Abstract
Autophagy is a conserved cellular process for degrading aggregate proteins and dysfunctional organelle. It is still debatable if autophagy and mitophagy (a specific process of autophagy of mitochondria) play important roles in myogenic differentiation and functional regeneration of skeletal muscle. We tested the hypothesis that autophagy is critical for functional regeneration of skeletal muscle. We first observed time-dependent increases (3- to 6-fold) of autophagy-related proteins (Atgs), including Ulk1, Beclin1, and LC3, along with reduced p62 expression during C2C12 differentiation, suggesting increased autophagy capacity and flux during myogenic differentiation. We then used cardiotoxin (Ctx) or ischemia-reperfusion (I/R) to induce muscle injury and regeneration and observed increases in Atgs between days 2 and 7 in adult skeletal muscle followed by increased autophagy flux after day 7. Since Ulk1 has been shown to be essential for mitophagy, we asked if Ulk1 is critical for functional regeneration in skeletal muscle. We subjected skeletal muscle-specific Ulk1 knockout mice (MKO) to Ctx or I/R. MKO mice had significantly impaired recovery of muscle strength and mitochondrial protein content post-Ctx or I/R. Imaging analysis showed that MKO mice have significantly attenuated recovery of mitochondrial network at 7 and 14 days post-Ctx. These findings suggest that increased autophagy protein and flux occur during muscle regeneration and Ulk1-mediated mitophagy is critical for recovery for the mitochondrial network and hence functional regeneration.
Keywords: mitophagy, Unc-51-like autophagy activating kinase 1, torque, muscle repair
skeletal muscle has a remarkable ability to regeneration after damage, and this regenerative capacity is conferred by myogenic stem cells located between the basal lamina and the sarcolemma referred to as satellite cells. Satellite cells when activated undergo proliferation, differentiation, and fusion to the existing myofiber to provide new myonuclei (9, 22). This myogenic process typically follows myofiber damage that can happen throughout the human life span but may also play a functional role in the maintenance of uninjured muscle myonuclei and satellite cell populations (36, 45). At the same time, a portion of the proliferating cells will exit the cell cycle to resume quiescence and repopulate the satellite cell population (27, 47). Recent studies show that the ability of satellite cells to enter myogenic differentiation or resume quiescence in the stem cell niches is negatively impacted by a poor cellular milieu (2, 3, 10) such as observed during aging or in obese people.
Macroautophagy is an evolutionally conserved cellular process responsible for degrading protein aggregates and damaged and/or dysfunctional organelles that are enveloped in a membrane structure called an autophagosome, which eventually merges with a lysosome (8, 31). After the cellular components are broken down by this process, the byproducts (amino and fatty acids) are used by the cell to synthesize new cellular components. Despite the importance, very little is known about the role of autophagy in skeletal muscle regeneration. Specifically, existing evidence suggests that stimulating autophagy in Duchenne muscular dystrophy promotes functional recovery; however, it is unclear if autophagy is activated and required for functional recovery after muscle injury (11, 35). Moreover, mitophagy, a form of macroautophagy specific for the degradation of damaged/dysfunctional mitochondria, and mitochondrial biogenesis both contribute significantly to mitochondrial quality control (30). Under normal conditions, mitophagy is believed to be a constantly ongoing process at a basal level in skeletal muscle necessary for proper turnover of mitochondria (18). Under stressed conditions, this process may be significantly enhanced to remove old/damaged mitochondria to maintain the metabolic homeostasis (13, 46). For example, Nichenko et al. (33) demonstrated that myotoxic skeletal muscle injury causes mitochondrial damage and is associated with increased expression of autophagy-related proteins and activation of autophagy 2 wk postinjury. Together, autophagy and mitophagy are generally accepted as being important for normal skeletal muscle function, but many facets of their functions remain unexplored. These include the cellular conditions that induce and/or activate autophagy and mitophagy and the functional role of their related proteins during skeletal muscle adaptation.
Unc-51-like kinase 1 (Ulk1) is a necessary kinase for initiating autophagy under the conditions of stress (17, 28). Ulk1 associates with autophagy gene 13 (Atg13) and FIP200, and this Ulk1-Atg13-FIP200 complex can be regulated by the mammalian target of rapamycin (mTOR) and AMP kinase (AMPK) signaling cascades that sense cellular energy status (20). Specifically, the Ulk1-Atg13-FIP200 complex is inhibited by mTOR in the presence of nutrients and activated by AMPK in low-nutrient states (16). Once activated, Ulk1 facilitates the formation of the autophagosome along with autophagy-related proteins Beclin1 (Atg6) and microtubule-associated proteins 1A/1B light chain 3A (LC3). Damaged proteins and organelles are targeted to the developing autophagosome by the polyubiquitin-binding protein p62/SQSTM1 (p62). Following the formation of autophagosome, fusion with a lysosome completes the degradation process. Importantly, cytosolic LC3 (LC3-I) is converted to LC3-II by conjugation to phosphatidylethanolamine (PE) during autophagosome formation. Upon fusion of the autophagosome with the lysosome, LC3-II and p62, which directly bind to LC3-II (34), are degraded. Therefore, LC3 and p62 represent two of the most ubiquitous autophagy-related proteins used to determine autophagy capacity and autophagy flux (i.e., ongoing autophagic degradation).
While it is clear that muscle injury results in damages to proteins and organelles, the extent to which autophagy participates in the clearing of the damage products is unclear. The objective of this study was to test if autophagy plays a role in muscle regeneration and functional recovery after injury. We hypothesized that inhibiting autophagy would compromise the functional regeneration of muscle (i.e., force-generating capacity).
MATERIALS AND METHODS
Animals and study design.
All mice were housed in temperature-controlled (21°C) cages in a specific-pathogen-free room with a 12:12-h light-dark cycle and free access to water and normal chow. Ulk1 muscle-specific knockout mice (MKO) were generated by crossbreeding myogenin-Cre mice (28) (generous gift from Dr. Eric Olsen) with loxP-flanked Ulk1 mice (ULK1fl/fl). Inducible Ulk1 muscle-specific knockout mice (iMKO) were generated by cross breeding human α-skeletal actin MerCreMer (HSA-MCM) mice with loxP-flanked Ulk1 mice (ULK1fl/fl). Wild-type (WT) littermates were used as controls. All animal protocols were approved by the University of Virginia Animal Care and Use Committee.
