Abstract
The serial analysis of gene expression (SAGE) method is used to study global gene expression in cells or tissues in various experimental conditions. However, its reproducibility has not yet been definitively assessed. In this study, we have evaluated the reproducibility of the SAGE method and identified the factors that affect it. The determination coefficient (R2) for the reproducibility of SAGE is 0.96. However, there are some factors that can affect the reproducibility of SAGE, such as the replication of concatemers and ditags, the number of sequenced tags and double PCR amplification of ditags. Thus, corrections for these factors must be made to ensure the reproducibility and accuracy of SAGE results. A bioinformatic analysis of SAGE data is also presented in order to eliminate these artifacts. Finally, the current study shows that increasing the number of sequenced tags improves the power of the method to detect transcripts and their regulation by experimental conditions.
INTRODUCTION
The recent developments in functional genomics allow the simultaneous analysis of all genes expressed in particular cell types or tissues of an organism. The aim of these analyses is to identify which genes are implicated in tissue or cell functions, and to understand the mechanisms by which variations in gene expression occur. Characterization of the differences in gene expression variations associated with diseases can also lead to the development of new therapies. Different methods are used to analyze global gene expression, such as differential display (DD) (1), cDNA microarrays (2) and serial analysis of gene expression (SAGE) (3). The SAGE method is based on two principles. First, a short nucleotide sequence, named tag, which is specifically localized at the last anchoring enzyme, such as NlaIII, restriction site contains enough information to identify specifically a transcript. Second, the concatemerization of tags allow to analyze 20–60 tags per sequencing reaction. Compared to DD and microarrays, SAGE has several advantages. The SAGE method gives the possibility of finding new transcripts since all cDNAs can be sequenced. Moreover, the evaluation of gene expression by SAGE is quantitative since the number of times a tag is sequenced in a library is representative of frequency, in the mRNA population.
SAGE libraries of several tissues are now available, such as pancreas (3), skeletal muscle (4), uterus (5), brain (6), kidney (7), oocyte (8), heart (9) and adipose tissues (10). The comparison of the genes expressed in these tissues during basal and experimental conditions reveals regulatory mechanisms and candidate genes for various diseases. Several studies have compared the results of the SAGE method with northern blot (11,12), real-time PCR (13), western blot (14) and proteomics (15). Although the validity of the SAGE method has been reported by several studies (3,4,11–14), the current report investigates, for the first time, the reproducibility of the SAGE method using more than 50 000 tags. Moreover, the factors affecting this reproducibility are studied and a new method designed to control for artifacts is reported.
MATERIALS AND METHODS
Sample preparation
Different experimental protocols and SAGE libraries were used in this study. C57BL6 mice (Charles River Canada Inc) were provided ad libidum access to Lab Rodent Diet No.5002 and water. In the first study, 14 male mice had a gonadectomy (GDX). One week later, 5α-dihydrotestosterone (DHT) (0.1 mg/mouse) was injected 1 h before sacrifice. The skin was sampled and stored at −80°C until analysis. For the second study, 24 male mice had an adrenalectomy (ADX) and were injected with cortisol (0.1 mg/mouse) 24 h before sacrifice. The gastrocnemius muscle was stored at −80°C until analysis. For the third study, 10 mice embryo had a sham surgery to the trachea 3 h before the sampling of lung. For another study, 58 female mice divided in two groups had GDX surgery and injection of vehicule 24 h before sacrifice. The uterus was sampled and stored at −80°C until analysis. Finally, 28 male mice had a GDX and were injected with vehicle or DHT, 24 or 6 h before death, respectively. The prostate was stored until analysis. The tissue samples of each group were pooled to eliminate inter-individual variations and to extract enough mRNA for the analysis.
