Abstract
Spermiogenesis entails a major biochemical and morphological restructuring of the germ cell involving replacement of the somatic histones by protamines packing the DNA into the condensed spermatid nucleus and elimination of the cytoplasm during the elongation phase. We describe H1T2, an histone H1 variant selectively and transiently expressed in male haploid germ cells during spermiogenesis. In round and elongating spermatids, H1T2 specifically localizes to a chromatin domain at the apical pole, revealing a polarity in the spermatid nucleus. Inactivation by homologous recombination shows that H1T2 is critical for spermiogenesis as male H1t2–/– mice have greatly reduced fertility. Analysis of spermiogenesis in H1t2 mutant mice shows delayed nuclear condensation and aberrant elongation. As a result, mutant spermatids are characterized by the presence of residual cytoplasm, acrosome detachment, and fragmented DNA. Hence, H1T2 is a protein required for proper cell restructuring and DNA condensation during the elongation phase of spermiogenesis.
Keywords: chromatin, mouse, protamine, acrosome
Mammalian spermatogenesis involves the differentiation of diploid spermatogonia into spermatocytes and then, through two successive meiotic divisions, into haploid round spermatids. During spermiogenesis, the haploid round spermatids undergo an elongation phase, transforming them into mature spermatozoa. This process entails a major biochemical and morphological restructuring of the germ cell where the majority of the somatic histones are replaced, first by transition proteins, then protamines packing the DNA into the sperm cell nucleus.
Differentiating male germ cells use specialized transcriptional regulatory mechanisms involving testis-specific paralogues of transcription factors such as TLF/TRF2, ALF, and TAF7L (1–3). In addition, male germ cells contain specialized chromatin components such as H1T, a histone H1 variant specifically expressed in pachytene spermatocytes and early haploid cells (4), and HILS1, a histone H1-related protein specifically expressed in elongating and elongated spermatids (5). However, no spermatogenesis abnormalities are observed in mice lacking the H1t gene, perhaps because of redundancy with the somatic H1.1, which is also expressed at this stage (6–9). This finding contrasts with the reduced fertility and defective spermiogenesis in mice lacking transition proteins 1 or 2 or haploinsufficient for protamines 1 or 2 (10–12). Thus, whereas H1 histones have been proposed to play a role in the formation of higher-order chromatin structure, the physiological functions of the tissue-specific variants remain obscure. Here, we describe a male germ cell-specific H1-related protein, H1T2, that plays a critical role during the elongation phase of spermiogenesis.
Materials and Methods
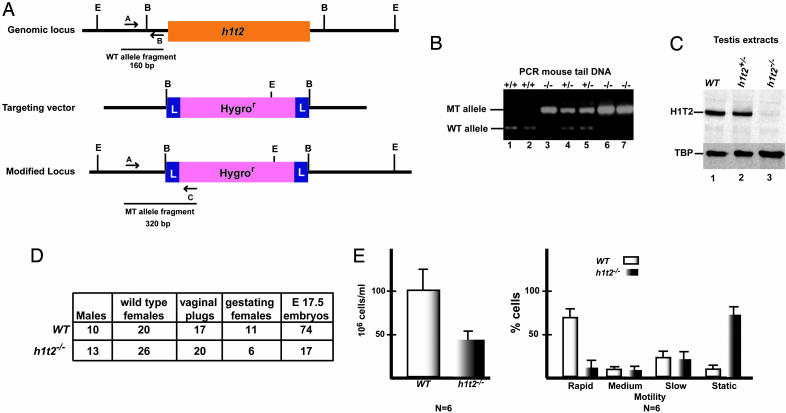
Cloning and Inactivation of H1t2. The H1t2 cDNA was cloned from a mouse testis cDNA library by screening with the short mouse TEST640 EST probe as described (2, 13). 5′ RACE using mouse testis RNA was performed to confirm the sequence of the full-length cDNA. The cloned cDNA was then used to screen a mouse genomic DNA library, and lambda phage encoding the H1t2 genomic locus were cloned and sequenced. To construct the targeting vector, an 8-kb genomic fragment spanning the gene was amplified by PCR using primers with NotI restriction sites and cloned into the corresponding site in pZERO (Invitrogen). This fragment was then cleaved with BglII to remove the H1t2 coding sequence along with short flanking 5′ and 3′ regions that were replaced by a cassette containing a hygromycin resistance gene flanked by Lox P sites (2) (see Fig. 4A). All steps were verified by restriction analysis and automated DNA sequencing. Homologous recombination in ES cells and generation of mutant mice by blastocyst injection were performed by standard procedures as described. (2, 14). Genotyping of ES cells and mice were performed by Southern blot and triplex PCR (see Fig. 3A for probes and primers used) (sequences are available upon request).
