Abstract
The type VI secretion system (T6SS) is a versatile molecular weapon used by many bacteria against eukaryotic hosts or prokaryotic competitors. It consists of a cytoplasmic bacteriophage tail‐like structure anchored in the bacterial cell envelope via a cytoplasmic baseplate and a periplasmic membrane complex. Rapid contraction of the sheath in the bacteriophage tail‐like structure propels an inner tube/spike complex through the target cell envelope to deliver effectors. While structures of purified contracted sheath and purified membrane complex have been solved, because sheaths contract upon cell lysis and purification, no structure is available for the extended sheath. Structural information about the baseplate is also lacking. Here, we use electron cryotomography to directly visualize intact T6SS structures inside Myxococcus xanthus cells. Using sub‐tomogram averaging, we resolve the structure of the extended sheath and membrane‐associated components including the baseplate. Moreover, we identify novel extracellular bacteriophage tail fiber‐like antennae. These results provide new structural insights into how the extended sheath prevents premature disassembly and how this sophisticated machine may recognize targets.
Keywords: bacterial molecular weapon, contractile bacteriophage tail, electron cryotomography, sub‐tomogram averaging, type VI secretion system
Subject Categories: Microbiology, Virology & Host Pathogen Interaction; Structural Biology
Introduction
The type VI secretion system (T6SS) is a dynamic nanomachine, widespread in Gram‐negative bacteria, that delivers effectors directly into target eukaryotic or bacterial cells for purposes of pathogenicity or competition 1, 2, 3. Its structure is known to comprise a membrane complex spanned the outer membrane (OM), periplasm and inner membrane (IM), a cytoplasmic baseplate associated with the membrane complex, an extended sheath anchored to the baseplate and loaded with an inner tube made of stacks of hexameric Hcp rings, and a spike complex on the tip of the inner tube 1, 2, 3. The extended sheath contracts to fire the inner tube/spike complex into target cells to deliver associated effectors 4, 5, 6. Bioinformatics and structural studies have provided strong evidence that this contractile mechanism is conserved with the contractile bacteriophage tail, R‐type pyocin, and phage tail‐like protein translocation structures 7.
Among these contractile machines, the T6SS is the only one that fires from inside the cell, and the only one known to recycle its components for multiple rounds of action 4, 8. The sheath proteins of the T6SS are equipped with a unique recycling domain recognized by the ATPase ClpV for rapid disassembly of the contracted sheath, allowing the sheath subunits to be reused to build other extended T6SS 6, 9, 10. High‐resolution cryo‐EM reconstructions have been obtained for purified R‐type pyocins in both the extended and contracted conformations, revealing how the energy for contraction is stored in the extended sheath and released upon contraction, and how the extended sheath interacts with the inner tube 11. For the T6SS, however, extended sheaths have not been reconstructed because cell lysis and purification trigger contraction. As a result, only the structure of the contracted sheath has been obtained by high‐resolution cryo‐EM 12, 13. Consequently, how the T6SS sheath subunits pack in the extended state, manage their recycling domains to prevent premature disassembly by ClpV, and interact with the Hcp inner tube is still unknown. In addition, the structural details of T6SS attachment to the membrane remain unclear.
Results and Discussion
Electron cryotomography (ECT) has proven powerful in visualizing native structures of bacterial nanomachines directly in their cellular context without purification 14. In fact, the structure of the T6SS was first discovered by ECT in Vibrio cholerae cells, wherein both extended and contracted conformations were seen, revealing the basic contractile mechanism 4. Following this discovery, we used correlated cryogenic photoactivated localization microscopy and ECT (cryo‐PALM/ECT) to identify the T6SS structure in vivo in Myxococcus xanthus 15, which was known to encode a T6SS in its genome (Fig EV1A). By fusing photoactivatable GFP with the sheath, localizing the signals in a frozen‐hydrated cell, and correlating the signals to electron densities in a cryotomogram of the same cell, we identified T6SS and confirmed that they formed extended and contracted structures just like their V. cholerae counterparts. We further noticed that in data taken close to focus, clear periodicities were visible in the M. xanthus T6SS sheaths (Fig EV2), opening the possibility of sub‐tomogram averaging to reveal structural details.
Figure EV1. Main and putative Myxococcus xanthus T6SS gene clusters.
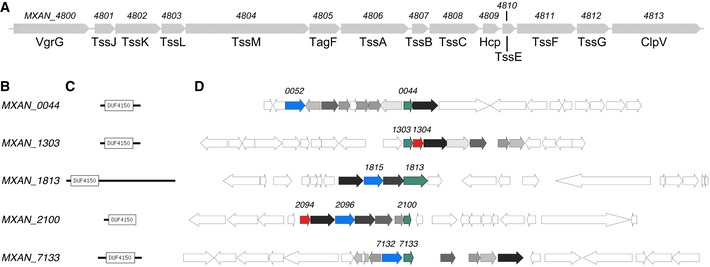
- The cluster of conserved T6SS genes (MXAN_4800–MXAN_4813) in the genome of M. xanthus.
- Locus number of putative PAAR‐repeat proteins.
- Domain architecture as predicted with PFAM 30.
- Gene neighborhood of putative PAAR‐repeat proteins (green). Distant homologs to proteins known to have a role in the function of T6SS in other organisms are highlighted: VgrG (red) and DUF2169‐containing proteins (blue). All gene clusters share similar set of homologs of less known proteins (shades of gray).
Figure EV2. Images of Myxococcus xanthus T6SS at different defoci.
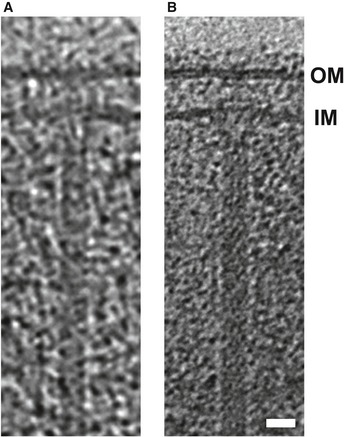
- A T6SS structure captured by correlated cryo‐PALM/ECT in our previous study 15 using a defocus of −10 μm during data collection.