SAGE and data analysis
Total RNA was isolated by Trizol (Invitrogen, Burlington, ONT) and polyadenylated RNA was extracted. The SAGE method was performed as previously described (4). The mRNA was annealed with biotin-5′T18-3′ primer and converted to cDNA using cDNA synthesis kit (Invitrogen, Burlington, Canada). The resulting cDNA library was digested with NlaIII (anchoring enzyme). The 3′ restriction fragments were isolated with streptavidin-coated magnetic beads (Dynal, Oslo, Norway) and split into two populations. Each population was ligated to one of the two annealed linker pairs and extensively washed to remove unligated linkers. Tags of eleven nucleotides were released from the last 3′ NlaIII restriction site (CATG) of each transcript by digestion with BsmFI (tagging enzyme). The two tags populations were blunted and ligated using the blunting kit from Takara Co. (Otsu, Japan). Ditags were amplified by PCR with an initial denaturation step of 1 min at 95°C followed by 24 cycles of 20 s at 94°C, 20 s at 60°C and 2 s at 72°C using 27 bp primers (4). The PCR products were digested with NlaIII and the band containing the ditags was isolated and extracted from the acrylamide gel. To produce concatemers, the purified ditags were self-ligated. The concatemers with length between 500 and 1800 bp were isolated by agarose gel and extracted with Gene-Clean Spin (BIO 101, Vista, USA). The resulting DNA fragments were ligated into SphI site of pUC19 and cloned into E.coli. White colonies were screened by PCR to select long inserts for automated sequencing. Finally, bioinformatics programs were used to process the data and compare the levels of gene expression between different samples.
SADE modification
The SADE method was developed by Virlon et al. (7). This method is a modification of the SAGE method, and is used when the mRNA content is too low. A second PCR amplification of 12 cycles was performed on the ditags purified from acrylamide gel after the first PCR amplification.
Microarray and data analysis
The samples were processed following the labeling protocol from Affymetrix (http://www.affymetrix.com/support/technical/technotes/smallv2_technote.pdf). Total RNA was converted to cDNA by incubation with 400 U SuperScript II reverse transcriptase (Invitrogen), by using a T7-oligo-d(T)24 as a primer, 1× first-strand buffer and 1 mM dNTPs at 42°C for 1 h. Second-strand synthesis was performed using 40 U DNA polymerase I, 10 U E.coli DNA ligase, 2 U RNAse H (Invitrogen, Burlington, ON), 1× reaction buffer and 0.2 mM dNTPs at 16°C for 2 h. The addition of 10 U of T4 polynucleotide kinase (Invitrogen) stops the reaction, and cDNAs were incubated for 10 min at 16°C. cDNAs were isolated by phenol–chloroform extraction. Then, cDNAs were transcribed in vitro by using the T7 BioArray High Yield RNA Transcript Labeling Kit (Enzo Diagnostics, Farmingdale, NY) to produce biotinylated cRNA. The mixture (20 μl final volume) was incubated at 37°C for 5 h, with gentle mixing every 30 min. Labelled cRNAs were purified by using an RNeasy Mini Kit (Qiagen, Valencia, CA). Purified cRNA was fragmented to 30–200mer cRNA using a fragmentation buffer, for 20 min at 94°C. The quality of cRNA amplification and cRNA fragmentation was monitored by micro-capillary electrophoresis (Bioanalizer 2100, Agilent Technologies, Mississauga, ON). The cRNA probes were hybridized to an MG-U74Av2 Genechip (Affymetrix, Santa Clara, CA). Fifteen micrograms of fragmented cRNA was incubated with 1× manufacturer-recommended hybridization buffer and 1× eukaryotic hybridization control for 16 h at 45°C with constant rotation (60 r.p.m.). Microarrays were processed by using the Affymetrix GeneChip Fluidic Station 400 (protocol EukGE-WS2Av4). Staining was made with streptavidin-conjugated phycoerythrin (SAPE) followed by amplification with a biotinylated anti-streptavidin antibody and by a second round of SAPE. Genechips were scanned using a GeneChip Scanner 3000 with autoloader (Affymetrix). The signal intensities for the β-actin and GAPDH genes were used as internal quality controls. The ratio of fluorescence intensities for the 5′ end and the 3′ end of these housekeeping genes was <2. Scanned images were analyzed with Microarray Suite 5.0 software (Affymetrix).
