Abstract
ApcMin mice have provided an example of a locus (Modifier of Min1; Mom1) modifying adenoma numbers in the intestines of inbred strains. Linkage analysis located Mom1 on chromosome 4, and further investigation identified secretory phospholipase A2 (Pla2g2a) as a candidate gene. Because of unknown variation introduced by a single founding male mouse, our Min stock, although Pla2g2aMom1-s, was not on a pure C57BL/6J background and exhibited several polymorphic loci, including a region on chromosome 18 distal to Apc. Through selective breeding for homozygosity for distal chromosome 18 markers, six recombinant lines that presented with limited intraline variation in adenoma numbers were established. One line (V) showed a particularly severe phenotype (mean adenoma number ± SEM, 370 ± 21) compared with the other lines that recorded significantly lower means (3- to 5-fold; P < 10–3, t test). Intercrosses between lines I and V showed suppression of the severe phenotype in the N1 generation. In N2 (and subsequent) backcrosses, tumor multiplicity depended on the origins of the WT and Min Apc alleles. Mice carrying both alleles from line V had a severe phenotype; others had mild disease very similar to line I (likelihood ratio statistic > 49.0; likelihood of odds > 10; P < 10–5). Frequency of allele loss at Apc was increased significantly in adenomas of mice with more severe disease. We propose that a modifier gene close to Apc or structural variation on chromosome 18 modifies polyp numbers in our mice, possibly by altering the frequency of WT Apc allele loss.
Keywords: MIN mouse, modifiers of MIN, tumor multiplicity
The two most common, well characterized, rare Mendelian predispositions to colorectal cancer (CRC) are hereditary nonpolyposis CRC [Online Mendelian Inheritance in Man database accession no. 114500] and familial adenomatous polyposis (FAP; Online Mendelian Inheritance in Man accession no. 175100). FAP patients inherit a mutated copy of the adenomatous polyposis coli (APC) gene (1–3), whereas hereditary nonpolyposis CRC is caused by inheritance of defective DNA mismatch repair genes such as MSH2 or MLH1 (4, 5). However, hereditary nonpolyposis CRC and FAP account for only a small fraction of the colorectal tumors presenting in the human population. The vast majority of CRCs (≈80%) do not result from a known inherited factor, are considered sporadic in origin, and demonstrate somatic mutation of the APC gene (6–10).
Many studies have suggested a role for uncharacterized genetic factors in predisposition to the common forms of colorectal tumors. Thus, relatives of CRC patients are at an increased risk of the disease, and segregation analysis has suggested dominant inheritance of CRC susceptibility (11, 12). In addition, an extensive analysis of twins showed that up to one-third of CRCs may have some inherited basis (13). The remaining, uncharacterized predisposition to CRC in humans is more likely to be the result of several genes of low-penetrance rather than high-penetrance mutations at single loci with large effects on risk (14). Much of the risk of CRC may result from a primary predisposition to colorectal adenomas. The number of colorectal adenomas presenting within FAP families or individuals with identical germ-line APC mutations has been shown to vary, suggesting that hereditary factors also may influence disease severity (15). The same genes that modify the phenotype of individuals with FAP also may influence the risk of CRC in the general population. A search for FAP modifier genes, either directly or through rodent models, therefore may lead to the identification of important susceptibility genes for human CRC.
The first mouse model used to study the involvement of the Apc gene in CRC is referred to as Min (Multiple intestinal neoplasia). ApcMin/+ (Min) mice are heterozygous for a truncating Apc mutation and develop numerous intestinal adenomas, thereby providing a good model of human FAP. The Min model has been used to provide an unambiguous example of a modifying locus in mice. A single locus was identified as a consequence of significant variation in the polyp number, depending on the inbred mouse strain harboring the Apc mutation. Linkage analysis located Mom1 on mouse chromosome 4, and further analysis identified the secretory phospholipase A2 (Pla2g2a) as a candidate gene (16–18). Unfortunately, studies in humans did not confirm PLA2G2A as a major modifier of colorectal cancer risk in humans because functional polymorphic variation did not exist (19, 20). A second locus, Mom2, on distal chromosome 18 was identified by Silverman et al. (21), although the underlying genetic defect has yet to be established. We now have direct evidence of a further Mom locus controlling susceptibility to a particularly severe form of intestinal disease in Min mice.
Materials and Methods
Husbandry. Mice were housed in conventional cages, and a standard maintenance diet was provided ad libitum. All procedures involving animals were carried out in accordance with the Animals (Scientific Procedures) Act 1986 and with guidance from the local Ethical Review Committee on animal experiments.
Preparation of the Intestinal Tracts of ApcMin/+ Min Mice for Tumor Counting. Mice were killed by CO2 asphyxiation when their quality of life was compromised, and the whole gastrointestinal tract was removed. The tract was separated into small intestine, cecum, and colon, with the small intestine divided further into four equal segments. The gut segments were cut along the line of the mesenteric attachment, flushed with 1× PBS, spread onto paper strips with villi uppermost, and then fixed for 24 h in phosphate-buffered formol saline before transfer to 70% ethanol for storage. Tumors were dissected from intestinal tracts as described in Sieber et al. (22).
