Abstract
Objective
O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT) catalyzes the addition of O-GlcNAc and GlcNAcylation has extensive crosstalk with phosphorylation to regulate signaling and transcription. Pig OGT is located near the region of chromosome X that affects follicle stimulating hormone level and testes size. The objective of this study was to find the variations of OGT between European and Chinese pigs.
Methods
Pigs were tested initially for polymorphism in OGT among European and Chinese pigs by polymerase chain reaction and sequencing at the U.S. Meat Animal Research Center (USMARC). The polymorphism was also determined in an independent population of pigs including European and Chinese Meishan (ME) breeds at the National Institute of Animal Science (NIAS, RDA, Korea).
Results
The intron 20 of OGT from European and Chinese pigs was 514 and 233 bp, respectively, in the pigs tested initially. They included 1 White composite (WC) boar and 7 sows (2 Minzu×WC, 2 Duroc [DU]×WC, 2 ME×WC, 1 Fengzing×WC) at USMARC. The 281-bp difference was due to an inserted 276-bp element and GACTT in European pigs. When additional WC and ME boars, the grandparents that were used to generate the 1/2ME×1/2WC parents, and the 84 boars of 16 litters from mating of 1/2ME×1/2WC parents were analyzed, the breeds of origin of X chromosome quantitative trait locus (QTL) were confirmed. The polymorphism was determined in an independent population of pigs including DU, Landrace, Yorkshire, and ME breeds at NIAS. OGT was placed at position 67 cM on the chromosome X of the USMARC swine linkage map.
Conclusion
There was complete concordance with the insertion in European pigs at USMARC and NIAS. This polymorphism could be a useful marker to identify the breed of origin of X chromosome QTL in pigs produced by crossbreeding Chinese and European pigs.
Keywords: Breeds of Origin, Pig, O-linked N-acetylglucosamine Transferase, Polymorphism, X chromosome
INTRODUCTION
O-GlcNAc transferase (OGT) catalyzes the addition of O-GlcNAc and β-N-acetylglucosaminidase (O-GlcNAcase, OGA) is responsible for O-GlcNAc removal, and only these two enzymes are known to regulate GlcNAcylation in mammals [1]. O-GlcNAc modifies numerous proteins and the GlcNAcylated proteins are involved in transcription, cell cycle, and stress responses. GlcNAcylation has extensive crosstalk with phosphorylation to regulate signaling, transcription and the cytoskeleton in response to nutrients and stress [1]. Nutrient-sensitive enzymes, OGT and AMP-activated protein kinase, cross-talk to mediate cellular metabolism [2]. OGT gene is highly conserved and is localized near the centromeres as a single X-linked gene in mammals [1]. The gene encoding OGT is evolutionarily conserved from plants to humans. In Arabidopsis, OGT is encoded by, SPINDLY (SPY) and SECRET AGENT (SEC) genes, and both genes are required during gametogenesis and embryogenesis [3]. O-GlcNAc is regulated throughout oogenesis and development in Xenopus laevis [4]. Intact OGT alleles are required for the completion of embryogenesis in mice [5]. OGT was identified as a promising placental biomarker of maternal stress in mice [6]. Hexosamine biosynthesis pathway and O-GlcNAcylation have important functions in pig preimplantation embryonic development and that inhibition of O-GlcNAcase is fatal for development [7]. The data suggests potential role of OGT and O-GlcNAcase in reproduction of many different species.
The Chinese pig breeds, including Meishan (ME), have been investigated for many unique reproductive traits both in females [8–10] and males [11–14]. While there are no reports of the role of OGT on male reproduction of Chinese pigs or breed differences in OGT expression, a genomic region located on chromosome X encompassing OGT was examined. In a previous study using ME×White composite (WC) crossbred boars, it was revealed that genomic region encompassing OGT on chromosome X affects follicle stimulating hormone (FSH) level and testes size [11]. As an initial step to investigate variations in OGT between European and Chinese pig breeds, attempts were made to amplify across several relatively short introns and a polymorphism was identified due to an inserted element in intron 20 of OGT [15]. The polymorphism was reported in the Korean native pig, but was not conclusive [16]. The objective of this study was to determine the OGT polymorphisms in European and Chinese pig breeds and evaluate whether this polymorphism could be an additional marker in identifying the breed of origin of X chromosomal region in either boars or sows produced for their reproductive characteristics by crossing ME or other Chinese breeds with the known polymorphism and European pig breeds.
