Abstract
Cell‐autonomous circadian oscillations strongly influence tissue physiology and pathophysiology of peripheral organs including the heart, in which the circadian clock is known to determine cardiac metabolism and the outcome of for instance ischemic stress. Human pluripotent stem cells represent a powerful tool to study developmental processes in vitro, but the extent to which human embryonic stem (ES) cell‐derived cardiomyocytes establish circadian rhythmicity in the absence of a systemic context is unknown. Here we demonstrate that while undifferentiated human ES cells do not possess an intrinsic functional clock, oscillatory expression of known core clock genes emerges spontaneously during directed cardiac differentiation. We identify a set of clock‐controlled output genes that contain an oscillatory network of stress‐related transcripts. Furthermore, we demonstrate that this network results in a time‐dependent functional response to doxorubicin, a frequently used anti‐cancer drug with known cardiotoxic side effects. Taken together, our data provide a framework from which the effect of oscillatory gene expression on cardiomyocyte physiology can be modeled in vitro, and demonstrate the influence of a functional clock on experimental outcome.
Keywords: cardiomyocytes, circadian rhythms, human embryonic stem cells
Subject Categories: Development & Differentiation, Stem Cells
Introduction
The circadian clock is a conserved time‐keeping system that regulates numerous body features such as behavior, metabolism, body temperature, tissue regeneration, and organ homeostasis in a diurnal manner 1. In the heart, the role of 24‐h rhythmicity is illustrated by oscillations in heart rate, blood pressure, and cardiac output 2, 3, 4, 5, 6. The circadian clock comprises a central clock in the brain (the suprachiasmatic nucleus (SCN)) and peripheral clocks that are present in almost all organs. The SCN is mainly entrained by light and subsequently synchronizes the peripheral clocks via neural and humoral factors 7. Interestingly, peripheral clocks function in a cell‐autonomous manner. When ablating the SCN, these clocks remain functional and even synchronized when subjected to a restricted feeding regime 8, 9. Autonomous rhythmicity is underscored by the persistence of circadian rhythms in in vitro cultured cells.
The molecular mechanism that underlies the core clock machinery consists of a transcriptional/translational feedback loop in which a heterodimer of BMAL1 and CLOCK drives rhythmic transcription of downstream genes. These include other core clock genes (period 1 (PER1), PER2, PER3, cryptochrome 1 (CRY1), CRY2, RORα/β, REV‐ERBα/β) as well as clock‐controlled genes (CCGs) that determine circadian organ physiology in a tissue‐specific manner. In the murine heart, ~6–12% of the expressed genes have a circadian expression pattern 10, 11, 12, 13, 14. Rhythmicity is essential for human tissue homeostasis as well, as highlighted by the fact that genetic or environmental (e.g., shift‐work) perturbation of the circadian clock results in a vast array of malignancies such as sleep disorders, inflammation, cancer 15, impairment of regenerative capacity 16, 17, metabolic disorders 18, 19, 20, and cardiovascular diseases 13, 21, 22, 23, 24. In addition, the onset of multiple malicious cardiac events is known to follow a diurnal pattern. Myocardial infarction 25, 26, arrhythmias 27, 28, and sudden cardiac death 29, 30 show a higher incidence in the sleep‐to‐wake transition in humans. The important role of circadian rhythmicity in cardiac injury and regeneration is further solidified by genetic experiments in mice in which a cardiomyocyte‐specific mutation of the Clock gene has been shown to blunt the heart's response to induced ischemic damage 31. Accordingly, clinical studies revealed that infarcts were larger and led to increased reduction in cardiac function when occurring in the sleep‐to‐wake transition 32, 33, 34.
Human pluripotent stem cell‐derived cardiomyocytes have emerged as a potential cellular source for replacement therapies. In addition, human ES cell‐derived as well as induced pluripotent stem cell‐derived cardiomyocytes are increasingly used for disease modeling and drug testing 35. While circadian rhythms play an essential role in cardiomyocyte function in vivo, nothing is known about circadian control of gene expression in pluripotent stem cell‐derived cardiomyocytes, which are often used to model cardiac function and disease.
Here we analyze temporal clock gene expression networks in human ES cells and ES cell‐derived cardiomyocytes. We demonstrate that circadian rhythmicity is absent in human ES cells and is established progressively during directed cardiac differentiation. The identified oscillatory networks are shown to significantly influence the function of human ES cell‐derived cardiomyocytes and determine their response to externally applied stressors. Our findings underscore that circadian rhythmicity can affect experimental outcome, which may have important ramifications for processes such as timed cell‐based therapy.
Results
Human embryonic stem cells express clock genes in a non‐oscillatory manner
Nearly all cells in the human body possess a functional clock as indicated by circadian rhythmicity of core clock gene expression. However, whether human embryonic stem (ES) cells display a functional circadian clock is unknown. Therefore, we compared global expression levels of six core clock genes ARNTL (coding for and henceforth referred to as BMAL1), PER2, CRY1, CRY2, CLOCK, and NR1D1 between pluripotent human ES cells and differentiated human osteosarcoma U2OS cells, a cell line known to possess a functional clock as confirmed here by anti‐phasic Bmal1‐dLuc and Per2‐dLuc signal after transduction of promoter‐based lentiviral luciferase constructs 36, 37 (Fig 1A and B). Transcripts of all genes could be detected in both cell types, with five out of six core clock genes having higher expression levels in U2OS cells compared to human ES cells (Student's t‐test, P < 0.05; Fig 1C). Only CRY1 showed a trend toward higher expression in human ES cells (Student's t‐test, P = 0.0506; Fig 1C), which is in line with previously reported observations in mouse ES and NIH3T3 cells 38. Protein levels of BMAL1, CRY1, and CLOCK were detected by Western blot (Fig 1D, left and Fig EV1) at corresponding levels to their mRNA transcripts (Fig 1D, right). From this, we conclude that while core clock genes are expressed and translated into proteins in human ES cells, this occurs with a different stoichiometry in comparison with U2OS cells and most likely also other differentiated cell types.
Figure 1. Non‐oscillatory expression of clock genes in human ES cells.
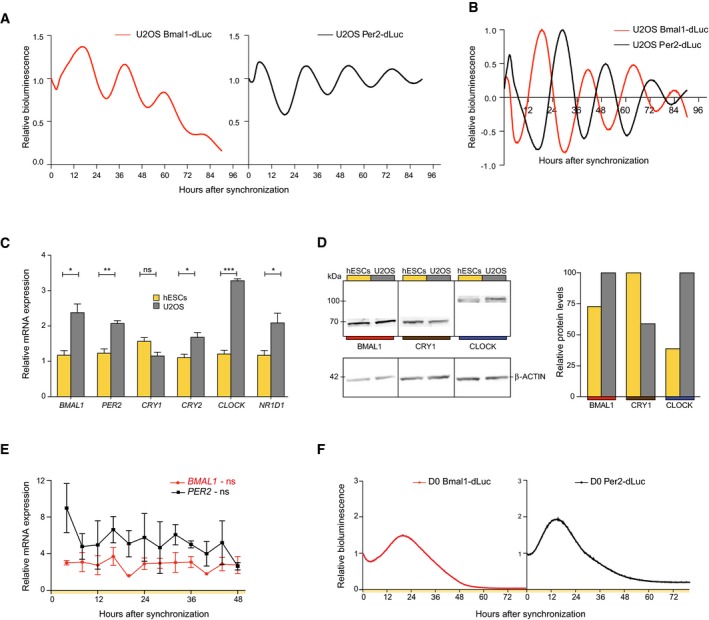
- Raw lentiviral promoter‐based luciferase reporter bioluminescence in U2OS cells after dexamethasone synchronization. Bioluminescence was measured with a LumiCycle32. Values are relative to T0.
- Detrended bioluminescent signals measured in (A).
- BMAL1, PER2, CRY1, CRY2, CLOCK, and NR1D1 expression levels in human ES cells compared to U2OS cells as determined by qRT–PCR. Expression levels were normalized to PPIA and compared between cell types using an unpaired two‐tailed Student's t‐test (ns: not significant, *P < 0.05, **P < 0.005, and ***P < 0.0005). Data are represented as mean ± s.e.m. of three independent replicates.
- Western blot for BMAL1, CRY1, and CLOCK. Protein levels were quantified and normalized to β‐ACTIN.
- qRT–PCR analysis of BMAL1 and PER2 expression over 48 h at a 4‐h interval in human ES cells. Circadian oscillations were analyzed using the RAIN algorithm, and the significance of rhythmicity across 48 h is indicated (ns: not significant). Data are represented as mean ± s.e.m. of three independent replicates.
- Bmal1‐dLuc and Per2‐dLuc values in synchronized human ES cells across 76 h measured by LumiCycle32. Representative tracks are shown. Values are relative to T0.
Figure EV1. Protein levels of clock genes in human ES cells versus U2OS cells.
Original Western blots for BMAL1, CRY1, and CLOCK protein.
The presence of core clock proteins in human ES cells led us to investigate their possible rhythmic expression over time. To this end, human ES cells were synchronized with forskolin 39, and BMAL1 and PER2 mRNA levels were measured every 4 h over a period of 48 h using qRT–PCR. Significance of 24‐h rhythmicity was assessed using RAIN, a nonparametric method detecting arbitrary wave forms in biological data 40. No apparent oscillatory expression pattern could be identified over the course of 2 days (RAIN, BMAL1, P > 0.99 and PER2, P = 0.97; Fig 1E). Additionally, to assess BMAL1 and PER2 transcription over time in human ES cells, we transduced human ES cells with Bmal1‐ and Per2‐promoter‐based lentiviral luciferase constructs 36, 37. After synchronization, no rhythmic bioluminescence was observed (Fig 1F). Therefore, clock genes are expressed in human ES cells, but in a non‐circadian manner.