To determine the association between muscle regeneration and autophagy, WT mice (C57BL/6) were injured using cardiotoxin (Ctx) or ischemia-reperfusion (I/R) injury, and skeletal muscle was harvested at several time points during regeneration. Ctx is widely used to induce muscle injury and regeneration in vivo (45) without significantly affecting the nerve or blood supply (21). Another cohort of WT mice was subjected to I/R injury to determine if the association between autophagy and muscle regeneration was conserved or limited to myotoxic injury. To determine the role of autophagy during muscle regeneration, two Ulk1 knockout approaches were utilized. Noninducible muscle-specific Ulk1 knockout mice (MKO) were used first to determine if muscle contractile function is impaired in uninjured adult skeletal muscle as a result of a developmental absence of Ulk1, in addition to impaired recovery during muscle regeneration. The inducible iMKO mice were used later to avoid compensatory or developmental alterations that may confound the dissection of the role of Ulk1 during muscle regeneration. All experiments were initiated when MKO, iMKO, and littermate controls were between 3 and 4 mo of age. Cre activity was induced in iMKO mice by intraperitoneal injection of tamoxifen (1 mg/day) for 10 days before the muscle injury, which allowed sufficient turnover of Ulk1 protein given consideration to its 16- to 24-h half-life (12, 24). This approach will result in deletion of the Ulk1 gene only in differentiated myofibers (see Fig. 5) but not prevent Ulk1 expression from the nuclei brought into the adult myofibers by the satellite cells. However, considering the short duration of the experiment, the contribution is limited, and even if it is true that some nuclei from satellite cells may restore Ulk1 expression partially, it would further support the conclusion that Ulk1-mediated autophagy is critical for functional muscle regeneration.
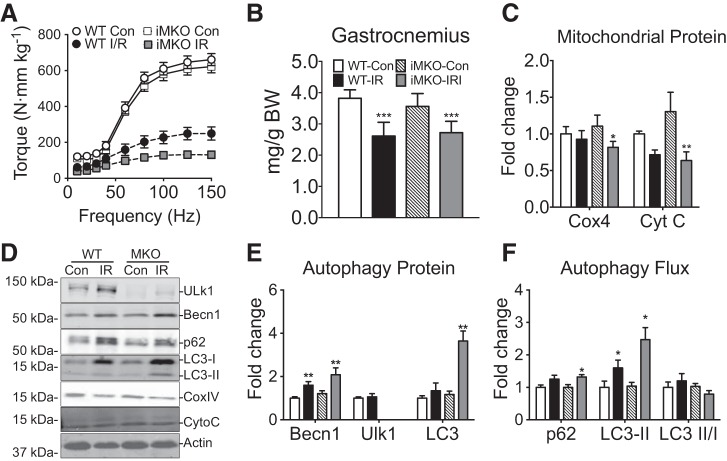
Fig. 5.
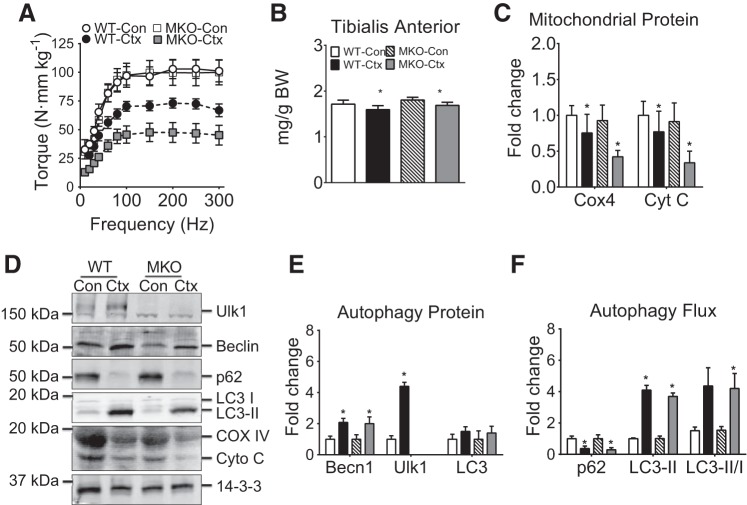
Muscle-specific conditional Ulk1 deficiency blunts functional muscle regeneration following I/R. A: quantification of gastrocnemius muscle mass and body mass for MKO and littermate WT mice at day 14 postinjury. B: torque frequency analysis of plantar flexors relative to WT Con. Mass and torque results are represented as means ± SE for n = 5–6 for each genotype/injury. C: quantification of COX4 and Cyt C at day 7 postinjury. All immunoblot results are represented as means ± SE for n = 5–6 for each genotype/injury. D: representative immunoblots of Ulk1, Beclin1, p62, LC3-I, LC3-II, COX4, Cyt C, and 14-3-3 for injured muscles (day 14 postinjury) and Con for MKO mice and littermate WT mice. E: quantification of Beclin1, Ulk1, and total LC3 indicant of autophagy capacity at day 14 postinjury. F: quantification of p62, LC3-II as an index of autophagy flux at day 14 postinjury.
Genotyping.
DNA was isolated using a phenol-chloroform-based DNA extraction protocol. PCR was performed with primers for the myogenin-Cre allele (5′-AGGTTCGTTCACTCATGGA-3′ and 5′-TCGACCAGTTTAGTTACCC-3′), loxP-flanked Ulk1 allele (5′-TCCTTCAGCACCAGCCGCATTA-3′ and 3′-GCAAACGCTAGTGTGAAGCGCA-5′), and the HSA-MCM allele (5′-GCATGGTGGAGATCTTTGA-3′ and 3′-CGACCGGCAAACGGACAGAAGC-5′). Cre-negative and iMKO mice that did not receive tamoxifen with genotype of ULk1+/+ or Ulk1fl/+ were used as WT mice, and mice with genotype of Ulk1fl/fl:Myog-Cre were used as MKO mice.
Muscle injury models.
We modified a previously described muscle injury model by injecting Ctx into the tibialis anterior (TA) or flexor digitorum longus (FDB) muscle of 3-mo-old mice (23). Briefly, Ctx (Naja nigricollis; 0.071 mg/ml diluted in saline; Calbiochem) was injected into the left TA muscle (0.1 ml) or left FDB muscle (0.03 ml), and an equal volume sterile saline was injected into the right hindlimb muscle for control. The TA muscle was selected because of its well-defined anatomical structure, our previous experience with Ctx-induced injury and regeneration (45), its contribution to dorsiflexion about the ankle joint, and our objective to assess its contractility during the repair phase after injury. The FDB muscle was selected because of our previous work with pMitoTimer and the ease to culture the muscle fibers for confocal microscopy imaging (29). Injection of Ctx or saline into the FDB was achieved by inserting a sterile needle at a point close to the heel and advancing the needle subcutaneously toward the base of the toes for 1/4 inch. Care was taken to ensure no solution leaked from the puncture site following injection. Muscles were harvested at 1, 3, 7, and 14 days postinjection. Contralateral limbs were used as controls.
I/R injury was induced in 3-mo-old mice as previously described (4). Briefly, while the mice were under anesthesia, the left hindlimb was shaved and depilated. The body temperature was maintained at 37°C on a stage via a circulating water bath. A 4.0 oz, 1/8 in. rubber band (Dentsply GAC International, Islandia, NY) was placed unilaterally above the femur using a McGivney hemorrhoidal ligator and kept in place for 1 h, a duration long enough to induce tissue damage and within clinical guidelines (14). Plantarflexor muscles were harvested at 0 (end of ischemic period), 3, and 12 h and 1, 2, 3, 7, and 14 days postinjury. Since posterior hindlimb muscles are the dominant muscles for hindlimb muscle function, plantarflexor muscles were selected for further analysis. Sham (Con) mice were subjected to the same protocol without addition of the rubber band.