MATERIALS AND METHODS
Experimental animals and primer design
Pigs tested initially for polymorphism in OGT among European and Chinese pigs included 1 WC boar and 7 sows (2 Minzu [MI] ×WC, 2 Duroc [DU]×WC, 2 ME×WC, 1 Fengzing [FE]×WC) at the U.S. Meat Animal Research Center (USMARC). WC population was comprised of equal contributions from Chester White, Landrace, Large White, and Yorkshire breeds [11]. Further WC and ME boars (n = 10 each), the grandparents that were used to generate the 1/2ME×1/2WC parents, and 84 boars of 16 litters from the mating of 1/2ME×1/2WC parents at USMARC were also studied. The polymorphism was also determined in 7 litters (n = 86) from 2 boars and 7 sows of the USMARC Swine Reference Population. The polymorphism was analyzed in an independent population of pigs (n = 122) including DU, Landrace, Yorkshire, and ME breeds at the National Institute of Animal Science (NIAS, RDA, Korea). The experimental protocol and standard operating procedures on experimental animals were reviewed and approved as exemption by the Institutional Animal Care and Use Committee of USMARC and the National Institute of Animal Science, RDA (Jeonju, Republic of Korea).
Pig OGT is consisted of 22 exons, and all exons are part of the coding sequence, which ranges from 182 to 3,322 base pairs. To find the variations in OGT between European and Chinese pig breeds, attempts were made to amplify across several relatively short introns, including intron 20 of OGT, based on the porcine OGT cDNA sequence (GenBank accession no. DQ400859) [15] and the OGT gene sequence in the Genome Browser for pig (http://genome.ucsc.edu/). Polymerase chain reaction (PCR) primer pairs were designed using the Primer 3 online-tool. The forward (OGT-2773F) and reverse (OGT-3083R) primer sequences to amplify across intron 20 corresponded to bases 2773–2793 and 3083–3059 of the porcine OGT cDNA sequence (GenBank accession no. DQ400859), respectively. The expected size of the PCR amplicon across intron 20 was 825 bp based on the gene sequence in the Genome Browser for pig (http://genome.ucsc.edu/).
PCR amplification and sequencing
PCR reactions were carried out in a 10 μL volume containing 50 ng genomic DNA, 1.5 mM MgCl2, 10 pmol of each primer (OGT-2773F: 5′-TCTGAAGCGTGTTCCCAATAG-3′ and OGT-3083R: 5′-GCTCAAGACAACC TAAACAAGTAAG-3′), 100 μM dNTP, and 0.35 U Taq polymerase. Amplification was performed under the following PCR conditions; 10 min at 95°C; 35 cycles of 30 s at 95°C, annealing for 30 s at 61°C, 1 min at 72°C; and a final extension of 5 min at 72°C. The amplified genomic DNA in the initial study from 1 WC boar and 7 sows [2MI×WC, 2DU× WC, 2ME×WC, 1FE×WC] at USMARC were cloned into pCRII vector (Invitrogen, Carlsbad, CA, USA) and sequenced in both directions. The amplified genomic DNA of European and ME pigs at NIAS were also sequenced. Agarose gel electrophoresis and sequencing of the PCR amplicons of porcine genomic DNA indicated that the size of the amplicon from White composite and Duroc pigs were 825 bp, while those of Chinese pigs were 544 bp.
RESULTS
OGT polymorphism and genotypes
Amplification of fragments containing intron 20 of OGT by PCR revealed a variation between the European and Chinese pig breeds. The size of the amplicons containing a part of exon 20, intron 20, and a part of exon 21 of OGT from a WC boar and two DU×WC sows was 825 bp (GenBank accession no. DQ404296), while those from MI×WC, ME×WC, and FE×WC sows were 825 bp and 544 bp. Thus, the size of amplicons from the Chinese pigs was 544 bp (GenBank accession no. DQ404295) (Figure 1A). The size of OGT intron 20 was determined by sequencing and it was 514 bp in DU and WC pigs, while it was 233 bp in Chinese pigs (Figures 1A, 1B). The sequence of OGT intron 20 from DU and WC pigs is identical to the OGT sequence that is in the Genome Browser for pig (NM_001039748.2, http://genome.ucsc.edu/).
Figure 1.