Genotyping and Loss of Heterozygosity Analysis at Apc. Genotyping of ApcMin/+ was conducted according to the procedure of Luongo et al. (23), and Pla2g2a was typed as described in Santos et al. (24). Microsatellite PCR was performed by using standard laboratory procedures. Loss of heterozygosity analysis of Apc alleles was conducted as described by Sieber et al. (22).
Statistical Analysis. The mapmanager qtx program (25) was used to perform interval mapping and permutation testing. stata statistical software (Version 8.0, StataCorp, College Station, TX) was used for other statistical analyses.
Results
Polyp Multiplicity in Our Colony of C57BL/6 ApcMin/+ Mice Indicates Presence of High- and Low-Scoring Subpopulations. An ApcMin/+ colony was established by crossing a single male ApcMin/+ mouse (Imperial Cancer Research Fund, Clare Hall, South Mimms, U.K.), reportedly on a C57BL/6J background, with two female C57BL/6J mice (Radiation and Genome Stability Unit, Medical Research Council, Harwell, U.K.). After 10 generations of inbreeding, mostly through sibling mating, the entire intestinal tracts of ApcMin/+ mice (n = 99) were scored for polyps. Although significant variation in multiplicity within the colony (median, 107; range, 32–494) was identified, the majority of mice (73%) with a lifespan of <125 days showed polyp numbers between 32 and 163. However, a subset of individuals (10%) showed notably higher polyp numbers (range 289–494) and shorter life spans (range 74–89 days). There was no evidence of a link between the sex of an individual and polyp number. To investigate the possibility of strain contamination, mice were tested for Mom1 by genotyping Pla2g2a and screened for chromosome 18 polymorphisms by using a panel of microsatellites spaced along the chromosome. As expected for C57BL/6, all animals were Pla2g2a–/–. However, seven microsatellites (D18Mit81, D18Mit184, D18Mit50, D18Mit9, D18Mit207, D18Mit186, and D18Mit188) were identified as polymorphic, all located to an interval on distal chromosome 18 between 26 and 32 centimorgans from the centromere; genetic maps place the Apc gene ≈15 centimorgans distal to the centromere on chromosome 18 and ≈11–17 centimorgans proximal to the polymorphic region (www.broad.mit.edu/cgi-bin/mouse/index). Each of the seven polymorphic markers gave two distinct allele sizes, one equivalent to those previously assigned to C57BL/6J and the other novel. The latter did not correspond to any known inbred strain of mouse, and genotyping of the original founding male ApcMin/+ mouse confirmed that it was homozygous for the novel alleles. We suspect that the founding male was derived from a stock of C57BL/6 animals held at the Imperial Cancer Research Fund (now Cancer Research UK) since the 1930s and originally derived from mice from The Jackson Laboratory.
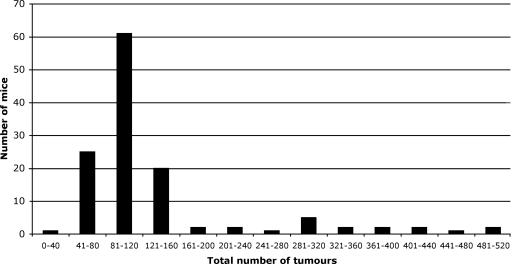
Establishment of Recombinant Lines Showing Interline Variation in Polyp Numbers. We next considered whether a modifier(s) of tumor multiplicity in ApcMin/+ mice mapped to the distal chromosome 18 segment encompassing the polymorphic microsatellite markers. The identified region is syntenic to human 18q and known to be involved in human CRC (26, 27). Accordingly, ApcMin/+ and Apc+/+ mice from the breeding colony were genotyped by using the seven microsatellites given above, and crosses were set up between individuals to produce progeny homozygous for each allele. Six lines then were maintained mostly by sibling mating within lines after genotyping of breeding pairs. After three subsequent generations, intestinal tracts of ApcMin/+ mice were assessed for polyp numbers. Line V showed a severe phenotype compared with the other lines, all of which recorded significantly lower means (P < 0.001, t tests with Bonferroni adjustments for multiple tests; see Table 1). There was no statistical difference in polyp multiplicity between males and females within each line, and the proportions of each gender were the same in all lines. Cluster analysis based on the number of adenomas presenting in the inbred lines suggested that severity of disease was a binary trait, with mild (<200 adenomas) and severe (>200 adenomas) forms (Table 1 and Fig. 1). Importantly, adenoma numbers within each line were stable through further generations (Table 2), demonstrating a genetic basis. Representative mice from each line were genotyped for Pla2g2a, and all were found to be Pla2g2aMom1-s, providing direct evidence that the effect was not due to Mom1 and indicating the presence of one or more additional modifiers. Any obvious mutation in the Apc gene in either line, in addition to the Min mutation, was ruled out after sequencing of the entire coding region, including splice sites and the 3′ and 5′ UTRs, of both WT and Min mice from lines I and V. However, this finding did not rule out the possibility of variants in the Apc introns and or in sequences flanking the gene.