- A similar structure captured in a cryotomogram of an M. xanthus cell with a defocus of −6 μm. Tomography slices through full 3D reconstructions of intact cells are shown in both cases.
Inspecting the more than 1,650 tomograms of M. xanthus cells in our tomography database 16, we identified 29 extended T6SS with an average length of 444 nm (an example is shown in Fig 1A), and 24 contracted T6SS with an average length of 282 nm (an example is shown in Fig 1B). Of the 29 extended T6SS structures, 16 were oriented close enough to horizontal (perpendicular to the electron beam) that we could align and average the membrane‐associated region (Fig 1C). The elongated structure of the sheath/Hcp tube was seen to end at a thin but distinct layer of density (L1). Another thicker layer of density (L2) was seen next to L1 with a gap in between. Between L2 and the IM, a cage‐like structure (L3) was revealed, which surrounds a thin axial density with a bulge close to the IM. The L1, L2, and most of the L3 densities together likely represent the baseplate structure in vivo, and the thin axial density is likely the spike complex. A distinct density spanning the periplasm with a wider part close to the IM and a narrower part close to the OM was seen. An extracellular cap‐like density was also seen on top of the OM outside of the cell. This extracellular cap together with the periplasmic density and a part of the cytoplasmic L3 density likely represents the membrane complex, which spans the OM, periplasm, and IM 17.
Figure 1. Visualization of the Myxococcus xanthus T6SS in vivo .
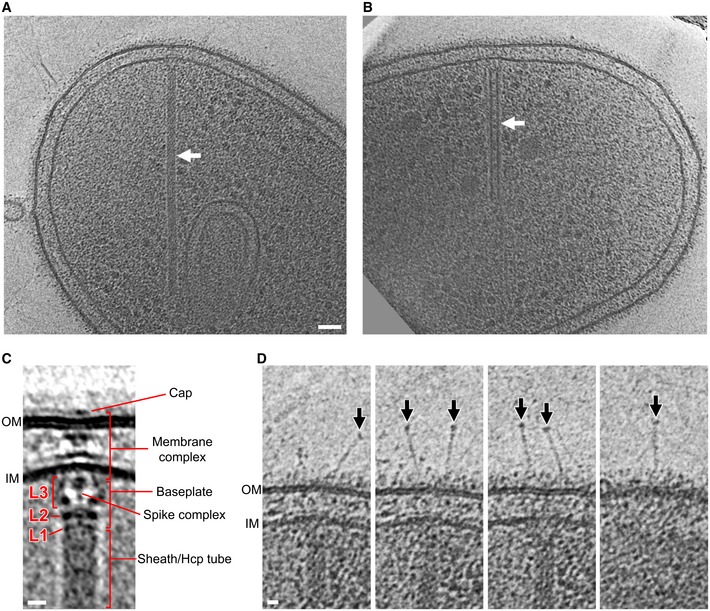
-
A, BTomographic slices through an extended T6SS (A) and a contracted T6SS (B) (arrows) in frozen‐hydrated M. xanthus cells.
-
CCentral slice of the sub‐tomogram average of the extended T6SS. L1–L3: different layers of densities in between the sheath and the IM.
-
DExtracellular bacteriophage tail fiber‐like antennae (arrows) captured in different slices of the tomogram shown in (A).
Interestingly, in individual tomograms of T6SS we found fiber‐like “antennae” densities originating from the OM and extending into the extracellular environment (Fig 1D). Of the 16 well‐orientated extended T6SS structures, 13 exhibited fiber‐like antennae. The number of antennae per T6SS varied from 1 to 6 with an average of 4. Since T6SS shares homologies with contractile bacteriophage, we note that these fiber‐like densities resemble bacteriophage tail fibers. The bulb‐like density on the tip of the antennae also bears a strong resemblance to similar structures on the bacteriophage T4 tail fibers that contain the putative receptor interaction sites 18, 19. However, the average length of the antennae seen here is ~60 nm, different from the bacteriophage T4 long (145 nm) and short (34 nm) tail fibers 20. Due to the crowded nature of the periplasm, we were unable to resolve whether a part of the antennae spans the periplasm to connect with the baseplate like a bacteriophage tail fiber, so the actual length of the antennae may be longer. We were also unable to identify genes in the M. xanthus genome with clear homology to bacteriophage T4 tail fiber proteins, so the protein identity of these extracellular antennae and whether they are related to bacteriophage tail fibers remain unclear. Since the gene of the antennae likely resides outside of the conserved T6SS gene cluster, it could be a specialized accessory component that varies among different bacteria, conferring different target specificity and firing behaviors 2.
To investigate the structure of the extended T6SS sheath, we computationally boxed the 29 extended sheaths observed in our cryotomograms into serial overlapping fragments and generated a sub‐tomogram average. This revealed clear structural features of the sheath surrounding the Hcp inner tube (Fig 2A–D). The extended sheath exhibits six‐fold symmetry along its long axis. In cross sections of the sheath, each of the six lobes was seen to contain a large and a small domain (Fig 2C and D; black arrowhead: large domain; white arrowhead: small domain). The large domains were connected to the inner tube by bridge densities; the small domains in turn connected to the large domains. A 3D volume of the sub‐tomogram average of the extended T6SS is shown in Fig 2E. By analyzing the autocorrelation of the extended sheath sub‐tomogram average through serial rotations and translations, a helical symmetry with a rise of 37 Å and a rotation of 22° for this six‐start right‐handed helix was revealed (Fig EV3A). We also generated a sub‐tomogram average of 24 contracted T6SS sheaths observed in our cryotomograms (Fig 2F–J). The cross section of the contracted sheath showed a cogwheel‐like structure with 12 distinct ridges (Fig 2H and I), consistent with previous in vivo observations in V. cholerae 4. Since there is no clear feature resolved along individual ridges at this resolution, we were not able to determine the helical symmetry of the contracted sheath by the autocorrelation method as we did for the extended sheath. Nonetheless, by the method we were able to determine the accurate pitch of the ridges (87.2°) (Fig EV3B), which is very similar to that of the contracted sheath from V. cholerae 4, 12.
Figure 2. Sub‐tomogram averages of the extended and contracted T6SS sheaths in intact Myxococcus xanthus cells.
