Abstract
We describe a physical mRNA mapping strategy employing fluorescent self-quenching reporter molecules (SQRMs) that facilitates the identification of mRNA sequence accessible for hybridization with antisense nucleic acids in vitro and in vivo, real time. SQRMs are 20–30 base oligodeoxynucleotides with 5–6 bp complementary ends to which a 5′ fluorophore and 3′ quenching group are attached. Alone, the SQRM complementary ends form a stem that holds the fluorophore and quencher in contact. When the SQRM forms base pairs with its target, the structure separates the fluorophore from the quencher. This event can be reported by fluorescence emission when the fluorophore is excited. The stem–loop of the SQRM suggests that SQRM be made to target natural stem–loop structures formed during mRNA synthesis. The general utility of this method is demonstrated by SQRM identification of targetable sequence within c-myb and bcl-6 mRNA. Corresponding antisense oligonucleotides reduce these gene products in cells.
INTRODUCTION
A major obstacle to employing antisense oligonucleotides for post-transcriptional gene silencing in mammalian cells is the apparent randomness with which they silence target mRNA expression. In addition to concerns related to intracellular delivery of nucleic acid molecules, oligonucleotide stability and hybridization kinetics, we have hypothesized that an extremely important element of this problem is our inability to reliably identify hybridization accessible nucleotides within the targeted mRNA. This failure arises as a result of mRNA secondary and tertiary structure which cannot be predicted in vivo, and is complicated by RNA-associated proteins.
Molecular beacons, containing a loop sequence flanked by complementary stems, were designed by Tyagi and Kramer (1) to identify target sequences with high specificity, while exhibiting a low background signal. We have reported that base pair sequence-specific fluorescent probes (designed after ‘molecular beacons’), we have called self-quenching reporter molecules (SQRMs) (Figure 1), can hybridize with an mRNA target in a living cell and reveal gene expression ‘real time’ (2). This has been confirmed by others (3). A logical extension would be to employ the intracellular signal as a means of locating single-stranded, hybridization accessible regions in any mRNA molecule in real time. Complementary nucleic acids directed to sites by this method should then reliably silence the expression of the encoding gene by a variety of mechanisms (4).
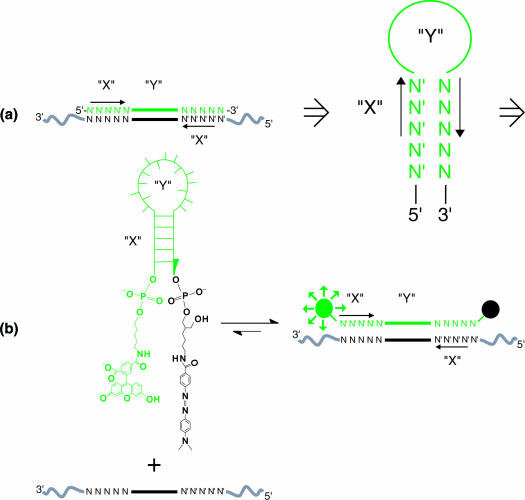
Figure 1.
SQRM design and reaction. (a) Concept: to exploit the traditional stem–loop structure of the SQRMs, a computer algorithm (‘AccessSearch’) searches an entire sequence of mRNA for complementary sequences of a desired length (stems) that are separated by a proscribed distance (loop). (b) Chemistry: the complementary sequences are synthesized as SQRM possessing 5′-fluorescein and 3′ DABCYL groups. In the absence of target, quenching of fluorescence occurs. Once hybridization of the loop sequence to a complementary target takes place, the moieties are separated and fluorescence can be detected.
A number of strategies have been developed to identify physically accessible regions within a given mRNA molecule. These include the use of random oligonucleotide libraries to probe for accessible sites, identified by subsequent RNase H cleavage fragments of the predicted size, mRNA hybridization to oligonucleotide arrays and algorithm-generated RNA structure models based on free energy predictions (4). The general utility of each of these approaches remains unclear, and none can be followed in real time, or in living cells.
Here, we report the successful use of SQRMs to identify hybridization accessible sites with the mRNA structure of three different targets: transcription factors c-myb (5,6) and bcl-6 (7,8) and tyrosine receptor kinase c-kit (9). All three targets were chosen because of their potential therapeutic utility in treating different forms of human carcinoma (10–13). Multiple SQRMs were developed for each of these targets and a small percentage of these SQRMs were found to have an adequate signal-to-noise ratio to indicate hybridization with mRNA (≥5:1). When introduced into living cells, antisense oligodeoxyribonucleotide (ODN) molecules directed to these sites allowed a decrease in mRNA and protein expression, as judged by real-time PCR and western blots.
MATERIALS AND METHODS
ODN and SQRM synthesis
ODNs were synthesized in the University of Pennsylvania Cancer Center Nucleic Acid Facility on an Expedite™ 8909 Nucleic Acid Synthesis System (Applied Biosystems) using standard phosphoramidite chemistry [bcl-6 antisense ODNs (AS ODNs) obtained from Integrated DNA Technologies]. The SQRM complements to the AccessSearch outputted RNA sequences are synthesized with 3′-C7-4-(4′-dimethylaminophenylazo)benzoic acid (DABCYL) Controlled Pore Glass (Glen Research) used as support and a fluorescein phosphoramidite (Cruachem) for 5′ end modification (Table 1). All ODNs and SQRMs were HPLC-purified. SQRMs were numbered according to the following convention: the SQRM's number designator corresponds to the 5′ base of mRNA to which the 3′ base of the SQRM complements, the adenine of the initiation codon is base (+1), and the base 5′ of the initiation A is designated (−1), no zero base. The full descriptor is characterized by the number of the first 5′ RNA base involved in a duplex with the probe, an arrow to indicate the direction of hybridization of the probe to the mRNA and a second number that corresponds to the 3′-most base involved in the helix.