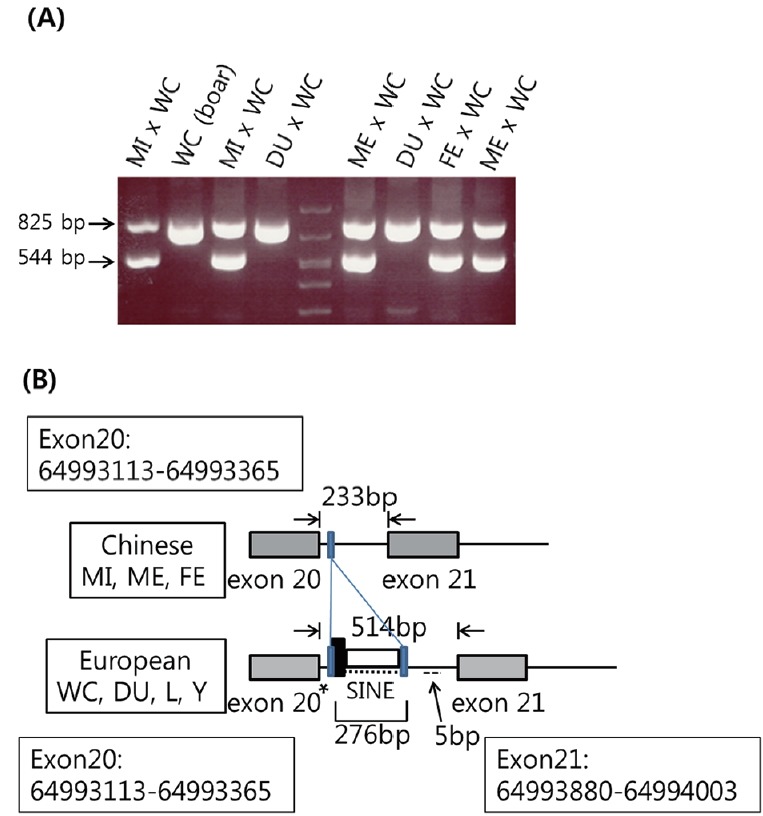
(A) Intron 20 of O-linked N-acetylglucosamine transferase (OGT) from European and Chinese pig breeds. Fragments containing intron 20 of OGT were compared by amplifying the region containing a part of exon 20, intron 20, and a part of exon 21 of OGT. All of polymerase chain reaction amplicons from Minzu (MI)×White composite (WC), Meishan (ME) ×WC, and Fengzing (FE)×WC sows had both 825-bp and 544-bp bands, while those from Duroc (DU)×WC sows and a WC boar had only the 825-bp band. (B) Schematic diagram of intron 20 of OGT in Chinese and European pigs. The intron 20 of Chinese pigs including MI, ME, and FE was 233 bp, while that of European pigs including WC, DU, Landrace (L), and Yorkshire (Y) was 514 bp, resulting in the 281-bp difference. The longer 514-bp intron 20 of OGT from European pigs contained an inserted 276-bp element (·····) near the beginning of the intron and GACTT (--) insertion at nucleotides 401–405 of the 514 bp intron 20. The 276-bp element (chrX:64993408–64993683) of the European pigs is located after the first 14-bp TGCTTGTTCTTTTT (chrX:64993394–64993407) sequence and the 3′-end of the element ends with the second 14-bp TGCTTGTTCTTTTT (chrX:64993670–64993683) sequence, shown as blue narrow rectangles and the Chinese pigs have only one such sequence. Black rectangle indicates the 33-bp (TTG)7 simple repeat (chrX:64993397–64993429) and it included the 4 to 14 bp of the 14-bp TGCTTGTTCTTTTT sequence and the (TTG)7 repeat sequence. The 240-bp SINE element (chrX:64993430–64993669), following the 33-bp (TTG)7 simple repeat, is shown by the white rectangle. The rs323323004 at chromosome position 64993390 on intron 20 is shown with *. The version of the swine genome assembly used to obtain sequence position information was the Swine Genome Sequencing Consortium Sscrofa10.2.
The 281-bp difference in the intron 20 between the European pigs and the Chinese pigs was due to an inserted 276-bp element near the beginning of the intron and GACTT insertion at nucleotides 401–405 of the 514 bp intron 20 in the European pigs (Figure 1B). The OGT genotypes of Chinese and European pigs were designated as A and B, respectively. The genotypes were determined in WC and ME boars (n = 10 each), the grandparents that were used to generate the 1/2ME×1/2WC parents, and the 84 boars of 16 litters from mating of 1/2ME×1/2WC parents at USMARC (Table 1; Figure 2). Consistently, insertions were only in WC boars, but not in ME boars. The OGT genotypes were determined in the 7 litters (n = 86) from 2 boars and 7 sows of the USMARC Swine Reference Population (Table 2). Consistently, insertions were only in European pigs. The OGT genotypes were also determined in an independent population of pigs including DU, Landrace, Yorkshire, and ME breeds (n = 122) at the National Institute of Animal Science (NIAS, RDA, Korea), and the results were from additional analysis of pigs from a previous study [16]. The insertions were only in European pigs (Table 3; Figure 3). There was complete concordance with the insertion in European pigs at USMARC and NIAS.
