Abstract
The crystal structure based model of the catalytic center of Ago2 revealed that the siRNA and the mRNA must be able to form an A-helix for correct positing of the scissile phosphate bond for cleavage in RNAi. This suggests that base pairing of the target mRNA with itself, i.e. secondary structure, must be removed before cleavage. Early on in the siRNA design, GC-rich target sites were avoided because of their potential to be involved in strong secondary structure. It is still unclear how important a factor mRNA secondary structure is in RNAi. However, it has been established that a difference in the thermostability of the ends of an siRNA duplex dictate which strand is loaded into the RNA-induced silencing complex. Here, we use a novel secondary structure prediction method and duplex-end differential calculations to investigate the importance of a secondary structure in the siRNA design. We found that the differential duplex-end stabilities alone account for functional prediction of 60% of the 80 siRNA sites examined, and that secondary structure predictions improve the prediction of site efficacy. A total of 80% of the non-functional sites can be eliminated using secondary structure predictions and duplex-end differential.
INTRODUCTION
RNAi has rapidly come to be recognized as a powerful tool for studies of gene function and a potential therapeutic modality. One of the challenges in applying small interfering RNAs is the identification of a potent siRNA/target sequence combination. The attempts, so far, have focused mainly on the occurrence of specific sequence motifs, such as predominance of A or U bases in the 5′ end of the antisense molecule. The reliance on observed nucleotide occurrence is due to an incomplete understanding of the factors involved in RNAi-mediated sequence-specific RNA degradation. Early on, rules for the selection of target sites were based mostly on practicality (1,2). Tuschel and co-workers applied the rules of 21mer targets, use of TT overhangs complementary to AA in the target sequence and low GC-content to select target sites. Drawing from criteria established for antisense oligodeoxyribonucleotide (ODN) applications, avoiding high GC-content was selected, in part, to avoid targets that might be bound in strong secondary structure.
Since the initial rules for siRNA design, numerous studies have been carried out to investigate the correct design of siRNAs. Each study, be it chemical, biochemical or in silico, has presented a justification for previous findings or given insight on a new aspect of RNAi mechanisms. Importantly, studies on the crystallographic structure of the Paz domain (3–7) have shown that it is not involved in protein–protein contacts in RNA-induced silencing complex (RISC), as once thought (8), but that the Paz domain is an RNA-binding domain. Specifically, the Paz domain binds to the 2 bp 3′ overhang characteristic of siRNAs (7). This corroborates the finding that siRNAs with overhangs are more functional than siRNAs without overhangs (1). Furthermore, an A-helical conformation of the target RNA–siRNA hybrid appears to be important for increased binding stability by the PAZ domain (7). This is consistent with the chemical modification studies on siRNAs and kinetic data, which demonstrate that inability of a duplex to form an A-helix is inhibitory for RNAi (9–13). Further, models of Ago2 ‘slicer’ activity (14,15) also predict that the target RNA–siRNA hybrid must be in an A-helical form for the scissile phosphate to be positioned correctly for the bond to be broken. Most importantly, the need for an A-helix conformation of the target RNA–siRNA hybrid suggests that the conformation of the bases involved is important, as the target site must be free of intermolecular base pairing. Thus, RISC-mediated RNAi would require the destabilization of secondary structures in the target.
The assumption that target and siRNA secondary structure can be involved in the RISC cleavage derives from the long-standing idea that antisense ODN paring using target mRNAs is inhibited by the secondary structure. In support of the importance of target mRNA secondary structure, several studies cited evidence, which suggested that secondary structure can indeed cause inhibition of RNAi (16,17). These types of observations have driven more recent attempts to predict secondary structures in order to guide selection of sites for RNAi targeting. In this regard, the two studies differ slightly in their approaches, although both are grounded in the nearest-neighbor rules of RNA folding, as in the Mfold program (18,19). While Ding et al. (20) rely on the single-stranded nature of sites based on a rigorous statistical sampling of the folded RNA, Luo and Chang (21) use a consistent predominance of unpaired nucleotides as a marker of a good site, as determined by the Mfold program, providing further evidence that the secondary structure is possibly a useful parameter.
In contrast to the idea that secondary structure plays a role in RNAi, Reynolds et al. (22) suggested that there is no correlation between secondary structure and RNAi. However, they found that the incorporation of GC-content in their algorithm for siRNA site prediction was important, a feature that would avoid areas of highly stable secondary structure. The major emphasis of their algorithm is on sequence-specific factors. For example, they use the difference in frequency of A/U bases in the 5′ and 3′ ends of the duplex; some refer to this as a difference in duplex-end stability. This relates to an essential aspect of RISC-mediated cleavage, the loading of the correct stand of the duplex into the complex. Previous results demonstrated that the thermodynamic stabilities of the ends of the siRNA duplexes influence the choice of the strand that is to be included in the RISC (23,24). Besides in vitro data, kinetic evidence obtained in Drosophila (10) and mammalian cell extracts (13) suggest that the RISC can engage in multiple rounds of mRNA target cleavage. Indeed, Haley and Zamore (10) observed that a 5′ mismatch of antisense with target mRNA can increase the ability of RISC to cleave a target in an ATP-independent fashion (10). Their data suggest that ATP is necessary for product release. Thus, a mismatch in the 5′ end may increase the ability of RISC to recycle by decreasing the stability of RISC and the cleaved product (reminiscent of ribozyme product release). This supports the motif of a weak binding base in the 5′ antisense position of an siRNA (22,25,26), and may tie into a more definitive explanation of the bias of the RISC complex for strands with a thermodynamically weak 5′ end. The effect of this phenomenon, a difference in duplex-end stabilities, is so great that it has been derived, or included, in the development of most motif-based siRNA selection algorithms (22,25,26), making it the most widely used feature in the siRNA design.