Human embryonic stem cell differentiation toward cardiomyocytes
Multi‐lineage differentiation of human ES cells has proven extremely valuable to understand developmental processes as well as to provide clinically relevant populations for cell‐based therapy and drug testing 41. To assess the presence of a functional clock upon differentiation, circadian rhythmicity was analyzed at two additional stages (D15 and D30) during directed differentiation of human ES cells toward cardiomyocytes (Fig 2A). To allow for the identification of early cardiac cells, we made use of a NKX2‐5‐eGFP (Homeobox protein NKX2‐5‐eGFP) reporter human ES cell line 42. Cardiac differentiation of human ES cells typically yields significant contribution of cardiomyocytes to the total population of cells 43, 44, 45, which was also seen here with ~50% cardiomyocytes around D15 as defined by FACS for cTNNT2 (cardiac Troponin T) (Fig EV2A). Different stages were characterized by clear changes in marker gene expression (Fig 2A). At day 0 (D0), cells expressed the pluripotency markers NANOG and POU5F1, both at the mRNA and protein level (Figs 2A and B, and EV2B). Upon differentiation, pluripotency factors quickly decreased and the expression of cardiac markers, such as NKX2‐5 and ACTC1 (actin alpha cardiac muscle 1), was observed in both (D15 and D30) spontaneously beating cultures as measured by qRT–PCR (Fig 2A). Immunostainings for α‐sarcomeric ACTIN and cTNNT2 confirmed sarcomeric structures at D15 and D30 (Fig 2B). Additionally, staining for MEF2C (Myocyte Enhancer Factor 2C) and GFP, to assess the presence of NKX2‐5‐eGFP‐positive cells, revealed the abundance of cardiomyocytes at D15 and D30. While the early cardiomyocyte progenitor marker ISL1 (insulin gene enhancer protein ISL‐1) was highly expressed at D15, maturation markers such as KCNJ2 (inward rectifier potassium channel 2) and SERCA2A (sarcoplasmic/endoplasmic reticulum Ca2+ ATPase) 44, 46 were the highest at D30 (Fig 2A). These results validate the in vitro transcriptomic maturation of these cells between D15 and D30, and confirm that the different stages represent distinct cardiac states that can be used to assess the presence of a functional clock across the transition from human ES cells to cardiomyocytes.
Figure 2. Characterization of distinct stages of cardiac differentiation.
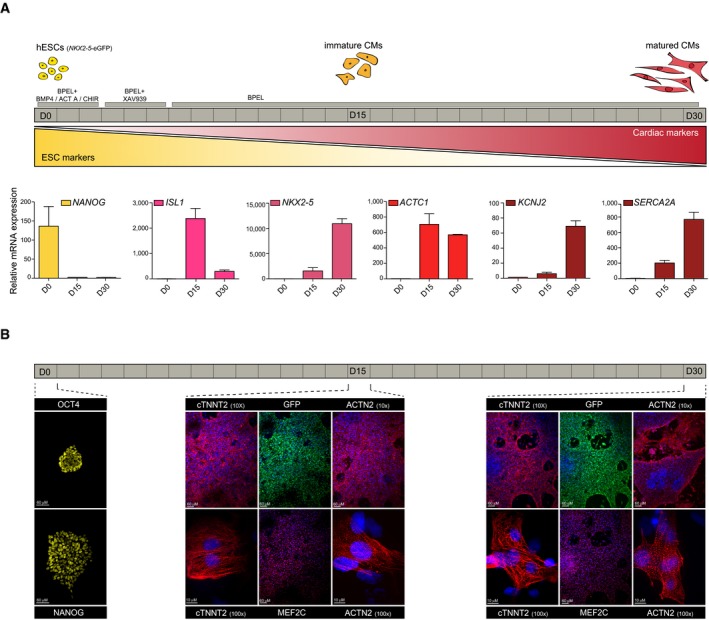
- Schematic of the directed cardiac differentiation and the three different stages used in this study. hESCs: human embryonic stem cells. CM: cardiomyocyte. Below are mRNA expression levels of pluripotency and cardiac markers as measured by qRT–PCR. Expression levels were normalized to a non‐oscillatory housekeeping gene (PPIA). Data are represented as mean ± s.e.m. of three independent replicates.
- Immunostaining for pluripotency markers OCT4 and NANOG (yellow) in human ES cells. cTNNT2 and sarcomeric actin (red) stainings reveal clear sarcomeric structures at all cardiac stages. Cardiomyocyte nuclei were stained for MEF2C (red), NKX2‐5‐eGFP‐positive cells were stained with anti‐GFP (green) and nuclei with Hoechst (blue).
Figure EV2. Cardiomyocyte quantification and characterization by FACS and qRT–PCR.
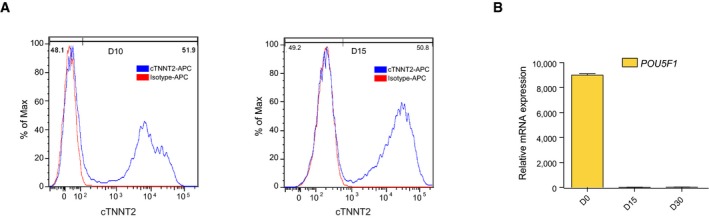
- FACS staining for cTNNT2 in D10 and D15 human ES cell‐derived cardiac cells. IgG1 isotype was used as a control.
- POU5F1 mRNA expression levels in human ES cells and D15 as well as D30 cardiac cells, as measured by qRT–PCR. Expression levels were normalized to a non‐oscillatory housekeeping gene (PPIA). Data are represented as mean ± s.e.m. of three independent replicates.
Rhythmic expression of clock genes emerges during cardiac differentiation
Whether and when human ES cells develop a functional clock upon differentiation, in the absence of systemic cues, is unknown. To investigate the possibility of an emerging clock, we compared mRNA levels of BMAL1, PER2, and CLOCK in D0 human ES cells and D15 and D30 human ES cell‐derived cardiac cells. Even though BMAL1, PER2, and CLOCK were expressed at all stages, their expression increased significantly from D0 to D15 and/or D30 (ANOVA with Bonferroni correction, P < 0.05; Fig 3A). This indicates that core clock gene expression levels gradually increase during directed cardiac differentiation.
Figure 3. Rhythmic clock gene expression emerges in (matured) cardiac cultures.
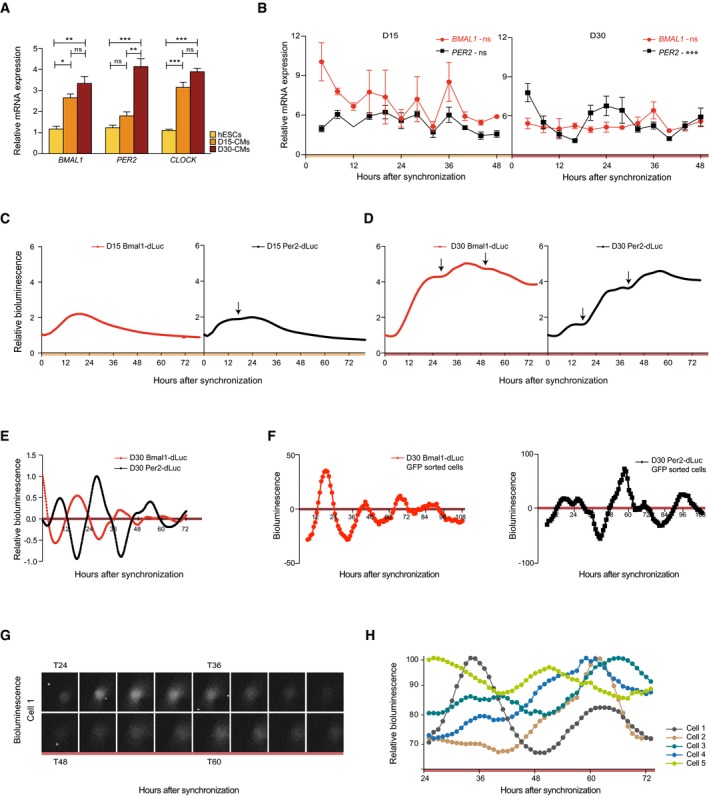
- BMAL1, PER2, and CLOCK expression levels at D0, D15, and D30 during directed cardiac differentiation as determined using qRT–PCR. Data are represented as mean ± s.e.m. of three independent replicates. Significant expression differences were tested by one‐way ANOVA, followed by a Bonferroni post hoc test (ns: not significant, *P < 0.05, **P < 0.005, ***P < 0.0005).
- qRT–PCR analysis of BMAL1 and PER2 expression over 48 h at a 4‐h interval in cardiac cells at D15. Expression levels in (A) and (B) were normalized to PPIA. Data are represented as mean ± s.e.m. of three independent replicates. Significance of rhythmicity across 48 h was analyzed using the RAIN algorithm and is indicated (ns: not significant, ***P < 0.0005).
- Promoter‐based destabilized luciferase (dLuc) reporter assay of the Bmal1 and Per2 promoter in synchronized cardiac cells at D15. Values are relative to T0. Measurements were performed using a LumiCycle32.
- Similar analysis as in (C) for D30.
- Detrended Bmal1‐dLuc and Per2‐dLuc luciferase signal measured in (D).
- Detrended Bmal1‐dLuc and Per2‐dLuc bioluminescence in NKX2‐5‐eGFP+ sorted and synchronized human ES cell‐derived cardiomyocytes at D30.
- Single‐cell analysis of Per2‐dLuc bioluminescence in sorted eGFP‐positive and synchronized D30 human ES cell‐derived cardiomyocytes.
- Representative Per2‐dLuc signal in five single D30 human ES cell‐derived cardiomyocytes over the course of 48 h.