Fig. 4.
H1t2 inactivation. (A) The structure of the mouse H1t2 genomic locus is schematized along with that of the targeting vector and the modified locus. The locations of diagnostic EcoRV (E) and BglII (B) restriction sites are shown. L designates the LoxP sites flanking the hygromycin resistance cassette. Arrows A, B, and C show the locations of oligonucleotide primers used for PCR genotyping. The resulting 160-bp (A-B) and 320-bp (A-C) fragments from the WT and mutated alleles are schematized. (B) Triplex PCR genotyping of mouse tail DNA with the primers A, B, and C shown in A. Ethidium bromide staining after agarose gel electrophoresis of PCR products is shown. The WT and mutant allele fragments are indicated to the left, and the genotypes of each mouse are indicated above each lane. (C) Immunoblot analysis of 10 μg of total testis extracts from the mice indicated above each lane with the anti-H1T2 mAb and anti-TATA binding protein (TBP) antibody as control. (D) Reduced fertility of male H1t2 mutant mice. Summary of data from crosses of H1t2–/– male mice and their WT littermates is shown. Each male was housed with two females until vaginal plugs were detected or up to 3 weeks in the absence of any obvious plugs. Females were killed at embryonic day 17.5, and the embryos were counted because mothers with small litters from the H1t2–/– crosses frequently ate their pups. (E) Sperm count and sperm motility. Sperm were recovered from dissected cauda epididymides of H1t2-null or WT mice. Sperm motion was assessed by the IVOS computer-assisted semen analysis system (version 10.7Q, Hamilton-Thorne Research, Beverly, MA). A drop of the sperm suspension was transferred to a cell-UV chamber, which was set at a temperature of 37°C. About 500 sperm cells from each of the six mice used were analyzed. The percentage of cells with rapid, medium, slow, or static motility is indicated.
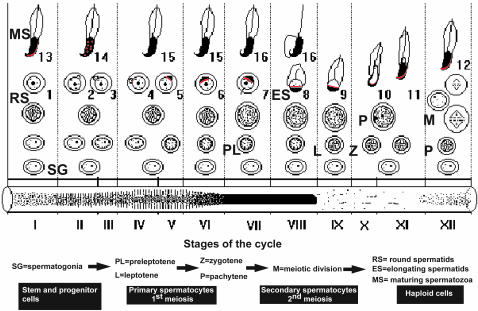
Fig. 3.
Summary of H1T2 expression during spermatogenesis. The spermatogenic differentiation program is schematized in relation to the transillumination pattern of the seminiferous tubule at the bottom. Representative cell types are indicated. Localized expression of H1T2 is indicated by red labeling.
Antibody Generation and Immunofluorescence. Monoclonal and polyclonal antibodies against the indicated H1T2 peptides coupled to ovalbumin were generated as described (15). Immunoblots were performed by standard techniques and revealed by chemiluminescence with an ECL kit (Amersham Pharmacia). Transillumination-assisted microdissection of seminiferous tubules from 6- to 10-week-old mice, squash preparations, and immunofluorescence were performed as described (3, 16, 17). Sperm were recovered from dissected cauda epididymides incubated in PBS for 30 min at 37°C. The sperm were then spotted on glass slides, air-dried, and fixed in 4% paraformaldehyde for 15 min before immunofluorescence and in situ TUNEL assay. TUNEL assays were performed with an ApopTag Red In situ apoptosis detection kit (Intergen, Purchase, NY) according to the manufacturer's instructions.