Immunoblot analysis.
Skeletal muscle sample preparation for immunoblot was described previously (5). Briefly, skeletal muscle were homogenized in extraction buffer [1% SDS, 5 mM EGTA, protease inhibitors (Sigma), and phosphatase inhibitor cocktails 2 and 3 (Sigma)] and boiled for 5 min in the presence of reducing agent β-mercaptoethanol, and supernatant was collected after 10,000 g spin for 10 min. Protein concentration was determined using a bicinchoninic acid kit (BCA; Pierce). Proteins were separated on 7, 10, or 15% SDS-PAGE gels before overnight transfer to PVDF membranes. Membranes were probed with the following antibodies for immunoblot analysis: Ulk1 (1:1,000; 150 kDa; Cell Signaling), Beclin1 (1:1,000; 60 kDa; Cell Signaling), LC3 I/II (1:1,000; 14 and 16 kDa; Cell Signaling), p62 (1:1,000; 62 kDa; Cell Signaling), cytochrome c oxidase 4 (COX4; 1:1,000; 17 kDa; Cell Signaling), cytochrome c (Cyt C; 1:1,000; 14 kDa; Cell Signaling), myogenin (1:1,000; 40 kDa; Abcam), tubulin (1:2,000; 52 kDa; Cell Signaling), β-actin (1:2,000; 40 kDa; Sigma), and 14-3-3 (1:2,000; 27 kDa; Cell Signaling). Quantification of the proteins was performed using Li-Cor Biosystems analysis software. For Ctx experiments, relative protein expression across time points from injured muscles was compared with the collective pool of contralateral control muscles. There was no significant difference among contralateral control muscles across the different time points (1–14 days, P ≥ 0.58). For I/R experiments, relative protein expression across time points from injured muscles was compared with the collective pool of sham control muscle. There was no significant difference among sham control muscles across the different time points (immediate: 14 days, P ≥ 0.73).
Mitochondrial imaging.
FDB muscles fibers were transfected with the pMitoTimer reporter gene as previously described (29). The MitoTimer encodes a mitochondria-targeted green fluorescent protein that was utilized to image the mitochondrial network using confocal microscopy (29).
Cell culture.
Mouse myoblast cell lines from American Type Culter Collection (Manassas, VA) with fewer than eight passages were used in all cell culture experiments. Myoblasts were cultured at subconfluent densities (100,000 cells/well, 6-well plate, 9-cm2 surface area) for 24 h in growth media [GM: DMEM (high glucose) with 20% fetal bovine serum and 1% penicillin/streptomycin]. Differentiation of myoblasts into fused myotubes was induced by serum withdrawal, i.e., incubating myoblasts in differentiation medium [DM: DMEM (high glucose) with 2% horse serum, 1% penicillin/streptomycin, 50 mM HEPES, 10 μg/ml transferrin, and 10 μg/ml insulin], which was replaced daily.
In vivo muscle function analysis.
In vivo maximal isometric torque of the ankle dorsiflexors (TA, extensor digitorum longus, and extensor hallucis longus muscle) and ankle plantarflexors (gastrocnemius, soleus, and plantaris muscle) was assessed as previously described (1, 7, 40). For dorsiflexors, mice were anesthetized using 1% isoflurane in oxygen, and then the left hindlimb was depilated and aseptically prepared, the foot was placed in a foot-plate attached to a servomotor (Model 300C-LR; Aurora Scientific, Aurora, ON, Canada), and Pt-Ir needle electrodes (Grass Technologies, West Warwick, RI) were inserted percutaneously on either side of the peroneal nerve that innervates the dorsiflexor muscles. Peak isometric torque was achieved by varying the current delivered to the nerve. Torque as a function of stimulation frequency was also assessed. Measurement of torque for plantarflexors was performed as outlined above with minor modifications. Briefly, Pt-Ir needle electrodes were inserted percutaneously on either side of the sciatic nerve that branches to the tibial nerve thus innervating the ankle plantarflexor muscles. Electrodes were placed ~1 cm proximal to the knee joint, which was secured with a knee clamp to maintain a knee joint angle of 90° (pictures detailing the dorsiflexor and plantarflexor electrode placement can be found in Ref. 6). To account for differences in body size among mice, torque (mN·m) was normalized by body mass (kg).
Statistics.
All results are presented as means ± SE. One-way ANOVA was used to compare protein expression during C2C12 cell differentiation (myoblasts to day 4 differentiation), during muscle regeneration following Ctx (TA muscle), and following I/R (plantaris muscle) injury. A two-way ANOVA was used to compare protein expression, muscle mass, and torque between knockout mice and littermate controls during muscle regeneration (genotype vs. injury). A significant interaction of 0.05 was required to perform between-variable post hoc analysis, and in such case a Tukey’s honestly significance difference was performed.
RESULTS
Activation of autophagy and increased autophagy flux during myogenic differentiation in vitro.
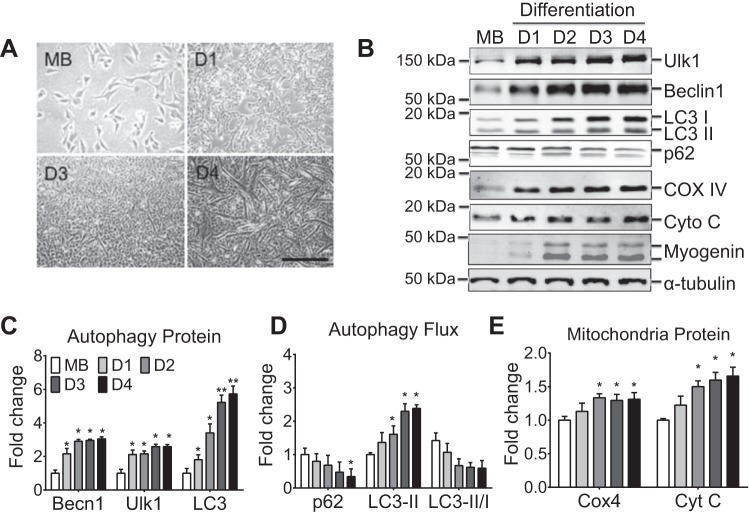
To determine if autophagy is associated with myogenic differentiation, an immortalized mouse cell line system was utilized, in which single nucleated C2C12 myoblasts fuse to form multinucleated myotubes in vitro (Fig. 1A). Protein expression of autophagy-related genes was determined during serum withdrawal-induced differentiation, including Ulk1, Beclin1, p62, and LC3. Both Ulk1 and Beclin1 were gradually induced to approximately threefold peaking at day 4 as myoblasts were fully differentiated into myotubes (Fig. 1, B–D, P < 0.05 relative to myoblasts). LC3 was induced approximately sixfold during C2C12 differentiation (Fig. 1, B–D, P < 0.05 relative to myoblasts and day 1). Ulk1, Beclin1, and LC3 represent the essential regulators of autophagy; therefore, these data suggest increased overall capacity for autophagy during myoblast differentiation.