To assess rhythmicity of clock gene expression at D15 and D30, cells were synchronized using dexamethasone 47 and three independent RNA samples were collected every 4 h over a period of 48 h (Fig EV3). BMAL1 and PER2 levels were analyzed by qRT–PCR to determine whether their expression oscillated in an anti‐phasic manner, a hallmark of a functional molecular circadian clock. Similar to undifferentiated human ES cells (Fig 1E), no clear circadian pattern was observed in the early stage D15 human ES cell‐derived cardiomyocytes (D15; RAIN, P = 0.095, P = 0.68 for BMAL1 and PER2, respectively; Fig 3B). Matured cardiac cells, however, showed significant oscillations for PER2 but not BMAL1 (D30; RAIN, P = 1.4E‐8 and P = 0.81; Fig 3B). To further validate the emergence of a functional clock, cultures were transduced with Bmal1‐dLuc and Per2‐dLuc lentiviral reporters. At D15, after synchronization, a small induction of oscillatory Per2‐based luciferase signal could be detected (Fig 3C), which is in line with previously described observations of Per2 as an early oscillator upon retinoic acid induced differentiation in mouse ES cells 48. In D30 synchronized populations, typical anti‐phasic oscillatory Per2‐ and Bmal1‐driven bioluminescence levels were observed, which confirms the presence of a clock at D30 (Figs 3D and E, and EV4A and B).
Figure EV3. Setup of CEL‐Seq experiment.
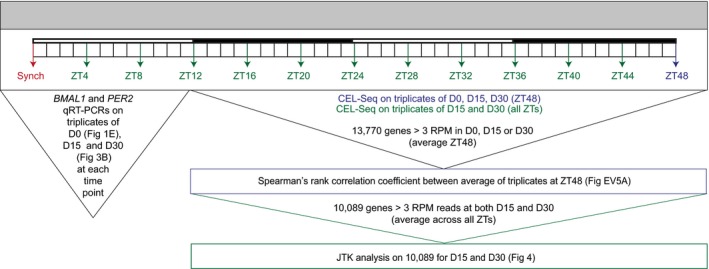
Schematic of RNA samples that were processed for qRT–PCR and/or CEL‐Seq. ZT: Zeitgeber and RPM: reads per million.
Figure EV4. Human ES cell‐derived cardiomyocytes show circadian oscillation in Bmal1‐ and Per2‐dLuc.
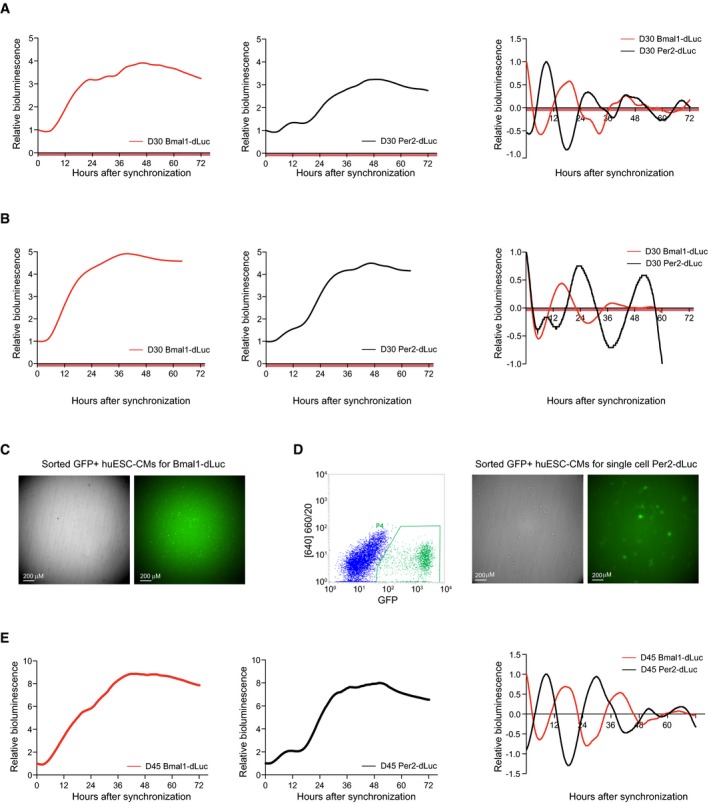
- Promoter‐based destabilized luciferase (dLuc) reporter assay of the Bmal1 (red) and Per2 (black) promoter in synchronized human ES cell‐derived cardiac cells at D30 (replicate for Fig 3D). Values are relative to T0. Right: Detrended Bmal1‐dLuc and Per2‐dLuc luciferase signal. Measurements were performed using a LumiCycle32.
- Similar analysis as in (A) on an additional independent replicate.
- Brightfield and fluorescent image of D30 GFP+ sorted human ES cell‐derived cardiomyocytes used for analysis in Fig 3F (left/red).
- Similar analysis as in (A) for D45 human ES cell‐derived cardiac cells.
In order to verify the contribution of cardiomyocytes to the observed oscillatory pattern in our cardiac cultures, NKX2‐5‐eGFP+ cells were purified via FACS. After sorting, strong Bmal1‐dLuc and Per2‐dLuc rhythmicity was detected in synchronized human ES cell‐derived cardiomyocytes (Figs 3F and EV4C and D). In addition to these observations in a sorted population, circadian Per2‐dLuc patterns were also found in bioluminescence recordings of single NKX2‐5‐eGFP+ human ES‐derived cardiomyocytes (Figs 3G and H, and EV4D), which further confirms the presence of a functional clock in D30 cardiomyocytes. To question whether circadian rhythmicity would persist during culture, 45 days old cardiac cultures were analyzed using real‐time reporter‐based luciferase measurements. Significant Per2‐ and Bmal1‐dLuc oscillations were observed (Fig EV4E). These results indicate that human ES cells develop a functional clock upon directed cardiac differentiation, with robust oscillations at D30 that persist in older in vitro cultures.
Human ES cell‐derived cardiomyocytes show a network of stress‐related clock output genes
A functional circadian clock translates into the oscillatory expression of clock‐controlled genes (CCGs). Gene expression profiling in numerous cell types and tissues has shown that around 3–16% of the transcriptome exhibits circadian rhythmicity 14. Conform differing physiological demands of organs, oscillating output genes vary per tissue. For the murine heart, ~6–12% of the expressed genes were shown to oscillate in a 24‐h manner 10, 11, 12, 13, 14. To identify CCGs during in vitro cardiomyocyte differentiation, genome‐wide mRNA levels were assessed by mRNA sequencing of purified RNA using CEL‐Seq, a previously described RNA profiling technique based on sequencing the 3′UTR of mRNAs, generating one read per transcript 49. We first compared the overall transcriptional profile of matured cardiac cells (D30) to that of human ES cells and D15 cultures 48 h post‐synchronization (Fig EV5A and Table EV1). Lowly expressed genes are typically not picked up robustly when analyzing highly multiplexed CEL‐Seq data at relatively low sequencing depth. To control for this, genes with an average of < 3 RPM (reads per million) across all time points were not used for further analysis. Based on ~14,000 genes with an expression level of more than 3 RPM at one of the stages, Spearman's rank correlation coefficients (ρ) showed that transcriptional programs were substantially different between cardiac cells at D15 and D30 (ρ = 0.53; Fig EV5A–C). Observed changes between states were consistent between our qRT–PCR and CEL‐Seq analyses, as indicated for several marker genes, which highlights the reliability of our sequencing datasets (Figs 2A and EV5D). Indeed, increased MYH7/MYH6 levels and multiple other markers (e.g., MYL2, PLN, and KCNJ2) confirm in vitro cardiomyocyte maturation as well as generally higher clock gene expression across differentiation (Fig EV5C and D).
Figure EV5. CEL‐Seq‐based characterization of transcriptome maturation and association between read coverage and oscillatory expression.
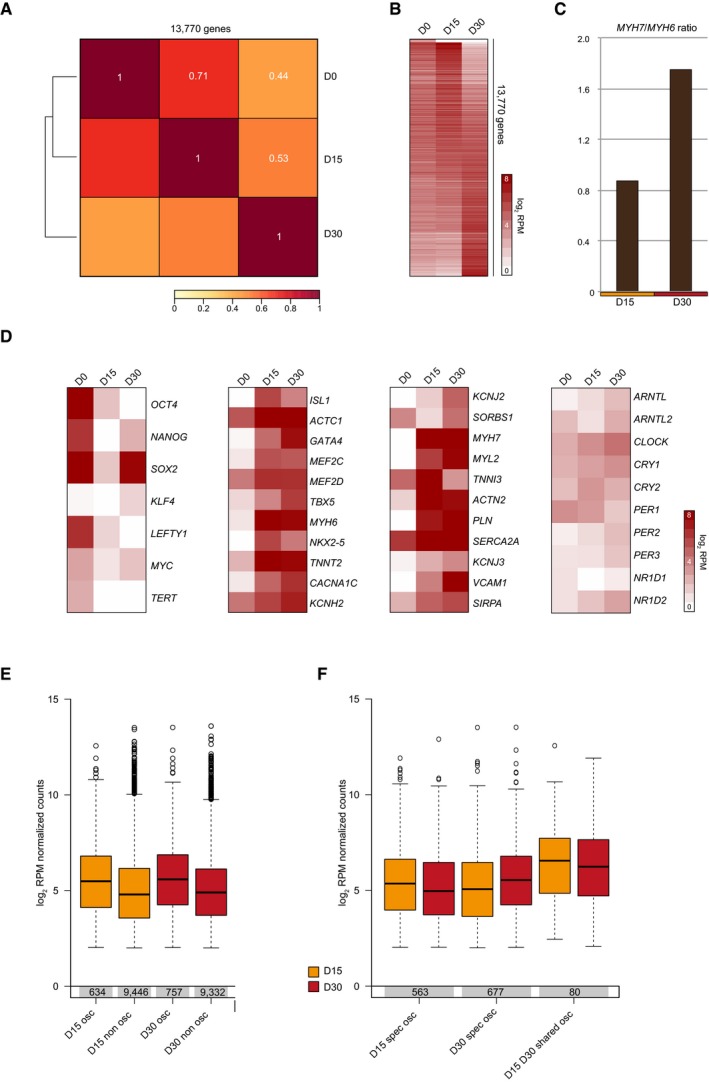
- Spearman's rank correlation with average linkage based on average RPM‐normalized CEL‐Seq read counts of three ZT48 replicates. Genes (n = 13,770) with an average ZT48 expression level of more than 3 RPM (reads per million) at one stage (either D0, D15, or D30) were used for analysis. Spearman's rank correlation coefficients are indicated in white.