RESULTS
Reproducibility
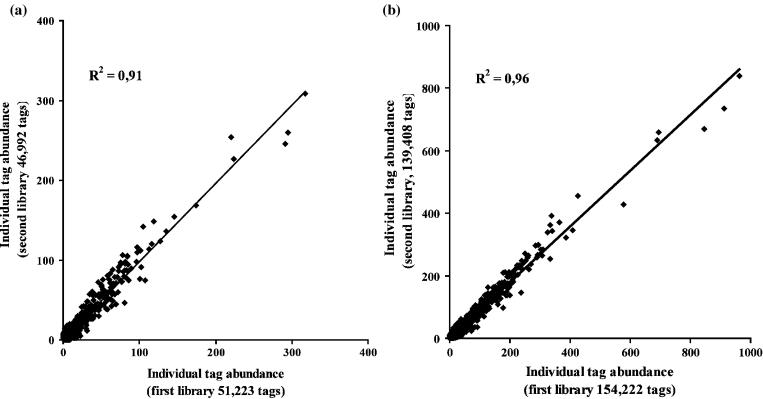
In order to investigate the reproducibility of SAGE, two libraries were generated in parallel from the same pool of total RNA. The libraries contained 51 223 and 46 992 tags representing 23 113 and 21 427 transcript species, respectively. Figure 1a shows the linear correlation (R2 = 0.91) between the gene expression level estimated in the two libraries. Using the comparative count display (CCD) test (16), no statistical difference was detected in the expression level of transcripts between these two libraries. The number of sequenced tags was increased in each library to determine the effect on the reproducibility of SAGE results. The libraries contained 154 222 and 139 408 tags, which represented 42 559 and 41 653 transcript species, respectively. Figure 1b shows the correlation between both libraries. The determination coefficient (R2) for this comparison is 0.96.
Figure 1.
Comparison of individual tag abundance estimated from two SAGE libraries of about 50 000 (a) or 150 000 (b) tags, each independently generated from the same pool of total RNA.
Bioinformatic analysis
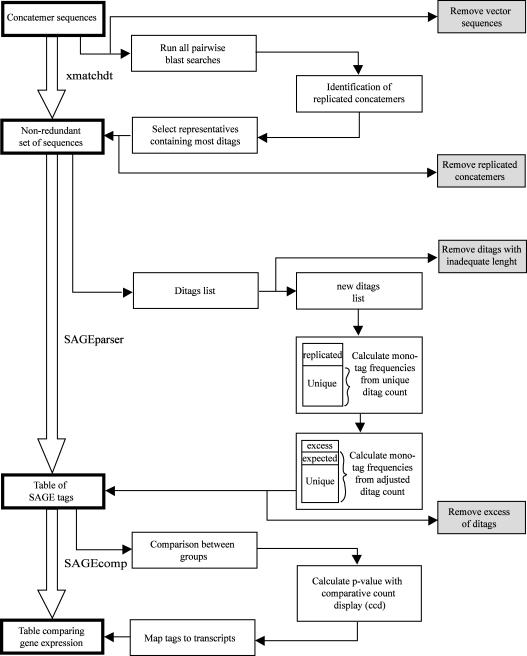
A new program package was developed to analyze SAGE results. This package, named SAGEparser, is a complete system to treat and analyze SAGE data (Figure 2). SAGEparser has been developed to eliminate the artifacts and to treat adequately the replicated ditags. The package contains three programs: xmatchdt, SAGEparser and SAGEcomp. They are publicly available on the web http://obesitygene.pbrc.edu/~eesnyder/sageparser.htm. This package is written in Perl using a minimum of external libraries and is portable to a variety of computer platforms. A SUN server was used for these analyses. Xmatchdt eliminates the replicated concatemers. Indeed, the probability that an identical series of tags occur in different concatemers is negligible. Replicate concatemers can occur when the clones are growing in SOC medium before plating on the LB-agar plates. Each concatemer sequence is blasted with all concatemer sequences and the similar sequences are clustered. The concatemer with the largest number of ditags is selected for each cluster, and the other replicated concatemers are eliminated. Table 1A shows the number of replicated concatemers. In the library containing 143 848 sequenced tags, 513 (11.13%) concatemers were replicated and eliminated by post-processing using the SAGEparser package.
Figure 2.
Overview of the SAGEparser software package.
Table 1.