Table 1. Tumor multiplicity in genetically segregating lines of ApcMin/+ mice.
| Location | Line I | Line II | Line III | Line IV | Line V | Line VI |
|---|---|---|---|---|---|---|
| No. of mice | 24 | 21 | 24 | 17 | 17 | 23 |
| Sm.ln. 1 | 10.6 ± 0.9 | 10.4 ± 0.8 | 14.8 ± 1.2 | 10.2 ± 0.5 | 25.8 ± 2.4 | 11.7 ± 0.7 |
| Sm.ln. 2 | 16.5 ± 1.5 | 10.5 ± 1.0 | 14.8 ± 1.0 | 10.8 ± 1.5 | 41.1 ± 2.8 | 11.6 ± 1.1 |
| Sm.ln. 3 | 47.5 ± 2.8 | 40.3 ± 1.9 | 50.6 ± 2.0 | 31.8 ± 3.1 | 172.5 ± 10.1 | 33.3 ± 1.7 |
| Sm.ln. 4 | 29.3 ± 1.4 | 32.0 ± 2.4 | 33.8 ± 1.7 | 21.9 ± 3.5 | 117.4 ± 9.3 | 24.5 ± 1.6 |
| Cecum | 1.6 ± 0.2 | 1.1 ± 0.2 | 1.3 ± 0.2 | 1.1 ± 0.4 | 1.0 ± 0.2 | 1.1 ± 0.2 |
| Colon | 6.4 ± 0.7 | 3.3 ± 0.4 | 4.8 ± 0.6 | 3.1 ± 0.4 | 12.1 ± 1.9 | 3.4 ± 0.5 |
| Total | 112.0 ± 4.9 | 97.7 ± 4.5 | 120.1 ± 4.4 | 78.8 ± 7.6 | 369.9 ± 20.5 | 85.6 ± 3.8 |
The table shows total number of adenomas in four segments of the small intestine (Sm.ln.), cecum, and colon. Small intestine segments 1 and 2 comprise all of the duodenum and jejeunum, and segments 3 and 4 comprise the proximal and distal ileum. Values are given as mean ± SEM.
Fig. 1.
Total numbers of adenomas in genetically segregating lines of C57BL/J mice.
Table 2. Total tumors presenting in the intestinal tracts of lines I and V Min over 13 generations.
| Generation* | Line I, total tumors (n) | Line V, total tumors (n) |
|---|---|---|
| N4 | 111.9 ± 8.2 (8) | 384.0 ± 53.2 (3) |
| N5 | 98.7 ± 3.8 (3) | 424.7 ± 24.4 (6) |
| N6 | 85.0 ± 3.2 (10) | 302.0 ± 28.4 (5) |
| N7 | 91.7 ± 10.1 (9) | 355.2 ± 28.3 (12) |
| N8 | 90.7 ± 7.6 (11) | 321.0 ± 22.0 (7) |
| N9 | 87.5 ± 8.7 (6) | 327.5 ± 29.9 (8) |
| N10 | 105.8 ± 11.5 (8) | 370.2 ± 26.3 (6) |
| N11 | 109.7 ± 21.8 (3) | 284.8 ± 22.1 (4) |
| N12 | 128.7 ± 6.1 (7) | 305.8 ± 21.1 (6) |
| N13 | 106.7 ± 7.2 (9) | 288.2 ± 19.9 (6) |
| N14 | 115.6 ± 7.4 (12) | 364.1 ± 16.5 (9) |
| N15 | 110.6 ± 8.5 (5) | 372 ± 13.3 (17) |
| N16 | 92.5 ± 6.9 (4) | 376.7 ± 15.4 (17) |
| Overall | 102 ± 25.5 (95) | 351.6 ± 72.1 (106) |
Values are given as means ± SEM, and the number of mice counted (n) is shown in parentheses. Lines were maintained by using standard protocols for inbred lines.
From original cross.
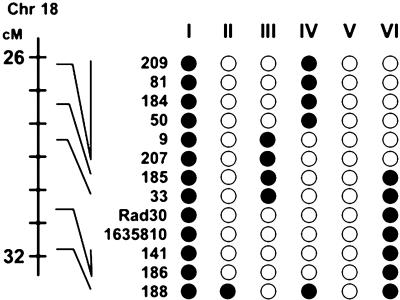
Do Potential Modifier(s) Map to Distal Chromosome 18? In addition to the original seven polymorphic markers used for genotyping the line, a further six informative markers, four microsatellites (D18Mit209, D18Mit185, D18Mit33 and D18Mit141), one polymorphic sequence-tagged site (1635810), and one SNP for Rad30, were identified and positioned within an ≈6-centimorgan region on distal chromosome 18 (Fig. 2). Again, two allele sizes were identified, one consistent with established data for C57BL6/J and the other introduced by the founding ApcMin/+ mouse. No other polymorphic markers were found from a screen by using an additional 60 markers evenly spaced along the length of chromosome 18, and no polymorphic markers were found in the Mom2 chromosomal region that lies distal to D18Mit188 (13).
