Abstract
The neuronal Na+/H+ exchanger NHE5 isoform not only resides in the plasma membrane but also accumulates in recycling vesicles by means of clathrin-mediated endocytosis. To further investigate the underlying molecular mechanisms, a human brain cDNA library was screened for proteins that interact with the cytoplasmic C-terminal region of NHE5 by using yeast two-hybrid methodology. One candidate cDNA identified by this procedure encoded β-arrestin2, a specialized adaptor/scaffolding protein required for internalization and signaling of members of the G protein-coupled receptor superfamily. Direct interaction between the two proteins was demonstrated in vitro by GST fusion protein pull-down assays. Sequences within the N-terminal receptor activation-recognition domain and the C-terminal secondary receptor-binding domain of β-arrestin2 conferred strong binding to the C terminus of NHE5. Full-length NHE5 and β-arrestin2 also associated in intact cells, as revealed by their coimmunoprecipitation from extracts of transfected CHO cells. Moreover, ectopic expression of both proteins caused a redistribution of β-arrestin2 from the cytoplasm to vesicles containing NHE5, and significantly decreased the abundance of the transporter at the cell surface. Comparable results were also obtained for the β-arrestin1 isoform. These data reveal a broader role for arrestins in the trafficking of integral plasma membrane proteins than previously recognized.
Keywords: endocytosis
Mammalian Na+/H+ exchangers (NHEs) constitute a family of at least nine twelve-membrane-spanning domain transporters that are differentially sorted to discrete membrane compartments where they engage in varied physiological processes (1, 2). For instance, the ubiquitous NHE1 isoform resides exclusively at the plasma membrane and is chiefly responsible for maintaining steady-state cytoplasmic pH and cell volume.
By comparison, two closely related isoforms, NHE3 and NHE5, fulfill more specialized roles. NHE3 is situated at the apical membrane of renal and intestinal epithelial cells and contributes prominently to systemic sodium, acid-base, and fluid-volume homeostasis (1, 3). In addition, NHE3 is internalized by a clathrin-dependent process into early endosomes, ultimately accumulating in a subapical or pericentriolar cluster of recycling endosomes (4–7). NHE5 is distinguished from NHE3 by its predominant expression in neuron-enriched regions of the brain (8), but otherwise is sorted in an analogous manner when ectopically expressed in neuronal and nonneuronal cells (9). This trafficking pathway is thought to serve as an efficient means of rapidly shuttling functional transporters to and from the cell surface in response to diverse signals. Moreover, in the case of NHE3, there is some indication that it also plays a crucial role in the rapid acidification of early/recycling endosomes; a condition that promotes efficient internalization and processing of cargo along the endocytic pathway (4, 10, 11). NHE5 conceivably fulfills an analogous function in neurons, but this function has yet to be demonstrated empirically.
At present, virtually nothing is known about the underlying molecular mechanisms that control the membrane sorting of NHE3 or NHE5. Only a limited number of sorting molecules have been identified that direct itinerant plasma membrane proteins along the endocytic pathway. These proteins include the heterotetrameric adaptor protein (AP)-2, which recognizes degenerate tyrosine-based motifs within the cytoplasmic tail of many integral plasmalemmal proteins (12, 13). Upon binding its target, AP-2 recruits clathrin to form coated pits followed by the binding of several accessory proteins required for vesicle formation and fusion along the endocytic pathway. Another class of more specialized adaptor proteins are the β-arrestins [β-arrestin1 (β-Arr1) and β-arrestin2 (β-Arr2)] that interact almost exclusively with specific phospho-serine/threonine residues of ligand-activated, seven-membrane-spanning, G protein-coupled receptors (GPCRs) (14, 15). The β-arrestins not only play crucial roles in desensitization of activated GPCRs but also promote their internalization by interacting spatially and temporally with components of the endocytic trafficking machinery, including clathrin (16), AP-2 (17), NSF (18), and ARF6 (19).
In this study, we report the unexpected finding that β-Arr1 and β-Arr2 interact directly with the neuronal NHE5 isoform and promote its accumulation in endocytic vesicles. These data support a role for β-arrestins in regulating the trafficking of integral plasma membrane proteins apart from receptor–ligand complexes.
Materials and Methods
Reagents. Carrier-free 22NaCl and [35S]methionine were obtained from NEN Life Science Products (PerkinElmer Canada, Woodbridge, ON, Canada). 5-(N-ethyl-N-isopropyl)amiloride was purchased from Sigma. α-MEM, FBS, penicillin/streptomycin, and trypsin-EDTA were purchased from Invitrogen. All other chemicals and reagents used in these experiments were obtained from BioShop Canada (Burlington, ON, Canada) or Fisher Scientific and were of the highest grade.