Fig. 1.
Autophagy-related protein content during myoblast differentiation. A and B: representative contrast light images of differentiating myoblast and immunoblot images of Ulk1, Beclin1, LC3-I, LC3-II, p62, cytochrome c oxidase complex IV (COX4), cytochrome c (Cyt C), myogenin (myog), and tubulin. C: quantification of Beclin1, Ulk1, and total LC3, collectively an index of autophagy capacity. D: quantification of p62, LC3-II, and LC3-II/I ratio collectively an index of autophagy flux. E: quantification of COX4 and Cyt C as a measure
of mitochondrial content. Results are represented as means ± SE from n = 4 separate cell culture experiments per day; n = 4 for each time point. *P < 0.05, significantly different from day 1. **P < 0.05, significantly different from day 2.
To assess ongoing autophagic degradation activity (i.e., autophagy flux) during differentiation, the PE-conjugated form of LC3 (LC3-II) and p62 were measured. Expression of p62 protein was reduced by 52% in myotubes at day 4 during differentiation compared with proliferating myoblasts (Fig. 1, B–D, P < 0.05). LC3-II protein content increased by approximately twofold at days 3 and 4 compared with proliferating myoblasts (Fig. 1, B–D, P < 0.05). These data suggest that autophagy flux is increased along with myogenic differentiation.
A more robust autophagy system during myogenic differentiation may reflect the development of the metabolic capacity of the myotube (15, 43). Consistently, the protein levels of the electron carrier cytochrome c (Cyt C) and electron transport chain protein in complex IV (COX4), which facilitates transfer of electrons to oxygen, were significantly greater at days 2–4 (35–50%) compared with those in the myoblasts (Fig. 1, B–E, P < 0.05). This suggests that the mitochondria reticulum is expanding during the differentiation process, which may demand greater autophagy for the maintenance of the growing mitochondrial network.
Early increase in autophagy proteins and late increase in autophagy flux during skeletal muscle regeneration in vivo.
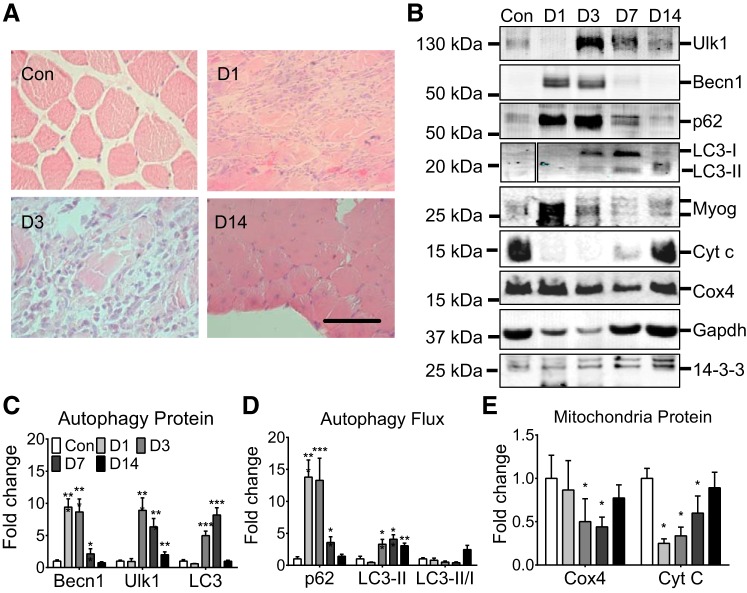
To determine if induction of autophagy proteins and activation of autophagy are associated with muscle injury and repair in vivo, Ctx was injected into the left hindlimb TA muscle of WT C57BL/6 mice. Beclin1 was profoundly induced (10-fold) at day 1 postinjury relative to the contralateral control TA muscle (P < 0.001), and the induction was reduced to basal level by day 14 postinjury (Fig. 2, B–D). Ulk1 and LC3 were induced with a peak at day 3 (P < 0.001) and day 7 (P < 0.001) postinjury relative to the contralateral control, respectively, and LC3 returned to the basal levels at day 14 while Ulk1 remained slightly elevated (2-fold, P < 0.01: Fig. 2, B–D). p62 was also profoundly induced at day 1 postinjury relative to the contralateral control (P < 0.001) but reduced significantly at day 7 and returned to the baseline by day 14. The rapid reduction of p62 at day 7 suggests an increase of autophagy flux (Fig. 2, B–D). This was also confirmed by an increase in LC3-II at days 7 and 14 postinjury (Fig. 2, B–D, P < 0.01). There was a significant reduction in COX4 and Cyt C content at days 3 and 7 relative to the contralateral control (P < 0.05), suggesting loss of mitochondria during muscle injury (Fig. 2E). Collectively, these data demonstrate 1) that autophagy proteins (enhanced autophagy capacity) increase transiently during early phase of muscle regeneration; and 2) that autophagy flux increases at around day 7, at the time after myogenic differentiation and during maturation in muscle regeneration (45).
Fig. 2.
Autophagy-related protein content during adult skeletal muscle regeneration. A: representative histological images of regenerating muscle at 1, 3, and 14 days postinjury relative to contralateral uninjured control: scale bar = 200 μm. B: representative immunoblot images of Ulk1, Beclin1, p62, LC3-I, LC3-II, myogenin, Cyt C, COX4, GAPDH, and 14-3-3- for cardiotoxin (Ctx)-injured muscles and contralateral control muscles at 1, 3, 7, and 14 days postinjury (D1, D3, D7, and D14). C: quantification of Beclin1, Ulk1, and total LC3, collectively an index of autophagy capacity, during muscle regeneration. D: quantification of p62, LC3-II, and LC3-II/I ratio collectively an index of autophagy flux, during muscle regeneration. E: quantification of COX4 and Cyt C as a measure of mitochondrial content. Results are represented as means ± SE from n = 5 mice for each day. *P < 0.05, **P < 0.01, ***P < 0.001, significantly different from contralateral control.