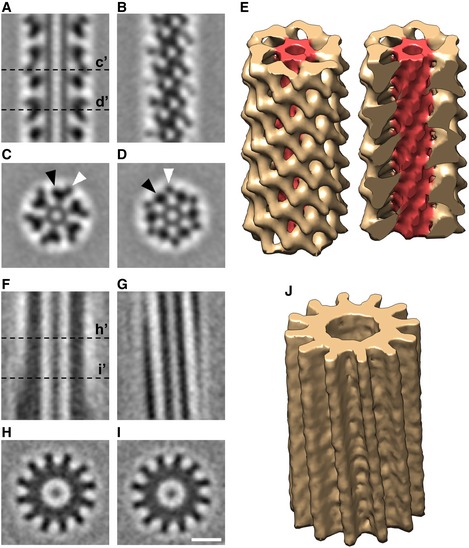
-
A, BSlices through the center (A) and surface (B) of the sub‐tomogram average of the extended T6SS sheath.
-
C, DSlices of cross sections C’ and D’ of the sub‐tomogram average shown in (A). Black and white arrowheads indicate the large and small domains of the sheath subunit, respectively.
-
E3D envelope of the extended T6SS sheath sub‐tomogram average. The sheath is colored in yellow and the Hcp inner tube in red. Left: the full molecule is displayed; right: a clipping plane is used to display the Hcp inner tube.
-
F–JThe same representations as in (A–E) for the contracted T6SS.
Figure EV3. Determination of the helical symmetries of Myxococcus xanthus T6SS extended and contracted sheaths by autocorrelation.
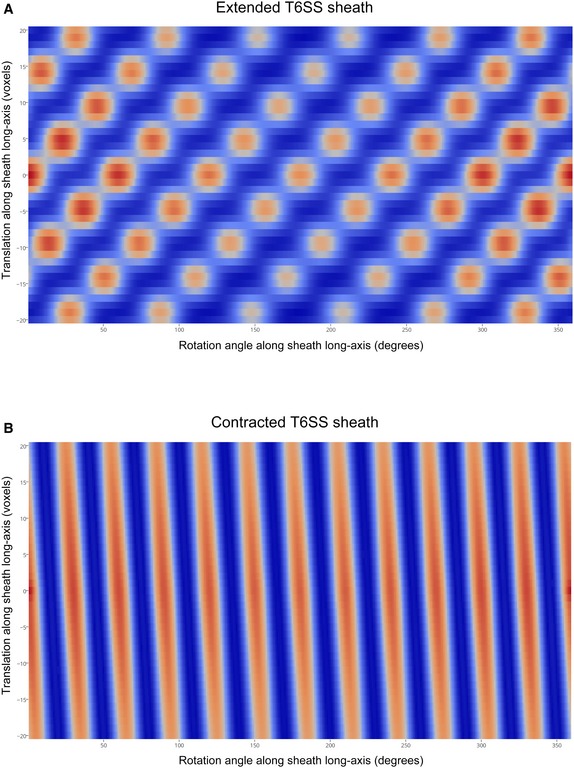
-
A, BAutocorrelation plots of the sub‐tomogram average of extended (A) and contracted (B) sheaths, respectively. Translational search: −20 to +20 voxels, with increments of 1 voxel (voxel dimensions: 7.8 Å × 7.8 Å × 7.8 Å); rotational search: 360°, with increments of 1°. Blue to red colors indicate low to high correlation.
Since it is unclear how a higher‐resolution structure of the extended T6SS sheath might be obtained directly, and since a wealth of structural information is available for the R‐type pyocin and contracted T6SS, we generated a pseudo‐atomic model of the extended T6SS sheath by applying this known information to our sub‐tomogram average. We started by superposing an atomic model of the extended R‐type pyocin (PDB 3J9Q) onto the sub‐tomogram average of the extended sheath using the Hcp inner tube as a reference (Fig 3A). In the extended R‐type pyocin structure, the sheath protein interacts with Hcp through an attachment α‐helix close to the C‐terminus 11, which is also present in the M. xanthus T6SS sheath protein TssC. In our sub‐tomogram average, we see a clear connection between the sheath and Hcp densities, which is well occupied by the attachment α‐helix (Fig 3A, arrow). The other parts of the pyocin sheath protein structure do not fit the density well, since the T6SS sheath is formed not by a single protein but by a TssB‐TssC heterodimer and contains an extra recycling domain. Near‐atomic‐resolution structures of the TssB‐TssC heterodimer were resolved previously in contracted T6SS sheaths purified from V. cholerae and Francisella novicida 12, 13. We used the more closely related V. cholerae structure (PDB 3J9G) as a template to generate a homology model of the M. xanthus TssB‐TssC heterodimer (MxTssBC) with which to replace the pyocin sheath protein structure. We superposed the MxTssBC model onto the pyocin sheath protein based on their structurally conserved domains 11 (Fig 3B). The result of this superposition suggests that the T6SS TssB‐TssC likely uses the same attachment α‐helix to interact with the Hcp protein in the inner tube (Fig 3B, arrow), but the MxTssBC model did not fit the density best in this orientation. It has been shown that upon pyocin sheath contraction, the sheath subunits rotate as rigid bodies. We therefore hypothesized that the structure of individual MxTssBC subunits remains the same in the extended sheath as in the contracted one. We then treated MxTssBC as a rigid body and slightly rotated it to best fit the density while maintaining the location of the attachment α‐helix for interaction with Hcp (Fig 3C and D). We also replaced the Hcp structure with a homology model of the M. xanthus Hcp hexamer (MxHcp) (constructed using PDB 3EAA 21 as a template) (Fig 3D). The conserved structural features at the interface of MxTssBC and MxHcp suggest that the sheath and Hcp tube in the T6SS likely interact in the same manner as in the R‐type pyocin (Fig 3A and D). We then replicated the MxTssBC model to populate the remaining five MxHcp subunits to create a hexamer of MxTssBC‐MxHcp, which constitutes one layer of the extended T6SS sheath (Fig 3D).
Figure 3. Generating pseudo‐atomic models of the Myxococcus xanthus T6SS sheath in extended and contracted conformations.
