Abstract
Analysing pooled DNA on microarrays is an efficient way to genotype hundreds of individuals for thousands of markers for genome-wide association. Although direct comparison of case and control fluorescence scores is possible, correction for differential hybridization of alleles is important, particularly for rare single nucleotide polymorphisms. Such correction relies on heterozygous fluorescence scores and requires the genotyping of hundreds of individuals to obtain sufficient estimates of the correction factor, completely negating any benefit gained by pooling samples. We explore the effect of differential hybridization on test statistics and provide a solution to this problem in the form of a central resource for the accumulation of heterozygous fluorescence scores, allowing accurate allele frequency estimation at no extra cost.
INTRODUCTION
DNA pooling is a well established method for reducing the cost and effort of large scale association studies (1–5). Samples from large numbers of individuals are pooled together before genotyping, thus reducing the workload from hundreds of samples per marker to a few per marker. Technologies also now exist to reduce the workload generated by large marker numbers to a single step (6–8). This means that a combination of the approaches could reduce thousands of genotypings of hundreds of individuals to a few tests. We have previously shown that one such method using Affymetrix GeneChips® to analyse DNA pools with a case-control design is feasible (6). This allows single nucleotide polymorphisms (SNPs) to be prioritized for individual genotyping by comparison of the fluorescence signal for an SNP from one pool with that from another.
A potential problem with this strategy is the differential hybridization of alleles, analogous to the differential amplification observed with more traditional methods of genotyping or sequencing (7). Equal allele doses do not correspond to equal fluorescence signals because of differences in chemistry and equipment response to fluorophores (2,9), so direct estimation of allele frequency is inaccurate. A solution is to use individuals heterozygous at an SNP to calibrate an observed fluorescence (2). This works because a heterozygote can be regarded as an exact 50:50 pool of each allele, allowing mathematical correction for differential hybridization and therefore accurate allele frequency estimation from pooled DNA (k-correction)(2,6). The only value required to enable allele frequency estimation from pools typed on DNA microarrays is the fluorescence score in a heterozygote. In order to reduce the standard error of the estimate of this calibration factor, data from several individuals heterozygous for a marker are needed (2). This is a significant problem for SNPs with low minor allele frequencies, as it means hundreds of individuals must be genotyped to be sure of having sufficient heterozygotes, negating the benefits of DNA pooling. We have examined the effect of differential hybridization on test statistics and report here a simple, free solution to this problem in the form of a website for accumulated heterozygote fluorescence scores.
MATERIALS AND METHODS
Statistical modelling
To investigate the need for a central resource of k-correction values, we modelled the effect of differential amplification or hybridization on a modified χ2-test statistic that takes into account the measurement error:
where is the estimated allele frequency of A, is the expected allele frequency of A, n is the number of individuals in the pool, V is the variance, ε is an error term and subscripts denote each pool. We estimated the error variance ε to be 0.0002 per pool (10).
Case and control pools were modelled as containing 225 individuals each. Allele frequency in case pools was varied with control allele frequencies set at 0.05 or 0.1 greater than case allele frequency. Levels of differential hybridization (Dh) were varied from 0.5 to 2 with . The test statistic was calculated in 0.05 increments.
The probability of finding at least x heterozygotes of frequency p in n samples was estimated using the tail probability of the binomial distribution, . The expected number of genotyped individuals needed to find a specified number of heterozygotes was calculated using the negative binomial distribution, .
Genotyping
Previous studies on other platforms (1,11–15) have suggested that several heterozygous values are needed for reliable allele frequency estimation from fluorescence scores. In order to explore this for the Affymetrix platform, we made three replicate pools using DNA from 100 individuals who had been previously genotyped individually for 104 SNPs. Each pool was genotyped in triplicate using Affymetrix 10K GeneChips® making a total of nine estimates for each SNP fluorescence score. Thirty-three other individuals were also genotyped using Affymetrix 10K GeneChips®. Data was obtained for 28 individuals genotyped by two other laboratories, six from one and twenty-two from the other, allowing comparison of correction using laboratory specific data, foreign data only, or all available data.