Replicated cloned concatemers and ditags
| Number of valid sequenced tags | 143 848 | |
| A | Total number of cloned concatemers | 4608 |
| Number of cloned concatemers eliminated as replica | 513 (11.13%) | |
| Number of unique concatemers | 4095 (88.87%) | |
| B | Total number of ditags | 82 711 |
| Number of excess ditags rejected | 4706 (5.69%) | |
| All ditags rejected | 9555 (11.55%) | |
| Difference | 49.25% |
The tagging enzyme BsmFI cuts usually 14 bp from its recognition site included in the linker. Since the last nucleotide of the BsmFI recognition site is also the first nucleotide of the NlaIII cutting site, a tag of 15 bp (CATG + 11 bp) is produced. For example, in a library of 143 848 sequenced tags, the use of a 14 bp tag identifies 73 474 tag species and 3252 (4.4%) multiple matched, whereas 15 bp tag reduces the latter to 2001 (2.5%) and increases the total number of transcript species to 81 501. Thus, using a 15 bp tag instead of a 14 bp tag decreases the number of multiple matches and increases the number of tags which uniquely identify a transcript. SAGEparser detects the presence of any vector sequence in the concatemer sequences and does not consider them further. SAGEparser also eliminates ditags of improper length, which is greater than or less than 22 bp in this study. Moreover, the SAGEparser program can be set for any ditag length.
Another important feature is the elimination of excess ditags. Since in a large library of several thousands of tags highly expressed, similar ditags can be expected to occur several times by simple ligation of the same two tag species. SAGEparser calculates the expected similar ditag count based on the frequency of their non-replicated component (mono) tags. Ditags in excess of this number are assumed to be due to preferential PCR amplification and are rejected. Table 1B shows the number of ditags eliminated. For a sample with 143 848 valid sequenced tags, from a total of 82 711 ditags, 9555 (11.55%) ditags have occurred more than once. On the other hand, only 4706 (5.69%) ditags in excess should be rejected as analyzed by SAGEparser.
SAGEcomp is used to find which transcripts are differentially expressed by a ratio 2 or greater between different samples according to the statistical CCD test (16). To associate the tags with the transcripts, each tag (CATG + 11 bp) is compared with UniGene and GenBank databases. The tag sequence has to be located at the last NlaIII site of a given transcript. The matching procedure is very restrictive to avoid the sequencing errors in expressed sequence tags (ESTs). The match of a tag with only one EST is not considered. Indeed, the possibility of matches with EST containing sequencing errors drops dramatically when at least two ESTs are identified in a UniGene cluster for a given tag sequence. Moreover, a minimum of one EST with a known polyA tail has to be in the UniGene cluster to identify the last NlaIII site on the corresponding mRNA.
Double PCR amplification
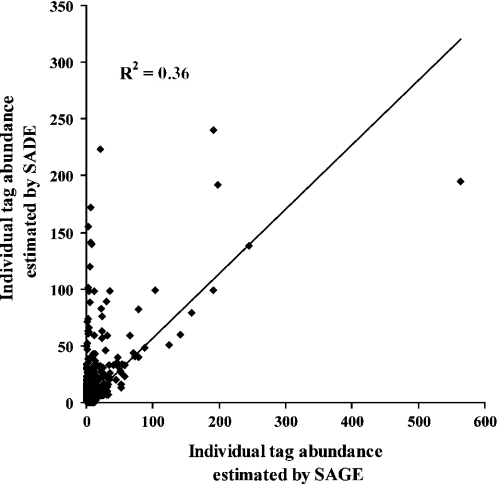
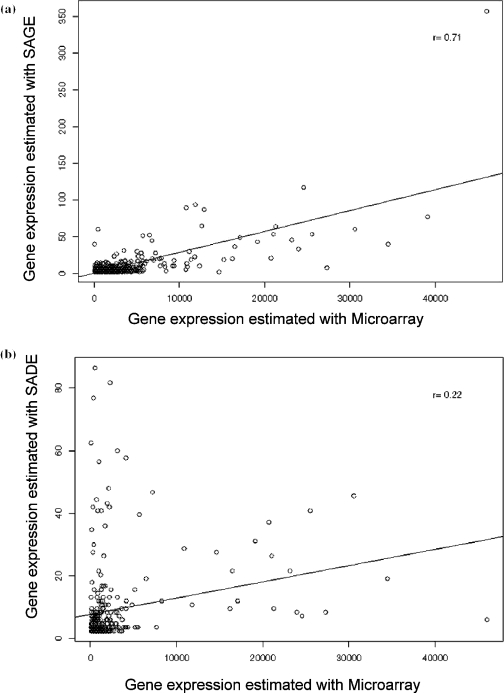
The last studied factor influencing the reproducibility is the use of the Virlon et al. (7) modification to the SAGE method with small amounts of tissue, known as SADE. The main modification of this method is a second PCR amplification to increase the quantity of ditags. To determine if reproducibility is affected by double PCR amplification, ditags were divided in two aliquots and a second round of PCR was performed only on one of them. The libraries contained 22 513 and 26 910 tags that represented 12 806 and 14 247 transcripts species, respectively. Figure 3 shows the low correlation between both methods (R2 = 0.36). A significant difference (p < 0.05) in the level of gene expression estimated by these two methods was observed for 53 transcripts. Moreover, oligonucleotides microarray results were compared with those from SAGE, with and without a second PCR amplification. Figure 4a shows that the correlation with results by microarray is 0.71 by SAGE, but only 0.22 for SADE (Figure 4b).