The factors influencing RNAi-mediated degradation of a target mRNA have not been deduced completely. Besides a length of 21 nt and 3′ overhangs, duplex-end energy difference is the only proven criteria. It is involved in the propensity of a strand to be incorporated into the RISC. Many other factors remain to be discovered. In this work, we introduce a novel approach for the determination of mRNA secondary structures and show that in combination with duplex-end energies the predicted strong secondary structures can account for 80% of non-functional siRNA target sites. These results offer an improvement over duplex-end energies alone. Our data also confirm that for RNAi targets, GC-content is truly linked to secondary structure potential. These studies outline the novel use of the most stable local substructure to determine the potential secondary structure at an siRNA site that differs from previous folding algorithms. Finally, the algorithm has the power to quickly screen through all the possible siRNA sites on an mRNA. This work not only highlights the use of our secondary structure predictions, but also provides mechanistic insights into RNAi and a direction for future work.
MATERIALS AND METHODS
siRNA
The siRNA duplexes targeting enhanced green fluorescent protein (EGFP) from pL-EGFP were in vitro transcribed using the Silencer Ambion kit as described previously (27). The sense sequences of the siRNAs 5′ to 3′ are A, GGUGAACUUCAAGAUCCGC; B, CAUCGAGGACGGCAGCGUG; C, GCAGAAGAACGGCAUCAAG; and D, GCUUCGAUAACUUCGUAUA. For 2a, 4a and 5a, the antisense strands were the same as the ones used by Kim and Rossi (28). The sense strands were GCAGACCCUGAAGUUCAUC, GUUCGAGGGCGACACUCUG and GCAGGAGUACAACUACAAC, respectively.
Computational time for the folding analysis program
On a Macintosh G4, with dual processors of 1.27 GHz and 1 GB of RAM, it took ∼20 min to analyze a file of 10 000 foldings, generated by Mfold program (18,19), for a sequence of 1000 nt.
Transfection protocol
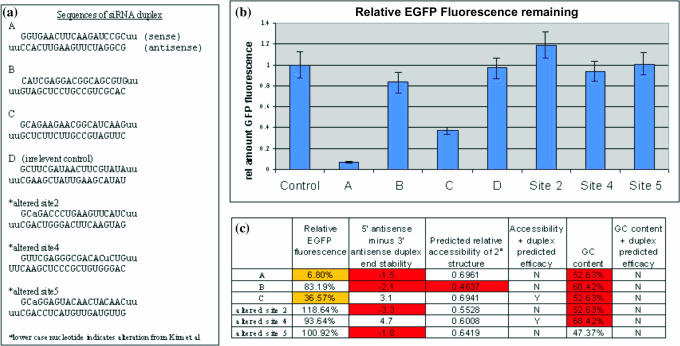
Three independent experiments, each in triplicate, were carried out in 24-well plates, for a total of 9 different tests for each sample. In brief, 0.1 μg EGFP plasmid, 10 pmol siRNA duplex and 8 μl of Lipofectamine 2000 were used. The Lipofectamine 2000 protocol of the manufacturer (Invitrogen) was followed with the noted concentrations. Cells were plated 24 h before transfection to achieve a density of 80% on the day of transfection. Twenty-four hours after transfection, cells were collected using phosphate-buffered saline and analyzed using fluorescent-activated cell sorting (FACS). A count of 50 000 cells per well was carried out. The histogram data were divided into four regions of intensity based on the level of the EGFP signal, and the two brightest regions were analyzed for the percentage of cells in the region. Averages were calculated for each plate and then compared across plates to confirm the results. The average of all the three 24-well plates is reported in Figure 3, with the resultant error.
Figure 3.
(a) The sequences of the siRNA duplexes used. (b) Average of FACS data for the amount of GFP signal relative to control. Data are the average from three 24-well plates with triplicate samples on each plate. Each plate was cotransfected with 0.1 μM EGFP vector and 20 nM in vitro transcribed siRNA. (c) Characteristics of sites. Comparing predicted versus knockdown. For example, site C was predicted to be an effective site as it had good duplex-end differential and predicted secondary structure accessibility. Site C produced 63.3% knockdown.
Accessibility predictions
To acquire a description of conformations of an mRNA, the mRNA was first folded using Mfold program with the parameters W = 0, MAXBP = 100, P = 100 and MAX = 9999 (Note: for sequences >1200 nt, it is often the case that the hard-wired max increment of 12 kcal/mol is used). This produces a set of 9999 confirmations of the mRNA that have a maximum distance of 100 nt between bases in a base pair, and where each folding is within 12 kcal/mol of the confirmation with the lowest free energy. The program next creates a list of all the possible n-mers (where n is the size of the target site) and calculates the theoretical maximum stability of a perfectly base-paired duplex of the n-mer and a complementary sequence. Next, its duplex-end energies are calculated using the nearest-neighbor rules for RNA–RNA hybrids. The stability of the 5′ end is equivalent to the free energy of the first four base parings [as in (24)] plus the energy of any overhanging bases. Our script that performs this calculation can be accessed from the City of Hope website. Finally, the folding analysis program analyzes the 9999 foldings to determine the most probable secondary structure for each of the n-mers.