Table 1.
Sequences of SQRMs identified using the AccessSearch program
| Sequence | |
|---|---|
| c-myb-targeted SQRMs | |
| SQRM 321←350 | 5′-F-ACAGACCAACGTTTCGGACCGTATTTCTGT-D-3′ |
| SQRM 772←799 | 5′ F-CATGTTCCATACCCTGTAGCGTTACATG-D-3′ |
| SQRM 1179←1207 | 5′ F-AGAAACACTCCAATTTATAGATTCTTTCT-D-3′ |
| SQRM 1454←1481 | 5′ F-AGATGCTACCTCAGACACCCTCTCATCT-D-3′ |
| SQRM 1649←1676 | 5′-F-GTCTGAATACCCAACTGTTCACGCAGAC-D-3′ |
| SQRM 1898←1915 | 5′ F-TCTCAGCCCGGACGCTGGTCATGTGAGA-D-3′ |
| SQRM 1965←1992 | 5′-F-GTTGAGGAATGATATATCCCAAGTCAAC-D-3′ |
| SQRM 1982←2011 | 5′-F-CATTCATGAAAAGTTTCATGTTGAGGAATG-D-3′ |
| bcl-6-targeted SQRMs | |
| SQRM 518←548 | 5′-F-CTGGGGGCAAAGGCTCTGCTCTCACACCCAG-D-3′ |
| SQRM 831←864 | 5′-F-GGCTGAGGGGGCAGCAGGTTTGAGGCCCTCAGCC-D-3′ |
| SQRM 832←863 | 5′-F-GCTGAGGGGGCAGCAGGTTTGAGGCCCTCAGC-D-3′ |
| SQRM 834←861 | 5′-F-TGAGGGGGCAGCAGGTTTGAGGCCCTCA-D-3′ |
| SQRM 1039←1073 | 5′-F-GCCTGGAGGATGCAGGCATTCTTACTGCTGCAGGC-D-3′ |
| SQRM 1190←1222 | 5′-F-AGGCTCGTGGGGAAAGGCGGCCCAGCTCAGCCT-D-3′ |
| SQRM 1367←1392 | 5′-F-GCTCTCGCTGCTGCTGCGGGGAGAGC-D-3′ |
| SQRM 1816←1841 | 5′-F-ACCTGTACAAATCTGGCTCCGCAGGT-D-3′ |
| SQRM 1821←1853 | 5′-F-CGGAGGTGGGCCACCTGTACAAATCTGGCTCCG-D-3′ |
| SQRM 2121←2141 | 5′-F-AAGCATCAACACTCCATGCTT-D-3′ |
| c-kit-targeted SQRMs | |
| SQRM 39←64 | 5′-F-TCTGGACGCGAAGCAGTAGGAGCAGA-D-3′ |
| SQRM 137←163 | 5′-F-TAATCTCGTCGCCCACGCGGACTATTA-D-3′ |
| SQRM 287←304 | 5′-F-CGTGTTTGTTGGTGCACG-D-3′ |
| SQRM 351←381 | 5′-F-TTTCCCATACAAGGAGCGGTCAACAAGGAAA-D-3′ |
| SQRM 580←607 | 5′-F-TCAGGATGAATTTTTCCGACAGCACTGA-D-3′ |
| SQRM 663←683 | 5′-F-TCTTCCCCTTCCCTAAGAAGA-D-3′ |
| SQRM 807←837 | 5′-F-TGAACTGATAGTCAACGTTGCCTGACGTTCA-D-3′ |
| SQRM 924←946 | 5′-F-AGATATTAATGAATCCTTTATCT-D-3′ |
| SQRM 962←994 | 5′-F-CTACATTTTCTCCATCGTTTACAAATACTGTAG-D-3′ |
| SQRM 1093←1116 | 5′-F-TCTGATATTACTTTCATTCTCAGA-D-3′ |
Stems are bold; F, fluorescein; and D, DABCYL.
The full descriptor of SQRM is characterized by the number of the 5′ base of mRNA to which the 3′ base of the SQRM complements RNA, an arrow indicating the direction of hybridization of the probe, and a second number that corresponds to the 3′-most base involved in the duplex [adenine of RNA initiation codon is base (+1), and the base 5′ of the initiation A is designated (−1), no zero base].
In vitro SQRM hybridization
The genes were cloned into pcDNA3 plasmids (Invitrogen; bcl-6 clone was a kind gift from R. Dalla-Favera, Columbia University). RNA was transcribed from linearized plasmids using the T7 RNA Polymerase RiboMax™ Kit (Promega). SQRMs (100 nM) were incubated for 30 min at 37°C with (1 μM) in vitro transcribed RNA, (10 μM) ODN (positive control) or (1 μM) scrambled ODN (negative control) target in SQRM buffer (100 mM Tris–HCl, pH 7.5, 2 mM MgCl2). Total RNA was isolated from Louckes cells (bcl-6 expressing human Burkitt lymphoma cell line kindly provided by Jill Lacy, Yale University) and K562 cells (non-expressing) with QIAgen RNeasy Kit (Qiagen). Fluorescence emission was monitored using a Packard FluoroCount Microplate Fluorometer (Packard Instrument Company).