Fig. 2.
Genotype and tumor multiplicity in genetically segregating lines of C57BL/6 Min mice. Distance from centromere of chromosome 18 is shown in centimorgans (cM), and the positions of D18Mit markers, one sequence-tagged site (1635810), and the SNP for Rad30 are shown to the right of the scale bar (18). The number identifying each of the recombinant lines is denoted by a Roman numeral. The open circles represent C57BL/6J alleles, and filled circles represent non-C57BL/6J alleles. None of the markers identified in the lines is consistent with linkage to a modifier that is distinct from APC.
The pattern of C57BL6/J and non-C57BL6/J alleles for the high-scoring (V) and the lower-scoring lines was inconsistent with a single modifier within the polymorphic region (Fig. 2). For example, if it were assumed that a modifier was closely linked to marker D18Mit188, then the absence of novel alleles for this marker for low-scoring line III would challenge this explanation (Fig. 2). It was not possible, however, to exclude the action of more than one modifier, and a number of potential modifiers map to the region, including Madh4 and Tcf4. Sequencing of the coding regions of these genes in the inbred lines did not, however, identify any variants predicted to have functional consequences. Before undertaking a more extensive analysis of this region for coding sequence variants, we attempted to verify the possible presence of any modifiers in the distal region of chromosome 18 by using backcrossing strategies.
Evidence for a Modifier Linked to ApcMin/+. By crossing low-scoring line I with the high-scoring line V, N1 hybrids were generated so that the Min was introduced from either line to give genotypes MinI/+V or MinV/+I at Apc [where I or V denotes the source of the mutated Apc (Min) or the WT (+/–) allele]. Overall, intestinal tracts from all hybrids (20 MinI/+V and 23 MinV/+I) showed polyp counts (mean ± SEM, 126.4 ± 6.3) entirely consistent with numbers expected for the lower-scoring line (112 ± 4.6; P = 0.15, t test). There was no apparent difference between males or females, and total polyp numbers were not dependent on whether the Min mutation was introduced from line I or V (MinI/+V, 123.3 ± 9.04, and MinV/+I, 135.4 ± 7.7, respectively; P = 0.28, t test). These data suggested that line I carried a dominant modifier (or possibly more than one) able to suppress tumor multiplicity in N1 mice.
Next, we backcrossed N1 animals (either MinI/+V or MinV/+I) with WT line V mice (+V/+V) and counted the polyps in N2 ApcMin/+ progeny (n = 76). A clear difference in phenotypic severity between crosses was observed (Table 3). With Min derived originally from line V, the majority of N2 mice (19 of 23, 83%) showed total tumor numbers of >200 (mean ± SEM, 408 ± 25; range, 264–637), and only 17% (4 of 23) had total numbers of <200 (139 ± 9, 124–163; Table 3). In contrast, when Min was donated from line I, most N2 mice (46 of 53, 87%) presented with <200 intestinal tumors (122 ± 4; 71–191) and a minority (7 of 53, 13%) had high tumor numbers (>200; 297 ± 31; 209–433; Table 3). Further backcrossing of +I/+V to MinI mice (MinI/+I or MinI/+V) over two more generations produced only two individuals (1%, 2 of 203; Table 3) with tumor numbers of >200. Furthermore, mice with the genotype MinI/+V showed predominately low tumor numbers (<200; 90%, 81 of 90; Table 3) in the two to five generations of a MinI/+V to +V/+V backcross. In contrast, N2–5 generation mice with a MinV/+V genotype showed predominantly high tumor numbers (>200; 46 of 55, 84%). There was no evidence to suggest that tumor multiplicity depended on imprinting or mitochondrial inheritance.
Table 3. Severity of tumor multiplicity in backcrossed ApcMin/+ mice.