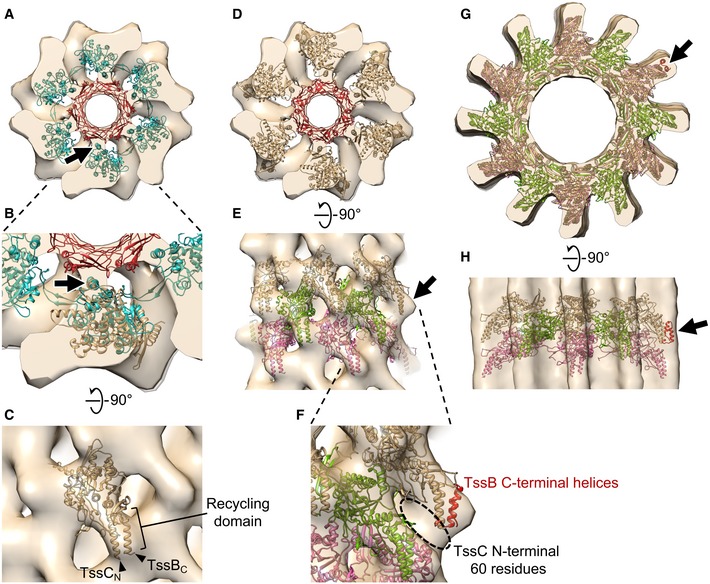
- An atomic model of the extended R‐type pyocin (PDB 3J9Q) placed in the envelope of the extended T6SS sheath sub‐tomogram average, fitting the Hcp inner tube (red) and the attachment α‐helix (arrow) in the sheath (cyan) to corresponding densities. The envelope shown here contains two layers of the extended sheath.
- Enlarged view of part of (A), with an MxTssBC model superposed on one sheath subunit of the R‐type pyocin structure. Arrow: the conserved attachment α‐helix in T6SS and R‐type pyocin sheath subunits.
- Side view of (B) after the MxTssBC model was rotated to best fit the density. TssCN and TssBC indicate the N‐terminus of TssC and C‐terminus of TssB, respectively.
- The model of hexameric MxTssBC‐MxHcp (one layer of the extended T6SS sheath) placed in the envelope of the extended M. xanthus T6SS sheath sub‐tomogram average. The envelope shown here contains two layers of the extended sheath.
- Side view of (D) after fitting three consecutive layers of the hexameric MxTssBC‐MxHcp model (shown in different colors) into the envelope. Arrow: density not fully occupied by the model.
- Enlarged view of part of (E), with the model of TssB C‐terminal helices (red; generated using PDB 4PS2) concatenated to the model in (E). Dashed circle: densities likely occupied by the TssC N‐terminal 60 amino acid residues, whose structure was absent in the MxTssBC model.
- The homology model of the contracted M. xanthus T6SS sheath (generated using PDB 3J9G) placed into the envelope of the contracted T6SS sheath sub‐tomogram average. Three consecutive layers of the hexameric MxTssBC model are shown in different colors.
- Side view of (G).
Next we propagated the MxTssBC‐MxHcp hexamer down the length of the sheath with a 37 Å translation and 22 degree rotation of each new layer to fit the density (Fig 3E). Most of the density was occupied, with the exception of the region surrounding the TssC N‐terminus and TssB C‐terminus in the MxTssBC model (Fig 3E, arrow). This structure comprises the recycling domain of the T6SS sheath (Fig 3C). A portion of this recycling domain was not resolved in the high‐resolution (< 3.5 Å) structures of contracted T6SS sheaths 12, 13 that we used to generate the MxTssBC model. An earlier lower‐resolution (6 Å) cryo‐EM reconstruction of the V. cholerae T6SS contracted sheath revealed that this missing part is composed of at least two helical elements and some additional densities 22. A crystal structure of the TssB C‐terminal domain is available (PDB 4PS2) that partially overlaps with the TssB C‐terminus in the MxTssBC model and contains the remaining two helices of the recycling domain. We concatenated this structure with the MxTssBC model and observed that it occupies the additional density well (Fig 3F). The additional ~60 residues of the TssC N‐terminus absent in the MxTssBC model likely occupy the remaining empty densities (Fig 3F, dashed circle).
We next constructed a pseudo‐atomic model of the contracted sheath by replicating MxTssBC subunits with the packing determined in the V. cholerae contracted sheath structure (PDB 3J9G) 12 and fitting the result into our sub‐tomogram average (Fig 3G and H). The inner diameter and dodecameric ridges of the model fit the density well. Interestingly, we observed additional densities at the tips of the ridges. As mentioned above, these densities were not observed in high‐resolution reconstructions of purified contracted sheaths 12, 13, but were seen in a lower‐resolution reconstruction 22. This suggests that the tip regions might exhibit greater flexibility than other parts. In our sub‐tomogram average, these additional densities could be partially occupied by the TssB C‐terminal helices (Fig 3G and H, arrows), as we saw for the extended conformation.
The pseudo‐atomic models of extended and contracted M. xanthus T6SS sheaths generated here support the hypothesis that the T6SS and R‐type pyocin share a similar architecture and mechanism (Fig 4). In our models, the protofilaments of the T6SS undergo similar motions to pyocin upon sheath contraction (Fig 4B, E, H and K), but when the same number of layers of the T6SS and pyocin is compared, the pyocin shows greater contraction (~33% shortening in the T6SS vs. ~56% in pyocin). However, since the length of extended T6SS can be more than 4X the length of pyocins 11, T6SS can still penetrate deeper into the target cell.
Figure 4. Comparing the models of Myxococcus xanthus T6SS sheath and R‐type pyocin in different conformations.