Description of mRNA folding analysis
Drawing upon statistical mechanics, we utilized a Boltzmann weighing of potential mRNA foldings to determine the most probable secondary structure of a position in an mRNA. Essentially, the free energy of a state determines the portion of molecules in a system adopting that state. Although Boltzmann weighing has been used by others for predicting RNA secondary structure (20,29–32), our algorithm is unique in that it uses local structural features to calculate the secondary structure of a target site. As described below, we use the stability of the most stable substructure within 100 nt of the target site to weigh the contribution of the folding to the most probable target site free energy (the interactions of the nucleotides that make up the target site).
The symbolic notation and mathematical equations used in the paper are as follows:
Let F be the set of foldings, where the maximum distance between bases in a base pair is 100 nt, and each folding is within 12 kcal/mol of the most stable folding.
Let Fi be the i-th element of F.
Let T be the set of all the 21 nt subsequences (target sites) of the target RNA, where Ti is the nucleotide subsequence from 1 to 21 of the sequence, T2 is the nucleotide subsequence from 2 to 22 and Tn is the nucleotide subsequence from n to (n + 20).
In Fi, for a specific Tj ε T, there is a 100 nt region centered at Tj. Denote Rij as the specific base parings for this 100 nt region.
For a given folding, Fi, and target site, Tj, find Rij. If the base pairings of Rij are the same as a previous Rpj, then the folding is not considered and we move onto the next folding, Fi+1. In Rij, find the most stable substructure and determine the total ΔG for that substructure, label it . Also determined , the structural stability, in Ri, of the nucleotides making up the target site, Tj. ΔG is determined using nearest-neighbor rules.
- For a particular Tj, Zj is
where n is the number of foldings considered, is the thermodynamic gas constant in kcal/(mol K), and T is 37°C. - For a Tj, the weighted stability of the target site secondary structure, called ‘str’, is
where n is the number of foldings considered. For each target site, Tj, there is an associated theoretical stability, , of a perfect helix between the target site nucleotides and a perfect complementary sequence.
- We define relative accessibility to be
The scaling factor, C, is simply (−60/21). This relates to the maximum possible ΔG for a 21 nt sequence, and is simply a way to scale the output between 0 and 1.
Site prediction
Target sites with unfavorable duplex-end energies or unfavorable accessibility are predicted to be ineffective sites. From a subset of siRNA effectivity data (33), a cut-off for predicted relative accessibility was set at 0.55. For duplex-end energy differences, we used −0.5 as the cut-off as this ensures that there is a difference in the end stability.
Origin of Pten and Icam siRNA sites evaluated
siRNA effectivity data comes from the work of Vickers et al. (33). This set of data was chosen due to its size, randomness and consistency of experimental method.
RESULTS
Predicting site effectiveness
Effective target sites are defined here as sequences susceptible to RNAi-mediated knockdown >55%. To identify such sequences, the algorithm combines the predicted strength of mRNA secondary structure and the relative stabilities of the corresponding siRNA duplex ends. Given an mRNA sequence, which includes the 5′-untranslated region (5′-UTR) and 3′-UTR, an exhaustive array of possible 21 base siRNA target sequences is generated. These sites are then used to create theoretical 21 bp siRNA duplexes, and the relative thermodynamic stabilities of the ends of the duplexes are determined (23,24). Those duplexes that have differential stabilities favoring antisense strand incorporation into the RISC are noted. Next, the potential target sites are examined by comparing the predicted secondary structure against the formation of a duplex between the target site and the siRNA antisense strand.
Secondary structure potential, or predicted secondary structure, is determined by the use of a nearest-neighbor RNA folding program [at present, we have been using the Mfold program of Zuker (18,19)] and a novel program that explores the sequence space of possible RNA foldings. First, a restricted set of RNA foldings is collected for the mRNA to be targeted. The nucleotide distance between paired bases and the thermodynamic stability of the total folding restricts the collection. These restrictions assume that the distance over which a base can find a pairing partner is limited by the kinetics of mRNA folding.
For each target, a subset of global foldings is then analyzed. This subset of foldings depends upon the local region centered on the target site. Within the region, there is a dominant substructure, meaning a most stable substructure. The stability of this substructure is used to determine the probability of the particular folding of the region. Each regional folding is measured only once. The thermodynamic stability of the most stable substructure in the region is used for a Boltzmann weight of the free energy of the secondary structure of the nucleotides that make up the target site. In this manner, the local folding around the target site is used to determine the likelihood that the bases of the target site will adopt the secondary structure represented in a particular folding (for more details see Materials and Methods).
In the final analysis, accessibility is the difference between the weighted average of the free energy of the target site secondary structure and the theoretical maximum stability of antisense binding. For the theoretical maximum stability of the antisense binding, the value of a perfect helix formed between the target site and the antisense sequence is utilized. This analysis provides a measure of the potential competition between an intramolecular secondary structure and the formation of an intermolecular duplex. For antisense ODNs, it is possible to calculate the change in mRNA free energy that occurs upon antisense hybridization [David Mathews program OligoWalk (34) is an excellent tool to perform this calculation]. However, RNAi incorporates the binding of an antisense RNA molecule that is complexed with one or more proteins. It is not clear how binding to the RISC will alter the mRNA structure. Using the difference between probable secondary structure and the theoretical maximum stability of a duplex containing the target sequence is a reasonable compromise between the exact calculation of the stability of RISC-antisense-mediated binding and the uncertainty of the effects that RISC binding to an mRNA might have upon the free energy. Furthermore, the difference described can be mathematically shown to be the ratio of the number of duplex molecules relative to the number of folded target sites.
Those target sequences that have good accessibility and for which there are favorable siRNA duplex-end stabilities for antisense sequence selection (23,24) (based upon empirically derived cut-offs) are predicted to be targets that will yield effective RNAi-mediated knockdown.