Website
A COM server constructed in Microsoft Visual Foxpro (VFP) was designed to manage and query a relational database containing data from DNA microarray analysis software. Visitors upload the output from chips used for individual genotyping, or download average heterozygote fluorescence scores as a tab-delimited text file, allowing the conversion of fluorescence scores from pooled DNA into allele frequency estimates.
Uploaded data undergoes a series of checks. A simple checksum is computed by summing the bytes or words of the data block ignoring overflow. This ensures that the file has not been uploaded before. Even minor modifications will change the checksum value, but if the data pass the first check it is imported into a temporary table and random records tested with their own individual checksums. This ensures that slightly modified files which have previously been uploaded are identified and discarded. If both checks are passed, the data are examined to determine whether it is output from the 10K or 100K chip so that it can be merged with the appropriate data set.
For each SNP, the heterozygote fluorescence score and variance is recalculated by a series of SQL queries using the new data, so that with each upload the estimate gradually converges to the ‘true’ population value.
RESULTS
Deriving allele frequency estimates from DNA microarrays
The original formula for k-correction was derived for correction of differential amplification using the ratio of independent readings for A and B alleles (2,6,11–14,16,17). In contrast, DNA microarray output is distorted by differential hybridization and for example, Affymetrix GeneChip® output data consists of two ‘relative allele scores’, RAS1 and RAS2, which are different measures of the same allele. Nevertheless, the k-correction formula still applies. The fluorescence score can be modelled as a value equal to the A allele frequency, scaled by some unknown factor x. For a heterozygous individual, where h is RASav in a heterozygote, the proportions of A allele (p) and B allele (q) are equal, so:
For pooled data, denoting the equivalent unknown allele frequencies as a (A allele) and b (B allele), and observed fluorescence score as f, the ratio of unknown proportions of pooled allele frequencies is:
Dividing the pooled ratio by the heterozygote ratio gives us:
Because a + b = 1 and a = bR, it follows that
and therefore that the corrected frequency of the B allele is
The only value required to enable data correction is therefore h, the fluorescence score in a heterozygote. The ratio R is related to k by k = Rh/(1 − h) and the use of either formula produces identical results.
Statistical modelling
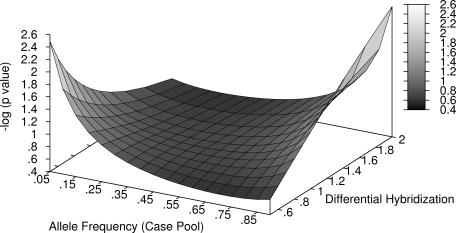
There was considerable distortion of the test statistic in our model (Figure 1). This was more marked the rarer the minor allele and the greater the degree of differential amplification or hybridization. Where there was no differential hybridization (i.e. differential hybridization of 1), the test statistic was higher when the difference between the allele frequencies of each pool was greater, as expected. However, when the minor allele was over hybridized, results were extremely liberal making a false positive call of association more likely. Conversely, when the major allele was over hybridized, the statistic was conservative. For example, with 225 individuals in each group, for an allele frequency of 0.1 in the cases and 0.05 in the controls, the P-value should be 0.048. The effect of differential hybridization in the range we studied produced p-values of between 0.228 and 0.003. With a 0.1 allele frequency difference between pools the effect was even more marked. For an allele frequency of 0.2 in cases and 0.1 in controls, the P-value should be 0.001. With differential hybridization the calculated P-value ranged between 0.024 and 10−5.
Figure 1.
The effect of differential hybridization and allele frequency on the test statistic. The control pool allele frequency was 0.05 greater than the case pool allele frequency shown on the x-axis.