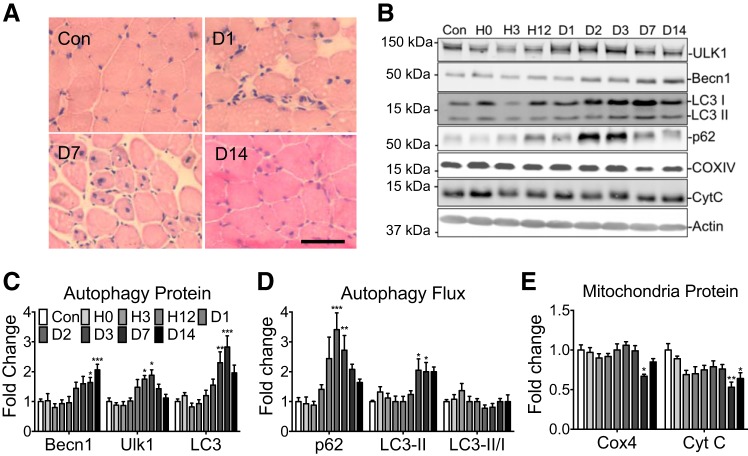
To further investigate the autophagic response during physiological muscle regeneration, we subjected mice to I/R by placing a rubber band tourniquet above the femur of the hindlimb to occlude the circulation for 1 h followed by release of the occlusion. It has been shown that I/R leads to prolonged recovery compared with myotoxic injury despite no difference in initial cell death (42). We observed clear evidence of muscle regeneration following I/R (Fig. 3A). Similar to Ctx-induced injury, I/R caused increased expression of Beclin1 (P < 0.001), Ulk1 (P < 0.05), and LC3 (P < 0.01), peaking between days 3 and 14 compared with the sham operated control (Con) (Fig. 3, B–D). p62 gradually increased following injury peaking at day 2 (P < 0.001) yet remained elevated through day 7 (P < 0.01) (Fig. 3, B–D) followed by a reduction to basal levels at day 14. The reduction of p62 is indicative of increased autophagy flux, which is further supported by increased LC3-II protein at days 3 and 7 during the later phases of regeneration (P < 0.05) (Fig. 3, B–D). There was a significant reduction in COX4 (P < 0.05) and Cyt C (P < 0.01) content at day 7, suggesting loss of mitochondria following I/R (Fig. 3E).
Fig. 3.
Autophagy-related protein content following hindlimb ischemia-reperfusion injury. A: representative histological images of regenerating plantaris muscle at 24 h and 7 and 14 days postinjury relative to uninjured control (Con): scale bar = 100 μm. B: representative immunoblot images of Ulk1, Beclin1, p62, LC3-I, LC3-II, Cyt C, COX4, and actin from the plantaris muscle at 0, 3, 12, 24, 48, and 72 h, and 7 and 14 days post-ischemia-reperfusion (I/R) injury and Con control. C: quantification of Beclin1, Ulk1, and total LC3, collectively an index of autophagy capacity, during muscle regeneration. D: quantification of p62, LC3-II collectively as an index of autophagy flux, during muscle regeneration. E: quantification of COX4 and Cyt C as a measure of mitochondrial content. Results are represented as means ± SE for n = 6 mice for each time point. *P < 0.05, **P < 0.01, ***P < 0.0001 significantly different from Con.
Ulk1 is required for mitochondrial remodeling in skeletal muscle regeneration in vivo.
To ascertain the role of Ulk1-mediated autophagy, we crossed myogenin-Cre mice with floxed-Ulk1 mice to generate skeletal muscle-specific Ulk1 knockout mice (MKO). Ulk1 has been shown to be essential for mitophagy (28). We first tested if MKO mice have normal muscle regeneration following Ctx injury. We chose muscle regeneration at day 7 postinjury because the most dramatic changes in autophagy proteins and autophagy flux occur at that time (Fig. 2). Although TA muscle mass was significantly lower than that of the uninjured muscles in both MKO and WT mice (main effect of injury, P < 0.001), there was no interaction among injured and uninjured MKO and WT mice (injury × genotype, Fig. 4A). We performed in vivo muscle functional analysis for ankle dorsiflexor muscles and detected a significant interaction between genotype and injury (P < 0.001). There was no difference in muscle strength between uninjured MKO and WT mice (Fig. 4B; P = 0.93). This suggests that inhibition of basal Ulk1-mediated autophagy does not have negative effects on muscle strength in the absence muscle injury. However, MKO mice generated significantly less muscle strength (48% recovery) compared with that of WT mice (70% recovery) 7 days after muscle injury during regeneration (Fig. 4B; P < 0.05). It is worth noting that the role of autophagy on neuromuscular junction formation has not been thoroughly investigated and was not the emphasis of this study and indeed strength data should be considered in the context that impaired autophagy may alter muscle fiber excitability during regeneration in the MKO mice. The expression of mitochondrial proteins COX4 and Cyt C was significantly reduced at day 7 following injury in both WT and MKO mice, independent of genotype (main effect of injury, P < 0.05), indicative of reduced mitochondrial content (Fig. 4, C and D). Ulk1 deficiency did not affect Beclin1 induction or p62 and LC3 protein accumulation with muscle injury compared with the WT littermate controls (main effect of injury, P < 0.01; Fig. 4, C–F).
Fig. 4.
Muscle-specific Ulk1 deficiency blunts functional muscle regeneration following Ctx. A: quantification of tibialis anterior muscle mass and body mass for muscle-specific Ulk1 knockout mice (MKO) mice and littermate wild-type (WT) mice at day 7 postinjury. B: torque-frequency analysis shown relative to uninjured WT (WT-Con) at day 7 postinjury. Mass and torque results are represented as means ± SE for n = 5 WT-Con and WT-Ctx and n = 7 MKO-Con and MKO-Ctx. C: quantification of COX4 and Cyt C at day 7 postinjury. All immunoblot results are represented as means ± SE for n = 5 mice for each genotype/injury. D: representative immunoblot images of Ulk1, Beclin1, p62, LC3-I, LC3-II, Cyt C, COX4, and 14-3-3 for injured muscles (day 7 postinjury) and contralateral control for MKO mice and littermate WT mice. E: quantification of Beclin1, Ulk1, and total LC3, collectively an index of autophagy capacity, at day 7 postinjury. F: quantification of p62, LC3-II collectively an index of autophagy flux, at day 7 postinjury.
To avoid any compensatory or developmental alterations in MKO, we obtained inducible muscle-specific Ulk1 knockout mice (iMKO), which allowed for deletion of the Ulk1 gene from adult skeletal muscle. We subjected iMKO and WT littermates to tourniquet-induced I/R and performed protein and functional analysis at day 14. Under basal conditions there was no difference in gastrocnemius (GA) wet weight between WT and iMKO; however, there was a significant reduction in weight in both WT (30%) and iMKO (25%) following I/R injury (main effect of injury, P < 0.001; Fig. 4A). Again, there was no difference in strength of posterior hindlimb muscles between WT and iMKO at baseline; however, I/R resulted in a dramatic loss of strength in WT (~30% recovery) that was exacerbated in iMKO (~10% recovery) compared with non-I/R control groups (significant interaction, P < 0.01; Fig. 5B). There was a significant interaction between genotype and injury (P < 0.05) showing that expression of COX4 and Cyt C was significantly reduced in iMKO following I/R (~40 and 50%, respectively), while there was no significant loss of COX4 and Cyt C expression in WT mice following I/R (Fig. 5, C and D). These findings raise the question with regard to the role of Ulk1, presumably mitophagy, in mitochondrial network remodeling during muscle regeneration, possibly suggesting impaired remodeling of the mitochondrial network and/or biogenesis due to deletion of the Ulk1 gene. Consistent with the time-course experiment, there was no difference in Ulk1 expression in WT I/R vs. WT Con at day 14 (Fig. 5, C–E; P = 0.36). Beclin1, p62, and LC3-II were induced in both iMKO and WT following I/R compared with respective Con (main effect of injury, P < 0.05), suggestive of increased autophagy capacity (Fig. 5, C–F).