Table 1.
Frequency of genotypes of OGT among ME and WC boars, and boars produced from 1/2 ME×1/2 WC parents1)
| A | B | Total | |
|---|---|---|---|
| ME* | 10 | - | 10 |
| WC* | - | 10 | 10 |
| Offspring | 37 | 47 | 84 |
| Total | 47 | 57 | 104 |
OGT, O-linked N-acetylglucosamine transferase; ME, Meishan; WC, White composite.
ME* and WC* boars were the grandparents. Genotypes of OGT from the Chinese and European boars at the U.S. Meat Animal Research Center were designated as A and B, respectively.
Figure 2.
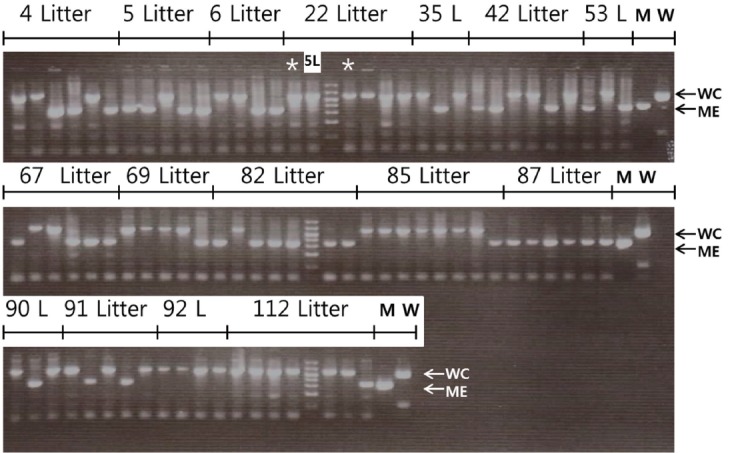
Confirmation of the breeds of origin of X chromosome by polymorphism in the intron 20 of porcine O-linked N-acetylglucosamine transferase (OGT). When boars produced by inter se mating of 1/2 Meishan (ME)×1/2 White composite (WC) parents, the breed of origin of X chromosome quantitative trait locus (QTL) was confirmed by the length polymorphism due to the presence of the inserted element in the intron 20 of OGT after polymerase chain reaction amplification. Out of 84 boars of 16 litters (L) analyzed, boars from 13 litters had both ME and WC alleles. They included ‘4, 5, 6, 35, 42, 53, 67, 69, 82, 85, 90, 91, and 112’ litters. Boars from the ‘22’ and ‘92’ litters had only WC allele, while boars from the ‘87’ litter had only ME allele. One of the ‘5’ litter (5L) boar was placed among the ‘22’ litter boars. Two boars (*) with ambiguous origin of X chromosome QTL from the ‘22’ litter were confirmed as having WC allele.
Table 2.
Frequency of genotypes of OGT among the seven litters produced by the 7 F1 sows and 2 WC boars1)
| A_ | AB | B_ | Total | |
|---|---|---|---|---|
| Du×WC litter | - | - | 15 | 15 |
| ME×WC litter | 2 | - | 3 | 5 |
| MI×WC litter | 4 | 3 | 7 | 14 |
| FE×WC litter | 5 | 3 | 4 | 12 |
| ME×WC litter | 1 | 3 | 10 | 14 |
| ME×WC litter | 4 | 2 | 7 | 13 |
| MI×WC litter | 2 | 3 | 8 | 13 |
| Total | 18 | 14 | 54 | 86 |
OGT, O-linked N-acetylglucosamine transferase; WC, White composite; Du, Duroc; ME, Meishan; MI, Minzu; FE, Fengzing.
One WC boar mated with 6 sows and the other boar WC mated with one MI×WC sow in the last column. Genotypes of OGT from Chinese and European pigs (male and female) were designated as A_ and B_, respectively.
Table 3.
Frequency of genotypes of OGT among the Duroc, Landrace, Yorkshire, and Meishan pigs1)
| A_ | B_ | |
|---|---|---|
| Duroc | - | 40 |
| Landrace | - | 36 |
| Yorkshire | - | 36 |
| Meishan | 10 | - |
| Total | 10 | 112 |
OGT, O-linked N-acetylglucosamine transferase.