Example of secondary structure predictions
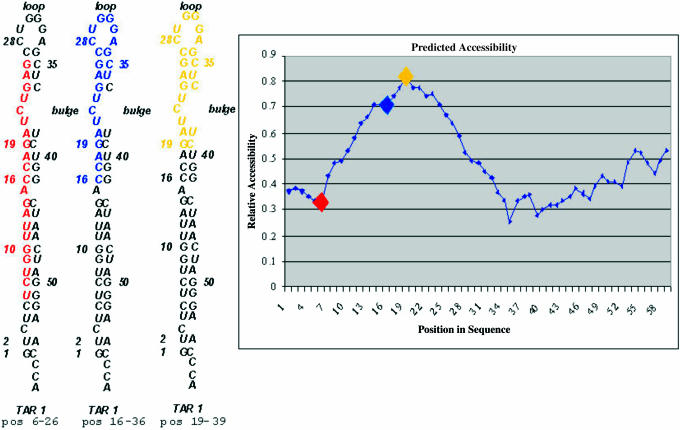
As an example of the algorithm's ability to predict secondary structure, the HIV-1 TAR element derived from the NL4-3 viral RNA sequence was folded and scanned using the algorithm. The 5′ end of all HIV transcripts contains the well-characterized and highly conserved TAR element (35,36). Figure 1 displays the results of the secondary structure predictions for this element based upon folding of the rev mRNA that contains TAR at its 5′ end. Each point in the graph represents a 21 nt subsequence, its x coordinate corresponding to its position in the mRNA, and its y coordinate corresponding to its relative accessibility. The greater the predicted accessibility and the less stable the secondary structure, the higher the sequence is on the y-axis. Figure 1 shows that in progressing from point 1 to point 43, the predicted accessibility correlates well with the known structure of TAR. For example, point 19 is predicted to be the most accessible site, and it corresponds to nucleotides 19–39, which includes a hairpin loop and a bulge, and has the fewest base pairs. The predicted accessibility is ∼0.8, which predicts that a perfectly matched 21mer to this sequence would have a ΔΔG that is 48 kcal/mol more stable than the predicted secondary structure of the TAR element for this 21mer. Thus, both the known structure and the predictions agree that the 21mer, at point 19, is predicted to be the most accessible site. The subsequence beginning at point 6 corresponds to a 21mer stretch in which most of the bases are paired. This site has a predicted accessibility of 0.33, which suggests that a 21mer duplex of this site and a complementary sequence have a ΔΔG that is only 19.8 kcal/mol more stable than the predicted secondary structure. The relative accessibility calculations in the other regions of TAR suggest that the predicted accessibilities to 21mer oligos are in close agreement with the known structure of TAR.
Figure 1.
The HIV TAR RNA was folded from a sequence derived from the pNL4-3 vector. The output of the secondary structure prediction is shown in the graph. The higher the relative accessibility score, the more accessible the site is predicted to be. Each point in the output graph represents a 21 nt stretch, with base 1 of the stretch corresponding to the position on the x-axis. For example, the red diamond represents nucleotide positions 6–26. The secondary structure predictions correspond to the known structure of TAR. For example, the 21 bp region starting from nucleotide 19 is the most accessible structure both in the graph of predicted accessibility and the known structure.
Further inclusions of different types of secondary structures create subtle and not-so subtle differences in predicted accessibility. The accessibility difference between sequences 1 and 6 is created by the inclusion of a single nucleotide bulge present at nucleotide 3. In contrast, the sharp differences in the predicted accessibilities of sequences 6 and 7 demonstrate the disruptive power of a hairpin loop. The G27–C34 base pair is destabilized by the presence of the hairpin loop, and lacks strong 3′ stacking interactions, as shown by the fact that C26–A33 is unpaired. Sequences 5 and 6 have similar accessibility predictions except that sequence 5 includes C5–G48 stacked on U6–G47, whereas sequence 6 includes G26–C35 stacked on C27–G34. The difference in predicted accessibility corresponds to the differences in the stabilities of a CG base pair stacked on an UG base pair and a GC base pair stacked on a CG base pair. Thus, these data demonstrate how even minor differences in the secondary structure of nucleotides involved in a region of base pairing are captured by the accessibility predictions in this algorithm. Importantly, the predicted accessibility plot strongly correlates with the known TAR structure, including the effects of nucleotide composition and the form of secondary structure.
Prediction of RNAi and published data
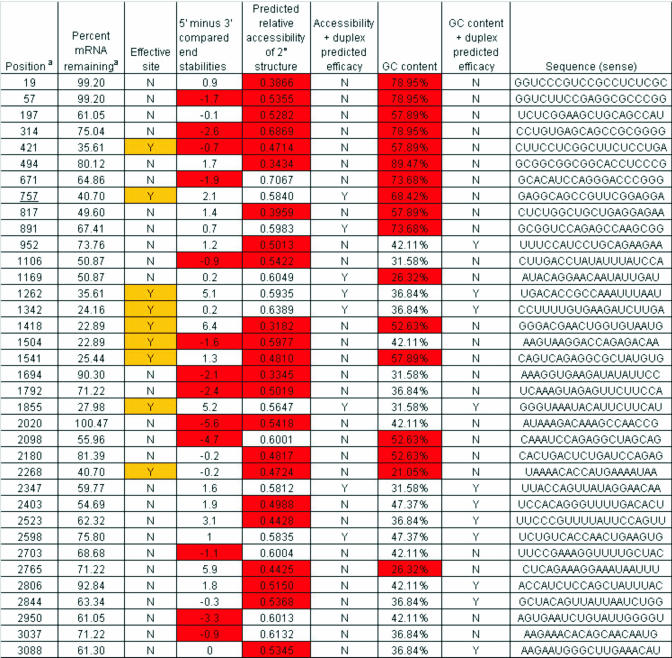
The ability of our algorithm to predict sites accessible to RNAi was analyzed using RNA knockdown data obtained from 80 published sites in 3 different mRNAs, Icam (33), Pten (33) and EGFP (28). Among these three data sets, a successful RNA knockdown is calculated to be 55% or greater reduction in the RNA relative to the untreated controls. Figure 2 contains the results of this analysis and a comparison of our predictions with experimentally determined knockdown efficiencies for sites for Pten. As described in Materials and Methods, target sequences for which the siRNA 5′ antisense end is −0.5 kcal/mol or more stable than the 3′ antisense end, and/or where the predicted accessibility value is <0.55, are predicted to be non-effective sites. The results indicate that the combined algorithm of duplex-end stability differences and accessibility predictions is capable of predicting the majority of RNAi sites, especially ineffective sites.