| Backcross, N1 genotype × parental line genotype (N2 genotype)
|
||||||
|---|---|---|---|---|---|---|
|
MinI/+V × +V/+V (MinI/+V)
|
MinV/+I × +V/+V (MinV/+V)
|
+I/+V × MinI/+I (MinI/+I or MinI/+V)
|
||||
| <200 tumors, %, | >200 tumors, %, | <200 tumors, %, | >200 tumors, %, | <200 tumors, %, | >200 tumors, %, | |
| Generation | (n), mean ± SEM | (n), mean ± SEM | (n), mean ± SEM | (n), mean ± SEM | (n), mean ± SEM | (n), mean ± SEM |
| N2 | 87 (46) 122 ± 4 | 13 (7) 297 ± 31 | 17 (4) 139 ± 9 | 83 (19) 408 ± 25 | 100 (32) 117 ± 4 | 0 (0) |
| N3 | 97 (22) 99 ± 4 | 3 (1) 447 | 16 (3) 138 ± 17 | 84 (16) 365 ± 23 | 96 (51) 122 ± 3 | 4 (2) 227 ± 10 |
| N4 | 93 (13) 92 ± 7 | 7 (1) 515 | 15 (2) 144 ± 49 | 85 (11) 339 ± 27 | 100 (63) 122 ± 3 | 0 (0) |
| N5 | — | — | — | — | 100 (55) 109 ± 3 | 0 (0) |
| Overall | 90 (81) 111 ± 3 | 10 (9) 338 ± 36 | 16 (9) 139 ± 10 | 84 (46) 377 ± 15 | 99 (201) 117 ± 2 | 1 (2) 227 ± 10 |
The percentage and number (n) of mice with greater or fewer than 200 tumors along with the mean ± SEM within each group is shown.
N2 mice (MinI/+V or MinV/+V) were genotyped for D18Mit184, D18Mit33, and D18Mit188, and the source of the donor Apc mutation was identified from breeding records (that is, in the MinI/+V ×+V/+V and +I/MinV ×+V/+V crosses in Table 3). Other markers found to be polymorphic between lines V and I on chromosomes 3 (D3Mit40), 4 (D4Mit144), 8 (D8Mit200), and 10 (D10Mit101) also were used to genotype the N2 mice. The mapmanager qtx program then was used to perform interval mapping and permutation testing. The following traits were considered: total tumor number, total small intestinal tumors, and intestine length. There was no evidence for any of these traits for a QTL linked to the chromosome 18 microsatellites D18Mit184, D18Mit33, or D18Mit188 (Table 4) and no evidence of linkage to other sites that were polymorphic between the lines (for D3Mit40, D4Mit144, D8Mit200, and D10Mit101; likelihood ratio statistic, 0.1; P > 0.5 in all cases). In contrast, with the exception of intestinal length, the values for all other traits were associated highly significantly (likelihood ratio statistic > 49.0, logarithm of odds > 10, P < 0.00001; Table 4) with the origin of the Min allele from line I or V, the latter tending to be present in animals with more severe disease. Given that the N2 animals used for linkage all carried the +V allele and that N1 animals (MinI/+V and MinV/+I) all had mild disease, we reasoned that the linkage data at Apc probably reflected the origins of both the Min and WT alleles from line V: only when both Apc alleles were of line V origin did mice develop severe polyposis. In support of this contention, we performed an additional cross of N1 WT animals (genotype +I/+V) to Min mice from line I (genotype MinI/+I). In this case, all of the N2 offspring (genotypes MinI/+I or MinI/+V) had mild disease (Table 3). Furthermore, in subsequent generations (N3–N5) in which these N2 mice were backcrossed to line I animals, polyp counts were very similar to those in N2 (Table 3).
Table 4. Likelihood ratio statistic (LRS) from interval mapping in N2 Min mice.
| QTL* | LRS (>13.9 highly significant) | P |
|---|---|---|
| Total tumors | ||
| Apc | 52.4 | >0.00001 |
| D18Mit184 | 1.6 | 0.20 |
| D18Mit33 | 0.3 | 0.57 |
| D18Mit188 | 0.1 | 0.72 |
| Total small intestine | ||
| Apc | 49.0 | >0.00001 |
| D18Mit184 | 1.1 | 0.28 |
| D18Mit33 | 0.1 | 0.71 |
| D18Mit188 | 0.0 | 0.93 |
| Age of death | ||
| Apc | 20.6 | 0.00001 |
| D18Mit184 | 0.1 | 0.1 |
| D18Mit33 | 0.0 | 0.0 |
| D18Mit188 | 0.1 | 0.1 |
| Int. length | ||
| Apc | 0.1 | 0.7 |
| D18Mit184 | 2.5 | 0.1 |
| D18Mit33 | 2.9 | 0.9 |
| D18Mit188 | 1.0 | 0.3 |
QTL, quantitative trait loci. Linkage with Apc was established by means of the Min lines I and V, and Apc genotype was defined as MinV/+V or MinI/+V using the breeding records for N2 mice.
Our data showed that although a modifier locus was linked to Apc, its position could not be refined further by using the available markers. We therefore evaluated a number of genes within 15 centimorgans of Apc that might act as phenotypic modifiers. Sequencing of the coding regions of Map3K8, Catna1, Egr1, Nrg2, Bin1, Ercc3, Dp1, and Cdc25c, however, failed to identify any variants between lines I and V.