Genotypes of OGT from the Chinese and European pigs (male and female) at the National institute of Animal Science (RDA, Korea) were designated as A_ and B_, respectively.
Figure 3.
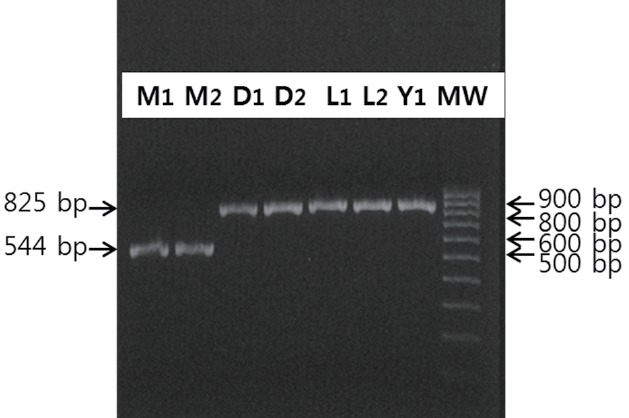
Intron 20 of O-linked N-acetylglucosamine transferase (OGT) from European and Chinese pigs in an independent population at the National Institute of Animal Science (NIAS, RDA, Korea). Fragments containing intron 20 of OGT were compared by amplifying the region containing a part of exon 20, intron 20, and a part of exon 21 of OGT. Polymerase chain reaction amplicons from Meishan (M1, M2) sows were 544-bp, while Duroc (D1, D2), Landrace (L1, L2), and Yorkshire (Y1) sows had only the 825-bp band.
Analysis of inserted element in the OGT polymorphism
The 276-bp element (chrX:64993408–64993683) in the intron 20 of the European pig is inserted after the first 14-bp TGCT TGTTCTTTTT (chrX:64993394–64993407) sequence and the 3′-end of the element ends with the second 14-bp TGCTTGTT CTTTTT (chrX:64993670–64993683) sequence (Figure 1B). In contrast, the Chinese pigs have only one of the 14-bp TGCTT GTTCTTTTT sequence. There are two nearby SNPs on exons 20 and 21. The rs81215029 at chrX:64993310 is located on exon 20 (nucleotide 2967 of cDNA DQ400859 and a.a. 929) and the rs335151398 at chrX:64993905 is located on exon 21 (nucleotide 3048 of cDNA DQ400859 and a.a. 956). Both of these A/G mutations results in synonymous codon. The rs323323004 at chrX: 64993390 is located on intron 20 and this A/G mutation is 4-bp before the first 14-bp TGCTTGTTCTTTTT sequence. Distribution of the SNPs among different pig breeds has not been reported.
DISCUSSION
Sequence analysis of the OGT polymorphism
When the longer 514-bp intron 20, the inserted 276-bp element of OGT from the WC pigs, or the shorter 233-bp intron 20 of Chinese pigs, were analyzed using the algorithm “Basic Local Alignment Search Tool (BLAST)”, there were numerous matches with different homologous regions. For the longer 514-bp intron 20 sequence, numerous homologous regions existed in the pig X chromosome and several other chromosomes including 2, 4, 6, 7, 9, 14, and 16. The homologous regions were mostly matching with the inserted 276-bp element of the intron 20. For the inserted 276-bp element of the intron 20, numerous homologous regions existed in the pig X chromosome and several other pig chromosomes similar to the longer 514-bp intron 20 sequence. The inserted 276-bp element of the intron 20 may contain a repetitive element. For the shorter 233-bp intron 20 sequence, there were only few matching pig and human sequences. The matching pig sequences included pig OGT and a few matching human sequences included human OGT (GenBank accession no. NG_015875) and Homo sapiens transposon SVA sequence (GenBank accession no. AB191243).
The 514-bp intron 20, including the inserted 276-bp element of OGT from the WC pigs, was analyzed by BLAT in the pig genome (http://genome.ucsc.edu/cgi-bin/hgBlat). There were two sequences that were recognized by the RepeatMasker program, which included a (TTG)n simple repeat and a short interspersed nuclear element (SINE). The position of 33-bp (TTG)n simple repeat was chrX:64993397–64993429. It consisted with the 4 to 14 bp of the 14-bp TGCTTGTTCTTTTT sequence and the (TTG)7 repeat sequence. The position of the 240-bp SINE element was chrX:64993430–64993669, following the 33-bp (TTG)7 simple repeat and it belongs to tRNA-Glu family. It is intriguing that the (TTG)7 repeat sequence, followed by the 240-bp tRNA-Glu family SINE element is located between the two 14-bp TGCTT GTTCTTTTT sequences. SINE repeats present near gene promoters may interfere negatively with gene expression [17]. However, there is only one SINE element present in the intron 20 of OGT of the European pig breeds. Twenty-six SINE and 16 long interspersed nuclear elements (LINE) are scattered within the introns of OGT in European pigs (http://genome.ucsc.edu/), while the numbers of SINE and LINE elements were not reported in Chinese pigs.
