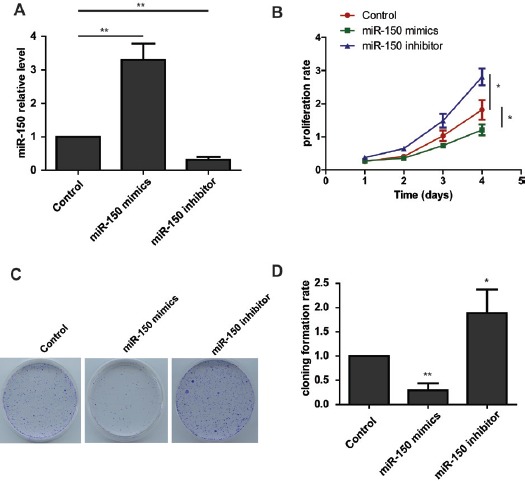
mir-150 Inhibits the Proliferation of Osteosarcoma Cells. (A) Real time PCR analysis of miR-150 levels in osteosarcoma cells transfected with the miR-150 mimics or the miR-150 inhibitor. (B) The CCK8 assay results show that the growth rate of miR-150-overexpressing osteosarcoma cells is decreased compared to control cells. Conversely, knockdown of miR-150 in osteosarcoma cells enhances cell growth. (C) and (D) The colony formation assays show that knockdown of miR-150 enhances osteosarcoma cells colony formation, while overexpression of miR-150 decreases colony formation in osteosarcoma cells.
Figure 3.
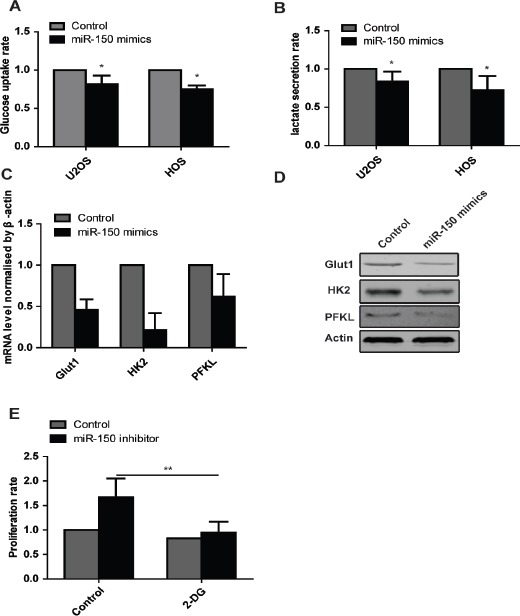
miR-150 Induces a Metabolic Shift in Osteosarcoma Cells. (A) Glucose uptake assays show that miR-150 decreases glucose uptake rate. (B) Lactate production assays show that miR-150 blocks lactate secretion. (C) Real time PCR show the mRNA level of Glut1, HK2, PFKL in cells transfected with miR-150 mimics. (D) Western blot show the protein level of Glut1, HK2, PFKL in cells transfected with miR-150 mimics. (E) The CCK8 assays show that inhibition of glycolysis with 2-DG blocks the enhanced proliferation rate induced by knockdown of miR-150.
Figure 4.
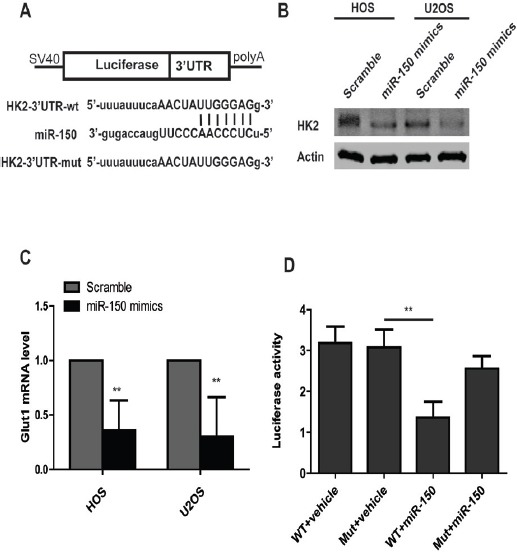
miR-150 Suppresses Expression of Glut1. (A) Sequences of miR-150 and the potential binding sites at the 3’UTR of Glut1. (B) Western blot assays show that overexpression of miR-150 suppresses the expression of Glut1. (C) Real time PCR show that overexpression of miR-150 increases Glut1 mRNA level. (D) Dual-luciferase assays show that miR-150 suppresses the luciferase activity in Glut1-3’UTR wild type group, but don’t in Glut1-3’UTR mutant group.