In vitro assays
Separate plasmids containing coding sequence for c-myb and luciferase are mixed in a 5:1 molar ratio and added to the TnT® rabbit reticulocyte lysate coupled transcription/translation system (Promega) with Redivue l-[35S]methionine (Amersham) and 1 U of RNase H (USB). Control ODN (5′-TGTCTGGTTGCAAAGCCTGGCATAAAGACA-3′) and AS ODN 321←350 were added to separate mixtures at 38- and 63-fold molar excess. Following incubation at 30°C for 90 min, aliquots were resolved on 10% SDS–PAGE and scanned on a Storm PhorsphorImager (Molecular Dynamics). In vitro transcribed bcl-6 mRNA (1 μM) was preincubated with (1 μM) SQRM 1190←1222 or two control sequences in SQRM buffer at 37°C for 10 min. RNase H buffer and enzyme were added, incubated at 37°C for 10 min, and the reaction was stopped by adding Proteinase K and run on a 1% agarose gel.
Western blotting
Adherent hamster fibroblast Tk−ts13 cells (American Type Culture Collection) engineered to express human c-myb from an exogenous vector were grown in DMEM supplemented with 10% heat-inactivated fetal calf serum, 1% l-Glutamine, 0.5% Penicillin/Streptomycin and Geneticin (G418; 1250 μg/ml) (Invitrogen). Cells were treated with AS ODN/Lipofectin® (Invitrogen) complex for 5 h, washed twice with cold PBS and incubated overnight at 37°C. Cells were incubated at 4°C for 30 min in lysis buffer (50 mM Tris–HCl, pH 7.6, 150 mM NaCl and 1% Triton X-100) containing a protease inhibitor cocktail tablet (Roche Diagnostics). Each sample contained 50 μg total protein and was resolved by 7.5% SDS–PAGE. Transfer to a polyvinyl difluoride membrane (Bio-Rad) was performed in glycine transfer buffer (192 mM glycine, 25 mM Tris, pH 8.8) for 1 h at 100 V and western blot was carried out with ECL Plus™ Western Blotting Detection Reagents Kit (Amersham). Antibodies used: 1:1000 dilution anti-c-Myb mAb (Upstate Biotechnology), peroxide-labeled anti-mouse antibody (1:3000 dilution; Upstate Biotechnology) and anti-actin mAb (1:7500 dilution, Oncogene Research Products).
Microinjection
Tk−ts13 cells were plated in 35 × 10 mm round dishes at 30–40% density 1 day before injection. Individual cells were viewed under an Olympus AX70 inverted microscope (Olympus America). Cytoplasmic SQRM injections (2 μM) were performed with a Micromanipulator 5171 and FemtoJet® Microinjector (EppendorfAG). Injection volume was regulated by injection pressure (260 hPa), injection time (0.3 s) and compensation pressure (86 hPa). Images were captured with a CoolSnap digital camera (Roper Scientific). IPLab Imaging software package (Scanalytics) was used for imaging and quantitation of the fluorescence intensity.
RESULTS
SQRM design considerations
The structural features of the SQRM suggest a rational strategy for locating target sequences. An algorithm that scans the cDNA sequence for complementary sequences of length (X), and separated by intervening sequence forming a loop of length (Y), was developed (Figure 1). Targeting SQRM to such locations provided two theoretical advantages. First, natural complements might be good targets of intra-molecular duplex formation within mRNA creating a loop available for hybridization. Second, if the SQRMs were fully complementary to the mRNA target, the possibility of the fluorophore and quencher contacting each other as a result of dangling from the non-hybridized DNA arms would be minimized. We had noted that some SQRMs failed to generate their maximal predicted fluorescence (defined as the fluorescence emitted by a fully DNase-digested SQRM), even when incubated with up to a 5-fold excess of target (data not shown). One explanation is that if the 5′ and 3′ ends of the SQRM are not complementary to the mRNA target, the two stem fragments (5 unpaired bases) and linkers (with 10 or more single bonds) provide enough length and flexibility to allow the fluorophore and quencher to contact each other while the loop portion is hybridized to the target. This was confirmed by building a molecular model using Insight II® (Biosym Technologies, San Diego, CA) (data not shown). This model suggests that the SQRM must be fully complementary to the mRNA target.
In vitro screening for hybridization accessible regions
To evaluate the utility of the SQRM design program, we tested its ability to identify accessible sequence in the mRNA of three therapeutically relevant genes: c-myb, bcl-6 and c-kit. The program was initially instructed to find complementary sequences 4–5 bases in length, separated by 18–20 nt in accord with original recommendations for designing molecular beacons (1). Later searches were broadened to find SQRMs with longer ‘stems’ and shorter ‘loop’ regions. Between 8 and 10 sequences were identified for targeting within each mRNA (Table 1).