Figure 2.
The excerpt is from our analysis of the published data. The percentage mRNA remaining is from Vickers et al. (33). As mentioned, effective sites are those which result in 55% or greater knockdown. For duplex energies, sites where the 5′ antisense end is 0.5 kcal/mol or more stable than the 3′ end are deemed unfavorable. The acceptable range for GC-content is 30–52%. And sites predicted to be accessible are those with a predicted relative accessibility of 0.55 or higher. The superscript letter ‘a’ represents the columns from Vickers et al. (33).
The algorithm had a positive predictive value of 55%, the best of the three methods tested. The algorithm had a specificity of 81%, correctly identifying 45 of the 55 ineffective sites. For effective sites (>55% knockdown), the algorithm had a sensitivity of 48%, correctly identifying 12 of the 25 experimental sites. Overall, the algorithm correctly predicted (effective or not effective) 71% of the sites tested.
We also investigated the predictive power of duplex-end stabilities. The studies of Khvorova et al. (23) and Schwarz et al. (24) have shown that a favorable differential in the stability of the 5′ and 3′ ends of the antisense strand in a siRNA duplex can lead to selective incorporation of the antisense strand into the RISC. Thus, the difference in duplex-end stabilities can be used to estimate the relative effectiveness of a given siRNA. Using duplex-end stability alone to predict site efficacy, only 60% of the sites (effective and ineffective) were correctly predicted as compared with 71% using the accessibility analyses. Although the sensitivity of this approach is good, the specificity is lower than for the combined accessibility duplex-end stability algorithm. By comparison, the approach of combining accessibility predictions and duplex-end stabilities resulted in an increase of specificity from 49 to 81%. Thus, duplex-end stabilities alone do not determine ineffective sites as well as the algorithm combining duplex-end stabilities and accessibility predictions. Looking at prevalence—in this case, the percentage of sites predicted to be effective—it can be seen that duplex-end energies alone (61%) acts as a weak filter for ineffective sites. The prevalence of effective sites in the sites tested was 31%.
Several sources have indicated that GC-content is useful for predicting effective sites. Essentially, those siRNAs harboring GC-contents outside of a specified range are predicted to be non-effective. Here we use the range 30–52%, as suggested by Reynolds et al. (22), and report the ability of a combined algorithm using GC-content and duplex-end stabilities to predict siRNA effectiveness. The results in Table 1 indicate that GC-content and our accessibility predictions behave similarly. However, the positive predictive value for the GC-content (36%) was much lower than for the combined accessibility, duplex-end stability algorithm, indicating that the combined algorithm is more reliable for effective site prediction than GC-content.
Table 1.
Comparison of predictions
| Methods | Sensitivity (percentage of effective sites correctly predicted) | Specificity (percentage of ineffective sites correctly predicted) | Positive predictive value (%) | Negative predictive value (%) | Prevalence (percentage predicted to be effective) |
|---|---|---|---|---|---|
| Duplex + GC | 32 | 75 | 36 | 71 | 28 |
| Accessibility + duplex | 48 | 81 | 55 | 78 | 28 |
| Duplex | 84 | 49 | 43 | 87 | 61 |
Duplex-end energies had the best sensitivity, but this was offset by a low selectivity. Overall, the best predictions were made using secondary structure accessibility predictions and duplex-end energy differential. This method had the highest selectivity and the best positive predictive value (reliability of positive predictions).
To further test the algorithm, we analyzed data obtained from RNAi-mediated knockdown of the mRNA for EGFP. A total of six siRNAs were tested. Of these, three novel siRNAs were designed based on the predictions for accessibility and duplex-end energies: siRNA A was predicted to have good accessibility but unfavorable duplex-end energy differences; siRNA B was predicted to have poor accessibility and unfavorable duplex-end energy differences; and siRNA C was predicted to have good accessibility and favorable duplex-end energy differences. Site D, an irrelevant control, was chosen for its lack of homology to known human mRNAs and the EGFP used in this study. The functional assay incorporated FACS to quantify EGFP expression. A greater than 55% decrease in fluorescence was observed for siRNAs A and C, but not for B (Figure 3). Thus, site C revealed that our prediction of an effective site was correct. Site B confirmed our prediction of an ineffective site. The result for site A showed that as with the published data, some sites with good predicted accessibility and bad duplex energies are still effective sites.