Germ Cell Chromatin Isolation. Chromatin isolation was performed essentially as described (18). Germ cells from 10-week-old mice were separated from seminiferous tubules by collagenase digestion, centrifugation at 2,000 × g for 5 min at 4°C, and passage through four layers of gauze until all residual tubules were retained. After centrifugation, the cells were resuspended in 15 mM Tris·HCl buffer, pH 7.5, containing 60 mM KCl, 15 mM NaCl, 10 mM MgCl2, 1 mM CaCl2, 1 mM DTT, and 250 mM sucrose (buffer N250). An equal volume of buffer N250 containing 0.6% Nonidet P-40 was added to the cells, and the suspension was gently mixed and incubated on ice for 15 min. After centrifugation at 2,000 × g for 5 min at 4°C, the supernatant cytoplasm was separated from the pelleted nuclei. Nuclei were recovered and washed three times with buffer N250 before lysis in 10 mM Pipes buffer, pH 6.5 containing 10 mM EDTA. After centrifugation at 6,000 × g for 20 min at 4°C in a microfuge, the nucleoplasmic fraction was recovered, and the chromatin pellet was recovered in SDS/PAGE loading buffer.
Results
Mouse TEST640 (mTEST640) Encodes a Histone H1 Variant. mTEST640 was one of a series of testis-specific ESTs isolated by Yuan et al. (19). In situ hybridization on sectioned testis and Northern blot analysis showed that mTEST640 expression was restricted to haploid cells with low-level expression in step-2 to step-4 round spermatids, which strongly increases through steps 5–12 (13). The full-length cDNA for mTEST640 was cloned from a testis library, revealing an ORF encoding a 399-aa protein. Database searches revealed that the mTEST640 N-terminal domain shows high similarity to the histone H1 proteins (Fig. 1) and contains a conserved winged three-helix bundle domain. Thus, mTEST640 is a unique haploid cell-specific histone H1 variant, which we designate as Hist1h1t2 (abbreviated as H1t2).
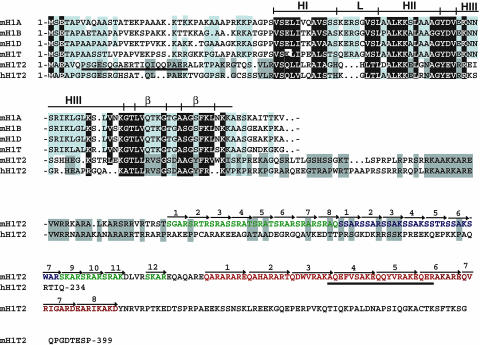
Fig. 1.
H1T2 is a novel member of the H1 linker histone family. The amino acid sequences of mouse H1T2 (mHlT2) and human H1T2 (hH1T2) are shown and are compared with those of known members of the mouse H1 family. Identical and highly conserved amino acids are shown in white on a black background. Positions with conserved amino acids are boxed in light gray. Amino acids conserved only in mH1T2 and hH1T2 are boxed in dark gray. Amino acids were classified as follows: small residues, P, A, G, S, and T; hydrophobic, L, I, V, A, F, M, C, Y, and W; polar/acidic, D, E, Q, and N; and basic, R, K, and H. The full-length amino acid sequences for H1T2 are shown, whereas only the amino acids of the conserved winged helix domain are shown for the other family members. The predicted locations of the α-helices (HI–HIII), the loops (L), and the β-sheets (β) forming the β-hairpin are also indicated based on the structure of histone H5 and H1 (20, 21). The arrows indicate the SR and heptapeptide repeats in mH1T2, which are shown in different colors. The peptides used for antibody production are underlined.
The H1 protein family has poorly conserved basic N- and C-terminal regions flanking the conserved three-helix bundle domain (20, 21). The C-terminal domain of H1T2 is highly basic, containing multiple serine-arginine-type repeats and eight copies of a heptapeptide sequence (Fig. 1). H1T2 is encoded by an intronless gene present on chromosome 15 (F2), whereas the other histone H1 genes are on chromosome 13 (22). Database searches identified a putative 234-aa human H1T2 orthologue encoded by chromosome 12 (locus q13.11). At several positions in the winged helix domain, human H1T2 shares amino acid identity with mouse H1T2, but not other H1 proteins (boxed in dark gray in Fig. 1). For example, in mouse and human H1T2, the loop connecting α-helices I and II is shorter and exhibits specific substitutions in the β-hairpin. However, human H1T2 does not contain an extended C-terminal domain with SR repeats. Aside from these minor differences, the H1T2 subfamily is highly related to the other histone H1 proteins, much more so than other members of the winged helix superfamily such as HILS1 (5).