To carry out the in vitro screening assay, SQRMs were added to (1 μM) in vitro transcribed mRNA. Each SQRM assay included complementary and scrambled DNA target controls. A 10 μM concentration of complementary oligonucleotide, a positive control, was used to ensure that the SQRM would achieve a high signal-to-noise ratio. The scrambled ODN was incubated with SQRMs at a 1 μM concentration, since this negative control was compared directly to the SQRM results with in vitro transcribed RNA. ‘Scrambled’ ODNs were designed by replacing each base with its complementary base in the same 5′ to 3′ direction. The fluorescence was monitored in a plate reader equilibrated to 37°C. Signal-to-noise ratios for all reactions can be found in Table 2.
Table 2.
SQRM signal-to-noise ratios
| Signal-to-noise ratios | RNA | ODN | SCR |
|---|---|---|---|
| c-myb-targeted SQRMs | |||
| SQRM 321←350 (n = 20) | 12.0 ± 3.8 | 32.9 ± 9.8 | 1.4 ± 0.4 |
| SQRM 772←799 | 1.3 ± 0.4 | 34.8 ± 10.6 | 1.1 ± 0.3 |
| SQRM 1179←1207 | 1.3 ± 1.2 | 3.1 ± 1.4 | 1.2 ± 0.4 |
| SQRM 1454←1481 | 0.9 ± 0.4 | 4.5 ± 1.4 | 1.3 ± 0.4 |
| SQRM 1649←1676 | 0.8 ± 0.2 | 7.8 ± 1.2 | 0.9 ± 0.2 |
| SQRM 1898←1915 | 0.9 ± 0.2 | 16.2 ± 1.9 | 0.9 ± 0.2 |
| SQRM 1965←1992 (n = 3) | 1.3 ± 0.6 | 12.4 ± 5.4 | 0.9 ± 0.5 |
| SQRM 1982←2011 (n = 3) | 1.2 ± 0.4 | 10.4 ± 2.3 | 5.1 ± 6.8 |
| bcl-6-targeted SQRMs | |||
| SQRM 518←548 | 2.3 ± 0.8 | 70.6 ± 16.0 | 2.0 ± 0.6 |
| SQRM 831←864 | 1.8 ± 0.2 | 33.0 ± 4.2 | 1.3 ± 0.2 |
| SQRM 832←863 | 1.1 ± 0.1 | 57.6 ± 6.6 | 1.6 ± 0.2 |
| SQRM 834←861 | 4.4 ± 2.4 | 150.6 ± 61.2 | 1.9 ± 0.8 |
| SQRM 1039←1073 | 2.2 ± 0.5 | 39.2 ± 8.6 | 2.7 ± 0.6 |
| SQRM 1190←1222 | 14.5 ± 1.6 | 57.6 ± 5.6 | 2.2 ± 0.4 |
| SQRM 1367←1392 | 1.1 ± 0.3 | 51.1 ± 13.7 | 2.0 ± 0.4 |
| SQRM 1816←1841 | 1.2 ± 0.3 | 17.8 ± 3.5 | 2.5 ± 0.5 |
| SQRM 1821←1853 | 3.2 ± 1.2 | 63.0 ± 7.8 | 3.3 ± 2.3 |
| SQRM 2121←2141 | 3.8 ± 1.9 | 207.0 ± 22.9 | 2.9 ± 1.0 |
| c-kit-targeted SQRMs | |||
| SQRM 39←64 | 2.5 ± 0.3 | 54.2 ± 15.8 | 4.5 ± 0.8 |
| SQRM 137←163 | 1.3 ± 0.1 | 4.6 ± 1.0 | 1.1 ± 0.2 |
| SQRM 287←304 | 4.5 ± 1.3 | 6.6 ± 2.2 | 1.2 ± 0.2 |
| SQRM 351←381 | 3.0 ± 1.1 | 11.5 ± 4.4 | N.D. |
| SQRM 580←607 | 1.8 ± 0.4 | 16.4 ± 3.4 | N.D. |
| SQRM 663←683 | 7.4 ± 3.0 | 4.5 ± 2.2 | 0.8 ± 0.3 |
| SQRM 807←837 | 6.4 ± 1.0 | 16.5 ± 3.0 | 1.4 ± 0.3 |
| SQRM 924←946 | 3.1 ± 0.6 | 7.2 ± 01.2 | 0.8 ± 0.1 |
| SQRM 962←994 | 5.4 ± 0.5 | 7.3 ± 0.7 | 1.6 ± 0.2 |
| SQRM 1093←1116 | 4.1 ± 0.4 | 4.7 ± 1.7 | 1.5 ± 0.4 |
N.D., no data.
All values ± SD are the result of six trials unless otherwise noted.
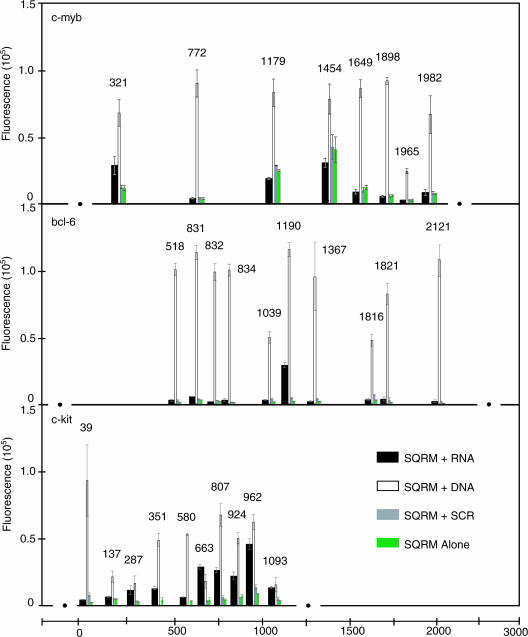
Eight SQRM sequences were identified for testing with the full-length [1923 nt plus ∼100 bases of the 3′-untranslated region (3′-UTR)] in vitro transcribed c-myb mRNA (Figure 2). SQRM 321←350 exhibited a signal-to-noise ratio of >5:1 (12.0 ± 3.8:1). The other c-myb SQRM failed to hybridize with their target sequence as revealed by their baseline, or lower, fluorescence. SQRMs 1965←1992 and 1982←2011 were directed to the 3′-UTR region of the c-myb sequence and showed a low signal-to-noise ratio (1.3 ± 0.6:1 and 1.2 ± 0.3:1, respectively). Whether targeting to the 3′-UTR would be unfavorable in general was not established.