Tumors from ApcMin/+ Line V Mice Show a Higher Frequency of Allele Loss of WT Apc than Line I Mice. Tumor formation in ApcMin/+ C57BL/6J mice generally occurs by somatic recombination and consequent allele loss of the WT Apc copy (28, 29). It was possible that the increased severity in intestinal disease in line V Min mice was the result of a higher frequency of allele loss in these mice compared with line I. Consequently, we tested adenomas from the parental lines, N1s and N2s, for allele loss. The results (Table 5) indicated that the frequency of allele loss was significantly higher in line V (96%) than in line I (77%; P = 0.003, Fisher's exact test). These results were in accordance with data on tumor multiplicity (Table 1). In N1 ApcMin/+ mice, the origin of the Min allele from line I or V had no detectable effect on polyp number or on the frequency of allele loss (P = 0.37, Fisher's exact test; Table 5). N1 mice had similar polyp numbers (Table 1) and frequency of loss to line I mice (P = 0.85, Fisher's exact test; Table 5) and a significantly decreased frequency of polyps and allele loss compared with line V mice (P = 0.001, Fisher's exact test; Table 5). In N2s, MinI/+V animals had similar polyp numbers and similar frequencies of allele loss to N1s and line I (P > 0.30 for both cases, Fisher's exact test; Table 5). N2 MinV/+V mice had more severe disease and a significantly higher frequency of loss than N2 MinI/+V animals (P = 0.029, Fisher's exact test; Table 5). N2 MinV/+V mice also had a higher frequency of loss than N1 and line I mice, although these differences did not reach statistical significance (P = 0.17 and P = 0.36, respectively, Fisher's exact test). Overall, mice with a MinV/+V genotype showed WT Apc loss in 90% (107 of 119; Table 5) of their polyps, whereas the frequency was 76% (170 of 233; Table 5) for other mice (MinI/+I, MinI/+V and MinV/+I combined). This difference was highly significant (P < 0.0001, Fisher's exact test).
Table 5. Frequency of allele loss of WT Apc in lines, N1 hybrids, and N2 backcrossed Min Mice.
| Type | Number | Frequency, % |
|---|---|---|
| Line | ||
| MinI/+I | 46/60 | 77 |
| MinV/+V | 52/54 | 96 |
| N1s | ||
| MinI/+V | 33/47 | 70 |
| MinV/+I | 40/51 | 78 |
| N2s | ||
| MinI/+V | 51/75 | 68 |
| MinV/+V | 55/65 | 85 |
Number of adenomas with loss of WT Apc allele/total number assayed.
In summary, our results indicate that mice with both the Min and WT Apc alleles derived from line V had higher frequencies of allele loss and higher polyp numbers than mice with both or one Apc allele(s) derived from line I. Overall, these data suggest that the differences in disease severity between mice might result, at least in part, from differences in the frequency of allele loss.
Discussion
We have provided evidence for a previously undescribed modifying locus for the severity of the intestinal phenotype in ApcMin mice. Our data show, in summary, that parental line I can be defined with respect to Apc and a modifier locus D by haplotypes ApcMin D/Apc+ D and line V as ApcMin d/Apc+ d (where d is the recessive allele associated with increased polyp numbers, which probably came from the founding mouse on a non-C57Bl6/J background). Thus, a cross between a line I Min mouse and a WT line V mouse is between ApcMin D/Apc + D and Apc+ d/Apc+ d; the affected offspring are then all ApcMin D/Apc+ d with low number polyps. The cross of these mice with WT mice from line I is effectively a coupling backcross where nonrecombinant, affected mice in the N2 are ApcMin D/Apc+ d with low polyp numbers; the few mice in this cross with high polyp numbers could either be due to variation in the phenotype or recombinants between Apc and the modifier locus D/d in the N1 mouse. The equivalent N2 backcross of N1 animals with line V WT mice generally produces nonrecombinants of type ApcMin d/Apc+ d with high polyp numbers and a few low-scoring mice owing to either variation in the phenotype or recombinants in the N1 parent. The nominal, estimated recombination fraction between the presumed modifier and Apc is consistent from the two sets of crosses, and the lack of evidence of linkage to markers distal to Apc, suggests that the modifier locus lies centromeric to Apc. The last N2 cross is between Apc+ D/Apc+ d N1 mice and ApcMin D/Apc+ D line I animals, thus producing ApcMin D/Apc+ D or ApcMin D/Apc+ d animals, all with low polyp numbers.
In summary, our data strongly suggest that the cause of our observations is that the modifier locus is linked to Apc and therefore mostly cosegregates with the Min mutation. The modifier model explains why the more severe phenotype generally results when both the WT and mutant copies of Apc are derived from line V, whereas the mild phenotype results if both Apc copies are from line I or if one copy of Apc (mutant or WT) is from line I and the other from line V. It is still possible that Apc sequence variants, either intronic or adjacent to the gene, influence the Min phenotype in our mice by altering gene or protein expression. More investigation about this possibility is needed, including more extensive sequencing around Apc and analyzing gene expression at the mRNA and protein levels.