Ulk1 is required for mitochondrial network remodeling in skeletal muscle during regeneration in vivo.
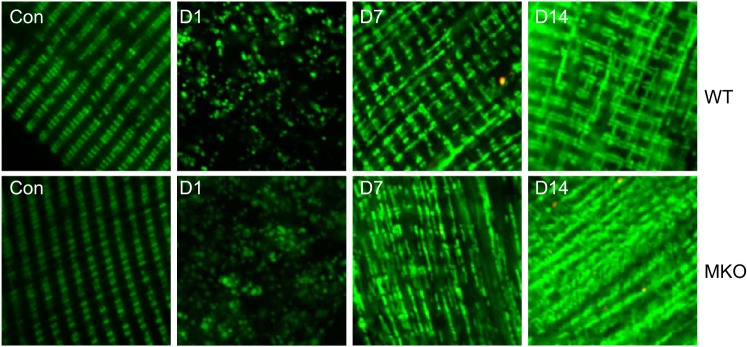
Our findings in muscle regeneration in vivo suggest that Ulk1-mediated autophagy, presumably mitophagy, is important for recovery of mitochondrial protein expression, presumably mitochondrial remodeling (less recovery of mitochondrial protein expression in MKO mice), and most importantly for functional regeneration (impaired muscle contractile function in MKO mice after injury). To address the importance of Ulk1 in the remodeling of mitochondrial network during muscle regeneration, we performed somatic gene transfer of a novel mitochondrial reporter gene, pMitoTimer (29), in MKO mice and WT littermate mice followed by Ctx-induced injury. This mitochondrial reporter gene was recently established by us for assessment of mitochondrial content, structure, oxidative stress, and mitophagy (29). Ten days before muscle injury, left hindlimb FDB muscles from MKO and WT littermates were transfected with pMitoTimer. At 1, 3, 7, and 14 days postinjury, confocal microscopy was performed to observe the mitochondrial network in injured and uninjured FDB muscles (Fig. 6). Normal striated alignment of intramyofibrillar mitochondria along Z-lines was observed with no noticeable difference in mitochondrial networks in uninjured FDB muscle between MKO and WT mice. Ctx-induced injury resulted in profound mitochondrial fragmentation at days 1 and 3, but a decent recovery of striated alignment of mitochondria was observed at day 7 and a nearly normal structure at day 14 in WT mice. This mitochondrial network recovery was significantly delayed in MKO mice (Fig. 6). These data strongly suggest that Ulk1 is necessary for proper mitochondrial remodeling during the maturation phase of muscle regeneration, which may account for impaired functional recovery after Ctx injury in MKO mice.
Fig. 6.
Mitochondrial network organization during muscle regeneration. Confocal microscopy analysis of the MitoTimer reporter gene was performed for somatic gene transfer-transfected flexor digitorum longus muscle following muscle injury at 1, 7, and 14 days of recovery and compared with the contralateral uninjured muscle (n = 4 mice per time point).
DISCUSSION
Autophagy is an important cellular process for maintaining organelle quality in skeletal muscle (30, 38), but little is known about its induction and functional role under conditions of stress and injury. We have previously demonstrated that endurance exercise training enhances autophagy protein expression and basal autophagy and that autophagy, specifically the expression of Beclin1, is necessary for mitochondrial biogenesis in skeletal muscle and for improved physical performance with exercise training (32). Herein, we tested the hypothesis that autophagy plays an important role during muscle regeneration, which typically occurs following trauma or strenuous bouts of exercise. We observed a clear induction of autophagy protein expression in a cell culture model of myogenic differentiation (Fig. 1). We then showed pronounced, transient induction of autophagy proteins early during muscle regeneration followed by increased autophagy flux in the late phase of maturation during muscle regeneration (Figs. 2 and 3). Importantly, mice with muscle-specific deletion of the Ulk1 gene, which has been shown to be essential for mitophagy (mitochondrial-specific autophagy) (39), displayed significantly impaired functional recovery with attenuated recovery of mitochondrial electron transport chain protein expression (Fig. 5). Using a novel mitochondrial reporter gene, pMitoTimer, we have obtained clear evidence that remodeling of mitochondrial network during the late stage of muscle regeneration is dependent on functional Ulk1 in skeletal muscle (Fig. 6). We conclude that autophagy, presumably mitophagy, is necessary for the mitochondrial remodeling and timely recovery of muscle function after injury.
We can approximate two phases of mitochondrial remodeling after injury: 1) mitochondrial degradation, and 2) mitochondrial network reorganization. Mitochondrial degradation is noticeable by fragmented mitochondria and a clear absence of mitochondria localized around the contractile units in both WT and MKO mice at 24 h postinjury (Fig. 6). At days 7 and 14 postinjury, the mitochondrial network becomes noticeable again in WT mice, even though total mitochondrial content has not yet returned to normal (Cyt C and COX4 in Fig. 4). In contrast, the mitochondrial network is much less developed in MKO mice at days 7 and 14 postinjury, and total mitochondrial content is even less compared with WT mice (Fig. 5). This suggests that Ulk1-mediated autophagy is important for the reorganization of the mitochondrial network after muscle damage, which is critical for optimal functional recovery.
Muscle regeneration is a metabolically demanding process that may depend on a well-developed mitochondrial network. Wagatsuma et al. (43) used chloramphenicol to inhibit mitochondrial protein synthesis after muscle damage and showed a reduction in muscle fiber cross-sectional area at 7 days, suggesting that an impaired mitochondrial network can negatively affect muscle repair. Nichenko et al. (33) advanced this narrative and connected mitochondrial maintenance to autophagy by chronically treating mice with an autophagy inhibitor (3-methyladenine) and showing less recovery of mitochondrial enzyme activities and muscle strength at 14 days after injury. Herein, we used a gene deletion method to precisely examine how a healthy mitochondrial network may be linked to muscle regeneration through autophagy. Indeed, the absence of Ulk1 unequivocally was associated with less recovery of muscle strength (Figs. 4 and 5) and delayed mitochondrial remodeling (Figs. 5 and 6). A true causal link between delayed mitochondrial remodeling (protein expression and/or proper alignment) and muscle weakness cannot be drawn from this study, but this work does highlight the importance of autophagy in functional regeneration in skeletal muscle.