Figure 2.
mRNA fluorescence assay. (a) Eight SQRMs were identified to target the full-length, 1923 nt human c-myb RNA; two were targeted to the ∼100 bases of the 3′-UTR region and overlap each other by a few bases. One of these SQRMs, 321←350, possesses a signal-to-noise ratio of >5:1, indicating that it opens when presented with a full-length RNA target. (b) Six SQRMs were hybridized with in vitro transcribed human bcl-6 targeted mRNA (2.4 kb). Of these SQRMs, 1190←1222 was shown to open, exhibiting a signal-to-noise ratio of ∼15:1. (c) Ten SQRMs were identified for a 1249 base fragment of the ∼2900 nt sequence of human c-kit mRNA. When hybridized with in vitro transcribed RNA, three (SQRMs 663←683, 807←837 and 962←994) have a signal-to-noise that is >5:1. All c-kit experiments were performed in triplicate; correction for background was not made.
Ten SQRMs each were synthesized to map the full-length, 2.4 kb, bcl-6 mRNA and a fragment of the human c-kit mRNA (−21 to 1228 of the 2931 nt coding sequence). In the case of bcl-6, only SQRM 1190←1222 showed a favorable signal-to-noise ratio (14.5 ± 1.5:1; Figure 2). The c-kit mapping SQRMs had arms of either 4 or 5 bases and intervening sequences ranging from 10 to 23 nt. Three of the ten SQRMs gave sufficient signal to indicate that they had hybridized with the mRNA. These were c-kit SQRMs 663←683, 807←837 and 962←994 (Figure 2). The ‘loop’ lengths of these molecules were 11, 21 and 13 nt, respectively, suggesting that this parameter at least was quite permissive with respect to mapping utility.
Evaluation of predicted sequence accessibility: in vitro assays
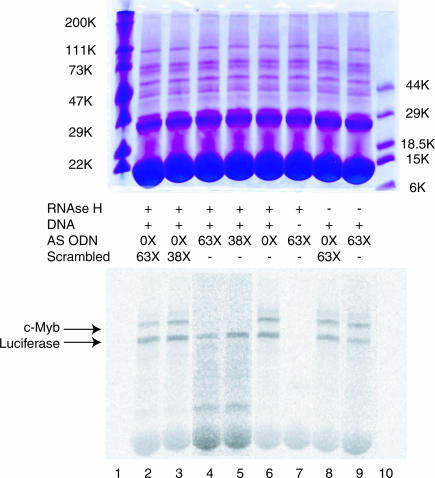
To evaluate the utility of SQRM mapping for the identification of hybridization accessible sequence, ODNs complementary to sequence identified as being available for hybridization were synthesized and targeted to c-myb and bcl-6 mRNAs. We first evaluated the ability of a specifically targeted ODN to direct RNase H-mediated cleavage of the mRNA to which it had been directed in a rabbit reticulocyte lysate transcription/translation system. This system was employed to better model the effect that cellular proteins might have on ODN hybridization. In the first series of experiments, we examined the ability of an AS ODN corresponding to c-myb SQRM 321←350 to direct RNA cleavage in comparison with a scrambled sequence not homologous to the c-myb or luciferase genes. Separate plasmids expressing full-length c-myb cDNA and luciferase mRNA (positive control) were mixed in a 5:1 molar ratio. This mixture was then added to the reticulocyte lysate along with AS ODN 321←350, or a scrambled ODN, at 38- and 63-fold molar excess to the predicted amount of protein the system would produce (14). Equal amounts of post-translation product from each reaction mixture were analyzed by electrophoresis in a single polyacrylamide gel. A PhosphorImage of this gel indicated that AS ODN 321←350, in the presence of RNase H, completely abolished c-Myb protein expression (Figure 3). When added at the 63-fold molar excess, the highest ODN concentration employed, luciferase protein was reduced ∼5-fold in the reaction mixture containing AS ODN 321←350, but this is in comparison with an ∼14-fold reduction in c-Myb protein. At the highest scrambled ODN concentration (63-fold), some non-specific decrease of the c-Myb and luciferase proteins was also noted (2.5- and 1.5-fold reductions, respectively).
Figure 3.
In vitro transcription and translation assay. Incubation of rabbit reticulocyte lysate with a c-myb AS ODN that corresponds to SQRM 321←350 decreases in vitro production of protein. Lanes 4 and 5 have been normalized to the luciferase (Luc) control band to show that there is no detectable presence of c-Myb protein. In the absence of exogenous DNA, there are no radiolabeled proteins at the molecular weights expected for c-Myb and luciferase.