H1T2 Expression Reveals Polarity in Spermatid Nuclei. Expression of mouse H1T2 was studied with a mAb generated against a C-terminal domain peptide and a rabbit polyclonal against a N-terminal domain peptide (underlined in Fig. 1). These antibodies recognize a single 48-kDa protein in testis extracts (see Fig. 4C, lane 1 and Fig. 6, which is published as supporting information on the PNAS web site) and were used for immunofluorescence on sequentially microdissected squash preparations of mouse seminiferous tubules where the exact stage of the cycle was identified and on sectioned testis.
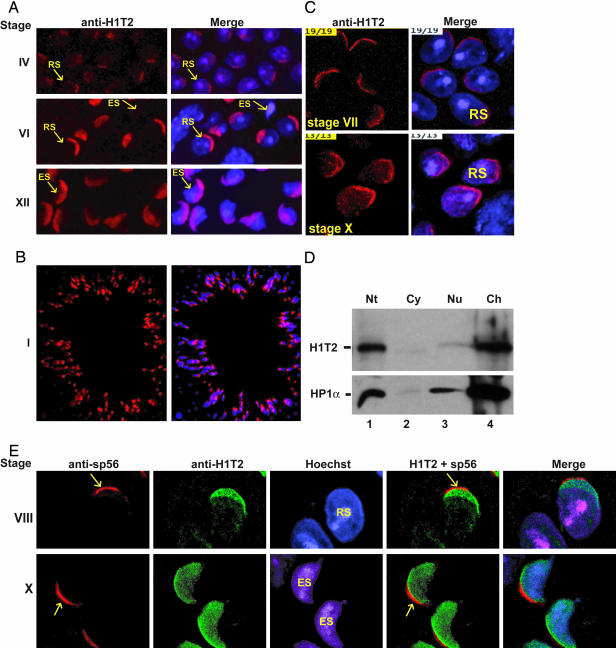
Weak H1T2 expression was first detected in round spermatids at stage IV, but expression strongly increased through stages V-VIII (Fig. 2 A and B, Fig. 7A, which is published as supporting information on the PNAS web site, and data not shown; summarized in Fig. 3: note that specificity of this signal is shown by the loss of labeling in H1T2-mutant spermatids, Fig. 7B and see below). This pattern of expression mirrors that previously described for HlT2's mRNA (13). Strikingly, the localization of H1T2 within the nucleus is highly polar, the protein being concentrated in a cap-like structure at the inner periphery of the nuclear membrane. This asymmetric localization of H1T2 in the nucleus of round spermatids is confirmed by confocal microscopy (Fig. 2C and see Movie 1, which is published as supporting information on the PNAS web site, showing a 3D reconstruction of H1T2 expression).
Fig. 2.
Immunolocalization of H1T2 in developing spermatids. (A) Immunofluorescence on squash preparations. (Left) The stages of the cycle are shown. (Right) The merge of the anti-H1T2 mAb image and the Hoechst-stained nuclei is shown. Representative examples of different cell types are indicated by the arrows. RS, round spermatids; ES, elongating spermatids. (B) Immunodetection of H1T2 in sectioned tubules from WT mice at stage I. The immunolabeling and the merge with the Hoechst-stained nuclei are shown. (C) Confocal images of staged squash preparations stained with mAb against H1T2 (Left) and merged with the Hoechst (Right). A single representative focal plane is shown. RS, round spermatid. (D) H1T2 is assocaited with chromatin. Western blot analysis of cytoplasmic (Cy), total nuclear (Nt), nucleoplasmic (Nu), and chromatin (Ch) fractions from purified germ cells is shown. Each fraction was subjected to 15% SDS/PAGE, and H1T2 and HP1α were revealed by using specific antibodies. (E) Confocal images of double-labeling of spermatids with polyclonal anti-H1T2 (green) and monoclonal anti-sp56 (red) antibodies. (Right) Both signals are superimposed. A single representative focal plane is shown. (Magnifications: ×40, A; ×20, B; and ×400, C and E.)