We observed higher frequencies of allele loss in adenomas from mice with more severe disease (Table 5). Although we cannot exclude the possibility that the higher frequency of allele loss is in some way the consequence of more severe disease, it is possible that higher rates of mitotic recombination caused the higher polyp numbers in our line V mice. Allele loss at Apc is known, for example, to be suppressed in mice that carry a Robertsonian translocation of chromosome 18, and these mice tend to have mild polyposis (28). Furthermore, crosses of distantly related strains often show lower overall levels of mitotic recombination, possibly as a result of multiple, large-scale chromosomal polymorphisms (29). We have not, to date, identified variation close to Apc that could explain differences in the frequencies of allele loss (data not shown). However, because other local chromosome factors, such as centromeric and telomeric repeats, also may influence chromosome pairing and crossing over in mitosis and because Apc lies only ≈15 centimorgans from the centromere/p-arm telomere, a search for differences between lines I and V needs to be conducted.
Overall, our results and data from other studies (30–32) may have important implications for human disease susceptibility once the specific modifiers and their functional consequences have been identified. To date, there has been considerable focus on SNPs as determinants of cancer risk, including genes involved in DNA repair, chromosome segregation, and recombination. It is possible, however, that some susceptibility alleles take the form of polymorphisms, or lower frequency variation, on a much larger scale than single base pair differences or small deletions. Variation would include, for example, low copy number repeats and variation in centromeres, telomeres, and other regions that influence chromosome structure or are prone to breakage (31, 32). Such sequence variants may well lead to susceptibility to specific tumor types that depends on the critical tumor suppressor loci that reside on the chromosome close to the variants and, as a result, may influence tumor initiation or progression.
Acknowledgments
We thank Kevin Whitehill, Rachael Bartram, and Pat Hillier (all of the National Radiological Protection Board) and staff of the Cancer Research Clare Hall laboratories for technical assistance. This work was supported in part by Commission of European Communities Contract Grant F14P-CT95-0008 and Cancer Research UK.
Abbreviations: CRC, colorectal cancer; FAP, familial adenomatous polyposis.
References
- 1.Bodmer, W. F., Bailey, C. J., Bodmer, J. Bussey, H. J., Ellis, A., Gorman, P., Lucibello, F. C., Murday, V. A., Rider, S. H., Scrambler, P., et al. (1987) Nature 328, 614–616. [DOI] [PubMed] [Google Scholar]
- 2.Groden, J., Thliveris, A., Samowitz, W., Carlson, M., Gelbert, L., Albertsen, H., Joslyn, G., Stevens, J., Spirio, L., Robertson, M., et al. (1991) Cell 66, 589–600. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler, K. W., Nilbert, M. C., Vogelstein, B., Bryan, T. M., Levy, D. B., Smith, K. J., Preisinger, A. C., Hamilton, S. R., Hedge, P., Markham, A., et al. (1991) Science 251, 1366–1370. [DOI] [PubMed] [Google Scholar]
- 4.Fishel, R., Lescoe, M. K., Rao, M. R. S., Copeland, N. G., Jenkins, N. A., Garber, J., Kane, M. & Kolodner, R. (1993) Cell 75, 1027–1038. [DOI] [PubMed] [Google Scholar]
- 5.Bronner, C. E., Baker, S. M., Morrison, P. T., Warren, G., Smith, L. G., Lescoe, M. K., Kane, M., Earabino, C., Lipford, J., Lindblom, A., et al. (1994) Nature 368, 258–260. [DOI] [PubMed] [Google Scholar]
- 6.Miyoshi, Y., Nagase, H., Ando H., Horii, A., Ichii, S., Nakatsuru, S., Aoki, T., Miki, Y., Mori, T. & Nakamura, Y. (1992) Hum. Mol. Genet. 1, 229–233. [DOI] [PubMed] [Google Scholar]
- 7.Miyaki, M., Tanaka, K., Kikuchi-Yanoshita, R., Muraoka, M. & Konishi, M. (1995) Crit. Rev. Oncol. Hematol. 19, 1–31. [DOI] [PubMed] [Google Scholar]
- 8.Powell, S. M., Zilz, N., Beazer-Barclay, Y., Bryan, T. M., Hamilton, S. R., Thibodeau, S. N., Vogelstein, B. & Kinzler, K. W. (1992) Nature 359, 235–237. [DOI] [PubMed] [Google Scholar]