On the other hand, insertion of the SINE element sequence may affect gene expression or insert a putative constitutive acceptor at chrX:64993713 and alternatively splicing the mRNA before the constitutive acceptor at chrX:64993880, and therefore altering the mRNA sequence with the premature stop codon (Alternative Splice Site Predictor: http://wangcomputing.com/assp/) [18]. Different number of inserted repeat elements may affect OGT gene expression between European and Chinese pig breeds.
Gene expression, evolution, and mapping
When breed-specific X-chromosome regional gene expression differences were determined, the chromosomal band Xq13, which contains 13 genes including OGT and X inactivation-specific transcript (XIST), ranked highest by criteria of placental gene expression and chromosomal location [19]. XIST, a long non-coding RNA that facilitates X-chromosome inactivation and also located on the chromosomal band Xq13, was highly expressed in the WC placenta, but was not detected in ME placenta during fetal development [19]. Since OGT is located on the X-chromosome near the Xist locus suggesting that tight dosage regulation is necessary for normal development [20], breed specific expression of OGT may differentially affect reproductive tissues and alter fetal development during pregnancy in Chinese or European pigs. Bischoff et al [19] reported the expression of OGT in MS and WC placentae using the pigs with the same genetic backgrounds from the same research center as the current report. Expression of OGT was higher in WC than MS pigs. Though the difference in the expression of OGT could be due to the inserted element in the intron 20 of OGT, specific experiments may be needed to ratify the effect of the inserted element. Differential expression of OGT may regulate the GlcNAcylation of proteins differentially in MS and WC placentae. However, expression of O-GlcNAcase (OGA) was not shown. Whether XIST expressed highly in the WC placenta and the presence of inserted element in the intron 20 of OGT in European pigs affected OGT expression and the levels of GlcNAcylated proteins in oviduct and uterus, and thus influenced preimplantation embryonic development needs further investigation.
OGT intron 20 from human and cattle were 240 and 243 bp (http://genome.ucsc.edu/), respectively. Therefore, they are comparable to 233-bp OGT intron 20 from the Chinese pigs, rather than 514-bp intron from the European pigs. The sequence homologies of the 233-bp OGT intron 20 from the Chinese pigs with the human and cattle were 76.3% and 69.5%, respectively. This suggests that the 276-bp element of the OGT intron 20 in European pigs may have been inserted later in evolution, after the speciation of Sus scrofa. The recent phylogenetic analyses of complete genome sequences from unrelated six European and four Asian wild boars, and six domestic pigs, revealed distinct Asian and European lineages that split during the mid-Pleistocene 1.6–0.8 Myr ago [21] and the data further supports our finding that the insertion may have taken place after the split of Asian and European lineages. Alternatively, the insertion of 276-bp element of the OGT intron 20 in European pig breeds may have taken place during the domestication process of these pigs.
In a previous study using ME×WC crossbred boars, it was revealed that genomic region located on chromosome X affects FSH level and testes size [11]. Further, boars with ME alleles at the X chromosome quantitative trait locus (QTL), among which were the first generation of inter se mating of 1/2 ME×1/2 WC parents, had smaller testicles, darker colored parenchyma, and lower total daily sperm production than boars with WC alleles [22]. When 84 boars of 16 litters were analyzed for their breeds of origin from inter se mating of 1/2 ME×1/2 WC parents, the breeds of origin of X chromosome QTL were confirmed, including two boars of ambiguous origin (Figure 2). Using the length polymorphism identified in 7 litters from 2 boars and 7 sows from the USMARC Swine Reference Population [23], OGT was mapped within the QTL for FSH, testes weight, and backfat depth (Figure 4). It requires further studies to determine whether OGT and other genes within the QTL are responsible for the effect.
Figure 4.