Yeast Two-Hybrid Screening. The human NHE5 cDNA encoding amino acids G544-L896 of the cytoplasmic C terminus was amplified by PCR methodology and subcloned into the pAS2–1 yeast expression vector (Clontech) in-frame with the GAL4 DNA-binding domain. The pAS2–1/NHE5 (amino acids 544–896) plasmid was cotransformed with a human brain cDNA library subcloned into the pACT2 vector in-frame with the GAL4 activation domain (Clontech) into the Saccharomyces cerevisiae strain AH109 by using a modified lithium acetate method (20). More than 2 × 106 clones were screened on the synthetic complete media containing 0.67% bacto-yeast nitrogen base without amino acids, 2% glucose, 1.5% bacto-agar, and 0.2% Leu-Trp-His-Ade drop-out mix. Colonies positive for β-gal activity were isolated and library-derived cDNAs in pACT2 that code for putative NHE5-interacting proteins were rescued from yeast cells and directly transformed into DH5α cells. The identities of the clones were determined by DNA sequencing. One of the clones identified from this screen was B1–7, which comprises the C-terminal half of β-Arr2, encompassing amino acids H211-C409.
Gene Construction and Plasmids. Full-length cDNAs of β-Arr2 and its paralog β-Arr1 were cloned from human brain mRNA by RT-PCR methodology, subcloned into a pCMV-based mammalian expression vector, and sequenced to verify their fidelity. For each β-arrestin, the myc-epitope was inserted at their extreme C terminus (β-Arr1myc and β-Arr2myc), and expression was verified by immunofluorescence microscopy. Dominant-negative constructs of β-Arr1/V53D and β-Arr2/V54D were generously provide by J. Staňková (University of Sherbrooke, Sherbrooke, QC, Canada) and S. S. Ferguson (University of Western Ontario, London, ON, Canada), respectively. The mammalian expression plasmid containing the human NHE5 cDNA tagged with a triple influenza virus hemagglutinin (HA) epitope in its first exomembranous loop (NHE5HA) was described (9).
Cell Culture and Transfection. CHO cells deficient in plasmalemmal NHE activity (AP-1 cells) (21) were maintained in complete α-MEM supplemented with 10% FBS/100 units/ml penicillin/100 μg/ml streptomycin/25 mM NaHCO3, pH 7.4, and incubated in an humidified atmosphere of 95% air and 5% CO2 at 37°C. The mammalian expression plasmids were transfected into AP-1 cells by using LipofectAMINE-2000 (Invitrogen).
GST Pull-Down Assay. For producing GST fusion proteins (22), different parts of the β-Arr1 and β-Arr2 cDNAs were amplified by PCR and inserted in-frame with GST using the bacterial pGEX-2T expression vector (Amersham Biosciences). After sequence verification, plasmids were transformed into the Escherichia coli BL21 strain. Overnight cultures of clonal BL21 cells were grown, then diluted 1:10, and protein expression was induced by further incubation in the presence of 0.4 mM isopropyl-1-thio-β-d-galactopyranoside at 30°C for 3 h. The cells were lysed, and the GST-fusion proteins were purified as described (22).
The bulk of the C-terminal cytoplasmic region of NHE5 (amino acids 491–896) was transcribed and translated in vitro in the presence of [35S]methionine by using the TNT-coupled reticulocyte lysate system (Promega). After three washes in 0.5% Nonidet P-40 buffer (0.5% Nonidet P-40/1 mM EDTA/proteinase inhibitor mixture in PBS), 200 μl of the purified GST-fusion proteins bound to the Sepharose beads were incubated with in vitro-translated 35S-labeled NHE5 C terminus (35S-NHE5-C; 2–3 μl of TNT reaction mixture) overnight at 4°C. After six washes with 0.5% Nonidet P-40 buffer, proteins were eluted in SDS sample buffer (50 mM Tris·HCl, pH 6.8/1% SDS/50 mM DTT/10% glycerol/0.1% Bromophenol blue) and analyzed by SDS/PAGE. The gels were dried, and the signals were detected by using a PhosphorImager (Molecular Dynamics).