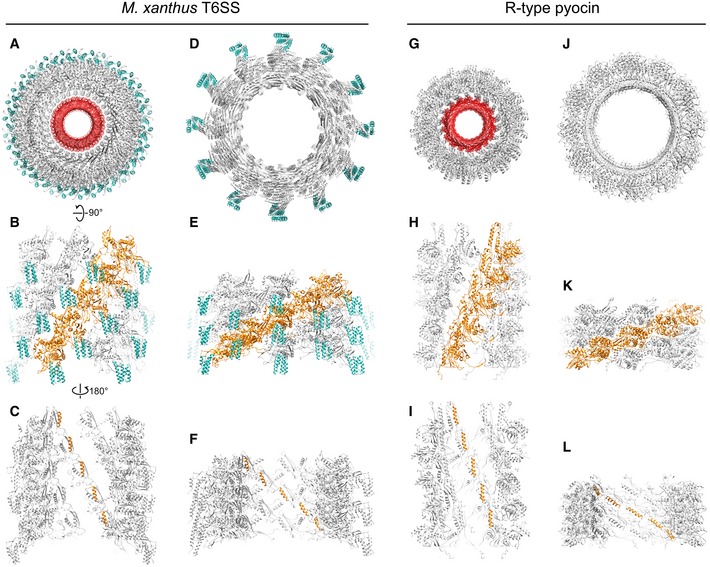
-
ATop view of the extended M. xanthus T6SS sheath model containing five layers of MxTssBC‐MxHcp hexamers. The sheath is colored gray, with the recycling domain colored cyan. The Hcp inner tube is colored red.
-
BSide view of the model in (A), with only the front half shown for clarity. One protofilament of the sheath is colored orange.
-
CBack view (from inside the tube) of model shown in (B). Here, only the attachment α‐helices in one protofilament of the sheath are colored orange.
-
D–FThe same representations as in (A–C) but for the contracted M. xanthus T6SS sheath.
-
G–LThe same representations as in (A–F) but for the R‐type pyocin.
The extended pyocin structure reported in a previous study revealed details of the interactions between the sheath and Hcp tube and showed that the Hcp tube packs in a helical fashion like the sheath 11. In the T6SS, however, how the Hcp hexamers stack in the inner tube is still in debate 7. Although our sub‐tomogram average of the extended T6SS is of insufficient resolution to reveal clear features of the Hcp hexamers in the inner tube, the helical arrangement of the density for the putative Hcp‐interacting α‐helix on the inside of the T6SS sheath reveals that Hcp hexamers in the T6SS are also packed helically with the symmetry of the sheath (Fig 4C, F, I and L).
While many aspects of the T6SS structure and contraction mechanism are clearly conserved with the R‐type pyocin, a key difference is the presence of recycling domains on the T6SS sheath (Fig 4A, B, D and E). As discussed above, in the contracted T6SS sheath, the recycling domain is exposed at the tip of the sheath ridges and is less conformationally rigid, possibly allowing access to the ATPase ClpV for sheath disassembly. Here, we show that in the extended sheath, the recycling domain is partly obstructed by interaction with the neighboring protofilament (Fig 4B). We speculate that this protection of the recycling domain prevents premature disassembly of the extended sheath by ClpV. In addition, in the extended pyocin structure, most of the interactions among sheath subunits are confined to within a single protofilament. Only the extended flexible arms on the N‐ and C‐termini of sheath subunits link neighboring protofilaments 11 (Fig 4H). Here, we observed that the recycling domain of the T6SS sheath is sandwiched in between two neighboring protofilaments (Fig 4B), suggesting that this domain might also be involved in assembly and stabilization of the extended sheath.
Finally, we placed our new sub‐tomogram average and the previously solved structures of individual components into the overall density (Fig 5A–C). The baseplate has been proposed to consist of TssA, TssE, TssF, TssG, and TssK 1. While TssA is initially recruited near the membrane, it has recently been shown to remain at the growing end (away from the membrane) of extending sheaths as new sheath and Hcp subunits are incorporated 23, and so is likely not present in the mature baseplate. It has been proposed that the T6SS sheath associates with the baseplate through TssE, because TssE is required for TssBC assembly 4 and shows clear homology to gp25 in the bacteriophage baseplate, which is known to interact with the bacteriophage sheath 24. Since TssE only comprises 131 residues and was proposed to interact with the sheath through a continuation of the three‐way “handshake” interactions of subunit domains in the assembled sheath 12, we reasoned that the density of TssE would be small and directly connected to the sheath density without any gap, similar to the gp25 location in the reported bacteriophage structure 24. We therefore proposed that the small but distinct density layer L1 is TssE and placed the average of the extended sheath/Hcp tube abutting it (Fig 5C). It is also possible that a part of the L1 density is contributed by another baseplate component TssK, which has been shown to directly interact with Hcp, TssC of the sheath, and TssL of the membrane complex, and therefore, it was proposed to connect the sheath/inner tube to the membrane complex 25. Purified TssK was previously shown to form a trimeric structure in solution with dimensions of around 100 × 100 × 90 Å 25. Based on our sub‐tomogram average, in order to span through the L1, L2, and L3 density layers (~300 Å) to interact with both the membrane complex and the sheath/Hcp tube, TssK must exhibit a more extended conformation or form a higher‐order oligomer in the assembled baseplate in vivo than in the isolated structure in vitro.
Figure 5. Placing available component structures into the Myxococcus xanthus T6SS structure in vivo .
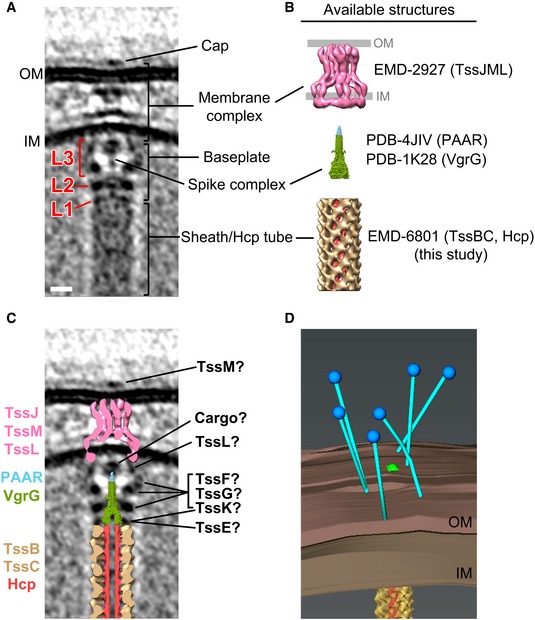
- Central slice of the sub‐tomogram average of M. xanthus T6SS membrane‐associated region.