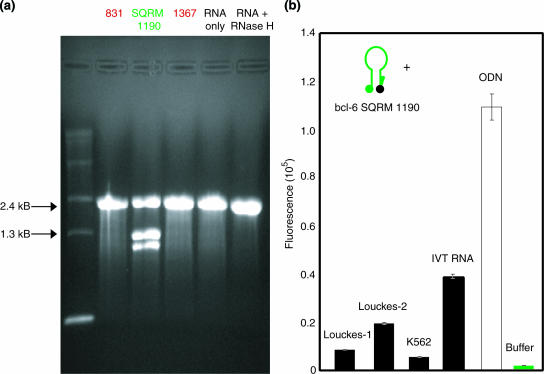
The efficacy of targeting the bcl-6 transcript was evaluated in vitro using two different assays. We first evaluated the ability of a SQRM to direct simple RNase H-mediated cleavage of an mRNA. In this experiment, in vitro transcribed mRNA was incubated with SQRM 1190←1222, or with two sense SQRM corresponding to non-opening locations, 831–864 and 1367–1392 (Figure 4a). As expected, bcl-6 SQRM 1190←1222 directed cleavage of the 2.4 kb bcl-6 transcript, yielding two fragments of the expected sizes, ∼1.1 and ∼1.3 kb, in an agarose gel. Specificity of cutting was demonstrated by the fact that neither of the control sequences had any effect on the mRNA.
Figure 4.
bcl-6 in vitro assays. (a) bcl-6 RNase H assay using SQRM 1190←1222. RNA was incubated with each SQRM and run on an agarose gel. The SQRM 1190 successfully recruits RNase H to cleave the RNA of the RNA/DNA hybrid. (b) bcl-6 SQRM 1190 was incubated with various targets and the fluorescence signal was measured. Louckes-1 (20 μg), Louckes-2 (40 μg) and K562 are RNA samples isolated from cell extracts. SQRM was also incubated with in vitro transcribed RNA (IVT RNA; a positive control for SQRM/RNA hybridization) and an ODN (positive control for SQRM function).
Next, we incubated SQRM 1190←1222 with total cellular RNA derived from Louckes Burkitt's Lymphoma cells, which express the bcl-6 gene, and K562 BCR/ABL-1 (+) myeloid leukemia cells, which do not. Fluorescence was monitored using a plate reader (Figure 4b). An ODN completely complementary to the SQRM served as the arbitrary 100% fluorescence control. As expected, SQRM fluorescence increased in the Louckes cell RNA when compared with that observed in the K562 RNA. Specificity was further demonstrated by the observation that fluorescence was proportional to the amount of RNA in which the SQRM was incubated. The signal-to-noise ratio for SQRM 1190←1222 when incubated in 40 μg of Louckes RNA was 2-fold higher than when incubated in 20 μg of RNA.
Evaluation of predicted sequence accessibility: in vivo assays
The results of the in vitro assays were encouraging, but it was required to demonstrate that they would accurately predict a targeted AS ODN ability to diminish mRNA expression in living cells. This was carried out in TK−ts13 cells engineered to express c-myb, and the bcl-6 expressing Louckes cells.
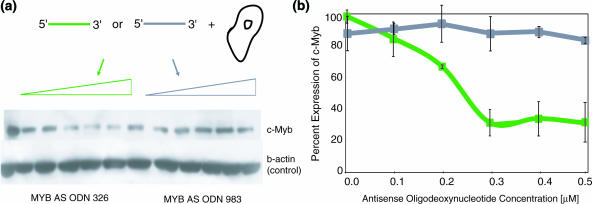
A c-myb targeted AS ODN corresponding to opening SQRM 321 (complementary to nucleotides 326–345) and an AS ODN corresponding to non-opening SQRM 970 (complementary to nucleotides 983–1000) were transfected into the TK−ts13 cells. Levels of protein expression were monitored by western blot and quantitated (Figure 5). Treatment with AS ODN 326←345 caused a decrease in the protein expression in direct proportion to the amount of the AS ODN added to the cultures. Concentrations of 0.3 μM were capable of reducing protein expression ∼70% compared with the level of untreated cells. In contrast, AS ODN 983←1000 did not bring about more than a 20% inhibition of the c-Myb protein. The results are in complete accord with the results obtained from the in vitro mapping assays.
Figure 5.
c-myb AS ODNs in vivo. An AS ODN corresponding to the SQRM 321 was synthesized and transfected into hamster fibroblast Tk−ts13 cells engineered to express human c-myb. (a) The western blot shows a decrease in protein expression following treatment with the AS ODN 326–345 as compared with AS ODN 983–1000 (negative control). (b) Graphical representation of the western blot data: AS ODN 326←345 (green); AS ODN 983–1000 (gray).
Bcl-6 targeted AS ODNs protected by phosphorothioate linkages between the five outermost bases were nucleoporated into the Louckes cells. Delivery efficiency was assessed using fluorescein-labeled ODNs and was judged to be ∼95%, with preservation of cell viability at ∼90% (data not shown). Control cells were subjected to the nucleofection procedure but in the absence of ODN. Quantitative real-time PCR was used to measure any resulting changes in mRNA expression; reaction was performed in triplicate each time for bcl-6 and 18S rRNA. When treated with the AS ODN 1190←1222 (corresponding to an opening SQRM), the cellular bcl-6 mRNA exhibited a 7-fold decrease in expression (data not shown). These results were confirmed by western blot analysis. The Bcl-6 protein expression was decreased by 80% compared with control cell values following treatment with AS ODN 1190←1222 (data not shown). There were no significant differences in the bcl-6 mRNA expression levels in the control populations. The scrambled control ODN, and AS ODN 1816←1841, corresponding to a non-opening SQRM, also decreased bcl-6 protein, but the decrease in expression (∼30%) was relatively insignificant compared with the AS ODN 1190←1222.