Figure 3.
Reproducibility of the SAGE method with one or two PCR amplifications.
Figure 4.
Comparison of the results obtained by microarrays and (a) the SAGE method or (b) the SADE method with two PCR amplifications.
Power of the SAGE method
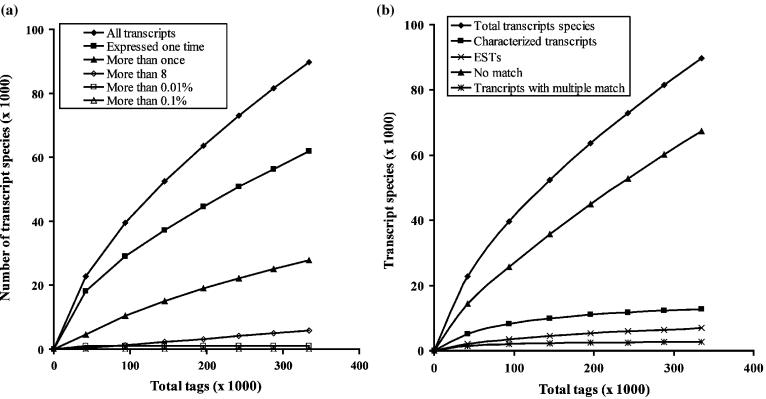
Figure 5a shows the influence of the number of sequenced tags on the power to detect transcript species. The number of high abundance tags (more than 0.1% and more than 0.01%) does not change when the number of sequenced tags is increased. However, the majority of transcript species are detected only once when the number of sequenced tags is low, whereas the percentage of tags detected more than once increased when the number of sequenced tags is increased. Figure 5b shows the type of transcript species detected with increasing number of sequenced tags. Increasing the number of sequenced tags increases the total number of transcripts assessed, as well as the number and proportion of those which are less characterized. Most of the highly expressed transcripts are well characterized and sequenced early in the SAGE library.
Figure 5.
(a) Influence of the number of sequenced tags on the detection of transcript species. (b) Type of transcript species detected according to the number of tags sequenced.
In order to evaluate the influence of the number of sequenced tags on the statistical power to detect differentially expressed transcripts, two libraries were made from mouse prostate. The first group was GDX and the other was GDX injected with DHT. As seen in Table 2, the increase in the number of sequenced tags allows the observation of more regulated transcripts.
Table 2.
Power of the SAGE method to detect significant changes according to the number of sequenced tags
| Number of sequenced tags in the first group | Number of sequenced tags in the second group | Number of transcript species after comparison | Number of transcripts differentially expressed |
|---|---|---|---|
| 30 387 | 47 376 | 29 203 | 22 |
| 79 884 | 96 075 | 52 936 | 52 |
| 130 729 | 144 438 | 73 040 | 83 |
DISCUSSION
The current study has investigated the reproducibility of the SAGE method and means to control for potential artifacts. Two libraries were made in parallel, from the same pool of total RNA. The comparison of the level of expression estimated by these two libraries has given a determination coefficient (R2) of 0.91. The current results show clearly that the SAGE method is reproducible. Yamamoto et al. (17) have assessed the reproducibility of SAGE with the use of two aliquots from a single library of 80 000 tags. The authors concluded that SAGE results are reproducible, but they are affected by the number of sequenced tags. However, they did not use any statistical analysis and did not take into account the first steps in the construction of the libraries which could be a potential source of variation. The present study shows that the first steps in the production of a SAGE library are also reproducible. Trendelenburg et al. (18) have compared two libraries constructed from a single sample. They have also concluded that the SAGE method is reproducible. However, only 15 000 tags have been sequenced from each library in that study, whereas 50 000 tags are commonly analyzed in SAGE studies.