- Available structures and models of individual components. The membrane complex structure (EMD‐2927) is shown with the OM and IM locations defined by previous work 17. The spike complex model was generated by concatenating the structure of VgrG from bacteriophage T4 (PDB 1K28) with non‐homologous parts removed with the crystal structure of the T6SS PAAR‐repeat protein (PDB 4JIV). The extended sheath/Hcp tube structure was generated in this study (EMD‐8601).
- Placing the central slices of available structures into the average shown in (A).
- 3D segmentation of the bacteriophage tail fiber‐like antennae shown in Fig 1D (blue: bacteriophage tail fiber‐like antennae; brown: inner and outer membranes; yellow: sheath; red: Hcp inner tube).
We next generated a model of the spike complex by concatenating the structure of VgrG from bacteriophage T4 (PDB 1K28) 26 with the crystal structure of the T6SS PAAR‐repeat protein (PDB 4JIV) 27 (Fig 5B). VgrG was shown to interact directly with the Hcp tube 28. We therefore placed the model of the spike complex adjacent to the tip of the fitted Hcp tube (Fig 5C). The model fits its potential density well in the center of baseplate, but the tip portion containing the PAAR‐repeat protein is clearly smaller than its corresponding density in the sub‐tomogram average. Interestingly, there is no gene of the PAAR‐repeat protein found in the M. xanthus T6SS gene cluster (Fig EV1A). There are, however, five genes (MXAN_0044, MXAN_1303, MXAN_1813, MXAN_2100, and MXAN_7133) outside of the T6SS gene cluster that encode different‐sized proteins with the PFAM protein domain model DUF4510 (Fig EV1B and C), which shares similarity with the PAAR‐repeat protein. It is therefore possible that one or more of these proteins form the spike tip with different sizes. All these five genes containing DUF4510 are present in gene clusters (Fig EV1D). Interestingly, two of the candidates (MXAN_2100 and MXAN_1303) also have neighboring genes encoding proteins related to the VgrG protein (i.e., proteins show sequence similarity with the TIGRFAM domain TIGR001646 and the PDB structure 4MTK). All five candidates but one (MXAN_1303) have at least one DUF2169‐containing protein within eight genes in the gene neighborhood. DUF2169‐containing proteins are not required for T6SS activity but seem to be important for toxin delivery in Agrobacterium tumefaciens 29. In the neighborhood of all five candidates but MXAN_7133, we found proteins with predicted 3‐oxoacyl‐(acyl carrier protein) synthase activity (due to their sequence similarity with the PRK06147 domain model). A protein with the same predicted activity was also found to be involved in the function of T6SS in Proteus mirabilis 30. These five gene clusters also share a number of other less well‐characterized genes (Fig EV1D, genes colored with different shades of gray). Based on these findings, we favor the gene cluster MXAN_2094–MXAN_2100 as the most plausible to be involved in the T6SS spike tip in M. xanthus. However, the similarity, but not identity, of the other four gene clusters begs the speculation that the T6SS in M. xanthus might be multifunctional, with several PAAR‐repeat tips to deliver different effectors. Because it has been shown that some T6SS cargo effectors associate with the PAAR‐repeat protein 2, 27, 31, we hypothesize that cargo effectors contribute to the bulge density on the spike tip (Fig 5C), similar to the observation in the in vitro structure of an effector‐loaded spike complex 32.
Recently, the T6SS membrane complex, composed of TssJ and TssM in the periplasm and TssL mainly in the cytoplasm, was purified and visualized by single particle cryo‐EM 17. The overall shape and dimensions of this single particle structure fit our in vivo densities well. However, the potential TssL density in the cytoplasm is more extended in our sub‐tomogram average, perhaps due to better structural preservation in the native environment and intact association with baseplate proteins (Fig 5C). Biochemical data in the same study also revealed that the C‐terminus of TssM is exposed to the cell surface during sheath assembling and contraction 17, which suggests that the extracellular cap structure seen here likely involves TssM. The remaining unassigned baseplate densities in L2 and L3 are likely contributed by the baseplate proteins TssF, TssG, and TssK (Fig 5C). The extracellular bacteriophage tail fiber‐like antennae which were seen in individual T6SS (Fig 1D) is shown by a 3D segmentation in Fig 5D.
In summary, here we have presented the first sub‐tomogram average of an extended T6SS sheath structure, revealing new insights into how its recycling domains are regulated. We also present the first visualization of the baseplate structure, revealing how it associates with the membrane complex and anchors the sheath. This structure also provides evidence of how cargo effectors might associate with the PAAR‐repeat protein of the spike complex. Our observation of extracellular antennae opens a new path for future research into how this important nanomachine recognizes targets and triggers firing. In the future, it will also be of great interest to locate all the components in the T6SS structure by imaging mutants (for instance as we did for the type IV pilus machines 33, 34). Myxococcus xanthus is not a good system for such mutant analysis, however, because of the low number of T6SS in the cells (we only obtained 16 T6SS structures showing clear views of the membrane‐associated components from > 1,650 tomograms) and the large size of the cells. New model systems such as T6SS‐containing minicells should therefore be sought for further dissection and higher‐resolution analysis of the T6SS in vivo.