Immunoprecipitation. AP-1 cells were transiently transfected with NHE5HA singly or in combination with β-Arr1myc or β-Arr2myc (DNA ratio of 1:1). After transfection (24 h), cells were lysed with ice-cold buffer (150 mM NaCl/10 mM EDTA/0.5% Nonidet P-40/0.2% sodium deoxycholate/20 mM Tris·HCl, pH 7.5/protease inhibitors). For immunoprecipitation, cell lysates (≈250 μg of protein) were incubated with either a rabbit polyclonal anti-myc antibody A-14 (Santa Cruz Biotechnology) or mouse monoclonal anti-HA antibody HA.11 (Covance, Richmond, CA) at 4°C for 2 h, followed by incubation with protein A-Sepharose beads overnight. After washing the beads in buffer four times, immune complexes were eluted in 2× SDS sample buffer for 1 h at room temperature, and analyzed by SDS/PAGE and Western blot. Membranes were blocked in PBS containing 5% nonfat milk overnight and then incubated with either the primary mouse monoclonal anti-HA antibody (1:5,000 dilution) followed by the secondary horseradish peroxidase (HRP)-conjugated anti-mouse IgG (The Jackson Laboratory, 1:10,000 dilution), or, for the reciprocal experiment, with an HRP-conjugated anti-myc monoclonal antibody, 9E10-HRP (Roche), or alternatively with a primary rabbit anti-myc polyclonal antibody followed by a secondary HRP-conjugated anti-rabbit IgG. The immunoreactive bands were visualized by an enhanced chemiluminescence detection system (Amersham Biosciences).
Immunofluorescence Confocal Microscopy. AP-1 cells grown on glass coverslips were transiently transfected with 0.5 μg of each of the NHE5HA and β-Arr2myc cDNAs. At 24 h posttransfection, the cells were fixed in 2% paraformaldehyde/PBS for 20 min and then blocked with 5% nonfat skim milk in PBS-0.1% Triton X-100 (PBS-TX) for 15 min. Cell monolayers were incubated with anti-HA mouse monoclonal antibody and anti-myc rabbit polyclonal antibody for 1 h. After washes in PBS-TX, cells were incubated with Cy3-conjugated anti-mouse IgG (The Jackson Laboratory) and FITC-conjugated anti rabbit IgG (Molecular Probes) for 1 h. All antibodies were diluted in PBS-TX. After extensive washes with PBS-TX, cover slips were mounted onto glass slides and analyzed by confocal laser scanning microscopy by using a Zeiss inverted microscope. Images were processed by using corel photo-paint v.12.
Measurement of Cell-Surface Expression of NHE5. AP-1 cells were cultured in 10-cm dishes to 50% confluence and then transiently cotransfected with a fixed amount (2 μg) of the NHE5HA cDNA and increasing amounts of β-Arr1myc or β-Arr2myc (0–8 μg). In all cases, the total DNA per transfection was constant at 10 μg per dish by adjusting with empty parental vector pCMV. After transfection (24 h), the cells were placed on ice and washed three times with ice-cold PBS buffer containing 0.1 mM CaCl2 and 1 mM MgCl2, pH 8.0 (PBSCM). Plasma membrane proteins were isolated by using a cell-surface biotinylation assay as described (23). Briefly, plasmalemmal proteins were indiscriminately labeled with membrane-impermeable N-hydroxysulfosuccinimydyl-SS-biotin (0.5 mg/ml, Pierce) for 30 min at 4°C. The solution was then discarded and unreacted biotin was quenched three times with ice-cold PBSCM containing 20 mM glycine. The cells were then lysed in PBS buffer containing 0.2% deoxycholic acid, 1% Triton X-100, and proteinase inhibitor mixture for 30 min on ice, and then centrifuged at 12,000 × g for 30 min at 4°C to remove insoluble cellular debris. A portion of the resulting supernatant was removed and represents the total fraction. The remaining supernatant was incubated overnight with streptavidin-Sepharose beads to extract biotinylated surface-membrane proteins according to manufacturers instructions. The proteins were then resolved by SDS/PAGE followed by Western blotting. The intensity of the bands was quantified by densitometry of films exposed in the linear range and then digitally imaged and analyzed with the FluorChem system (Alpha Innotech, San Leandro, CA).
Measurement of NHE Activity. Subconfluent AP-1 cells (cultured in 12-well plates) were transiently transfected with vectors containing NHE5HA alone or together with β-Arr1myc, β-Arr2myc, β-Arr1/V53D, or β-Arr2/V54D, as indicated. Transfection efficiency reached 40–60% of total cells, and was monitored by Western blots and immunofluorescence microscopy. The protein content in each transfected sample set was measured to ensure that equal numbers of cells were analyzed. Twenty-four hours posttransfection, the cells were assayed for plasmalemmal NHE5 activity as defined by measuring the rate of H+-dependent 22Na+ influx that is inhibited by the selective NHE antagonist 5-(N-ethyl-N-isopropyl)amiloride as described (9).