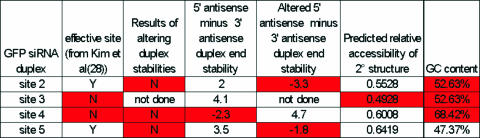
We also tested duplex modifications of three previously used siRNAs (28). For siRNA duplexes 2a and 5a, the sense strand was modified by replacing the U in position 3 with an A, creating an A–A mismatch that destabilized the 5′ sense strand. This made a duplex with end stabilities that favor the sense strand incorporation over the antisense strand into the RISC (i.e. this in effect turned a good duplex into a bad duplex). Figure 4 compares the results of the altered duplexes with the original duplexes. The sites for 2a and 5a both exhibit predicted accessibilities somewhat lower than for site A. In the functional assays, sites 2a and 5a were ineffective. The difference between sites for duplex 2a, 5a and A suggests that if a site is predictively more accessible due to secondary structure, there is a less stringent requirement for siRNA duplex instability at the 5′ end of the antisense strand. In site 4a, a bad duplex was changed into a good duplex by changing a C to an U in the sense strand, in a position corresponding to the base pairing partner of the fourth nucleotide from the 5′ end of the antisense strand (for 2a and 5a, the altered nucleotide was at the third position from the 5′ end). This change resulted in a GU wobble base pair in the siRNA duplex between the sense and antisense strands. Contrary to our predictions, site 4a was still an ineffective site. Thus, our predictions were correct in four out of six experimental sites (one out of two for effective, and three out of four for ineffective), a result corresponding to the frequency with which we correctly predicted the published sites, further demonstrating that the predicted secondary structure can be used to help pre-evaluate sites for siRNA efficacy.
Figure 4.
For sites 2, 4 and 5, we altered the sense strand of the siRNA duplex to reverse the duplex-end stabilities. For example, sites 2 and 5 had an A substituted for a U in the third position of the sense strand. This created an A–A mismatch in the duplex, weakening the stability of the 5′ antisense end (see Figure 3). Our results show that the changes in duplex-end stability reduced the effectiveness of the siRNA duplexes. However, for site 4, the change from unfavorable duplex energies to favorable duplex energies did not result in an increase in the effectiveness of the siRNA.
DISCUSSION
The factors involved in RNAi-mediated sequence-specific mRNA degradation are not completely understood. Although there have been numerous discoveries, RNA binding by the Paz domain for example, the best supported determinant, outside of length and the presence of overhangs, of a good siRNA duplex is the difference in duplex-end thermodynamics. The data obtained in this study show that duplex-end thermodynamics can account for the activity of 60% of the sites tested. Furthermore, the data obtained in the present study show that siRNA efficacy predictions based upon a combination of the most probable mRNA secondary structure and siRNA duplex-end stabilities are useful in the selection of target sites, improving the number of correct predictions to 70%. Most importantly, using these predictions, it is possible to quickly predict 80% of the ineffective sites for any given siRNA/target combination, thereby focusing upon a smaller subset of sequences to identify more optimal siRNA/target sequences.
Many algorithms use GC-content in their assessment of potential site efficacy. Here, we show that there is a similarity in the results for GC-content and secondary structure predictions. This is due to a usually strong correlation between high GC-content and stable secondary structures. However, relying on GC-content alone is not as effective as the accessibility predictions, and the two methods have many discrepancies in their predictions. Notably, the GC-content algorithm results in more false negatives (fewer predicted good sites) than accessibility predictions, which are based on the secondary structure. To elaborate on this point, if we were to randomly predict that 60 of the published sites are ineffective, 27% of the time the random prediction would be better than the GC-content algorithm (based on a binomial distribution). For the algorithm using accessibility predictions, the random sampling would be better only 5% of the time. In addition, the random prediction would have fewer false negatives than the GC-content prediction 27% of the time. For accessibility predictions, the probability that a random prediction would have a better result is only 5% or 5-fold better than for GC-content. The probability that a random prediction of 22 effective sites would produce a more accurate result, then using the GC-content algorithm is 37%, much higher than for accessibility predictions (2%). Thus, GC-content is a more restrictive parameter than accessibility predictions.
Recent studies (10,13) investigated the in vitro kinetics of the RISC cleavage of mRNA targets. Both studies demonstrated an increase in the ability of RISC to cleave the target RNA upon ATP addition. Haley and Zamore (10) reported that mismatches between the antisense 5′ end and the target increased the rate of cleavage in the absence of ATP. Both studies concluded that the RISC activity is dependent upon product release. In searching for other factors to resolve false negatives, we did not see a simple correlation with the 5′ end energies and increased efficiency suggesting that in vivo product release is not rate limiting for RISC.
In the present study, we have shown that duplex-end energies and predicted accessibility can aid in the design of effective siRNA duplexes. This suggests that secondary structure can act as a barrier to RNAi, but how strong this effect is in the presence of other factors remains to be determined. It is already known that duplex-end energy determines which strand of an siRNA duplex will be preferentially loaded into the RISC, but it is not known if there is a quantitative correlation of a particular energy bias with the preference for strand loading. It is attractive to hypothesize that the more favorable the energy difference, the more RISC will be loaded with the antisense strand, such that there will be enough active enzyme to overcome other barriers to activity. For example, a site on Pten corresponding to position 1418 has a duplex-end energy difference of 6.4 kcal. Despite a low-predicted accessibility, Vickers et al. (33) reported that this site, 5′-GGGACGAACUGGUGUAAUG-3′, corresponded to a knockdown of >70%. However, it may be that our secondary structure predictions are in error for this site (due to imperfect nature of secondary structure predictions), or that other factors are contributing to the efficacy of the site. As an example, site A from our studies with EGFP, 5′-AAGGUGAACUUCAAGAUCCGC-3′, was able to knockdown EGFP by >90%, despite having unfavorable duplex-end energies. Its high-predicted accessibility leads us to hypothesize that high accessibility is related to a lowering of the activation energy needed to form a stable siRNA/target duplex and hence cleavage at this site. However, we cannot rule out other factors not taken into account in our computational approach.