Furthermore, the number of sequenced tags affects the reproducibility of the SAGE method. The current study shows that the reproducibility also increases with the number of sequenced tags. The determination coefficient goes from 0.91 to 0.96 when the number of sequenced tags increases from 50 000 to 150 000. This study is the first to sequence such a high number of tags from the same pool of RNA.
The computer program used to analyze SAGE data has a direct influence on the results. SAGEparser has been developed to improve the processing of SAGE data and control for the artifacts. Two important operations are necessary. The first one is to eliminate replicated concatemers, since the probability that a group of ditags is found in the same order in several concatemers is negligible. Several replicated concatemers have been observed. In addition, replicated concatemers have to be eliminated first in order to estimate the number of replicated ditags produced by the PCR amplification. Second, the analysis of SAGEparser is important to preserve a just proportion of the similar ditags. The necessity to eliminate the replicated ditags has already been discussed in the original SAGE study by Velculescu et al. (3). Since, they sequenced only 1000 SAGE tags, these authors suggested that the probability of any the two tags being coupled together to form the same ditags more than once is small, even for abundant transcripts, and that repeated ditags potentially produced by biased PCR could be excluded from analysis without substantially altering the final results. However, most of the other SAGE studies usually sequence around 50 000 tags. The SAGEparser program does not eliminate all replicated ditags since two tags of high expression level are expected to produce the same ditag many times. For example, for two tags having a frequency of 2% each, the probability of occurrence of these two tags to form a unique ditag is 0.02 * 0.02 = 0.0004 equivalent to 1/2500. Thus, in a library of 25 000 ditags, the ditag should be observed 10 times. SAGEparser is designed to eliminate only the replicated ditags which are in excess based on this model. The mono-tag frequencies are calculated by considering only one count for each ditag species. Then, the program calculates the frequency of each ditag with mono-tags frequencies, and compares the calculated and observed frequencies. When the observed frequency of the ditags is lower than the calculated frequency, all similar ditags are conserved. Otherwise, the ditags in excess are eliminated. With this analysis, the right number of replicated ditags is eliminated and results are more representative of the reality. Generally, SAGE studies use tags of 14 bp length. However, the present results show the advantage of using a tag of 15 bp. The percentage of multiple matches is lower with a tag of 15 bp. For example, the 15th basepair allows to specifically match rat myosin heavy chain I, IIa or IIx whereas 14 bp cannot distinguish these isoforms.
A last factor identified to affect the reproducibility of SAGE is the use of pre-large scale PCR described by Virlon et al. (7). SADE has been suggested when the quantity of mRNA is too small. In SADE, ditags are amplified by PCR twice rather than only once. However, the effects of this modification on the quantitative aspect of SAGE have not been investigated in detail and statistically. In the current study, ditags were separated in two aliquots, only one of which was subjected to a pre-large scale. The determination coefficient (R2) between SAGE and SADE was only 0.36. Thus, the reproducibility of SAGE is affected by the SADE modifications. A further comparison with microarray studies also shows the detrimental effect of double amplification of ditags on the estimated level of gene expression. The correlation coefficient between microarray and the SAGE method decreased from 0.71 without pre-large scale PCR to 0.22 with the SADE modification. Therefore, the quantitative aspect of the results is affected by the second PCR amplification. Moreover, several transcripts had significant differences in expression levels estimated by the SAGE and SADE methods. Velculescu et al. (19) have also previously suggested that PCR amplification can affect the accuracy of SAGE to estimate the level of gene expression. It has also been mentioned that an increase in the number of PCR cycle can potentially distort the quantification of gene expression level by SAGE (20).
An increase in the number of sequenced tags enhances the reproducibility and the power of the SAGE strategy to detect known and novel transcripts as well as their differential regulation by experimental conditions. When the number of sequenced tags is low, the majority of tags are observed only once and correspond to known transcripts. When the number of sequenced tags is increased, the proportion of tags expressed more than once increases and the statistical power to detect changes between experimental conditions is greater. The novel transcripts are generally expressed at low abundance level and more tags must be sequenced to detect them. Indeed, the majority of transcripts have only a very small number of copies per cell. For instance, it has been estimated that about 80% of transcripts are present at less than 5 copies per cell (21) or that 83% of transcripts have only one copy per cell (22). To detect these very low abundance transcripts, a very powerful method is necessary.