Polarized H1T2 expression persists in steps 9–14 elongating spermatids (stages IX-II), before rapidly disappearing by stage III (Figs. 2 A and C, 3, and 7A). At these later stages, the distinctive morphology of the spermatids shows that H1T2 is principally localized at the apical pole (Fig. 2 A and C). Polar localization of H1T2 in elongating spermatids is confirmed by immunolabeling of sectioned tubules (Fig. 2B). Confocal analysis of double-labeling with antibodies against H1T2 and sp56, a component of the acrosomal matrix whose antibodies specifically label this compartment of the acrosome (23, 24), showed that H1T2 was also localized at the apical pole of round spermatids (Fig. 2E and Fig. 8, which is published as supporting information on the PNAS web site). H1T2 overlaps with the Hoeschst-stained DNA and is adjacent to, but does not overlap with, sp56, indicating that it is localized uniquely in the nucleus under the acrososme. Hence, H1T2 is localized in a specific domain at the apical pole, revealing an inherent asymmetry in the round spermatid nucleus.
To determine whether H1T2 was tightly associated with the chromatin, extracts were prepared from the nucleoplasmic and chromatin fractions of purified germ cell nuclei. H1T2 could be detected in total extracts from nuclei, but not in the cytoplasmic fraction (Fig. 2D, lanes 1 and 2). Little H1T2 was extracted in the nucleoplasmic fraction (Fig. 2D, lane 3), whereas the vast majority of the protein was present in the chromatin fraction (Fig. 2D, lane 4). As a control, the same extracts were also probed for the presence of HP1α, a known chromatin component. H1T2 and HP1α are extracted in a similar manner, showing that H1T2 is a chromatin-associated protein.
H1T2 Is Essential for Normal Male Fertility. Developmentally, H1T2 expression only partially overlaps with that of H1T, which disappears early in the elongation phase (25), but is concomitant with transition proteins and protamines as somatic histones are removed and the DNA is condensed into the nucleo-protamine complex. Both the timing of its expression and its polarized localization in the germ cell nucleus suggest that H1T2 may be required to organize DNA condensation during the elongation phase. To test this possibility, the H1t2 gene was inactivated in mice by homologous recombination (Fig. 4 A and B). Testicular H1T2 protein levels were not significantly affected in H1t2+/– animals, but were absent from H1t2–/– testis (Fig. 4C).
Adult H1t2–/– mice appeared normal and females were fertile (data not shown). In contrast, whereas fertility of H1t2 heterozygous male mice was comparable to WT (data not shown), that of H1t2–/– male mice was strongly reduced (Fig. 4D). Although the litter size was small, the pups were normal and viable. Moreover, H1t2–/– mice showed a reduced sperm count and severely reduced sperm motility compared with WT (Fig. 4E). The majority of the mutant sperm were slow moving or static with almost no rapidly moving cells. Thus, H1T2 is essential for normal spermatogenesis and male fertility.
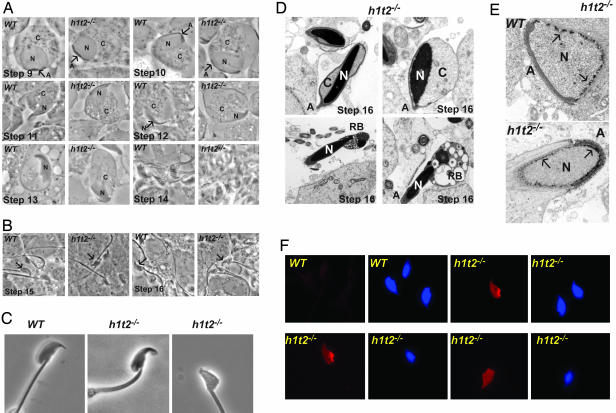
Loss of H1T2 Leads to Abnormal Elongation and DNA Condensation. Histological analysis of seminiferous tubules, sectioned testes, and epididymides revealed no major morphological changes in the mutant animals (Fig. 9, which is published as supporting information on the PNAS web site). However, closer examination of the elongation phase when H1T2 is strongly expressed by live cell cytology (16) on squash preparations from sequential staged dissected seminiferous tubules from 6- to 8-week-old male H1t2–/– mice or their WT littermates did reveal abnormalities. In WT step-11 elongating spermatids, nuclear condensation and protrusion of the nuclear apex are observed, whereas in H1t2-mutant mice, nuclear condensation is reduced and the nucleus remains within the cytoplasm (Fig. 5A). This phenomenon is more evident in step-12 spermatids, which normally show the most elongated nucleus with fully condensed DNA seen during mouse spermiogenesis. In step-12 spermatids from H1t2-mutant mice, the nucleus is only partially condensed and its protrusion from the cytoplasm is diminished (Fig. 5A).