- Heatmap of average ZT48 CEL‐Seq read counts for 13,770 genes used in (A). Genes were sorted according to human ES cell expression from high (top) to low (bottom). Log2 RPM values were plotted.
- MYH7/MYH6 CEL‐Seq read count ratio in D15 and D30 human ES cell‐derived cardiac cultures (average of three ZT48 replicates) as a measure for maturation.
- Heatmaps of average ZT48 CEL‐Seq read counts in human ES cells (D0) and human ES cell‐derived cardiac cells (D15 and D30). The left heatmap shows pluripotency markers, the two middle heatmaps depict cardiomyocyte and cardiomyocyte maturation markers, and the right panel comprises core circadian clock genes. Log2 RPM values were plotted.
- Boxplots depict average log2 RPM‐normalized CEL‐Seq counts across all time points for D15 and D30 oscillatory and non‐oscillatory genes. 10,089 genes with average RPM values above 3 across all time points (ZT4‐ZT48) on which JTK analysis was done in both D15 and D30 were used. Bottom and top of the boxes are the first and third quartiles. The line within the boxes represents the median and whiskers denote the interval within 1.5? the interquartile range from the median. Outliers are plotted as dots.
- Boxplots depict average log2 RPM‐normalized CEL‐Seq counts across all time points (ZT4‐ZT48) for D15‐specific, D30‐specific, and shared cardiac oscillators. Boxplot representation is as described in (E).
To assess the possible presence of oscillatory transcripts at D15 and the identity of CCGs in D30 cardiac cells, in which a functional clock was found (Fig 3), three independent RNA samples were collected every 4 h over a period of 48 h and sequenced using CEL‐Seq (Fig EV3 and Table EV1). Around 10,000 genes had an average expression of more than 3 RPM in both D15 and D30 (Fig EV3) and were screened for oscillatory expression over 48 h as determined by JTK‐cycle 50. This revealed 643 and 757 oscillating transcripts (P < 0.05) at D15 and D30, respectively (Fig 4A and Table EV2). The oscillatory transcripts of D15 could result from a starting clock as indicated by small circadian Per2‐dLuc signals at this time point (Fig 3C), but are mostly distinct from the CCGs that were found at D30 (Fig 4B). This limited fraction of overlap might be an underrepresentation, as detecting oscillatory transcription of genes has been shown to rely strongly on sequencing depth 51. Relatively low expression also explains the absence of core clock genes from the rhythmic transcripts (Fig EV5D). Indeed, in our data for both D15 and D30, oscillatory genes had on average more coverage than non‐oscillatory transcripts (Fig EV5E) and shared oscillators between D15 and D30 (n = 80) had higher expression levels than stage‐specific oscillators (Fig EV5F). A fraction of the oscillators (D15 only, D30 only and shared) was found to overlap known mouse cardiac CCGs 14 (Fig 4C) including genes with a known important role in cardiomyocytes (COL4A1, SPON2, SLC23A2, AQP1, and STC1; Fig 4D) 52, 53, 54, 55. These data thus contain common rhythmically expressed clock‐controlled genes between mouse hearts and human ES cell‐derived cardiomyocytes.
Figure 4. Identification of oscillatory transcripts at D15 and D30.
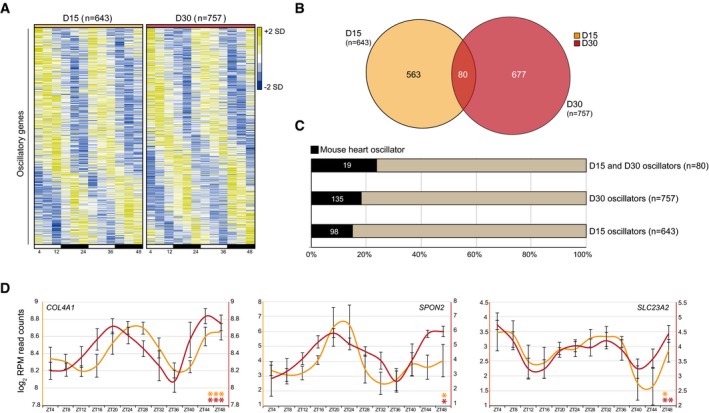
- 643 and 757 oscillators for D15 and D30 cultures, as analyzed using JTK‐cycle (adj. P < 0.05). Heatmaps represent z‐normalized RPM values of the average of three independent replicates. Oscillatory genes were ranked by their phase of expression and visualized using Java TreeView.
- Venn diagram of JTK‐cycle detected oscillators for D15 and D30.
- Fraction of JTK‐cycle detected oscillators that were previously found to be rhythmically expressed in mouse hearts 14.
- Examples of overlapping oscillators between D15, D30 cardiac cells and mouse hearts. Average log2 RPM read counts of three replicates, smoothened over 2 time points ± s.e.m., were plotted. Significance of rhythmicity across 48 h was analyzed using the JTK‐cycle algorithm (* JTK P < 0.05, ** JTK P < 0.005, and *** JTK P < 0.0005).
STRING protein–protein analysis 56 on D30 oscillators that were also found to have circadian expression in mouse hearts (n = 135; Table EV2) revealed a putative, highly interactive network (Fig 5A). D15 rhythmic transcripts that overlap mouse heart oscillators (n = 98), however, did not show such interactions (Fig 5B). Gene ontology (GO) analysis for these oscillators showed enrichment for extracellular matrix formation terms at D15, while D30 oscillators were enriched for cardiac development and stress response terms (Fig 5A and B). Interestingly, the D30 interaction network was centered around UBC (ubiquitin C) (Fig 5A), one of the four genes encoding for ubiquitin in mammals and one of the most abundant proteins in eukaryotic cells 57. Although Ubc is expressed in multiple tissues in mice (http://biogps.org/) 58, it has only been shown to oscillate in the murine heart 52 and (skeletal) muscle 14 (JTK, P = 3.32E‐6 and P = 5.97E‐7, respectively; Fig 5C). This suggests that Ubc is a heart‐ and muscle‐specific CCG in vivo, and concurs with our identification of UBC as a circadian CCG in in vitro D30 human ES cell‐derived cardiomyocytes (D30, JTK, P = 0.0032; Fig 5D). Among the putative UBC interacting partners, several genes were known oscillators in the murine heart according to the CircaDB database (http://circadb.hogeneschlab.org/) 11, 14, 59. Interestingly, many of the oscillating UBC interaction partners in D30 human ES cell‐derived cardiomyocytes were involved in cardiac function (PLN) 60, stress response (BNIP3, RRAGA, DNAJA1 and HSPH1) 61, hypertrophy (RGS2) 62, and even contained therapeutic targets such as TSPO (Translocator protein) 63 (Fig 5D). This indicates that the oscillators that were identified here possibly contribute to multiple molecular mechanisms with a circadian clock dependency, but could also suggest a role for circadian processes in pathophysiological events such as ischemic damage after myocardial infarction.
Figure 5. Circadian stress network results in time‐dependent apoptotic response of human ES cell‐derived cardiac cultures.
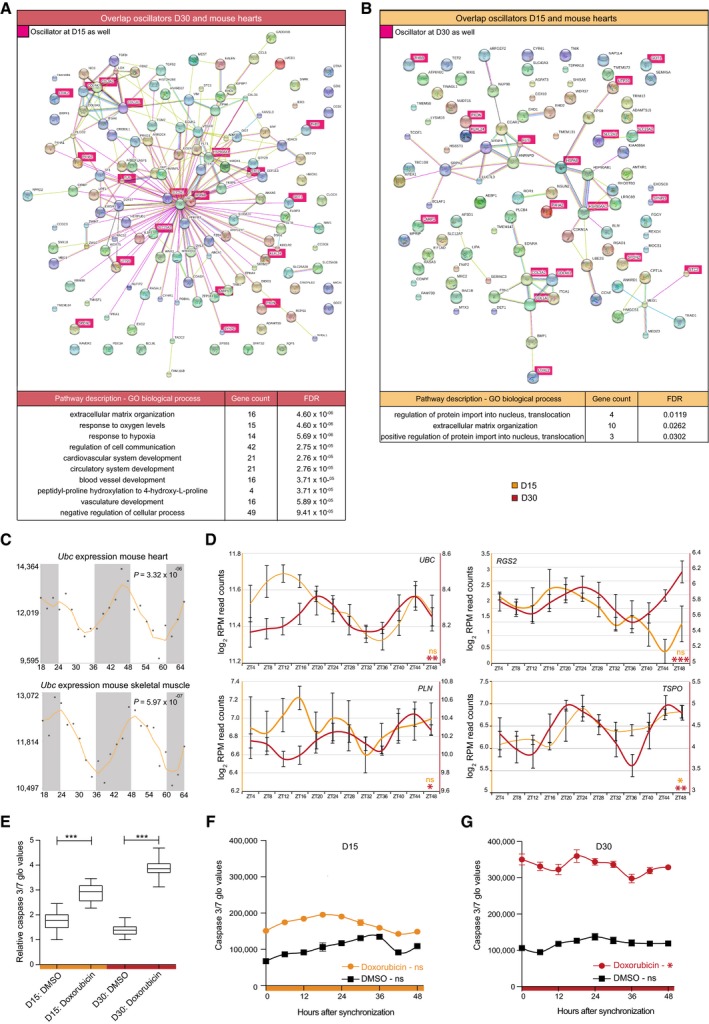
- STRING interaction network of overlapping oscillators between D30 cardiac cells and mouse hearts with corresponding GO analysis. Genes that oscillate at both D15 and D30 are depicted in pink. Mouse heart oscillators were deducted from Zhang et al 14.