To explore whether the enhanced glycolysis induced by knockdown of miR-150 was required for the increased proliferation rate, we treated cells transfected with miR-150 inhibitor with 2-deoxyglucose (2-DG), a glucose analog that inhibits glycolysis by acting on hexokinase. We used CCK8 assay to measure cell proliferation rate. The data showed that inhibition of glycolysis by 2-DG was sufficient to reduce osteosarcoma cell growth (Figure 3. C). In conclusion, these data showed that enhanced glycolysis induced by miR-150 is required for the increased cell proliferation.
miR-150 regulates Glut1 expression in osteosarcoma cells.
To further elucidate the molecular mechanism by which miR-150 regulates glycolysis, we used miRanda and targetscan to predict potential downstream targets of miR-150 based on the 3’UTR. We found that the region in the Glut1 (HK2) 3’UTR was a predicted target of miR-150. In order to verify Glut1 as the target of miR-150, we transfected two osteosarcoma cell lines HOS and U2OS with miR-150 mimics, after 48h post-transfection, western blot and RT-PCR analysis were performed. The results showed that overexpression of miR-150 led to a marked decrease of endogenous Glut1 protein level and mRNA level compared to osteosarcoma cells transfected scrambles. Next, to examine whether miR-150 directly binds to the seed region of Glut1, we performed a luciferase reporter assay with plasmids containing the 3’UTR of Glut1. As expected, there was reduced luciferase activity in osteosarcoma cells co-transfected with pGL-Glut1-3’UTR vector and miR-150 mimics, however, the suppression of the luciferase activity was rescued in the 3’UTR mutant group. Taken together, the results indicated that miR-150 directly downregulates Glut1 expression through binding the 3’UTR.
Discussion
MicroRNAs have emerged as key regulators of many biological events, including cell proliferation, cell differentiation, cell apoptosis and cell metabolism (Cho, 2011; Lages et al., 2012). Previous reports showed that aberrant expression of miR-150 was associated with several cancers. increased miR-150 expression promoted prostate cancer cell development by suppressing p27Kip1 (Liu et al., 2015); downregulation of miR-150 promotes pertuzumab resistance in ovarian cancer cells via AKT activation (Wuerkenbieke et al., 2015). Li X et al., (2015a) reported that miR-150 expression was significantly decreased in osteosarcoma cell lines and miR-150 inhibits cell proliferation, invasion by regulating the expression of Sp1, however the effect of miR-150 on glycolysis in osteosarcoma remains unknown.
In our study, we provide important evident in support of miR-150 acting as tumor suppressor by targeting the expression of Glut1 in osteosarcoma. We detected the reduced expression of miR-150 in osteosarcoma, which is consistent with the published microRNA assay data and the results of Li X et.al, which found that miR-150 is down-regulated in osteosarcoma cell lines, and overexpression of miR-150 inhibits cell proliferation by targeting Sp1. Along with our findings, these data indicate the tumor suppressor role of miR-150. More extensively effort is needed to investigate the mechanism underline the dysregulation of miR-150in osteosarcoma.
The molecular mechanism underlining enhanced glycolysis in cancer cells are complicated and remains largely unclear (Kroemer and Pouyssegur, 2008). In our study, we found that miR-150 plays key role in regulating glycolysis in osteosarcoma. Gain of miR-150 function decreases glucose uptake and lactate secretion, while loss of miR-150 function increases glucose uptake and lactate secretion. The enhanced glycolysis induced by downregulation of miR-150 is required for osteosarcoma cell proliferation. In addition, we identified Glut1 as a downstream target of miR-150. We are the first to report the glycolysis role of miR-150 in osteosarcoma through directly targeting Glut1 expression.