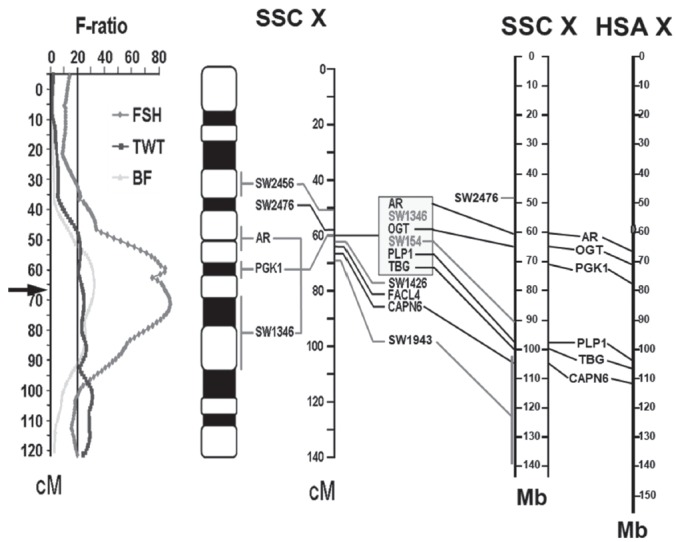
Localization of O-linked N-acetylglucosamine transferase (OGT) on porcine X chromosome. Using the length polymorphism identified in the intron 20 of the OGT gene, OGT was mapped to 67 cM marked by an arrow (→) on the porcine chromosome X within the quantitative trait locus (QTL) for follicle stimulating hormone (FSH), testes weight (TWT), and backfat depth (BF). OGT is located between SW1346 and SW154, which is about 65 Mb on the physical map of SSC X. AR and PGK1 are located near the OGT, and PLP1, TBG, and CAPN6 are located around 100 Mb on SSC X. The corresponding genes are shown on HSA X. Gray lines are pig-specific sequences and black lines indicate human-pig homologues. The gray oval at 60 Mb on HSA X is the centromere. SSC, Sus scrofa chromosome, AR, androgen receptor, PGK1, phosphoglycerate kinase 1, PLP1, proteolipid protein 1, TBG, thyroxine-binding globulin, CAPN6, calpain 6.
We report a polymorphism due to an insertion in the intron 20 of OGT in European pigs, but not in Chinese pigs. There was complete concordance with the insertion in the European pigs at USMARC and NIAS. When 84 boars of 16 litters were analyzed from inter se mating of 1/2 ME×1/2 WC parents, the breeds of origin of X chromosome QTL for FSH, testes weight, and backfat depth were confirmed. This polymorphism could be an additional marker in identifying the breed of origin in sows produced by crossing ME or other Chinese breeds with European pig breeds for their reproductive characteristics and in boars for their X chromosomal region.
ACKNOWLEDGMENTS
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The U.S. Department of Agriculture (USDA) prohibits discrimination in all its programs and activities on the basis of race, color, national origin, age, disability, and where applicable, sex, marital status, familial status, parental status, religion, sexual orientation, genetic information, political beliefs, reprisal, or because all or part of an individual’s income is derived from any public assistance program. (Not all prohibited bases apply to all programs.) Persons with disabilities who require alternative means for communication of program information (Braille, large print, audiotape, etc.) should contact USDA’s TARGET Center at (202) 720-2600 (voice and TDD). To file a complaint of discrimination, write to USDA, Director, Office of Civil Rights, 1400 Independence Avenue, S.W., Washington, D.C. 20250-9410, or call (800) 795-3272 (voice), or (202) 720-6382 (TDD). USDA is an equal opportunity provider and employer.
This paper was supported in part by research funds of Chonbuk National University in 2009 to Kim, Jong Gug.