- Same analysis as in (A) for D15 oscillators.
- Ubc mRNA oscillation in mouse hearts and skeletal muscle. Data were obtained from CircaDB (http://circadb.hogeneschlab.org/, deducted from 14, 83). Corresponding CircaDB P‐values are indicated.
- Expression levels of four D30 oscillatory genes (UBC, RGS2, PLN, and TSPO) across 48 h. Average log2 RPM CEL‐Seq counts of three replicates were plotted for D15 and D30 and smoothened over two time points ± s.e.m. Significance of rhythmicity across 48 h was analyzed using the JTK‐cycle algorithm (ns: not significant; * JTK P < 0.05, ** JTK P < 0.005, and *** JTK P < 0.0005).
- Apoptosis, as measured by caspase 3/7 activity, after doxorubicin administration in D15 and D30 human ES cell‐derived cardiac cells across all samples from (F) and (G). Bottom and top of the boxes are the 25th and 75th percentiles. The line within the boxes represents the median and whiskers denote the minimum and maximum values. The effect of doxorubicin versus DMSO was tested using a Mann–Whitney U‐test (***P < 0.0005).
- Apoptosis measured with 6‐h intervals in synchronized D15 cultures after administration of doxorubicin (orange) and DMSO as control (black) for 6 h. Data are represented as the mean ± s.e.m. of three independent replicates. Significance of rhythmicity across 48 h was analyzed using the RAIN algorithm and is indicated (ns: not significant).
- Same as in (F) for D30 cultures with the doxorubicin response depicted in red (RAIN, ns: not significant and *P < 0.05). Data are represented as the mean ± s.e.m. of three independent replicates.
Human ES cell‐derived cardiomyocytes show rhythmicity in doxorubicin‐induced apoptosis
Mouse hearts show circadian rhythmicity in their tolerance to ischemia and reperfusion after myocardial infarction 31. In humans, a similar time of the day pattern in the onset and severity of myocardial infarction has been described 25, 32, 33, 34, 64, 65. The combination of time‐dependent pathophysiology and the enrichment of oscillating stress‐associated genes (around UBC) in our CEL‐Seq datasets prompted us to assess whether in vitro derived cardiac cells would show a functional circadian reaction to induced stress. The anthracycline doxorubicin is a widely used anti‐cancer drug that is often administered in the clinic, but is also known to have severe cardiotoxic side effects 66, 67. These effects are recapitulated in in vitro human ES cell‐derived cardiomyocytes in which doxorubicin is known to induce apoptosis and has proven to be a good model for induced cardiotoxicity 68, 69. To determine whether the sensitivity of human ES cell‐derived cardiac cells to doxorubicin‐induced apoptosis displays an oscillatory response, we synchronized cultures at D15 and D30 and administered doxorubicin (10 μM) every 6 h over the course of 48 h (see Materials and Methods). A marked induction of apoptosis was found at both stages, as indicated by elevated active caspase 3/7 levels over control DMSO‐treated samples (Mann–Whitney U‐test, P < 0.0005 for D15 and D30; Fig 5E), with matured D30 cells being more sensitive to doxorubicin than D15 cultures (P < 0.05). Interestingly, the strength of the apoptotic response demonstrated a significant circadian pattern at D30, but not D15 (RAIN, P < 0.05 and P = 0.85 for D30 and D15, respectively; Fig 5F and G), which reveals the functional consequences of a circadian clock in cardiomyocytes. These results highlight the potential of reducing cardiotoxic side effects by the use of time‐based cancer therapy, but also indicate that taking diurnal rhythmicity into account could possibly improve other treatment strategies.
Discussion
Circadian rhythmicity is crucial to heart function, but also influences pathophysiology as indicated by, for instance, diurnal rhythmicity of cardiac damage after infarction 25, 31, 32, 33, 34, 64, 65. As human ES cell‐derived cardiomyocytes are emerging as a powerful tool to model developmental and disease processes as well as being a potential cellular source for regenerative therapies, we examined the presence and possible implications of a functional clock in human ES cells and their cardiac derivatives. While human ES cells do express core clock genes, no circadian clock was observed. Upon differentiation toward cardiomyocytes however, a functional core clock pathway was gradually established (Fig 6) as determined by robust anti‐phasic oscillations of BMAL1 and PER2. This work is the first demonstration of a functional clock in human ES cell‐derived (cardiac) cells and may serve as a paradigm for the emergence of diurnal rhythms in other human pluripotent stem cell‐derived cell types. At D30, 757 CCGs were identified, 18% of which are known to oscillate in the murine heart. Importantly, our data uncover additional transcripts with specific oscillatory behavior in human ES cell‐derived cardiomyocytes. As some of these newly identified oscillators are known to play an important role in human heart physiology (PLN, KCNE4, TSPO, CAV1, RGS2), this stresses the importance of using human cells for modeling cardiovascular processes and disease.
Figure 6. Schematic representation of the emergence and consequences of circadian rhythmicity during directed cardiac differentiation of human ES cells.
hESCs: human ES cells. CMs: cardiomyocytes.
Importantly, a defined set of the oscillators could clearly be linked to stress response, which was confirmed by a time‐dependent response to doxorubicin administration. This highlights the possible beneficial effects of drug administration at a specific time of the day to decrease cardiotoxic side effects. Notably, next to explicit clock synchronization steps, such as with forskolin or dexamethasone, simple medium changes can also reset the internal clock of cell cultures 70. Our results demonstrate that circadian mechanisms can influence cellular response to external stressors and thus is an important factor to consider when interpreting experimental results. Our data stress the importance of testing compounds in a time‐controlled manner when using in vitro cultured cardiomyocytes, and may also extent to other ES cell‐based disease models.
Materials and Methods
ESC culture and cardiomyocyte differentiation
NKX2‐5‐eGFP human ES cells 42 (stable reporter line generated from wild‐type HES3 cells 71) were cultured in Essential 8™ medium (Gibco) on Matrigel (BD, Corning) without penicillin/streptomycin. Cells were differentiated in a monolayer toward cardiomyocytes as previously described 45. In short, human ES cells were cultured in E8 until 60% confluent. Cells were then supplemented with 1% DMSO enriched E8 medium for 24 h. On D0, cells were put on BPEL medium supplemented with Activin A (20 ng/ml, R&D Systems), BMP4 (20 ng/ml, R&D Systems), and CHIR99021 (1.5 μM, Axon Medchem). At D3, medium was changed to BPEL with XAV939 (5 μM, Tocris), and on D6, BPEL without any supplements was used. BPEL medium: IMDM, no phenol red (Gibco) and F12 Ham's F12 nutrient Mix (Gibco) in a 1:1 ratio supplemented with 5% (v/v) PFHM‐II (Gibco), 0.25% (w/v) BSA, 1% (v/v) Chemically Defined Lipid Concentrate (Gibco), 0.1% ITS‐X (Gibco), 450 μM α‐MTG (Sigma), 2 mM GlutaMax, 50 μg/ml L‐ascorbic acid 2‐phosphate (Sigma), and 0.25% penicillin/streptomycin (10,000 U/ml, Gibco).
Lentiviral constructs and transduction
Lentiviral plasmids harboring luciferase reporters of the Per2 and Bmal1 promoters were described previously and kindly provided by Prof. Dr. Liu 36, 37, 72. Viral particles were concentrated via ultracentrifugation after three harvests in HEK293T cells. Cells were transduced with concentrated Bmal1‐dLuc or Per2‐dLuc lentivirus 2 days before circadian bioluminescent measurements.
Bioluminescent recording and data analysis
Human ES cell‐derived cardiac cells were differentiated for up to 45 days and transduced with lentiviral reporters at described time points after synchronization with 100 nM dexamethasone 47 for 2 h. Subsequently, medium was changed to recording medium [BPEL, 10 mM HEPES, 100 μM D‐Luciferin Potassium Salt (Promega)]. Human ES cells were cultured in E8 medium and synchronized for 2 h with forskolin 39. Forskolin was chosen as a synchronizing agent for human ES cells, since dexamethasone has been implemented in multiple stem cell differentiation protocols and might therefore induce premature differentiation 73, 74, 75, 76, 77, 78. Subsequently, medium was changed to ES recording medium (E8, 10 mM HEPES, 100 μM D‐Luciferin Potassium Salt (Promega)). Culture dishes were sealed with high vacuum grease (Dow Corning) and monitored via the use of a LumiCycle32 device (Actimetrics) at 37°C. Bioluminescence from each dish was continuously recorded (integrated signal of 70 s with intervals of 10 min). Raw data (counts/s) were baseline subtracted (polynomial order 3).
Microscopic real‐time bioluminescence analysis
Human ES cells were differentiated for up to 30 days and transduced with Bmal1‐or Per2‐dLuc lentivirus 2 days before recording. Bioluminescence was assessed with an LV200 microscope (Olympus) in a humidified chamber under 5% CO2, at 37°C. Bioluminescence was detected for multiple consecutive days, using an EM CCD camera (Hamamatsu), with exposure times of 1 h. Image series were analyzed in ImageJ. Cells were synchronized with 100 nM dexamethasone for 2 h and changed to normal BPEL medium, containing 100 μM D‐Luciferin Potassium Salt (Promega). For pure cardiomyocyte population experiments, human ES cell‐derived cardiac populations were sorted with a FACS Jazz flow cytometer (BD Biosciences) based on GFP positivity, replated on Matrigel‐coated dishes, and bioluminescence was assessed 7 days later.
Immunostaining
Cells were fixed with 4% paraformaldehyde (PFA) for 15 min, blocked for 1 h in blocking buffer (5% FBS, 0.25% Triton X‐100 in PBS), and stained for OCT4 (SantaCruz, #5279), NANOG (Cell Signaling, #3580S), TNNT2 (ThermoFisher Scientific, #MA5‐12960), ACTN2 (Sigma, #A7811), MEF2C (Cell Signaling, #sc‐13266), GFP (Abcam, #ab6556) in staining buffer (1% BSA, 0.25% Triton X‐100 in PBS). Nuclei were stained with Hoechst for 15 min. Images were made using a spinning disk microscope (PerkinElmer).