In conclusion, our data show that miR-150 was obviously downregulated in osteosarcoma, thus leading to the enhanced cell glycolysis. The altered glycolysis is required for the cell rapid growth. These results identify miR-150 as a potential therapeutic target for osteosarcoma.
References
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Cho WC. Promises and challenges in developing miRNA as a molecular diagnostic tool for lung cancer. Expert Rev Mol Diagn. 2011;11:763–6. doi: 10.1586/erm.11.71. [DOI] [PubMed] [Google Scholar]
- Davalos A, Goedeke L, Smibert P, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–7. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson WS, Goorin AM. Current treatment of osteosarcoma. Cancer Invest. 2001;19:292–315. doi: 10.1081/cnv-100102557. [DOI] [PubMed] [Google Scholar]
- Geller DS, Gorlick R. Osteosarcoma:a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705–18. [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism:Warburg and beyond. Cell. 2008;134:703–7. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Hu JW, Sun P, Zhang DX, et al. Hexokinase 2 regulates G1/S checkpoint through CDK2 in cancer-associated fibroblasts. Cell Signal. 2014;26:2210–6. doi: 10.1016/j.cellsig.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Pouyssegur J. Tumor cell metabolism:cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–82. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Lages E, Ipas H, Guttin A, et al. MicroRNAs:molecular features and role in cancer. Front Biosci (Landmark Ed) 2012;17:2508–40. doi: 10.2741/4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hu L, Tian C, et al. microRNA-150 promotes cervical cancer cell growth and survival by targeting FOXO4. BMC Mol Biol. 2015a;16:24. doi: 10.1186/s12867-015-0052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen L, Wang W, et al. MicroRNA-150 inhibits cell invasion and migration and is downregulated in human osteosarcoma. Cytogenet Genome Res. 2015b;146:124–35. doi: 10.1159/000437379. [DOI] [PubMed] [Google Scholar]
- Liu DZ, Zhang HY, Long XL, et al. MIR-150 promotes prostate cancer stem cell development via suppressing p27Kip1. Eur Rev Med Pharmacol Sci. 2015;19:4344–52. [PubMed] [Google Scholar]
- Luetke A, Meyers PA, Lewis I, et al. Osteosarcoma treatment - where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–32. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Mitra R, Sun J, Zhao Z. microRNA regulation in cancer:One arm or two arms? Int J Cancer. 2015;137:1516–8. doi: 10.1002/ijc.29512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Li K, Huang Y, et al. miR-495 mediates metabolic shift in glioma cells via targeting Glut1. J Craniofac Surg. 2015;26:e155–8. doi: 10.1097/SCS.0000000000001385. [DOI] [PubMed] [Google Scholar]
- Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and function. Thromb Haemost. 2012;107:605–10. doi: 10.1160/TH11-12-0836. [DOI] [PubMed] [Google Scholar]
- Wei Z, Cui L, Mei Z, et al. miR-181a mediates metabolic shift in colon cancer cells via the PTEN/AKT pathway. FEBS Lett. 2014;588:1773–9. doi: 10.1016/j.febslet.2014.03.037. [DOI] [PubMed] [Google Scholar]
- Wu P, Liang J, Yu F, et al. miR-145 promotes osteosarcoma growth by reducing expression of the transcription factor friend leukemia virus integration 1. Oncotarget. 2016;7:42241–51. doi: 10.18632/oncotarget.9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerkenbieke D, Wang J, Li Y, et al. miRNA-150 downregulation promotes pertuzumab resistance in ovarian cancer cells via AKT activation. Arch Gynecol Obstet. 2015;292:1109–16. doi: 10.1007/s00404-015-3742-x. [DOI] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Shi Z, et al. Metabolic reprogramming of cancer-associated fibroblasts by IDH3alpha downregulation. Cell Rep. 2015;10:1335–48. doi: 10.1016/j.celrep.2015.02.006. [DOI] [PubMed] [Google Scholar]