Fig. 5.
Phenotype of H1t2 mutant spermatids. (A) Light microscope images of staged squash preparations from WT or H1t2–/– seminiferous tubules. Close-up images of representative individual elongating spermatids from the indicated steps of spermiogenesis are shown. N, C, and A indicate the nucleus, cytoplasm, and acrosome, respectively. (B) Close-up images of elongating spermatids from the indicated steps of spermiogenesis. (C) Abnormalities in sperm from cauda epididymides. Light microscope images of representative individual spermatozoa from WT and H1t2-mutant cauda epididymides. (D) Electron microscopy of late-stage elongated spermatids. The nucleus (N) and abnormal residual cytoplasm (C) are indicated along with the detached acrosome (A) and the residual body (RB). (E) Electron microscopy of early elongating spermatids. The arrows indicate the representative foci of perinuclear heterochromatin. (F)(Left) In situ TUNEL assay detects fragmented DNA. (Right) The corresponding Hoeschst stain is shown. (Magnifications: ×100, A and B; ×150, C; ×5,000, D and E; and ×40, F.)
At step 13, nuclear condensation is not yet completed in H1t2–/– spermatids and residual cytoplasm remains around the nucleus, whereas the WT nuclei have assumed their distinctive morphology and the cytoplasm has been eliminated (Fig. 5A). At later stages, the spermatid head is clearly deformed and of irregular shape showing bent or blunted apex (Fig. 5B). In agreement with these observations, severe morphological abnormalities were observed in >80% of spermatozoa isolated from cauda epididymides of mutant mice (Fig. 5C). Electron microscopy shows abnormal retention of residual cytoplasm around the nucleus of late-stage spermatids and the abnormal presence of the residual body (Fig. 5D). The presence of residual cytoplasm may impair tight association of the acrosome with the nuclear membrane, leading to its detachment. None of these abnormalities are observed in the H1t2 heterozygous animals (data not shown). Hence, in the absence of H1T2, spermatids fail to polarize correctly, leading to defective and incomplete elongation and retention of cytoplasm or residual bodies as well as retarded DNA condensation.
Electron microscopy on early elongating spermatids further reveals that in WT spermatids perinuclear heterochromatin foci are excluded from the region beneath the acrosome where H1T2 is localized (Fig. 5E and Fig. 10, which is published as supporting information on the PNAS web site). In contrast, in spermatids lacking H1T2 heterochromatin foci are all around the nuclear membrane, including the region beneath the acrosome (Figs. 5E and 10). These observations provide direct evidence suggesting that H1T2 is necessary for the polarity in nuclear chromatin organization.
Defective DNA condensation within the sperm nucleus, such as is observed in mice haploinsufficient for protamines 1 or 2, is known to provoke DNA fragmentation (12). The nuclei of >80% of the sperm from cauda epididymides of mutant mice could be labeled by using an in situ TUNEL assay to detect fragmented DNA (Fig. 5F). In addition to a generalized labeling, more intense localized labeling could be observed in some nuclei (Fig. 5F Upper Right and Lower Left). In contrast, no labeling was observed in sperm from WT animals (Fig. 5F Upper Left). These results confirm that the loss of H1T2 leads to defective DNA condensation, supporting the idea that formation of an asymetrically localized H1T2 chromatin domain is essential for organizing proper DNA condensation.
Taken together, the analyses of the H1T2 mutant spermatids show that this protein is essential for proper elongation and DNA condensation during spermiogenesis.