It is estimated that a cell generally contains about 300 000 mRNA molecules (23). However, the number of transcript species has not yet been exactly quantified since the whole transcriptome has not been fully characterized in mammalian cells. Although the genome may contain approximately between 27 000 and 120 000 genes (24) many genes produce more than one transcript species due to various mechanisms such as alternative splicing, mRNA editing, as well as multiple promoters and polyadenylation sites. The current study shows that the sequencing of 350 000 tags from one library gives the possibility to detect around 90 000 transcript species. The majority of these transcripts are expressed at low levels and are novel transcripts. Some of these transcripts can be sequencing errors. However, the experimental data of Wang (25) show that the error rate for sequenced SAGE tags is lower than 2% per tag and that 67% of novel SAGE tags are derived from novel transcripts. Chen et al. (26) also suggest that the sequencing error rate is lower than 2% by tag. These authors also observed that all of erroneous SAGE tags had only a single-base error. However, the lower expressed tags should be considered since a major part of the transcripts are expressed with less than 5 copies per cell (21). Increasing the number of sequenced tags in a SAGE study can improve the reproducibility as well as the power to detect the less abundant tags. However, this increase can be made only according to the budget and the time-consuming limitation of the SAGE method.
The current study has shown that the SAGE method has very good reproducibility. However, several factors can affect this reproducibility. First, a suitable way to treat the replicated concatemers and ditags was investigated, and a new software package was developed. Second, the influence of the number of sequenced tags was studied. When the number of sequenced tags is increased, the SAGE method is more powerful for the identification of regulated genes and the reproducibility is also improved. Finally, the current results have clearly demonstrated that pre-large scale PCR importantly reduces the reproducibility of the SAGE method and the ability to accurately quantify gene expression levels.
Acknowledgments
This work was supported by Genome Québec and Genome Canada. Funding to pay the Open Access publication charges for this article was provided by Genome Québec and Genome Canada.
REFERENCES
- 1.Liang P., Bauer D., Averboukh L., Warthoe P., Rohrwild M., Muller H., Strauss M., Pardee A.B. Analysis of altered gene expression by differential display. Methods Enzymol. 1995;254:304–321. doi: 10.1016/0076-6879(95)54022-9. [DOI] [PubMed] [Google Scholar]
- 2.Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 3.Velculescu V.E., Zhang L., Vogelstein B., Kinzler K.W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 4.St-Amand J., Okamura K., Keitaro M., Shimizu S., Sogowa Y. Characterization of control and immobilized skeletal muscle: an overview from genetic engineering. FASEB J. 2001;15:684–692. doi: 10.1096/fj.00-0150com. [DOI] [PubMed] [Google Scholar]
- 5.Larose M., St-Amand J., Yoshioka M., Belleau P., Morissette J., Labrie C., Raymond V., Labrie F. Transcriptome of mouse uterus by serial analysis of gene expression (SAGE): comparison with skeletal muscle. Mol. Reprod. Dev. 2004;68:142–148. doi: 10.1002/mrd.20065. [DOI] [PubMed] [Google Scholar]
- 6.Datson N.A., van der Perk-de Jong J., van den Berg M.P., de Kloet E.R., Vreugdenhil E. MicroSAGE: a modified procedure for serial analysis of gene expression in limited amounts of tissue. Nucleic Acids Res. 1999;27:1300–1307. doi: 10.1093/nar/27.5.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virlon B., Cheval L., Buhler J.M., Billon E., Doucet A., Elalouf J.M. Serial microanalysis of renal transcriptomes. Proc. Natl Acad. Sci. USA. 1999;96:15286–15291. doi: 10.1073/pnas.96.26.15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neilson L., Andalibi A., Kang D., Coutifaris C., Strauss J.F., III, Stanton J.A., Green D.P. Molecular phenotype of the human oocyte by PCR–SAGE. Genomics. 2000;63:13–24. doi: 10.1006/geno.1999.6059. [DOI] [PubMed] [Google Scholar]