A role for autophagy in muscle regeneration can be drawn from recently findings in satellite cells. Tang and Rando (41) reported muscle fibers and isolated satellite cells treated with the autophagy inhibitors 3-methyladenine and chloroquine had less DNA replication, indicating autophagy is critical for satellite cell activation. They concluded that satellite cell activation is metabolically demanding and that autophagy played an important role in facilitating satellite cell transition from quiescence to activation. Garcia-Prat et al. (19) demonstrated that defective autophagy was responsible for satellite cell senescence, a feature of muscle aging, and that rescuing autophagy induction led to greater fiber cross-sectional area 8 days after injury in skeletal muscle from aged mice. Here, our genetic model did not affect satellite cells, which allowed us to gain insight the role of autophagy in differentiated/mature myofibers in skeletal muscle (Figs. 1–3).
To explore the activity of autophagy and the requirement of Ulk1-mediated autophagy during recovery from injury, we utilized two established models of muscle injury, Ctx and I/R. A side-by-side functional and histological comparison has been previously reported (42). Ctx and I/R injury cause temporary functional deficits due to cellular damage to critical contractile, cytoskeletal, and metabolic proteins (25, 37, 44). I/R injury, however, is more heterogeneous leading to regional differences in regeneration within the injured muscle compared with Ctx injury. The difference between these two models is supported by differences in central nucleated fibers (marker of ongoing regeneration) and inflammatory cell infiltration (42). This notion is further supported by our analysis of autophagy flux.
Autophagy flux defines the rate of ongoing autophagic degradation activity within a cell. The most ubiquitous markers of autophagy flux are expression and localization of the structural proteins LC3 and p62 (26). Although not part of our intended analysis, there were clear differences in p62 and LC3-II protein content during regeneration following Ctx and I/R injuries. Specifically, peak p62 and LC3-II expression was much greater following Ctx compared with I/R injury (Figs. 2 and 3), likely reflecting the difference in heterogeneity of the injury. Additionally, p62, which is responsible for targeting damaged proteins to the autophagosome, peaked at day 1 after Ctx injury compared with day 2 after I/R injury (Figs. 2 and 3), demonstrating a different timing of maximal protein damage and a need for degradation processes. This finding supports that there is a difference of pathophysiology between the two injuries models and that therefore the timing and magnitude of the autophagy process are different. That said, current technologies to assess autophagy flux are limited to multiple static measurements of autophagy gene transcript levels as well as protein expression and localization performed over a time course (26). To better distinguish assess autophagy flux, it is best to conduct additional experiments, in which a drug (such as bafilomycin or chloroquine) is used target the final step of autophagy (26). While our data support the overall hypothesis that autophagy capacity and flux are greater during muscle regeneration, more precise experiments are required to further elucidate autophagy flux.
In conclusion, using ex vivo and in vivo approaches that span the cellular to whole tissue, we demonstrate that increased autophagy capacity and flux occur during the muscle regenerative process. Skeletal muscle-specific Ulk1 deficiency causes impaired functional muscle regeneration and mitochondrial network reorganization. It is currently unclear what cellular responses to injury (e.g., inflammatory cells migration) are leading to autophagy induction, but it is clear that autophagy, presumably mitophagy, plays an important role during muscle regeneration.
GRANTS
This work was supported by National Institutes of Health (NIH) Grant R01-AR-0050429 (to Z. Yan), American Heart Association (AHA) Postdoctoral Fellowship (14POST20450061; to R. C. Laker), NIH Grant T32-HL-007284-38 and American Heart Association Postdoctoral Fellowship (12POST12030231; to J. A. Call), and NIH Grant T32-HL-007284-37 and AHA Grant 114PRE20380254 (to R. J. Wilson).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
J.A.C., M.K., and Z.Y. conceived and designed research; J.A.C., R.J.W., R.C.L., and M.Z. performed experiments; J.A.C., R.J.W., R.C.L., and M.Z. analyzed data; J.A.C., R.J.W., M.Z., and Z.Y. interpreted results of experiments; J.A.C., R.J.W., R.C.L., and Z.Y. prepared figures; J.A.C. and R.J.W. drafted manuscript; J.A.C., R.J.W., M.K., and Z.Y. edited and revised manuscript; J.A.C., R.J.W., R.C.L., M.Z., M.K., and Z.Y. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Karyn Esser for the generous gift of HAS-MerCreMer mice. We also thank Dr. David F. Kashatus and the members of the Zhen Yan and Kyle L. Hoehn laboratories for critical feedback and discussion.
REFERENCES
- 1.Baltgalvis KA, Call JA, Cochrane GD, Laker RC, Yan Z, Lowe DA. Exercise training improves plantar flexor muscle function in mdx mice. Med Sci Sports Exerc 44: 1671–1679, 2012. doi: 10.1249/MSS.0b013e31825703f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernet JD, Doles JD, Hall JK, Kelly Tanaka K, Carter TA, Olwin BB. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med 20: 265–271, 2014. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blau HM, Cosgrove BD, Ho AT. The central role of muscle stem cells in regenerative failure with aging. Nat Med 21: 854–862, 2015. doi: 10.1038/nm.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonheur JA, Albadawi H, Patton GM, Watkins MT. A noninvasive murine model of hind limb ischemia-reperfusion injury. J Surg Res 116: 55–63, 2004. doi: 10.1016/S0022-4804(03)00232-4. [DOI] [PubMed] [Google Scholar]
- 5.Call JA, Chain KH, Martin KS, Lira VA, Okutsu M, Zhang M, Yan Z. Enhanced skeletal muscle expression of extracellular superoxide dismutase mitigates streptozotocin-induced diabetic cardiomyopathy by reducing oxidative stress and aberrant cell signaling. Circ Heart Fail 8: 188–197, 2015. doi: 10.1161/CIRCHEARTFAILURE.114.001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Call JA, Lowe DA. Eccentric contraction-induced muscle injury: reproducible, quantitative, physiological models to impair skeletal muscle’s capacity to generate force. Methods Mol Biol 1460: 3–18, 2016. doi: 10.1007/978-1-4939-3810-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Call JA, Warren GL, Verma M, Lowe DA. Acute failure of action potential conduction in mdx muscle reveals new mechanism of contraction-induced force loss. J Physiol 591: 3765–3776, 2013. doi: 10.1113/jphysiol.2013.254656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell 15: 344–357, 2008. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chargé SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev 84: 209–238, 2004. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 10.Conboy IM, Conboy MJ, Wagers AJ, Girma ER, Weissman IL, Rando TA. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature 433: 760–764, 2005. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- 11.De Palma C, Morisi F, Cheli S, Pambianco S, Cappello V, Vezzoli M, Rovere-Querini P, Moggio M, Ripolone M, Francolini M, Sandri M, Clementi E. Autophagy as a new therapeutic target in Duchenne muscular dystrophy. Cell Death Dis 3: e418, 2012. doi: 10.1038/cddis.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorsey FC, Rose KL, Coenen S, Prater SM, Cavett V, Cleveland JL, Caldwell-Busby J. Mapping the phosphorylation sites of Ulk1. J Proteome Res 8: 5253–5263, 2009. doi: 10.1021/pr900583m. [DOI] [PubMed] [Google Scholar]