- 9.Miyaki, M., Konishsi, M., Kikuchi-Yanoshita, R., Enomoto, M., Igari, T., Tanaka, K., Muraoka, M., Takahashi, H., Amada, Y., Fukayama, M., et al. (1994) Cancer Res. 54, 3011–3020. [PubMed] [Google Scholar]
- 10.Rowan, A. J., Lamlum, H., Ilyas, M., Wheeler, J., Straub, J., Papadopoulou, A., Bicknell, D., Bodmer, W. F. & Tomlinson, I. P. (2000) Proc. Natl. Acad. Sci. USA 97, 3352–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cannon-Albright, L. A., Skolnick, M. H., Bishop, D. T., Lee, R. G. & Burt, R. W. (1988) N. Engl. J. Med. 319, 533–537. [DOI] [PubMed] [Google Scholar]
- 12.Johns, L. E. & Houlston, R. (2001) Am. J. Gastroenterol. 96, 2992–3003. [DOI] [PubMed] [Google Scholar]
- 13.Lichtenstein, P., Holm, N. V., Verkasalo, K., Iudou, A., Kaprio, J., Koskenvuo, M., Rykkala, T., Skytthe, A. & Hemminki, K. (2000) N. Engl. J. Med. 343, 78–85. [DOI] [PubMed] [Google Scholar]
- 14.Fearnhead, N. S., Wilding, J. L., Winney, B., Tonks, S., Bartlett, S., Bicknell, D. C., Tomlinson, I. P., Mortensen, N. J. & Bodmer, W. F. (2004) Proc. Natl. Acad. Sci. USA 101, 15992–15997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crabtree, M. D., Tomlinson, I. P., Hodgson, S. V., Neale, K., Phillips, R. K. & Houlston, R. S. (2002) Gut 51, 420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietrich, W. F., Lander, E. S., Smith, J. S., Moser, A. R., Gould, K. A., Luongo, C., Borenstein, N. & Dove, W. F. (1993) Cell 75, 631–639. [DOI] [PubMed] [Google Scholar]
- 17.MacPhee, M., Chepenik, K. P., Liddell, R. A., Nelson, K. K., Siracusa, L. D. & Buchberg, A. M. (1995) Cell 81, 957–966. [DOI] [PubMed] [Google Scholar]
- 18.Cormier, R. T., Hong, K. H., Halberg, R. B., Hawkins, T. L., Richardson, P., Mulherkar, R., Dove, W. F. & Lander, E. S. (1997) Nat. Genet. 17, 88–91. [DOI] [PubMed] [Google Scholar]
- 19.Riggins, G. J., Markowitz, S., Wilson, J. K., Vogelstein, B. & Kinzler, K. W. (1995) Cancer Res. 55, 5184–5186. [PubMed] [Google Scholar]
- 20.Tomlinson, I. P., Beck, N. E., Neale, K. & Bodmer, W. F. (1996) Ann. Hum. Genet. 5, 369–376. [DOI] [PubMed] [Google Scholar]
- 21.Silverman, K., Koratkor, R., Siracusa, L. D. & Buchberg, A. M. (2002) Genome Res. 12, 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sieber, O. M., Howarth, K. M., Thirlwell, C., Rowan, A., Mandir, N., Goodlad, R. A., Gilkar, A., Spencer-Dene, B., Stamp, G., Johnson, V., et al. (2004) Cancer Res., 64, 8876–8881. [DOI] [PubMed] [Google Scholar]
- 23.Luongo, C., Moser, A. R., Gledhill, S. & Dove, W. F. (1994) Cancer Res. 54, 5947–5952. [PubMed] [Google Scholar]
- 24.Santos, J., Herranz, M., Perez de Castro, I., Pellicer, A. & Fernandez-Piqueras, J. (1998) Oncogene 17, 924–929. [DOI] [PubMed] [Google Scholar]
- 25.Manly, K. F., Cudmore, R. H., Jr., & Meer, J. M. (2001) Mamm. Genome 12, 930–932. [DOI] [PubMed] [Google Scholar]
- 26.Cho, K. R., Oliner, J. D., Simons, J. W., Hedrick, L., Fearon, E. R. Preisinger, A. C., Hedge, P., Silverman, G. A. & Vogelstein, B. (1994) Genomics 19, 525–531. [DOI] [PubMed] [Google Scholar]
- 27.Kern, S. E., Fearon, E. R. Termette, K. W. F., Enterline, J. P., Leppert, M., Nakamura, T., White, R., Vogelstein, B. & Hamilton, S. R. (1998) J. Am. Med. Assoc. 261, 3099–3103. [DOI] [PubMed] [Google Scholar]
- 28.Haigis, K. M. & Dove, W. F. (2003) Nat. Genet. 33, 33–39. [DOI] [PubMed] [Google Scholar]
- 29.Haigis, K. M., Caya, J. G., Reichelderfer, M. & Dove, W. F. (2002) Proc. Natl. Acad. Sci. USA 99, 8927–8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haigis, K. M., Hoff, P. D., White, A., Shoemaker, A. R., Halberg, R. B. & Dove, W. F. (2004) Proc. Natl. Acad. Sci. USA 101, 9769–9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abeysinghe, S. S., Chuzhanova, N., Kraeczak, M., Ball, E. V. & Cooper, D. N. (2003) Hum. Mutat. 21, 229–244. [DOI] [PubMed] [Google Scholar]
- 32.Chuzhanova, N., Abeysinge, S. S., Krawczak, M. & Cooper D. N. (2003) Hum. Mutat. 22, 245–251. [DOI] [PubMed] [Google Scholar]