Footnotes
CONFLICT OF INTEREST
We certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
REFERENCES
- 1.Butkinaree C, Park K, Hart GW. O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta. 2010;1800:96–106. doi: 10.1016/j.bbagen.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bullen JW, Balsbaugh JL, Chanda D, et al. Cross-talk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK) J Biol Chem. 2014;289:10592–606. doi: 10.1074/jbc.M113.523068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartweck LM, Scott CL, Olszewski NE. Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics. 2002;161:1279–91. doi: 10.1093/genetics/161.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dehennaut V, Lefebvre T, Leroy Y, et al. Survey of O-GlcNAc level variations in Xenopus laevis from oogenesis to early development. Glycoconj J. 2009;26:301–11. doi: 10.1007/s10719-008-9166-0. [DOI] [PubMed] [Google Scholar]
- 5.Shafi R, Iyer SP, Ellies LG, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci USA. 2000;97:5735–9. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howerton CL, Morgan CP, Fischer DB, Bale TL. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc Natl Acad Sci USA. 2013;110:5169–74. doi: 10.1073/pnas.1300065110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibutani M, Mori T, Miyano T, Miyake M. Removal of O-GlcNAcylation is important for pig preimplantation development. J Reprod Dev. 2015;61:341–50. doi: 10.1262/jrd.2014-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohrer GA, Ford JJ, Wise TH, Vallet JL, Christenson RK. Identification of quantitative trait loci affecting female reproductive traits in a multigeneration Meishan-White composite swine population. J Anim Sci. 1999;77:1385–91. doi: 10.2527/1999.7761385x. [DOI] [PubMed] [Google Scholar]
- 9.Jiang Z, Robinson JAB, Verrinder Gibbins AM, et al. Mapping of QTLs for prolificacy traits on SSC8 using a candidate gene approach. 7th World Congress on Genetics Applied to Livestock Production; 2002 August 19–23; Montpellier, France. 2002. [Google Scholar]
- 10.King AH, Jiang Z, Gibson JP, Haley CS, Archibald AL. Mapping quantitative trait loci affecting female reproductive traits on porcine chromosome 8. Biol Reprod. 2003;68:2172–9. doi: 10.1095/biolreprod.102.012955. [DOI] [PubMed] [Google Scholar]
- 11.Rohrer GA, Wise TH, Lunstra DD, Ford JJ. Identification of genomic regions controlling plasma FSH concentrations in Meishan-White Composite boars. Physiol Genomics. 2001;6:145–51. doi: 10.1152/physiolgenomics.2001.6.3.145. [DOI] [PubMed] [Google Scholar]
- 12.Kim JG, Vallet JL, Rohrer GA, Christenson RK. Characterization of porcine uterine estrogen sulfotransferase. Domest Anim Endocrinol. 2002;23:493–506. doi: 10.1016/s0739-7240(02)00172-8. [DOI] [PubMed] [Google Scholar]
- 13.Kim JG, Vallet JL, Christenson RK. Molecular cloning and endometrial expression of porcine amphiregulin. Mol Reprod Dev. 2003;65:366–72. doi: 10.1002/mrd.10314. [DOI] [PubMed] [Google Scholar]
- 14.Thorson JF, Desaulniers AT, Lee C, et al. The role of RFamide-related peptide 3 (RFRP3) in regulation of the neuroendocrine reproductive and growth axes of the boar. Anim Reprod Sci. 2015;159:60–5. doi: 10.1016/j.anireprosci.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Kim JG, Ford JJ, Rohrer GA, Nonneman DJ. Molecular cloning of porcine OGT cDNA and mapping to the X chromosome. Biology of Reproduction, Society for the Study of Reproduction Meeting. 2006;(Supplement):154. [Google Scholar]
- 16.Nam YS, Kim DW, Kim MJ, Cho KH, Kim JG. Length polymorphism in OGT between Korean native pig, Chinese Meishan, and the Western pig breeds. J Anim Sci Technol. 2015;57:12. doi: 10.1186/s40781-015-0045-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Estecio MR, Gallegos J, Dekmezian M, et al. SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters. Mol Cancer Res. 2012;10:1332–42. doi: 10.1158/1541-7786.MCR-12-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang M, Marin A. Characterization and prediction of alternative splice sites. Gene. 2006;366:219–27. doi: 10.1016/j.gene.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 19.Bischoff SR, Tsai SQ, Hardison NE, et al. Differences in X-chromosome transcriptional activity and cholesterol metabolism between placentae from swine breeds from Asian and Western origins. PLoS One. 2013;8:e55345. doi: 10.1371/journal.pone.0055345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olivier-Van Stichelen S, Abramowitz LK, Hanover JA. X marks the spot: does it matter that O-GlcNAc transferase is an X-linked gene? Biochem Biophys Res Commun. 2014;453:201–7. doi: 10.1016/j.bbrc.2014.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groenen MA, Archibald AL, Uenishi H, et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393–8. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ford JJ, Wise TH, Lunstra DD, Rohrer GA. Interrelationships of porcine X and Y chromosomes with pituitary gonadotropins and testicular size. Biol Reprod. 2001;65:906–12. doi: 10.1095/biolreprod65.3.906. [DOI] [PubMed] [Google Scholar]
- 23.Rohrer GA, Alexander LJ, Keele JW, Smith TP, Beattie CW. A microsatellite linkage map of the porcine genome. Genetics. 1994;136:231–45. doi: 10.1093/genetics/136.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]