- 13.Drake JC, Wilson RJ, Yan Z. Molecular mechanisms for mitochondrial adaptation to exercise training in skeletal muscle. FASEB J 30: 13–22, 2016. doi: 10.1096/fj.15-276337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drolet BC, Okhah Z, Phillips BZ, Christian BP, Akelman E, Katarincic J, Schmidt ST. Evidence for safe tourniquet use in 500 consecutive upper extremity procedures. Hand (NY) 9: 494–498, 2014. doi: 10.1007/s11552-014-9667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duguez S, Féasson L, Denis C, Freyssenet D. Mitochondrial biogenesis during skeletal muscle regeneration. Am J Physiol Endocrinol Metab 282: E802–E809, 2002. doi: 10.1152/ajpendo.00343.2001. [DOI] [PubMed] [Google Scholar]
- 16.Egan D, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy 7: 643–644, 2011. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461, 2011. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank M, Duvezin-Caubet S, Koob S, Occhipinti A, Jagasia R, Petcherski A, Ruonala MO, Priault M, Salin B, Reichert AS. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys Acta 1823: 2297–2310, 2012. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 19.García-Prat L, Martínez-Vicente M, Perdiguero E, Ortet L, Rodríguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, Sandri M, Muñoz-Cánoves P. Autophagy maintains stemness by preventing senescence. Nature 529: 37–42, 2016. doi: 10.1038/nature16187. [DOI] [PubMed] [Google Scholar]
- 20.Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510, 2008. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris JB. Phospholipases in snake venoms and their effects on nerve and muscle. Pharmacol Ther 31: 79–102, 1985. doi: 10.1016/0163-7258(85)90038-5. [DOI] [PubMed] [Google Scholar]
- 22.Hawke TJ, Garry DJ. Myogenic satellite cells: physiology to molecular biology. J Appl Physiol (1985) 91: 534–551, 2001. [DOI] [PubMed] [Google Scholar]
- 23.Hochreiter-Hufford AE, Lee CS, Kinchen JM, Sokolowski JD, Arandjelovic S, Call JA, Klibanov AL, Yan Z, Mandell JW, Ravichandran KS. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 497: 263–267, 2013. doi: 10.1038/nature12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiao H, Su GQ, Dong W, Zhang L, Xie W, Yao LM, Chen P, Wang ZX, Liou YC, You H. Chaperone-like protein p32 regulates ULK1 stability and autophagy. Cell Death Differ 22: 1812–1823, 2015. doi: 10.1038/cdd.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalogeris T, Bao Y, Korthuis RJ. Mitochondrial reactive oxygen species: a double edged sword in ischemia/reperfusion vs preconditioning. Redox Biol 2: 702–714, 2014. doi: 10.1016/j.redox.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12: 1–222, 2016. [Erratum in Autophagy 12: 443, 2016.] 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell 129: 999–1010, 2007. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 112: 1493–1502, 2008. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laker RC, Xu P, Ryall KA, Sujkowski A, Kenwood BM, Chain KH, Zhang M, Royal MA, Hoehn KL, Driscoll M, Adler PN, Wessells RJ, Saucerman JJ, Yan Z. A novel MitoTimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J Biol Chem 289: 12005–12015, 2014. doi: 10.1074/jbc.M113.530527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res 8: 3–5, 2005. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 31.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6: 463–477, 2004. doi: 10.1016/S1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 32.Lira VA, Okutsu M, Zhang M, Greene NP, Laker RC, Breen DS, Hoehn KL, Yan Z. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. FASEB J 27: 4184–4193, 2013. doi: 10.1096/fj.13-228486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichenko AS, Southern WM, Atuan M, Luan J, Peissig KB, Foltz SJ, Beedle AM, Warren GL, Call JA. Mitochondrial maintenance via autophagy contributes to functional skeletal muscle regeneration and remodeling. Am J Physiol Cell Physiol 311: C190–C200, 2016. doi: 10.1152/ajpcell.00066.2016. [DOI] [PubMed] [Google Scholar]
- 34.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145, 2007. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 35.Pauly M, Daussin F, Burelle Y, Li T, Godin R, Fauconnier J, Koechlin-Ramonatxo C, Hugon G, Lacampagne A, Coisy-Quivy M, Liang F, Hussain S, Matecki S, Petrof BJ. AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am J Pathol 181: 583–592, 2012. doi: 10.1016/j.ajpath.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Pawlikowski B, Pulliam C, Betta ND, Kardon G, Olwin BB. Pervasive satellite cell contribution to uninjured adult muscle fibers. Skelet Muscle 5: 42, 2015. doi: 10.1186/s13395-015-0067-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramadasan-Nair R, Gayathri N, Mishra S, Sunitha B, Mythri RB, Nalini A, Subbannayya Y, Harsha HC, Kolthur-Seetharam U, Srinivas Bharath MM. Mitochondrial alterations and oxidative stress in an acute transient mouse model of muscle degeneration: implications for muscular dystrophy and related muscle pathologies. J Biol Chem 289: 485–509, 2014. doi: 10.1074/jbc.M113.493270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, Scorrano L, Rudolf R, Sandri M. Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J 29: 1774–1785, 2010. doi: 10.1038/emboj.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 15: 741–750, 2013. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Southern WM, Nichenko AS, Shill DD, Spencer CC, Jenkins NT, McCully KK, Call JA. Skeletal muscle metabolic adaptations to endurance exercise training are attainable in mice with simvastatin treatment. PLoS One 12: e0172551, 2017. doi: 10.1371/journal.pone.0172551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang AH, Rando TA. Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J 33: 2782–2797, 2014. doi: 10.15252/embj.201488278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vignaud A, Hourde C, Medja F, Agbulut O, Butler-Browne G, Ferry A. Impaired skeletal muscle repair after ischemia-reperfusion injury in mice. J Biomed Biotechnol 2010: 724914, 2010. doi: 10.1155/2010/724914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagatsuma A, Kotake N, Yamada S. Muscle regeneration occurs to coincide with mitochondrial biogenesis. Mol Cell Biochem 349: 139–147, 2011. doi: 10.1007/s11010-010-0668-2. [DOI] [PubMed] [Google Scholar]
- 44.Weiser MR, Williams JP, Moore FD Jr, Kobzik L, Ma M, Hechtman HB, Carroll MC. Reperfusion injury of ischemic skeletal muscle is mediated by natural antibody and complement. J Exp Med 183: 2343–2348, 1996. doi: 10.1084/jem.183.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, Hart R, Lin L, Thurmond FA, Williams RS. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem 278: 8826–8836, 2003. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- 46.Yan Z, Lira VA, Greene NP. Exercise training-induced regulation of mitochondrial quality. Exerc Sport Sci Rev 40: 159–164, 2012. doi: 10.1097/JES.0b013e3182575599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev 93: 23–67, 2013. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]