Materials and Methods
Electron cryotomography
Myxococcus xanthus DK 1622 strain was grown in 10 ml CTT medium at 32°C with 250 rpm shaking. The cells were mixed with 10‐nm colloidal gold (Sigma‐Aldrich, St. Louis, MO) pretreated with bovine serum albumin and subsequently applied to freshly glow‐discharged Quantifoil copper R2/2 200 EM grids (Quantifoil Micro Tools GmbH, Jena, Germany). The grids were plunge‐frozen in a liquid ethane propane mixture 35 using an FEI Vitrobot Mark III (Thermo Fisher Scientific, Waltham, MA). The frozen grids were imaged in an FEI Polara 300 keV field emission gun transmission electron microscope (Thermo Fisher Scientific, Waltham, MA) equipped with a Gatan energy filter (Gatan, Pleasanton, CA) and a Gatan K2 Summit direct detector (Gatan, Pleasanton, CA). Energy‐filtered tilt series of images were collected automatically from −60° to +60° at 1° intervals using the UCSF tomography data collection software 36 with a total dosage of 160 e−/Å2, a defocus of −6 μm, and a pixel size of 3.9 Å. The images were subsequently binned by 2, aligned, and contrast‐transfer‐function‐corrected using the IMOD software package 37. SIRT reconstructions were then produced using the TOMO3D program 38. T6SS structures in the tomograms were located by visual inspection and manually rotated to similar orientation for subsequent sub‐tomogram averaging processes. To generate the sub‐tomogram averages of the T6SS sheaths, model points were distributed along the sheaths with 5 voxels apart. Overlapping boxes centered on the model points with dimensions of 60 × 60 × 60 voxels were cropped out, aligned, and averaged using the PEET program 39. Five iterations of alignment were performed using maximum angular searches of 24°, 12°, 6°, 3°, and 1.5° along the sheath long axis, with search steps of 8°, 4°, 2°, 1°, and 0.5°, and translation of 3, 3, 3, 2, and 2 voxels, respectively. Cylindrical masks with outer radii of 18 voxels (extended sheath) and 24 voxels (contracted sheath) were applied along the long axis of the sheath during the PEET processes. Six‐fold and 12‐fold axial symmetries were applied for the final extended and contracted sheath averages, respectively. To generate the sub‐tomogram averages of the T6SS membrane‐associated region, model points were put on the intersection of the inner membrane and the putative baseplate density. Boxes centered on the model points with dimensions of 100 × 200 × 100 voxels were cropped out, aligned, and averaged using PEET. Three iterations of alignment were performed using maximum angular searches of 3°, 1°, and 0.3°, with search steps of 1°, 0.25°, and 0.1°, and translation of 3, 2, and 2 voxels, respectively. A two‐fold symmetry along the particle long axis was applied to the final average.
Bioinformatics analysis
To find distant homologs of the PAAR repeat in the M. xanthus, we identify the PSSM models of the PAAR_like superfamily (cl21497) from the Conserved Domain Database (CDD) 40: PAAR_1 (cd14737), PAAR_2 (cd14738), PAAR_2 (cd14739), PAAR_4 (cd14740), PAAR_5 (cd14741), PAAR_RHS (cd14742), PAAR_CT_1 (cd14743), PAAR_CT_2 (cd14744); and the two imported conserved domains: PAAR_motif (pfam05488), DUF4150 (pfam13665). We built a library of these domains to use with RPS‐BLAST v2.2.36 41 to search proteins similar to these models in the M. xanthus genome. We found five hits with E‐value below the cutoff of 1E‐30: MXAN_0044 (4E‐53), MXAN_7133 (2E‐49), MXAN_1303 (1E‐47), MXAN_2100 (1E‐47), MXAN_1813 (8E‐31). To further investigate these genes, we used MiST2.2 42 and SeqDepot 43 databases to fetch information about the gene and their neighborhood. We used CDVist 44 (cdvist.utk.edu) for domain architecture prediction skipping HMMER and only one search against CDD database and another independent search with the PDB database. All other parameters were kept as default.
Data availability
The sub‐tomogram averages of the M. xanthus T6SS that support the findings of this study have been deposited in the Electron Microscopy Data Bank with accession codes EMD‐8600 (membrane‐associated region), EMD‐8601 (extended sheath), EMD‐8602 (contracted sheath). The coordinates of the extended and contracted sheath models have been deposited in the Protein Data Bank with accession code 5URW and 5URX, respectively.
Author contributions
Y‐WC and LAR collected, processed, and analyzed the electron cryotomography data. Y‐WC built the models. DRO performed the bioinformatics analysis. Y‐WC, DRO and GJJ wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank Prof. Hong Z. Zhou and Dr. Peng Ge for providing the initial concept of how to place the MyTssBC model in the sub‐tomogram average of extended T6SS sheath and Dr. Catherine Oikonomou for discussions and editorial assistance. This work was supported by NIH grant R01 AI127401 to G.J.J.
EMBO Reports (2017) 18: 1090–1099
References
- 1. Cianfanelli FR, Monlezun L, Coulthurst SJ (2016) Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol 24: 51–62 [DOI] [PubMed] [Google Scholar]
- 2. Ho BT, Dong TG, Mekalanos JJ (2014) A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zoued A, Brunet YR, Durand E, Aschtgen MS, Logger L, Douzi B, Journet L, Cambillau C, Cascales E (2014) Architecture and assembly of the Type VI secretion system. Biochim Biophys Acta 1843: 1664–1673 [DOI] [PubMed] [Google Scholar]
- 4. Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ (2012) Type VI secretion requires a dynamic contractile phage tail‐like structure. Nature 483: 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Basler M, Ho BT, Mekalanos JJ (2013) Tit‐for‐tat: type VI secretion system counterattack during bacterial cell‐cell interactions. Cell 152: 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Basler M, Mekalanos JJ (2012) Type 6 secretion dynamics within and between bacterial cells. Science 337: 815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kube S, Wendler P (2015) Structural comparison of contractile nanomachines. AIMS Biophysics 2: 88–115 [Google Scholar]
- 8. Vettiger A, Basler M (2016) Type VI secretion system substrates are transferred and reused among sister cells. Cell 167: 99–110.e112 [DOI] [PubMed] [Google Scholar]
- 9. Pietrosiuk A, Lenherr ED, Falk S, Bonemann G, Kopp J, Zentgraf H, Sinning I, Mogk A (2011) Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J Biol Chem 286: 30010–30021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kapitein N, Bonemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A (2013) ClpV recycles VipA/VipB tubules and prevents non‐productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol 87: 1013–1028 [DOI] [PubMed] [Google Scholar]