Genotyping
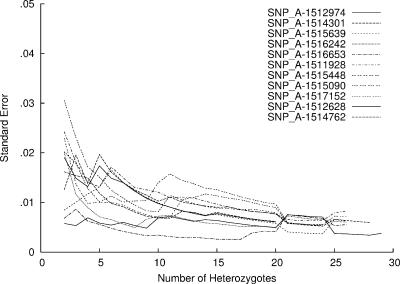
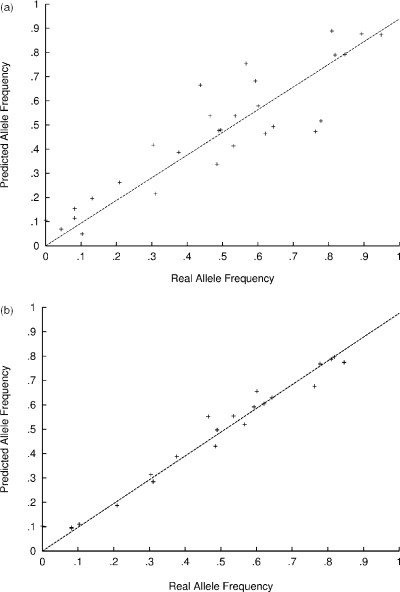
The 33 individuals genotyped to generate heterozygous RASav scores, were heterozygous for 9757 markers in at least 1 chip, 7787 in more than 6 chips and 4370 in more than 12 chips. The standard error of the heterozygote RASav was <0.02 averaged over 6 heterozygotes (Figure 2). With at least 20 heterozygotes, the standard error was <0.01. When the predicted allele frequency was compared with the observed, even a small number of heterozygotes contributing to k improved the allele frequency estimate of pooled samples (Figure 3). Without correction, the correlation coefficient r, between actual and predicted allele frequency was 0.892 but k-correction with data from 10 to 14 heterozygotes increased r to 0.987.
Figure 2.
Standard error of mean RASav for a random selection of SNPs showing that at least 20 estimates of RASav are needed for a standard error <0.01.
Figure 3.
Correlation between real and predicted allele counts using data from 100 individually genotyped samples compared with allele frequency estimates from pooled data. (a) No correction; (b) corrected with RASav data from 10 to 14 heterozygotes.
An assumption of this central resource is that accumulated heterozygote RASav scores can correct pool data regardless of the laboratories from which the correction values were obtained. The worst case scenario is that the only heterozygote RASav scores available are from foreign laboratories. Correcting our data using available heterozygote RASav scores from our own laboratory only, the correlation coefficient r was 0.984. Using foreign heterozygote RASav scores it was still excellent but a little lower at 0.972. Using all available heterozygote data from any laboratory produced marginally the best outcome, with r = 0.985.
Website
The website for collecting and distributing accumulated heterozygote fluorescence scores is at http://cogent.iop.kcl.ac.uk/rcorrection.cogx. There is currently data for 61 individuals for the Affymetrix 10K mapping chip v1.0. Data for the 100K GeneChip® will be available soon and we welcome data from other platforms.
DISCUSSION
We have shown that distortion of test statistics by differential hybridization is an important factor for those performing association studies by DNA pooling using DNA microarrays. This is particularly true for rare SNPs, and as real differences between pools increase. Correction of RASav scores from the Affymetrix GeneChip® using heterozygous individuals generates an accurate estimate of allele frequencies and we can expect this to apply to other platforms on which DNA pooling is possible. Such correction is therefore desirable but two things confound the investigator trying to use this method. First, rarer SNPs for which k-correction is more important are also those least likely to be heterozygous. Second, multiple measures of the heterozygous fluorescence score are required for accurate correction. The Affymetrix 100K Mapping Set contains 35 312 SNPs with a minor allele frequency of ≤0.1, which is about 30% of the entire chip, and it is likely that chips from other manufacturers will be similar. One can expect to type 10 individuals to find just one heterozygous value for an SNP with a minor allele frequency of 0.1, but to be 99% sure requires the genotyping of 24 individuals. At least six heterozygotes are needed to reduce the standard error to <0.02, but even this requires 69 individual genotypes. For a more stringent standard error of 0.01, sampling at least 20 heterozygotes would be required, which would need 171 extra chip genotypings, and for rarer SNPs the numbers rise rapidly. Fortunately, the allele frequency estimate is quite robust to the estimates of the correction factor (data not shown), and even one heterozygote score is better than none. Nevertheless, this means that any investigator attempting to use DNA microarrays for DNA pooling faces the dilemma of ignoring the effect of differential hybridization on the test statistic or adding several hundreds of thousands of dollars to the project.