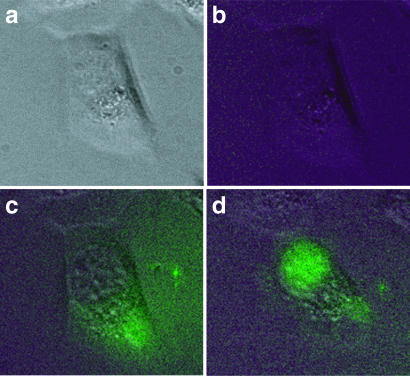
To demonstrate that SQRM could identify hybridization accessible sequence in living cells real-time, we microinjected SQRM 321←350 and a scrambled SQRM control into the hamster fibroblast cells (TK−ts13) engineered to express c-myb. SQRMs were end-protected by phosphorothioate linkages between the five outermost bases. The development of fluorescence was monitored post-injection every 10 min for a total of 30 min. Representative photomicrographs are shown for a cell injected with SQRM 321 (Figure 6). There was no increase in fluorescence detected 30 min after the injection of scrambled SQRM (data not shown). In accord with previous reports that oligonucleotides rapidly diffuse from cytoplasm into nucleus (15), nuclear fluorescence intensity increased as cytoplasmic fluorescence decreased. These results correlate well with the fluorescence levels observed in the in vitro assays with RNA targets. Of significant interest, they also suggest that real-time visualization of specific mRNA expression is being detected with these probes.
Figure 6.
Microinjections into cells. Hamster fibroblast Tk−ts13 cells engineered to express c-myb were injected with SQRM 321. Images were captured pre-injection (a) phase (b) fluorescence; post-injection (c) T0 and (d) at 30 min.
DISCUSSION
The difficulty involved in identifying hybridization accessible sequence within RNA molecules in vivo has been a significant impediment to the development of AS ODN for either informatic or therapeutic purposes. The widespread adoption of RNA interference has ameliorated this problem to some degree, but increasing experience with this methodology has shown that it is often necessary to make numerous molecules before one with considerable gene silencing activity is found. We believe that those data presented in this manuscript allow us to state that we have now made significant progress in solving this problem, at least in the case of AS ODN. The c-myb SQRM 321←350, for example, exhibited an acceptable signal-to-noise ratio and outperformed all other c-myb SQRMs tested (Figure 2). These promising results were further supported by the cell-free in vitro coupled transcription–translation system assay (Figure 3). When an antisense ODN that corresponds to this SQRM was incubated with the cell-free mixture, a dramatic decrease in the target protein was seen. This was also seen in vivo, where treatment of human c-myb-expressing hamster fibroblast cells with increasing concentrations of AS ODN affected a decrease in protein expression after 24 h (Figure 5). Furthermore, the hybridization was seen in vivo, in real time, following microinjection of these cells with SQRM 321←350 (Figure 6).
To further demonstrate the utility of the strategy we present in this paper, two other mRNAs, the transcription factor bcl-6 and the PDGF family receptor c-kit, were also mapped using the same procedures. In the case of bcl-6, efficient silencing was shown with SQRM 1190←1222, which exhibited the greatest fluorescence in vitro (Figure 2). The AS ODN corresponding to this SQRM sequence also yielded the best gene silencing activity (an ∼7-fold mRNA reduction) as demonstrated using real-time PCR. A western blot revealed a significant decrease in BCL-6 protein expression with AS ODN 1190←1222, but no significant decrease in the case of all remaining molecules. Interestingly, AS ODN 1190←1222 contains a G quartet in its sequence, which has been cited as a cause of non-specific inhibition of cell function. However, use of the appropriate controls, including a scrambled molecule (33mer) with a G quartet and the same base content as AS ODN 1190←1222, ruled this out because none had any effect on bcl-6 expression (data not shown).
Nucleic acid-mediated gene silencing, employing various types of targeting ‘vectors’ including AS ODN, ribozymes, DNAzymes and short interfering RNAs, has been used with considerable success in the laboratory (16–18). Many also hope to employ nucleic acid-based gene silencing for therapeutic purposes, but important technical issues remain to be solved before this approach will become reliable enough for routine treatment of disease. These issues include the problem of inefficient delivery of molecules to targeted cells (19), inability to intracellularly ‘address’ molecules to their mRNA targets (20,21), and a limited ability to predict secondary and tertiary mRNA structure in vivo (22). Localizing hybridization accessible sequences within any given mRNA is complex since many unpredictable factors govern how these molecules fold (23). Off-target silencing by siRNAs has become a concern (24), particularly when used for functional analysis of genomic/array data in the laboratory. As antisense DNA does not enter the siRNA pathway, it may be more specific than siRNA, while exhibiting similar levels of attenuation. We believe that the work presented here provides a useful approach to this problem and one that might, when combined with efficient methods for delivering these molecules into cells, result in the development of truly useful nucleic acid molecules for informatic and therapeutic usages.