Discussion
During spermiogenesis, nuclear structure undergoes a dramatic reorganization unique to male germ cells that packs the DNA in a specific topology within the sperm nucleus (26, 27). How the specificity of this process is achieved is at present not well understood. Here, we describe the variant histone H1T2 that, in addition to the transition proteins and protamines, is necessary for proper DNA condensation during the elongation phase.
An intriguing property of H1T2 is that it is specifically localized at the apical pole, hence revealing an inherent polarity within the spermatid nucleus. It remains to be determined whether H1T2 is imparting this polarity by itself or whether its localization reflects a polarity induced by other proteins. Indeed, the mechanism by which H1T2 is localized in a polar fashion within the spermatid nucleus also awaits elucidation. In this respect, however, it is interesting to note that there exists a family of high molecular weight proteins, thecins, whose identities are as yet unknown, that also have a polar expression profile analogous to that of H1T2 (28). These proteins are components of the perinuclear theca beneath the acrosome, a region also depleted in nuclear pores and comprising the specialized acroplaxome structure (29). The specialization of nuclear membrane structure beneath the acrosome suggests that there may be communication between the acrosome and the nucleus that directly or indirectly positions H1T2, although H1T2 is not essential for acrosome formation and positioning that initially appear normal in the mutant spermatids. Our observations do, however, show that there is a polarity in the spermatid nucleus, and they open up possibilities for identification of additional proteins involved in nuclear polarization during spermiogenesis.
Supplementary Material
Acknowledgments
We thank I. Michel for excellent technical assistance; H. Tournaye for the spermogram; A. Dierich, E. Metzger, and the Institut de Génétique et de Biologie Moléculaire et Cellulaire ES and animal facilities; M. Oulad-Abdelghani and the mAb facility; G. Duval and the polyclonal antibody facility, and J. L. Vonesch and the staff of the confocal microscopy facility and DNA sequencing and peptide synthesis facilities. I.M. was supported by a fellowship from the Fondation pour la Recherche Médicale, and S.B was supported by a fellowship from the Association pour la Recherche contre le Cancer. This work was supported by grants from the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Hôpital Universitaire de Strasbourg, the Ministère de la Recherche et de la Technologie, the European Union (RTN-00026), the Association pour la Recherche Contre le Cancer, the Ligue Nationale Contre le Cancer, and Organon (Akzo/Nobel).
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY496853 and AY496854).
References
- 1.Sassone-Corsi, P. (2002) Science 296, 2176–2178. [DOI] [PubMed] [Google Scholar]
- 2.Martianov, I., Fimia, G. M., Dierich, A., Parvinen, M., Sassone-Corsi, P. & Davidson, I. (2001) Mol. Cell 7, 509–515. [DOI] [PubMed] [Google Scholar]
- 3.Pointud, J. C., Mengus, G., Brancorsini, S., Monaco, L., Parvinen, M., Sassone-Corsi, P. & Davidson, I. (2003) J. Cell Sci. 116, 1847–1858. [DOI] [PubMed] [Google Scholar]
- 4.Drabent, B., Bode, C., Bramlage, B. & Doenecke, D. (1996) Histochem. Cell Biol. 106, 247–251. [DOI] [PubMed] [Google Scholar]