A solution to this problem is a central collection of individual heterozygous genotype results. This is possible because the markers used are a fixed set in a standardized system with replicable results (6), and any investigator using DNA microarrays will therefore generate useful k-correction data as a by-product. For example, the output from microarrays used for linkage or loss of heterozygosity studies in which the genotype call data is collected, but for which the fluorescence data is discarded by the investigator, falls into this category. Data from different laboratories can be merged and used to correct allele frequency estimates. We have therefore designed a central resource at http://cogent.iop.kcl.ac.uk/rcorrection.cogx for the accumulation of heterozygous fluorescence scores from such experiments.
Data available for download includes SNP identity, map position, current estimate of the calibration factor, variance of the calibration factor and the number of heterozygous individuals contributing to the factor, for each marker. The website can currently handle data for the Affymetrix 10K Mapping GeneChip® Array versions 1.0 and 2.0 and the new 100K Mapping Set, but in principle this resource could be used for any standard chipset for which DNA pooling is possible. This means that an excellent estimate of the heterozygous fluorescence score and therefore calibration factor for k-correction can be obtained even for rare SNPs, and this will steadily improve with time. Such a resource could not easily exist before the advent of DNA microarray technology because the marker sets used by investigators, and the platforms used for genotyping, were all variable.
This resource will significantly assist those planning association studies by DNA pooling, and will also allow the accurate estimation of population allele frequencies, quickly, easily and cheaply.
Acknowledgments
We are grateful to Geoff Scopes and Affymetrix UK for advice, Professor Robert H. Brown Jr, Dr Jing Xi and Dr Betsy Hosler of the Day Neuromuscular Laboratory, Massachusetts General Hospital for advice and sample data, and Nick Leaves and the MRC Gene Service for help. At the time of this work, A.A.-C. was a UK Medical Research Council (MRC) Clinician Scientist Fellow. C.L.S. is funded by a GKT Medical School Prize PhD Studentship. P.C.S. is funded by NEI grant EY-12662 and MRC grant G9700821 and a Distinguished Visiting Professorship from the University of Hong Kong. This work was funded in part by UK MRC grant G9424799. Funding to pay the Open Access publication charges for this article was provided by the Department of Neurology, Institute of Psychiatry.
REFERENCES
- 1.Breen G., Harold D., Ralston S., Shaw D., St Clair D. Determining SNP allele frequencies in DNA pools. Biotechniques. 2000;28:464–466. doi: 10.2144/00283st03. 468, 470. [DOI] [PubMed] [Google Scholar]
- 2.Le Hellard S., Ballereau S.J., Visscher P.M., Torrance H.S., Pinson J., Morris S.W., Thomson M.L., Semple C.A., Muir W.J., Blackwood D.H., Porteous D.J., Evans K.L. SNP genotyping on pooled DNAs: comparison of genotyping technologies and a semi automated method for data storage and analysis. Nucleic Acids Res. 2002;30:e74. doi: 10.1093/nar/gnf070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jawaid A., Bader J.S., Purcell S., Cherry S.S., Sham P.C. Optimal selection strategies for QTL mapping using pooled DNA samples. Eur. J. Hum. Genet. 2002;10:125–132. doi: 10.1038/sj.ejhg.5200771. [DOI] [PubMed] [Google Scholar]
- 4.Norton N., Williams N.M., Williams H.J., Spurlock G., Kirov G., Morris D.W., Hoogendoorn B., Owen M.J., O'Donovan M.C. Universal, robust, highly quantitative SNP allele frequency measurement in DNA pools. Hum. Genet. 2002;110:471–478. doi: 10.1007/s00439-002-0706-6. [DOI] [PubMed] [Google Scholar]