Acknowledgments
Supported in part by grants from the NIH PO1-CA72765 (A.M.G.), the Doris Duke Charitable Foundation (A.M.G.), and a grant from the Polish Research Council 2PO5A 12326 (J.B.O.). Funding to pay the Open Access publication charges for this article was provided by the NIH. A.M.G. is a Distinguished Clinical Scientist of the Doris Duke Charitable Foundation. V.P., M.R., P.G. and K.V. were members of the Roy and Diana Vagelos Scholars Program.
REFERENCES
- 1.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 2.Sokol D.L., Zhang X., Lu P., Gewirtz A.M. Real time detection of DNA:RNA hybridization in living cells. Proc. Natl Acad. Sci. USA. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perlette J., Tan W.H. Real-time monitoring of intracellular mRNA hybridization inside single living cells. Anal. Chem. 2001;73:5544–5550. doi: 10.1021/ac010633b. [DOI] [PubMed] [Google Scholar]
- 4.Gewirtz A.M., Sokol D.L., Ratajczak M.Z. Nucleic acid therapeutics: state of the art and future prospects. Blood. 1998;92:712–736. [PubMed] [Google Scholar]
- 5.Gewirtz A.M., Calabretta B. A c-myb antisense oligodeoxynucleotide inhibits normal human hematopoiesis in vitro. Science. 1988;242:1303–1306. doi: 10.1126/science.2461588. [DOI] [PubMed] [Google Scholar]
- 6.Mucenski M.L., McLain K., Kier A.B., Swerdlow S.H., Schreiner C.M., Miller T.A., Pietryga D.W., Scott W.J., Jr, Potter S.S. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 7.Baron B.W., Nucifora G., McCabe N., Espinosa R., III, Le Beau M.M., McKeithan T.W. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc. Natl Acad. Sci. USA. 1993;90:5262–5266. doi: 10.1073/pnas.90.11.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang C.C., Ye B.H., Chaganti R.S., Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl Acad. Sci. USA. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yarden Y., Kuang W.J., Yang-Feng T., Coussens L., Munemitsu S., Dull T.J., Chen E., Schlessinger J., Francke U., Ullrich A. Human proto-oncogene c-kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987;6:3341–3351. doi: 10.1002/j.1460-2075.1987.tb02655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calabretta B., Sims R.B., Valtieri M., Caracciolo D., Szczylik C., Venturelli D., Ratajczak M., Beran M., Gewirtz A.M. Normal and leukemic hematopoietic cells manifest differential sensitivity to inhibitory effects of c-myb antisense oligodeoxynucleotides: an in vitro study relevant to bone marrow purging. Proc. Natl Acad. Sci. USA. 1991;88:2351–2355. doi: 10.1073/pnas.88.6.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris N.L., Jaffe E.S., Stein H., Banks P.M., Chan J.K., Cleary M.L., Delsol G., De Wolf-Peeters C., Falini B., Gatter K.C. A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood. 1994;84:1361–1392. [PubMed] [Google Scholar]
- 12.Ye B.H., Chaganti S., Chang C.C., Niu H., Corradini P., Chaganti R.S., Dalla-Favera R. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. EMBO J. 1995;14:6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratajczak M.Z., Luger S.M., DeRiel K., Abrahm J., Calabretta B., Gewirtz A.M. Role of the KIT protooncogene in normal and malignant human hematopoiesis. Proc. Natl Acad. Sci. USA. 1992;89:1710–1714. doi: 10.1073/pnas.89.5.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelham H.R.B., Jackson R.J. Efficient messenger-RNA-dependent translation system from reticulocyte lysates. Eur. J. Biochem. 1976;67:247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- 15.Leonetti J.P., Mechti N., Degols G., Gagnor C., Lebleu B. Intracellular distribution of microinjected antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1991;88:2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jen K.-Y., Gewirtz A.M. Suppression of gene expression by targeted disruption of messenger RNA: available options and current strategies. Stem Cells. 2000;18:307–319. doi: 10.1634/stemcells.18-5-307. [DOI] [PubMed] [Google Scholar]
- 17.Hannon G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 18.Sun L.Q., Cairns M.J., Saravolac E.G., Baker A., Gerlach W.L. Catalytic nucleic acids: from lab to applications. Pharmacol. Rev. 2000;52:325–347. [PubMed] [Google Scholar]
- 19.Juliano R.L., Yoo H. Aspects of the transport and delivery of antisense oligonucleotides. Curr. Opin. Mol. Ther. 2000;2:297–303. [PubMed] [Google Scholar]
- 20.Sullenger B.A. Colocalizing ribozymes with substrate RNAs to increase their efficacy as gene inhibitors. Appl. Biochem. Biotechnol. 1995;54:57–61. doi: 10.1007/BF02787911. [DOI] [PubMed] [Google Scholar]
- 21.Rossi J.J. Ribozymes in the nucleolus. Science. 1999;285:1685. doi: 10.1126/science.285.5434.1685. [DOI] [PubMed] [Google Scholar]
- 22.Tinoco I., Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 23.Dreyfuss G., Kim V.N., Katoaoka N. Messenger-RNA-binding proteins and the messages they carry. Nature Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 24.Scacheri P.C., Rozenblatt-Rosen O., Caplen N.J., Wolfsberg T.G., Umayam L., Lee J.C., Hughes C.M., Shanmugam K.S., Bhattacharjee A., Meyerson M., et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]