RNA isolation and CEL‐Seq
Cardiac cells were derived from human ES cells in 48‐well plates. After synchronization, biological triplicates (independent wells) with comparable cardiac purity were collected every 4 h over the course of 48 h (ZT4‐ZT48) (Fig EV3). RNA was extracted using the standard TRIzol (Invitrogen) protocol, and 10 ng of total RNA per sample was used for library preparation and sequencing. RNA was processed as described previously 49, 79, and paired‐end sequencing was performed on the Illumina Nextseq platform with a read length of 75 base pairs. Read 1 was used to identify the sample barcode and library index, while read 2 was aligned to the hg19 human RefSeq transcriptome (downloaded from the UCSC genome browser) using BWA 80. CEL‐Seq only sequences the most 3′ end of a transcript, generating one read per transcript. Reads that mapped equally well to multiple locations were discarded. Around 500,000 reads were sequenced per sample. Samples were reads per million (RPM) normalized (Table EV1).
Quantitative RT–PCR
Purified RNA was treated with DNAse (Promega) and reversibly transcribed with Superscript III reverse transcriptase (ThermoFisher Scientific). qRT–PCR on biological triplicate samples was carried out in triplicate (technical replicates) in CFX‐384 Touch™ Real‐time PCR detection system (Bio‐Rad). PPIA was used as housekeeping gene, and fold changes were calculated to the lowest values among all replicates. Primer sequences: PPIA (fw): ttctgctgtctttgggacct, PPIA (rv): caccgtgttcttcgacattg, NANOG (fw): cagccctgattcttc, NANOG (rv): tgcatctgctggaggctgag, POU5F1 (fw): ctgaagcagaagaggat, POU5F1 (rv): gggccgcagcttacacat, ISL1 (fw): ctgcttttcagcaactggtca, ISL1 (rv): ggactggctaccatgctgtt, NKX2‐5 (fw): caagtgtgcgtctgcctttc, NKX2‐5 (rv): ctttcttttcggctctagggtcct, ACTC1 (fw): atgccatcatgcgtctggat, ACTC1 (rv): acgttcagcagtggtgacaa, KCNJ2 (fw): tgggtcttgggaattctggttt, KCNJ2 (rv): gaacatgtcctgttgctggcg, SERCA2A (fw): cgaacccttgccactcatct, SERCA2A (rv): ccagtattgcaggttccaggt, BMAL1 (fw): ggctcatagatgcaaaaactgg, BMAL1 (rev): ctccagaacataatcgagatgg, PER2 (fw): ggccatccacaaaaagatcctgc, PER2 (rv): gaaaccgaatgggagaatagtcg, CRY1 (fw): ctccatgggcactggtctcagtg, CRY1 (rv): tccccaccaatttcagctgcaac, CRY2 (fw): ccaagagggaagggcagggtagag, CRY2 (rv): aggatttgaggcactgttccgagg, CLOCK (fw): aagttagggctgaaagacgacg, CLOCK (rv): gaactccgagaagaggcagaag, NR1D1 (fw): acagctgacaccacccagatc, NR1D1 (rv): catgggcataggtgaagatttct.
Western blotting
Cells were lysed in RIPA buffer, and protein concentration was measured using a BCA assay (ThermoFisher Scientific). 12.5 μg protein lysate was loaded, separated by 10% SDS–PAGE, and transferred to a nitrocellulose membrane. Membranes were blocked with 5% milk powder (Nestlé) in T‐BST and probed with anti‐BMAL1 (1:1,000, #ab3350, Abcam), anti‐CRY1 (1:1,000, #13474‐1‐A, Proteintech), or anti‐CLOCK (1:250, #PA1‐520, ThermoFisher Scientific) antibodies, followed by a peroxidase‐conjugated antibody (1:5,000, #sc‐2004, Santa Cruz). ECL Plus Western blotting substrate (#32132, ThermoFisher Scientific) was used for chemiluminescence detection with an ImageQuant™ LAS 4000 imager (GE Healthcare). HRP‐coupled anti‐β‐ACTIN (1:5,000, #5125S, Cell Signaling) was used as a loading control. Band intensities were calculated with ImageJ.
Apoptosis measurements
Human ES cells were differentiated in 96‐well white walled plates for the course of 15 and 30 days. Cardiac cells were synchronized with 100 nM of dexamethasone for 2 h, and 10 μM of doxorubicin HCl (Sigma D1515) was administered at 6‐h intervals for a total time of 6 h. Apoptosis levels of three replicate wells (per condition), represented by active caspase 3 and caspase 7 levels, were measured using a CaspaseGlo 3/7 kit (Promega) following manufacturer's instructions. Bioluminescence was read out with a Centro microplate luminometer (Berthold Technologies).
JTK‐cycle analysis
RPM‐normalized read counts were obtained for each sample. As lowly expressed genes are typically not picked up robustly using CEL‐Seq, genes with an average of > 3 RPM across all time points (ZT4‐ZT48) as well as the replicates were selected for JTK‐cycle analysis. Around 10,000 genes reached this threshold in both D15 and D30, and these form the list on which JTK‐cycle was run (Table EV2). The following settings were used: jtkdist (12,3), periods (6:6), jtk.init (periods, 4). Significant oscillators with an adjusted P‐value of < 0.05 were selected for further analyses. To identify mouse heart oscillators, JTK was run with similar settings on normalized GC‐RMA intensity values of 24 samples (CT18‐CT62, sampled every 2 h) for 35,556 genes downloaded from the GEO‐database 81 (accession GSE54652) 14.
STRING and gene ontology analysis
STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database (www.string-db.org) was used to investigate the relationship between the overlap of known murine heart oscillators and identified D15 and D30 oscillators 56. Gene ontology terms were retrieved via www.string-db.org.
CircaDB gene expression website
The circadian expression database (CircaDB, http://circadb.hogeneschlab.org/) is an open access online platform 59 compiling circadian gene expression profiles from microarrays and RNA sequencing experiments 11, 14, 18, 82, 83, 84, 85, 86, 87, 88. The embedded JTK‐cycle algorithm defines the significance of rhythmic gene expression.
Statistics
All data were shown as means ± s.e.m. Student's t‐tests were carried out to assess differences between qRT–PCR mean values within the same experiments. One‐way ANOVA, followed by a Bonferroni post hoc test, was carried out to test increasing mRNA levels of clock factors during directed cardiac differentiation. A difference of P < 0.05 was considered significant. To calculate general induction of apoptosis upon doxorubicin, a Mann–Whitney U‐test was used. Differences in doxorubicin‐effect sizes between D15 and D30 cardiac cells were assessed via non‐overlapping 95% effect interval sizes. Statistical analyses to detect circadian oscillations in RNA levels (qRT–PCR) as well as doxorubicin‐based apoptosis were performed by RAIN 40.
Data availability
Primary data
Dierickx P, Vermunt MW, Muraro MJ, Creyghton MP, Doevendans PA, van Oudenaarden A, Geijsen N, Van Laake LW (2017) Circadian networks in human embryonic stem cell‐derived cardiomyocytes. Gene Expression Omnibus GSE97142.
Referenced data
Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB (2014) A circadian gene expression atlas in mammals: Implications for biology and medicine. Gene Expression Omnibus GSE54652.
Author contributions
PD, PAD, LWVL, and NG conceived and designed the research; PD conducted and analyzed the experiments; MJM supervised by AvO, made sequencing libraries, and processed CEL‐Seq data; PD and MWV supervised by MPC, analyzed and interpreted the CEL‐Seq data; PD, MWV, NG, and LWVL wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Acknowledgements
We thank Prof. Dr. Christine Mummery, Dorien Ward‐van Oostwaard (Leiden UMC) and Dr. David Elliot (Murdoch Childrens Research Institute) for providing us with the NKX2‐5‐eGFP human ES cell line, and Cathelijne W. van den Berg for the detailed differentiation protocol. We also thank Prof. Dr. Andrew Liu (University of Memphis) for sharing the lentiviral reporter plasmids. We thank Geert Geeven (Hubrecht Institute) for statistical assistance. This work was funded in part by NWO/ZonMw (Veni 91612147).
EMBO Reports (2017) 18: 1199–1212
Contributor Information
Pieterjan Dierickx, Email: p.dierickx@hubrecht.eu.
Linda W Van Laake, Email: l.w.vanlaake@umcutrecht.nl.