- 5.Sham P.C., Bader J.S., Craig I., O'Donovan M., Owen M. DNA pooling: a tool for large-scale association studies. Nature Rev. Genet. 2002;3:862–871. doi: 10.1038/nrg930. [DOI] [PubMed] [Google Scholar]
- 6.Butcher L.M., Meaburn E., Liu L., Hill L., Al-Chalabi A., Plomin R., Schalkwyk L., Craig I. Genotyping pooled DNA on microarrays: a systematic genome screen of thousands of SNPs in large samples to detect QTLs for complex traits. Behav. Genet. 2004;34:549–555. doi: 10.1023/B:BEGE.0000038493.26202.d3. [DOI] [PubMed] [Google Scholar]
- 7.Butcher L.M., Meaburn E., Dale P.S., Sham P., Schalkwyk L.C., Craig I.W., Plomin R. Association analysis of mild mental impairment using DNA pooling to screen 432 brain-expressed single-nucleotide polymorphisms. Mol. Psychiatry. 2004 doi: 10.1038/sj.mp.4001589. doi:10.1038/sj.mp.4001589. [DOI] [PubMed] [Google Scholar]
- 8.Uhl G.R., Lin Q.R., Walther D., Hess J., Naiman D. Polysubstance abuse-vulnerability genes: genome scans for association, using 1,004 subjects and 1,494 single nucleotide polymorphisms. Am. J. Hum. Genet. 2001;69:1290–1300. doi: 10.1086/324467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohlke K.L., Erdos M.R., Scott L.J., Fingerlin T.E., Jackson A.U., Silander K., Hollstein P., Boehnke M., Collins F.S. High-throughput screening for evidence of association by using mass spectrometry genotyping on DNA pools. Proc. Natl Acad. Sci. USA. 2002;99:16928–16933. doi: 10.1073/pnas.262661399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bader J.S., Bansal A., Sham P.C. Efficient SNP-based tests of association for quantitative phenotypes using pooled DNA. GeneScreen. 2001;1:143–150. [Google Scholar]
- 11.Germer S., Holland M.J., Higuchi R. High-throughput SNP allele-frequency determination in pooled DNA samples by kinetic PCR. Genome Res. 2000;10:258–266. doi: 10.1101/gr.10.2.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou G., Kamahori M., Okano K., Chuan G., Harada K., Kambara H. Quantitative detection of single nucleotide polymorphisms for a pooled sample by a bioluminometric assay coupled with modified primer extension reactions (BAMPER) Nucleic Acids Res. 2001;29:e93. doi: 10.1093/nar/29.19.e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki T., Tahira T., Suzuki A., Higasa K., Kukita Y., Baba S., Hayashi K. Precise estimation of allele frequencies of single-nucleotide polymorphisms by a quantitative SSCP analysis of pooled DNA. Am. J. Hum. Genet. 2001;68:214–218. doi: 10.1086/316928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoogendoorn B., Norton N., Kirov G., Williams N., Hamshere M.L., Spurlock G., Austin J., Stephens M.K., Buckland P.R., Owen M.J., et al. Cheap, accurate and rapid allele frequency estimation of single nucleotide polymorphisms by primer extension and DHPLC in DNA pools. Hum. Genet. 2000;107:488–493. doi: 10.1007/s004390000397. [DOI] [PubMed] [Google Scholar]
- 15.Buetow K.H., Edmonson M., MacDonald R., Clifford R., Yip P., Kelley J., Little D.P., Strausberg R., Koester H., Cantor C.R., Braun A. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc. Natl. Acad. Sci. USA. 2001;98:581–584. doi: 10.1073/pnas.021506298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsson C., Liljedahl U., Syvanen A.C. Quantitative analysis of SNPs in pooled DNA samples by solid-phase minisequencing. Meth. Mol. Biol. 2003;2:313–317. doi: 10.1385/1-59259-327-5:167. [DOI] [PubMed] [Google Scholar]
- 17.Ross P., Hall L., Haff L.A. Quantitative approach to single-nucleotide polymorphism analysis using MALDITOF mass spectrometry. Biotechniques. 2000;29:620–626. doi: 10.2144/00293rr05. [DOI] [PubMed] [Google Scholar]