- 9.Anisimov S.V., Tarasov K.V., Stern M.D., Lakatta E.G., Boheler K.R. A quantitative and validated SAGE transcriptome reference for adult mouse heart. Genomics. 2002;80:213–222. doi: 10.1006/geno.2002.6821. [DOI] [PubMed] [Google Scholar]
- 10.Bolduc C., Larose M., Lafond N., Yoshioka M., Rodrigue M.A., Morissette J., Labrie C., Raymond V., St-Amand J. Adipose tissue transcriptome by serial analysis of gene expression. Obes. Res. 2004;12:750–757. doi: 10.1038/oby.2004.90. [DOI] [PubMed] [Google Scholar]
- 11.Velculescu V.E., Zhang L., Zhou W., Vogelstein J., Basrai M.A., Bassett D.E., Jr, Hieter P., Vogelstein B., Kinzler K.W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhang L., Zhou W., Velculescu V.E., Kern S.E., Hruban R.H., Hamilton S.R., Vogelstein B., Kinzler K.W. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 13.Welle S., Bhatt K., Thomton C.A. High-abundance mRNAs in human muscle: comparison between young and old. J. Appl. Physiol. 2000;89:297–304. doi: 10.1152/jappl.2000.89.1.297. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Zulueta M., Ensz L.M., Mukhina G., Lebovitz R.M., Zwacka R.M., Engelhardt J.F., Oberley L.W., Dawson V.L., Dawson T.M. Manganese superoxide dismutase protect nNOS neurons from NMDA and nitric oxide-mediated neurotoxicity. J. Neurosci. 1998;18:2040–2055. doi: 10.1523/JNEUROSCI.18-06-02040.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gygi S.P., Rochon Y., Franza B.R., Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lash A.E., Tolstoshev C.M., Wagner L., Schuler G.D., Strausberg R.L., Riggins G.J., Altschul S.F. SAGEmap: a public gene expression resource. Genome Res. 2000;10:1051–1060. doi: 10.1101/gr.10.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto M., Wakatsuki T., Hada A., Ryo A. Use of serial analysis of gene expression (SAGE) technology. J. Immunol. Methods. 2001;250:45–66. doi: 10.1016/s0022-1759(01)00305-2. [DOI] [PubMed] [Google Scholar]
- 18.Trendelenburg G., Prass K., Priller J., Kapinya K., Polley A., Muselmann C., Ruscher K., Kannbley U., Schimitt A.O., Castell S., et al. Serial analysis of gene expression identifies metallothionein-II as major neuroprotective gene in mouse focal cerebral ischemia. J. Neurosci. 2002;22:5879–5888. doi: 10.1523/JNEUROSCI.22-14-05879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velculescu V.E., Vogelstein B., Kinzler K.W. Analysing uncharted transcriptomes with SAGE. Trends Genet. 2000;16:423–425. doi: 10.1016/s0168-9525(00)02114-4. [DOI] [PubMed] [Google Scholar]
- 20.Ye S.Q., Lavoie T., Usher D.C., Zhang L.Q. Microarray, SAGE and their applications to cardiovascular diseases. Cell Res. 2002;12:105–115. doi: 10.1038/sj.cr.7290116. [DOI] [PubMed] [Google Scholar]
- 21.Peters D.G., Kassam A.B., Yonas H., O'Hare E.H., Ferrell R.E., Brufsky A.M. Comprehensive transcript analysis in small quantities of mRNA by SAGE-lite. Nucleic Acids Res. 1999;27:e39. doi: 10.1093/nar/27.24.e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Velculescu V.E., Madden S.L., Zhang L., Lash A.E., Yu J., Rago C., Lal A., Wang C.J., Beaudry G.A., Ciriello K.M., et al. Analysis of human transcriptomes. Nature Genet. 1999;23:387–388. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 23.Hastie N.C., Bishop J.O. The expression of three abundance classes of messenger RNA in mouse tissues. Cell. 1976;9:761–774. doi: 10.1016/0092-8674(76)90139-2. [DOI] [PubMed] [Google Scholar]
- 24.Pennisi E. Human Genome Project. And the gene number is …? Science. 2000;288:1146–1147. doi: 10.1126/science.288.5469.1146. [DOI] [PubMed] [Google Scholar]
- 25.Wang S.M. Response: the new role of SAGE in gene discovery. Trends Biotechnol. 2003;21:57–58. doi: 10.1016/s0167-7799(02)00031-8. [DOI] [PubMed] [Google Scholar]
- 26.Chen J., Sun M., Lee S., Zhou G., Rowley J.D., Wang S.M. Identifying novel transcripts and novel genes in the human genome by using novel SAGE tags. Proc. Natl Acad. Sci. USA. 2002;99:12257–12262. doi: 10.1073/pnas.192436499. [DOI] [PMC free article] [PubMed] [Google Scholar]