References
- 1. Aschoff J (1983) Circadian control of body temperature. J Therm Biol 8: 143–147 [Google Scholar]
- 2. Ohishi K, Nagasato K, Aoi W, Nakamura T, Ichinose K, Nishiura Y, Satoh A, Tsujihata M, Shibata Y, Nagataki S (1993) Circadian rhythms of blood pressure and heart rate in patients with human T‐lymphotropic virus type‐I‐associated myelopathy. Tohoku J Exp Med 169: 67–75 [DOI] [PubMed] [Google Scholar]
- 3. Shea SA, Hilton MF, Hu K, Scheer FAJL (2011) Existence of an endogenous circadian blood pressure rhythm in humans that peaks in the evening. Circ Res 108: 980–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Richards AM, Nicholls MG, Espiner EA, Ikram H, Cullens M, Hinton D (1986) Diurnal patterns of blood pressure, heart rate and vasoactive hormones in normal man. Clin Exp Hypertens A 8: 153–166 [DOI] [PubMed] [Google Scholar]
- 5. Degaute JP, Van Cauter E, van de Borne P, Linkowski P (1994) Twenty‐four‐hour blood pressure and heart rate profiles in humans. A twin study. Hypertension 23: 244–253 [DOI] [PubMed] [Google Scholar]
- 6. Delp MD, Manning RO, Bruckner JV, Armstrong RB (1991) Distribution of cardiac output during diurnal changes of activity in rats. Am J Physiol 261: H1487–H1493 [DOI] [PubMed] [Google Scholar]
- 7. Cajochen C, Kräuchi K, Wirz‐Justice A (2003) Role of melatonin in the regulation of human circadian rhythms and sleep. J Neuroendocrinol 15: 432–437 [DOI] [PubMed] [Google Scholar]
- 8. Pezuk P, Mohawk JA, Yoshikawa T, Sellix MT, Menaker M (2010) Circadian organization is governed by extra‐SCN pacemakers. J Biol Rhythms 25: 432–441 [DOI] [PubMed] [Google Scholar]
- 9. Hara R, Wan K, Wakamatsu H, Aida R, Moriya T, Akiyama M, Shibata S (2001) Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells 6: 269–278 [DOI] [PubMed] [Google Scholar]
- 10. Storch K‐F, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ (2002) Extensive and divergent circadian gene expression in liver and heart. Nature 417: 78–83 [DOI] [PubMed] [Google Scholar]
- 11. Tsimakouridze EV, Straume M, Podobed PS, Chin H, LaMarre J, Johnson R, Antenos M, Kirby GM, Mackay A, Huether P et al (2012) Chronomics of pressure overload‐induced cardiac hypertrophy in mice reveals altered day/night gene expression and biomarkers of heart disease. Chronobiol Int 29: 810–821 [DOI] [PubMed] [Google Scholar]
- 12. Martino T, Arab S, Straume M, Belsham DD, Tata N, Cai F, Liu P, Trivieri M, Ralph M, Sole MJ (2004) Day/night rhythms in gene expression of the normal murine heart. J Mol Med 82: 256–264 [DOI] [PubMed] [Google Scholar]
- 13. Martino TA, Tata N, Belsham DD, Chalmers J, Straume M, Lee P, Pribiag H, Khaper N, Liu PP, Dawood F et al (2007) Disturbed diurnal rhythm alters gene expression and exacerbates cardiovascular disease with rescue by resynchronization. Hypertension 49: 1104–1113 [DOI] [PubMed] [Google Scholar]
- 14. Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB (2014) A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci USA 111: 16219–16224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eckel‐Mahan K, Sassone‐Corsi P (2013) Metabolism and the circadian clock converge. Physiol Rev 93: 107–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Janich P, Toufighi K, Solanas G, Luis NM, Minkwitz S, Serrano L, Lehner B, Benitah SA (2013) Human epidermal stem cell function is regulated by circadian oscillations. Cell Stem Cell 13: 745–753 [DOI] [PubMed] [Google Scholar]
- 17. Janich P, Pascual G, Merlos‐Suárez A, Batlle E, Ripperger J, Albrecht U, Cheng H‐YM, Obrietan K, Di Croce L, Benitah SA (2011) The circadian molecular clock creates epidermal stem. Nature 480: 209–214 [DOI] [PubMed] [Google Scholar]
- 18. Rudic RD, McNamara P, Curtis A‐M, Boston RC, Panda S, Hogenesch JB, FitzGerald GA (2004) BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol 2: e377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kennaway DJ, Varcoe TJ, Voultsios A, Boden MJ (2013) Global loss of bmal1 expression alters adipose tissue hormones, gene expression and glucose metabolism. PLoS ONE 8: e65255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bray MS, Ratcliffe WF, Grenett MH, Brewer RA, Gamble KL, Young ME (2013) Quantitative analysis of light‐phase restricted feeding reveals metabolic dyssynchrony in mice. Int J Obes 37: 843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Furlan R, Barbic F, Piazza S, Tinelli M, Seghizzi P, Malliani A (2000) Modifications of cardiac autonomic profile associated with a shift schedule of work. Circulation 102: 1912–1916 [DOI] [PubMed] [Google Scholar]
- 22. Martino TA, Oudit GY, Herzenberg AM, Tata N, Koletar MM, Kabir GM, Belsham DD, Backx PH, Ralph MR, Sole MJ (2008) Circadian rhythm disorganization produces profound cardiovascular and renal disease in hamsters. Am J Physiol Regul Integr Comp Physiol 294: R1675–R1683 [DOI] [PubMed] [Google Scholar]
- 23. Penev PD, Kolker DE, Zee PC, Turek FW (1998) Chronic circadian desynchronization decreases the survival of animals with cardiomyopathic heart disease. Am J Physiol 275: H2334–H2337 [DOI] [PubMed] [Google Scholar]
- 24. Dierickx P, Du Pré B, Feyen DAM, Geijsen N, van Veen T, Doevendans PA, van Laake LW (2015) Circadian rhythms in stem cell biology and function Stem cells and cardiac regeneration, pp 57–78. Cham: Springer International Publishing; [Google Scholar]
- 25. Muller JE, Stone PH, Turi ZG, Rutherford JD, Czeisler CA, Parker C, Poole WK, Passamani E, Roberts R, Robertson T (1985) Circadian variation in the frequency of onset of acute myocardial infarction. N Engl J Med 313: 1315–1322 [DOI] [PubMed] [Google Scholar]
- 26. Mukamal KJ, Muller JE, Maclure M, Sherwood JB, Mittleman MA (2000) Increased risk of congestive heart failure among infarctions with nighttime onset. Am Heart J 140: 438–442 [DOI] [PubMed] [Google Scholar]
- 27. Tofler GH, Gebara OC, Mittleman MA, Taylor P, Siegel W, Venditti FJ, Rasmussen CA, Muller JE (1995) Morning peak in ventricular tachyarrhythmias detected by time of implantable cardioverter/defibrillator therapy. The CPI Investigators. Circulation 92: 1203–1208 [DOI] [PubMed] [Google Scholar]
- 28. Maron BJ, Semsarian C, Shen W‐K, Link MS, Epstein AE, Estes NAM, Almquist A, Giudici MC, Haas TS, Hodges JS et al (2009) Circadian patterns in the occurrence of malignant ventricular tachyarrhythmias triggering defibrillator interventions in patients with hypertrophic cardiomyopathy. Heart Rhythm 6: 599–602 [DOI] [PubMed] [Google Scholar]
- 29. Arntz HR, Willich SN, Oeff M, Brüggemann T, Stern R, Heinzmann A, Matenaer B, Schröder R (1993) Circadian variation of sudden cardiac death reflects age‐related variability in ventricular fibrillation. Circulation 88: 2284–2289 [DOI] [PubMed] [Google Scholar]
- 30. Maron BJ, Kogan J, Proschan MA, Hecht GM, Roberts WC (1994) Circadian variability in the occurrence of sudden cardiac death in patients with hypertrophic cardiomyopathy. JAC 23: 1405–1409 [DOI] [PubMed] [Google Scholar]
- 31. Durgan DJ, Pulinilkunnil T, Villegas‐Montoya C, Garvey ME, Frangogiannis NG, Michael LH, Chow C‐W, Dyck JRB, Young ME (2010) Short communication: ischemia/reperfusion tolerance is time‐of‐day‐dependent: mediation by the cardiomyocyte circadian clock. Circ Res 106: 546–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reiter R, Swingen C, Moore L, Henry TD, Traverse JH (2012) Circadian dependence of infarct size and left ventricular function after ST elevation myocardial infarction. Circ Res 110: 105–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Seneviratna A, Lim GH, Devi A, Carvalho LP, Chua T, Koh T‐H, Tan H‐C, Foo D, Tong K‐L, Ong H‐Y et al (2015) Circadian dependence of infarct size and acute heart failure in ST elevation myocardial infarction. PLoS ONE 10: e0128526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suarez‐Barrientos A, Lopez‐Romero P, Vivas D, Castro‐Ferreira F, Nunez‐Gil I, Franco E, Ruiz‐Mateos B, Garcia‐Rubira JC, Fernandez‐Ortiz A, Macaya C et al (2011) Circadian variations of infarct size in acute myocardial infarction. Heart 97: 970–976 [DOI] [PubMed] [Google Scholar]
- 35. Dierickx P, Doevendans PA, Geijsen N, van Laake LW (2012) Embryonic template‐based generation and purification of pluripotent stem cell‐derived cardiomyocytes for heart repair. J Cardiovasc Trans Res 5: 566–580 [DOI] [PubMed] [Google Scholar]
- 36. Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM et al (2007) Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129: 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA (2008) Redundant function of REV‐ERBα and β and non‐essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet 4: e1000023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Umemura Y, Koike N, Matsumoto T, Yoo S‐H, Chen Z, Yasuhara N, Takahashi JS, Yagita K (2014) Transcriptional program of Kpna2/Importin‐α2 regulates cellular differentiation‐coupled circadian clock development in mammalian cells. Proc Natl Acad Sci USA 111: E5039–E5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yagita K, Okamura H (2000) Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat‐1 fibroblasts. FEBS Lett 465: 79–82 [DOI] [PubMed] [Google Scholar]
- 40. Thaben PF, Westermark PO (2014) Detecting rhythms in time series with RAIN. J Biol Rhythms 29: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Keller G (2005) Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev 19: 1129–1155 [DOI] [PubMed] [Google Scholar]
- 42. Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC et al (2011) NKX2‐5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods 8: 1037–1040 [DOI] [PubMed] [Google Scholar]
- 43. Bunda S, Liu P, Wang Y, Liu K, Hinek A (2007) Aldosterone induces elastin production in cardiac fibroblasts through activation of insulin‐like growth factor‐I receptors in a mineralocorticoid receptor‐independent manner. Am J Pathol 171: 809–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cao N, Huang Y, Zheng J, Spencer CI, Zhang Y, Fu J‐D, Nie B, Xie M, Zhang M, Wang H et al (2016) Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science 352: 1216–1220 [DOI] [PubMed] [Google Scholar]