- 5.Yan, W., Ma, L., Burns, K. H. & Matzuk, M. M. (2003) Proc. Natl. Acad. Sci. USA 100, 10546–10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drabent, B., Saftig, P., Bode, C. & Doenecke, D. (2000) Histochem. Cell Biol. 113, 433–442. [DOI] [PubMed] [Google Scholar]
- 7.Lin, Q., Sirotkin, A. & Skoultchi, A. I. (2000) Mol. Cell. Biol. 20, 2122–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fantz, D. A., Hatfield, W. R., Horvath, G., Kistler, M. K. & Kistler, W. S. (2001) Biol. Reprod. 64, 425–431. [DOI] [PubMed] [Google Scholar]
- 9.Nayernia, K., Drabent, B., Adham, I. M., Moschner, M., Wolf, S., Meinhardt, A. & Engel, W. (2003) Biol. Reprod. 69, 1973–1978. [DOI] [PubMed] [Google Scholar]
- 10.Yu, Y. E., Zhang, Y., Unni, E., Shirley, C. R., Deng, J. M., Russell, L. D., Weil, M. M., Behringer, R. R. & Meistrich, M. L. (2000) Proc. Natl. Acad. Sci. USA 97, 4683–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao, M., Shirley, C. R., Yu, Y. E., Mohapatra, B., Zhang, Y., Unni, E., Deng, J. M., Arango, N. A., Terry, N. H., Weil, M. M., et al. (2001) Mol. Cell. Biol. 21, 7243–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho, C., Willis, W. D., Goulding, E. H., Jung-Ha, H., Choi, Y. C., Hecht, N. B. & Eddy, E. M. (2001) Nat. Genet. 28, 82–86. [DOI] [PubMed] [Google Scholar]
- 13.Penttila, T. L., Yuan, L., Mali, P., Hoog, C. & Parvinen, M. (1995) Biol. Reprod. 53, 499–510. [DOI] [PubMed] [Google Scholar]
- 14.Martianov, I., Viville, S. & Davidson, I. (2002) Science 298, 1036–1039. [DOI] [PubMed] [Google Scholar]
- 15.Mengus, G., May, M., Jacq, X., Staub, A., Tora, L., Chambon, P. & Davidson, I. (1995) EMBO J. 14, 1520–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parvinen, M. & Hecht, N. B. (1981) Histochemistry 71, 567–579. [DOI] [PubMed] [Google Scholar]
- 17.Martianov, I., Brancorsini, S., Gansmuller, A., Parvinen, M., Davidson, I. & Sassone-Corsi, P. (2002) Development (Cambridge, U.K.) 129, 945–955. [DOI] [PubMed] [Google Scholar]
- 18.Remboutsika, E., Lutz, Y., Gansmuller, A., Vonesch, J. L., Losson, R. & Chambon, P. (1999) J. Cell Sci. 112, 1671–1683. [DOI] [PubMed] [Google Scholar]
- 19.Yuan, L., Liu, J. G. & Hoog, C. (1995) Biol. Reprod. 52, 131–138. [DOI] [PubMed] [Google Scholar]
- 20.Ramakrishnan, V., Finch, J. T., Graziano, V., Lee, P. L. & Sweet, R. M. (1993) Nature 362, 219–223. [DOI] [PubMed] [Google Scholar]
- 21.Cerf, C., Lippens, G., Muyldermans, S., Segers, A., Ramakrishnan, V., Wodak, S. J., Hallenga, K. & Wyns, L. (1993) Biochemistry 32, 11345–11351. [DOI] [PubMed] [Google Scholar]
- 22.Wang, Z. F., Sirotkin, A. M., Buchold, G. M., Skoultchi, A. I. & Marzluff, W. F. (1997) J. Mol. Biol. 271, 124–138. [DOI] [PubMed] [Google Scholar]
- 23.Bookbinder, L. H., Cheng, A. & Bleil, J. D. (1995) Science 269, 86–89. [DOI] [PubMed] [Google Scholar]
- 24.Kim, K. S., Cha, M. C. & Gerton, G. L. (2001) Biol. Reprod. 64, 36–43. [DOI] [PubMed] [Google Scholar]
- 25.Drabent, B., Bode, C., Miosge, N., Herken, R. & Doenecke, D. (1998) Cell Tissue Res. 291, 127–132. [DOI] [PubMed] [Google Scholar]
- 26.Zalensky, A. O., Allen, M. J., Kobayashi, A., Zalenskaya, I. A., Balhorn, R. & Bradbury, E. M. (1995) Chromosoma 103, 577–590. [DOI] [PubMed] [Google Scholar]
- 27.Meyer-Ficca, M., Muller-Navia, J. & Scherthan, H. (1998) J. Cell Sci. 111, 1363–1370. [DOI] [PubMed] [Google Scholar]
- 28.Bellve, A. R., Chandrika, R. & Barth, A. (1990) J. Cell Sci. 96, 745–756. [DOI] [PubMed] [Google Scholar]
- 29.Fawcett, D. W. & Chemes, H. E. (1979) Tissue Cell 11, 147–162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.