One way to increase the efficacy of site prediction is to find other factors involved in RISC activity. One possible factor is the presence of an A or U at the 5′ end of the antisense molecule, as described in the work of Ui-Tei et al. (25). A more kinetic argument comes from the work of Haley and Zamore (10), which shows that the presence of an AU base pair at the 5′ end of the antisense/target RNA correlates with an increase in the efficiency of RISC turnover. Specifically, they showed that a mismatch in the 5′ end position of the antisense increased the cleavage efficiency. In the sites we examined, we observed a slight predominance of effective sites with A/Us in the first portion of the antisense. However, there are sites among the most effective that have G or C in this position. Notably, the two sites are mentioned above in Pten and EGFP sequences.
In comparison to popular motif-based algorithms, our combination of predicted secondary structure and duplex-end stabilities performs quite well. Specifically, the published algorithms of Amarzguioui and Prydz (26) and Dharmacon (22) were capable of predicting ∼70% of the sites from Vickers et al. (33) correctly (55 and 54 of the 76 sites from Vickers et al.) [the comparison was done using a cut-off of four for Amarzaguioui and Prydz (26) and six for Reynolds et al. (22), as suggested by the authors, with functionality defined as the ability to knockdown 55% or more]. This indicates that the secondary structure predictions are as important as the motifs used. Notably, the greatest mechanical similarity of the three methods is the use of duplex-end thermodynamics. Amarzguioui and Prydz (26) and Reynolds et al. (22) use a count of the A/U nucleotides in either end of the siRNA. This feature, which we have shown to account for the effectivity of 60% of the sites, probably accounts for the similarities in results.
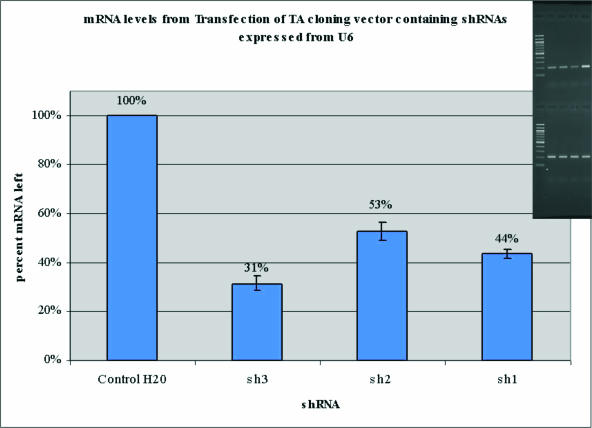
The small improvement gained by the three methods suggests that there are important factors that are yet to be discovered. A likely factor is the presence of proteins bound to the mRNA. For example, Figure 5 shows the knockdown of the three target sites by shRNA [transfections done as in Materials and Methods, but with a U6 shRNA expression vector (37) and limiting EGFP plasmid as transfection control]. Site 3 is the most effective, followed by 2 and 1. In cell extract accessibility assays, in which DNA oligo pairing to the native mRNA in cell lysates is measured by the extent of RNase H-mediated cleavage of the target (38), the trend was similar. An ODN complementary to site 3 directed the cleavage of almost all the target mRNA whereas site 2 ODN directed <80% cleavage. The influence of heterogeneous nuclear ribonucleoproteins binding along mRNAs on the RISC-mediated cleavage is not known. Studies with ribozyme/target interactions demonstrated that the hnRNPs are clearly important in both the association and dissociation steps (38–40). It is also possible, but less likely, that proteins bound to sites on an mRNA sterically inhibit RISC interactions within that region. Conversely, certain hRNPs may guide the RISC to target sites, as proposed for the human fragile X protein (41) or even aid in the destabilization of target mRNA secondary structures. Message marking by splicing/transport factors such as Y14 that binds to spliced RNAs several nucleotides upstream of 5′–3′ exon boundaries (42,43) could impede RISC binding. In this context, we examined the Pten mRNA for sites near exon–exon junctions and found that the distribution of effective and non-effective sites was similar to the mRNA as a whole. This suggests that if Y14 is bound to human mRNAs, it does not pose a serious hindrance for RISC-mediated cleavage. We are currently investigating further the potential role of RNA chaperones in RNAi site accessibility.
Figure 5.
Relative knockdown of target mRNA. GAPDH used to normalize amount of target. Percentage knockdown measured by semiquantitative RT–PCR, example of gel inset (lanes are 100 bp ladder, sh1, sh2, sh3 and control, respectively. Upper panel is for target mRNA and the lower panel is for GAPDH). RT showed no signal (data not shown).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Table 2.
Breakdown of predictions
| Methods | Number of published sites | Number of correctly predicted | Percentage of correctly predicted |
|---|---|---|---|
| GC content and duplex | |||
| Total sites | 80 | 49 | 61.25 |
| Effective sites | 25 | 8 | 32.00 |
| Ineffective sites | 55 | 41 | 74.55 |
| Accessibility + duplex | |||
| Total sites | 80 | 57 | 71.25 |
| Effective sites | 25 | 12 | 48.00 |
| Ineffective sites | 55 | 45 | 81.82 |
| Duplex | |||
| Total sites | 80 | 48 | 60.00 |
| Effective sites | 25 | 21 | 84.00 |
| Ineffective sites | 55 | 27 | 49.09 |
Secondary structure based accessibility predictions improved the number of correctly predicted sites, and the algorithm was able to screen out eighty percent of the ineffective sites.