- 45. van den Berg CW, Elliott DA, Braam SR, Mummery CL, Davis RP (2015) Differentiation of human pluripotent stem cells to cardiomyocytes under defined conditions. Methods Mol Biol 1353: 163–180 [DOI] [PubMed] [Google Scholar]
- 46. Liu J, Lieu DK, Siu C‐W, Fu J‐D, Tse H‐F, Li RA (2009) Facilitated maturation of Ca2 + handling properties of human embryonic stem cell‐derived cardiomyocytes by calsequestrin expression. Am J Physiol Cell Physiol 297: C152–C159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U (2000) Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289: 2344–2347 [DOI] [PubMed] [Google Scholar]
- 48. Yagita K, Horie K, Koinuma S, Nakamura W, Yamanaka I, Urasaki A, Shigeyoshi Y, Kawakami K, Shimada S, Takeda J et al (2010) Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro . Proc Natl Acad Sci USA 107: 3846–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hashimshony T, Wagner F, Sher N, Yanai I (2012) CEL‐seq: single‐cell RNA‐seq by multiplexed linear amplification. Cell Rep 2: 666–673 [DOI] [PubMed] [Google Scholar]
- 50. Hughes ME, Hogenesch JB, Kornacker K (2010) JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome‐scale data sets. J Biol Rhythms 25: 372–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li J, Grant GR, Hogenesch JB, Hughes ME (2015) Considerations for RNA‐seq analysis of circadian rhythms. Meth Enzymol 551: 349–367 [DOI] [PubMed] [Google Scholar]
- 52. Bray MS, Shaw CA, Moore MWS, Garcia RAP, Zanquetta MM, Durgan DJ, Jeong WJ, Tsai J‐Y, Bugger H, Zhang D et al (2008) Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol‐Heart C 294: H1036–H1047 [DOI] [PubMed] [Google Scholar]
- 53. Sheikh‐Hamad D, Bick R, Wu G‐Y, Christensen BM, Razeghi P, Poindexter B, Taegtmeyer H, Wamsley A, Padda R, Entman M et al (2003) Stanniocalcin‐1 is a naturally occurring L‐channel inhibitor in cardiomyocytes: relevance to human heart failure. Am J Physiol‐Heart C 285: H442–H448 [DOI] [PubMed] [Google Scholar]
- 54. Li L, Weng Z, Yao C, Song Y, Ma T (2015) Aquaporin‐1 deficiency protects against myocardial infarction by reducing both edema and apoptosis in mice. Sci Rep 5: 13807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yan L, Wei X, Tang Q‐Z, Feng J, Zhang Y, Liu C, Bian Z‐Y, Zhang L‐F, Chen M, Bai X et al (2011) Cardiac‐specific mindin overexpression attenuates cardiac hypertrophy via blocking AKT/GSK3β and TGF‐β1‐Smad signalling. Cardiovasc Res 92: 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta‐Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP et al (2015) STRING v10: protein‐protein interaction networks, integrated over the tree of life. Nucl Acids Res 43: D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Radici L, Bianchi M, Crinelli R, Magnani M (2013) Ubiquitin C gene: structure, function, and transcriptional regulation. Adv Biosci Biotechnol 04: 1057–1062 [Google Scholar]
- 58. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW et al (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol 10: R130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pizarro A, Hayer K, Lahens NF, Hogenesch JB (2013) CircaDB: a database of mammalian circadian gene expression profiles. Nucl Acids Res 41: D1009–D1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. MacLennan DH, Kranias EG (2003) Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol 4: 566–577 [DOI] [PubMed] [Google Scholar]
- 61. Chen EY, Xu H, Gordonov S, Lim MP, Perkins MH, Ma'ayan A (2012) Expression2Kinases: mRNA profiling linked to multiple upstream regulatory layers. Bioinformatics 28: 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nunn C, Zou M‐X, Sobiesiak AJ, Roy AA, Kirshenbaum LA, Chidiac P (2010) RGS2 inhibits beta‐adrenergic receptor‐induced cardiomyocyte hypertrophy. Cell Signal 22: 1231–1239 [DOI] [PubMed] [Google Scholar]
- 63. Motloch LJ, Hu J, Akar FG (2015) The mitochondrial translocator protein and arrhythmogenesis in ischemic heart disease. Oxid Med Cell Longev 2015: 234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Muller JE, Tofler GH, Stone PH (1989) Circadian variation and triggers of onset of acute cardiovascular disease. Circulation 79: 733–743 [DOI] [PubMed] [Google Scholar]
- 65. Peckova M, Fahrenbruch CE, Cobb LA, Hallstrom AP (1998) Circadian variations in the occurrence of cardiac arrests: initial and repeat episodes. Circulation 98: 31–39 [DOI] [PubMed] [Google Scholar]
- 66. Lefrak EA, Pitha J, Rosenheim S, Gottlieb JA (1973) A clinicopathologic analysis of adriamycin cardiotoxicity. Cancer 32: 302–314 [DOI] [PubMed] [Google Scholar]
- 67. Benjamin RS, Chawla SP, Ewer MS (1988) Adriamycin cardiac toxicity—An assessment of approaches to cardiac monitoring and cardioprotection. In Organ directed toxicities of anticancer drugs, Hacker MP, Lazo JS, Tritton TR (eds), Boston, MA: Martinus Nijhoff Publishing. Proceedings of the First International Symposium on the Organ Directed Toxicities of Anticancer Drugs, Burlington, VT, June 4–6, 1987
- 68. Maillet A, Tan K, Chai X, Sadananda SN, Mehta A, Ooi J, Hayden MR, Pouladi MA, Ghosh S, Shim W et al (2016) Modeling doxorubicin‐induced cardiotoxicity in human pluripotent stem cell derived‐cardiomyocytes. Sci Rep 6: 25333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Burridge PW, Li YF, Matsa E, Wu H, Ong S‐G, Sharma A, Holmström A, Chang AC, Coronado MJ, Ebert AD et al (2016) Human induced pluripotent stem cell‐derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin‐induced cardiotoxicity. Nat Med 22: 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Guenthner CJ, Luitje ME, Pyle LA, Molyneux PC, Yu JK, Li AS, Leise TL, Harrington ME (2014) Circadian rhythms of Per2: Luc in individual primary mouse hepatocytes and cultures. PLoS ONE 9: e87573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A (2000) Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro . Nat Biotechnol 18: 399–404 [DOI] [PubMed] [Google Scholar]
- 72. Ramanathan C, Khan SK, Kathale ND, Xu H, Liu AC (2012) Monitoring cell‐autonomous circadian clock rhythms of gene expression using luciferase bioluminescence reporters. J Vis Exp 67: e4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cai J, Zhao Y, Liu Y, Ye F, Song Z, Qin H, Meng S, Chen Y, Zhou R, Song X et al (2007) Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology 45: 1229–1239 [DOI] [PubMed] [Google Scholar]
- 74. Tropel P, Noël D, Platet N, Legrand P, Benabid A‐L, Berger F (2004) Isolation and characterisation of mesenchymal stem cells from adult mouse bone marrow. Exp Cell Res 295: 395–406 [DOI] [PubMed] [Google Scholar]
- 75. Kubo A, Shinozaki K, Shannon JM, Kouskoff V, Kennedy M, Woo S, Fehling HJ, Keller G (2004) Development of definitive endoderm from embryonic stem cells in culture. Development 131: 1651–1662 [DOI] [PubMed] [Google Scholar]
- 76. Jaiswal N, Haynesworth SE, Caplan AI, Bruder SP (1997) Osteogenic differentiation of purified, culture‐expanded human mesenchymal stem cells in vitro . J Cell Biochem 64: 295–312 [PubMed] [Google Scholar]
- 77. Mackay AM, Beck SC, Murphy JM, Barry FP, Chichester CO, Pittenger MF (1998) Chondrogenic differentiation of cultured human mesenchymal stem cells from marrow. Tissue Eng 4: 415–428 [DOI] [PubMed] [Google Scholar]
- 78. Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR (1999) Multilineage potential of adult human mesenchymal stem cells. Science 284: 143–147 [DOI] [PubMed] [Google Scholar]
- 79. Simmini S, Bialecka M, Huch M, Kester L, van de Wetering M, Sato T, Beck F, van Oudenaarden A, Clevers H, Deschamps J (2014) Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nat Commun 5: 5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li H, Durbin R (2010) Fast and accurate long‐read alignment with Burrows‐Wheeler transform. Bioinformatics 26: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M et al (2013) NCBI GEO: archive for functional genomics data sets–update. Nucl Acids Res 41: D991–D995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB (2009) Harmonics of circadian gene transcription in mammals. PLoS Genet 5: e1000442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB et al (2007) Circadian and CLOCK‐controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci USA 104: 3342–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, Campbell KS, Arbogast S, Reid MB, Walker JR et al (2010) CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci USA 107: 19090–19095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109: 307–320 [DOI] [PubMed] [Google Scholar]
- 86. Rudic RD, McNamara P, Reilly D, Grosser T, Curtis A‐M, Price TS, Panda S, Hogenesch JB, FitzGerald GA (2005) Bioinformatic analysis of circadian gene oscillation in mouse aorta. Circulation 112: 2716–2724 [DOI] [PubMed] [Google Scholar]
- 87. Hoogerwerf WA, Sinha M, Conesa A, Luxon BA, Shahinian VB, Cornélissen G, Halberg F, Bostwick J, Timm J, Cassone VM (2008) Transcriptional profiling of mRNA expression in the mouse distal colon. Gastroenterology 135: 2019–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk H‐D, Kramer A, Maier B (2009) A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA 106: 21407–21412 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Review Process File
Data Availability Statement
Primary data
Dierickx P, Vermunt MW, Muraro MJ, Creyghton MP, Doevendans PA, van Oudenaarden A, Geijsen N, Van Laake LW (2017) Circadian networks in human embryonic stem cell‐derived cardiomyocytes. Gene Expression Omnibus GSE97142.
Referenced data
Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB (2014) A circadian gene expression atlas in mammals: Implications for biology and medicine. Gene Expression Omnibus GSE54652.