- 11. Ge P, Scholl D, Leiman PG, Yu X, Miller JF, Zhou ZH (2015) Atomic structures of a bactericidal contractile nanotube in its pre‐ and postcontraction states. Nat Struct Mol Biol 22: 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kudryashev M, Wang RY, Brackmann M, Scherer S, Maier T, Baker D, DiMaio F, Stahlberg H, Egelman EH, Basler M (2015) Structure of the type VI secretion system contractile sheath. Cell 160: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clemens DL, Ge P, Lee BY, Horwitz MA, Zhou ZH (2015) Atomic structure of T6SS reveals interlaced array essential to function. Cell 160: 940–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oikonomou CM, Chang Y‐W, Jensen GJ (2016) A new view into prokaryotic cell biology from electron cryotomography. Nat Rev Microbiol 14: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang YW, Chen S, Tocheva EI, Treuner‐Lange A, Lobach S, Sogaard‐Andersen L, Jensen GJ (2014) Correlated cryogenic photoactivated localization microscopy and cryo‐electron tomography. Nat Methods 11: 737–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ding HJ, Oikonomou CM, Jensen GJ (2015) The Caltech Tomography Database and Automatic Processing Pipeline. J Struct Biol 192: 279–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Durand E, Nguyen VS, Zoued A, Logger L, Pehau‐Arnaudet G, Aschtgen MS, Spinelli S, Desmyter A, Bardiaux B, Dujeancourt A et al (2015) Biogenesis and structure of a type VI secretion membrane core complex. Nature 523: 555–560 [DOI] [PubMed] [Google Scholar]
- 18. Bartual SG, Otero JM, Garcia‐Doval C, Llamas‐Saiz AL, Kahn R, Fox GC, van Raaij MJ (2010) Structure of the bacteriophage T4 long tail fiber receptor‐binding tip. Proc Natl Acad Sci U S A 107: 20287–20292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thomassen E, Gielen G, Schutz M, Schoehn G, Abrahams JP, Miller S, van Raaij MJ (2003) The structure of the receptor‐binding domain of the bacteriophage T4 short tail fibre reveals a knitted trimeric metal‐binding fold. J Mol Biol 331: 361–373 [DOI] [PubMed] [Google Scholar]
- 20. Yap ML, Rossmann MG (2014) Structure and function of bacteriophage T4. Future Microbiol 9: 1319–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jobichen C, Chakraborty S, Li M, Zheng J, Joseph L, Mok YK, Leung KY, Sivaraman J (2010) Structural basis for the secretion of EvpC: a key type VI secretion system protein from Edwardsiella tarda. PLoS ONE 5: e12910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kube S, Kapitein N, Zimniak T, Herzog F, Mogk A, Wendler P (2014) Structure of the VipA/B type VI secretion complex suggests a contraction‐state‐specific recycling mechanism. Cell Rep 8: 20–30 [DOI] [PubMed] [Google Scholar]
- 23. Zoued A, Durand E, Brunet YR, Spinelli S, Douzi B, Guzzo M, Flaugnatti N, Legrand P, Journet L, Fronzes R et al (2016) Priming and polymerization of a bacterial contractile tail structure. Nature 531: 59–63 [DOI] [PubMed] [Google Scholar]
- 24. Taylor NM, Prokhorov NS, Guerrero‐Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG (2016) Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533: 346–352 [DOI] [PubMed] [Google Scholar]
- 25. Zoued A, Durand E, Bebeacua C, Brunet YR, Douzi B, Cambillau C, Cascales E, Journet L (2013) TssK is a trimeric cytoplasmic protein interacting with components of both phage‐like and membrane anchoring complexes of the type VI secretion system. J Biol Chem 288: 27031–27041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanamaru S, Leiman PG, Kostyuchenko VA, Chipman PR, Mesyanzhinov VV, Arisaka F, Rossmann MG (2002) Structure of the cell‐puncturing device of bacteriophage T4. Nature 415: 553–557 [DOI] [PubMed] [Google Scholar]
- 27. Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG (2013) PAAR‐repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500: 350–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin JS, Ma LS, Lai EM (2013) Systematic dissection of the agrobacterium type VI secretion system reveals machinery and secreted components for subcomplex formation. PLoS ONE 8: e67647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bondage DD, Lin JS, Ma LS, Kuo CH, Lai EM (2016) VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor‐effector complex. Proc Natl Acad Sci U S A 113: E3931–E3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alteri CJ, Himpsl SD, Pickens SR, Lindner JR, Zora JS, Miller JE, Arno PD, Straight SW, Mobley HL (2013) Multicellular bacteria deploy the type VI secretion system to preemptively strike neighboring cells. PLoS Pathog 9: e1003608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS (2013) Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110: 7032–7037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whitney JC, Quentin D, Sawai S, LeRoux M, Harding BN, Ledvina HE, Tran BQ, Robinson H, Goo YA, Goodlett DR et al (2015) An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang YW, Rettberg LA, Treuner‐Lange A, Iwasa J, Sogaard‐Andersen L, Jensen GJ (2016) Architecture of the type IVa pilus machine. Science 351: aad2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang YW, Kjaer A, Ortega DR, Kovacikova G, Sutherland JA, Rettberg LA, Taylor RK, Jensen GJ (2017) Architecture of the Vibrio cholerae toxin‐coregulated pilus machine revealed by electron cryotomography. Nat Microbiol 2: 16269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tivol WF, Briegel A, Jensen GJ (2008) An improved cryogen for plunge freezing. Microsc Microanal 14: 375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zheng SQ, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA (2007) UCSF tomography: an integrated software suite for real‐time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol 157: 138–147 [DOI] [PubMed] [Google Scholar]
- 37. Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three‐dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- 38. Agulleiro JI, Fernandez JJ (2011) Fast tomographic reconstruction on multicore computers. Bioinformatics 27: 582–583 [DOI] [PubMed] [Google Scholar]
- 39. Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR (2006) The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313: 944–948 [DOI] [PubMed] [Google Scholar]
- 40. Marchler‐Bauer A, Bo Y, Han L, He J, Lanczycki CJ, Lu S, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR et al (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45: D200–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ulrich LE, Zhulin IB (2010) The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res 38: D401–D407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ulrich LE, Zhulin IB (2014) SeqDepot: streamlined database of biological sequences and precomputed features. Bioinformatics 30: 295–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adebali O, Ortega DR, Zhulin IB (2015) CDvist: a webserver for identification and visualization of conserved domains in protein sequences. Bioinformatics 31: 1475–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Review Process File
Data Availability Statement
The sub‐tomogram averages of the M. xanthus T6SS that support the findings of this study have been deposited in the Electron Microscopy Data Bank with accession codes EMD‐8600 (membrane‐associated region), EMD‐8601 (extended sheath), EMD‐8602 (contracted sheath). The coordinates of the extended and contracted sheath models have been deposited in the Protein Data Bank with accession code 5URW and 5URX, respectively.