Acknowledgments
We thank Steven L. Johnson, L. Scherer, A. Ehsani, D. Kim, M. Robbins, M. Weinberg, S. Gu, M. Lee, D. Castanotto and M. Amarzguioui for their kind suggestions and discussion. This work was supported by NIH grant HL074704. Funding to pay the Open Access publication charges for this article was provided by NIH grant HL074704.
REFERENCES
- 1.Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elbashir S.M., Harborth J., Weber K., Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 3.Lingel A., Simon B., Izaurralde E., Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nature Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 4.Song J.J., Liu J., Tolia N.H., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L. The crystal structure of the Argonaute 2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nature Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 5.Lingel A., Simon B., Izaurralde E., Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 6.Yan K.S., Yan S., Farooq A., Han A., Zeng L., Zhou M.M. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 7.Ma J.B., Ye K., Patel D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tahbaz N., Kolb F.A., Zhang H., Jaronczyk K., Filipowicz W., Hobman T.C. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–194. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harborth J., Elbashir S.M., Vandenburgh K., Manninga H., Scaringe S.A., Weber K., Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 10.Haley B., Zamore P.D. Kinetic analysis of the RNAi enzyme complex. Nature Struct. Mol. Biol. 2004;11:599–606. doi: 10.1038/nsmb780. [DOI] [PubMed] [Google Scholar]
- 11.Amarzguioui M., Holen T., Babaie E., Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu Y.L., Rana T.M. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez J., Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 15.Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 16.Vickers T.A., Wyatt J.R., Freier S.M. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 2000;28:1340–1347. doi: 10.1093/nar/28.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bohula E.A., Salisbury A.J., Sohail M., Playford M.P., Riedemann J., Southern E.M., Macaulay V.M. The efficacy of small interfering RNAs targeted to the type 1 insulin-like growth factor receptor (IGF1R) is influenced by secondary structure in the IGF1R transcript. J. Biol. Chem. 2003;278:15991–15997. doi: 10.1074/jbc.M300714200. [DOI] [PubMed] [Google Scholar]
- 18.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathews D.H., Sabina J., Zuker M., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 20.Ding Y., Chan C.Y., Lawrence C.E. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo K.Q., Chang D.C. The gene-silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochem. Biophys. Res. Commun. 2004;318:303–310. doi: 10.1016/j.bbrc.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 23.Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 24.Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 25.Ui-Tei K., Naito Y., Takahashi F., Haraguchi T., Ohki-Hamazaki H., Juni A., Ueda R., Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amarzguioui M., Prydz H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- 27.Kim D.H., Longo M., Han Y., Lundberg P., Cantin E., Rossi J.J. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 28.Kim D.H., Rossi J.J. Coupling of RNAi-mediated target downregulation with gene replacement. Antisense Nucleic Acid Drug Dev. 2003;13:151–155. doi: 10.1089/108729003768247619. [DOI] [PubMed] [Google Scholar]
- 29.Mironov A.A., Dyakonova L.P., Kister A.E. A kinetic approach to the prediction of RNA secondary structures. J. Biomol. Struct. Dyn. 1985;2:953–962. doi: 10.1080/07391102.1985.10507611. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz M., Steger G. Base-pair probability profiles of RNA secondary structures. Comput. Appl. Biosci. 1992;8:389–399. doi: 10.1093/bioinformatics/8.4.389. [DOI] [PubMed] [Google Scholar]
- 31.Wu J.C., Shapiro B.A. A Boltzmann filter improves the prediction of RNA folding pathways in a massively parallel genetic algorithm. J. Biomol. Struct. Dyn. 1999;17:581–595. doi: 10.1080/07391102.1999.10508388. [DOI] [PubMed] [Google Scholar]
- 32.McCaskill J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29((6–7)):1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- 33.Vickers T.A., Koo S., Bennett C.F., Crooke S.T., Dean N.M., Baker B.F. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- 34.Mathews D.H., Burkard M.E., Freier S.M., Wyatt J.R., Turner D.H. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muesing M.A., Smith D.H., Capon D.J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 37.Castanotto D., Rossi J.J. Construction and transfection of PCR products expressing siRNAs or shRNAs in mammalian cells. Methods Mol. Biol. 2004;252:509–514. doi: 10.1385/1-59259-746-7:509. [DOI] [PubMed] [Google Scholar]
- 38.Lee N.S., Bertrand E., Rossi J.J. Enhancement of ribozyme function by RNA binding proteins. Methods Mol. Biol. 1997;74:275–279. doi: 10.1385/0-89603-389-9:275. [DOI] [PubMed] [Google Scholar]
- 39.Herschlag D., Khosla M., Tsuchihashi Z., Karpel R.L. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertrand E.L., Rossi J.J. Facilitation of hammerhead ribozyme catalysis by the nucleocapsid protein of HIV-1 and the heterogeneous nuclear ribonucleoprotein A1. EMBO J. 1994;13:2904–2912. doi: 10.1002/j.1460-2075.1994.tb06585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carthew R.W. RNA interference: the fragile X syndrome connection. Curr. Biol. 2002;12:R852–R854. doi: 10.1016/s0960-9822(02)01352-0. [DOI] [PubMed] [Google Scholar]
- 42.Le Hir H., Izaurralde E., Maquat L.E., Moore M.J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Custodio N., Carvalho C., Condado I., Antoniou M., Blencowe B.J., Carmo-Fonseca M. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA. 2004;10:622–633. doi: 10.1261/rna.5258504. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.