Results
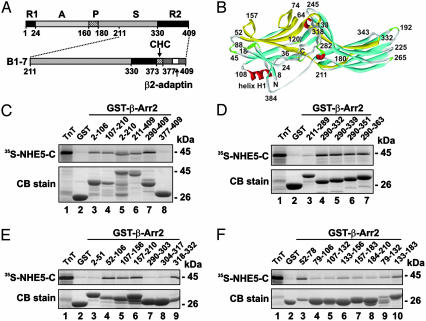
Identification of β-Arr2 as a Binding Partner of NHE5. Numerous studies have highlighted the central importance of the cytoplasmic C-terminal region of the NHEs as the principal regulatory domain of the transporter (1). Therefore, a series of bait fragments spanning the predicted cytoplasmic C terminus (residues 454–896) of human NHE5 fused to the GAL4 DNA-binding domain were used to probe a human brain cDNA library for interacting partners by using the yeast two-hybrid system (24). In this report, we describe results obtained with one of the fragments encompassing the last 353 aa (residues 544–896). More than 2 × 106 clones were screened, from which nine positive clones were obtained. One clone, B1–7, encoded the C-terminal half of β-Arr2 (residues 211–409), which contains elements involved in recognition of membrane receptors, clathrin and AP-2 (illustrated in Fig. 1 A and B). The yeast two-hybrid interaction was confirmed in mating assays by cotransforming yeast stains MATα strain Y187 and AH109 with pAS2–1/NHE5 (residues 544–896) and pACT2/β-Arr2 (residues 211–409). The NHE5 (residues 544–896) bait did not interact with the GAL4-activation domain encoded by pGAD10, nor did β-Arr2 (residues 211–409) interact with the GAL4 DNA-binding domains encoded by pAS2–1, suggesting that the interaction of these peptides was specific.
Fig. 1.
Identification of β-Arr2 as a NHE5-interacting protein. (A) Simplified diagram depicting the basic structure of β-Arr2 and the region (clones B1–7) identified by yeast two-hybrid screening of a human brain cDNA library using the C terminus of NHE5 as a probe. β-Arr2 is comprised of a receptor activation-recognition domain (A), a phosphate sensor domain (P), a secondary receptor-binding domain (S), and two regulatory domains located at the N (R1) and C (R2) termini (14). The R2 domain contains sites that interact with the clathrin heavy chain (CHC) (16) and the β2-adaptin subunit of the AP-2 adaptor complex (17). (B) Ribbon diagram of β-arrestin depicting β-strand (turquoise), α-helix (red), turns (green), and coil (gray) structures. Segments overlaid in yellow represent elements involved in binding NHE5. This figure generated with ds viewerpro 5.0 software (Accelrys; which can be accessed at www.accelrys.com) based on the 1.9-Å crystal structure of bovine β-arrestin (25). (C–F) Mapping of the interaction sites of β-Arr2 with the C terminus of NHE5 was demonstrated by protein-binding pull-down assays using purified GST–β-Arr2 fusion constructs incubated with 35S-NHE5-C (residues 491–896). Purified GST complexes were resolved by SDS/PAGE. Proteins were stained with Coomassie blue dye and radioactivity was analyzed by using a PhosphorImager. Data shown are representative of at least three independent experiments.
Delineation of the NHE5-Binding Domain of β-Arr2. To map elements within β-Arr2 that confer direct binding to the C terminus of NHE5, we performed in vitro protein-binding pull-down assays. GST-fusion proteins containing segments spanning the length of β-Arr2 were generated and incubated with 35S-NHE5-C (residues 491–896). Complexes of GST-βArr2 and 35S-NHE5-C were purified, subjected to SDS/PAGE, and analyzed by using a PhosphorImager. The strength of binding was evaluated semiquantitatively by comparing the signal intensity of bound 35S-NHE5-C in relation to the amount of GST-fusion protein (detected by Coomassie blue staining) loaded per sample. 35S-NHE5-C bound to the GST-β-Arr2 (residues 211–409) construct generated from the original yeast clone B1–7 (Fig. 1C, lane 6). Examination of further truncations of β-Arr2 (residues 211–409) (Fig. 1 C, lanes 7 and 8, D, lanes 3–7, and E, lanes 7–9) delineated the minimal interacting region to a positively-charged cluster comprising amino acids 318–332 (318ILVSYRVKVKLV-VSR332). 35S-NHE5-C also bound broadly to fragments encompassing the N-terminal half of β-Arr2 (amino acids 2–210) (Fig. 1C, lanes 3–5). Examination of progressively smaller fragments from this region (Fig. 1 E, lanes 3–6, and F, lanes 3–10) showed that high-affinity binding was conferred by amino acids 52–78, and to a lesser extent, by amino acids 133–183; considerably weaker interactions were also detected with other fragments from within this region. The locations of these three binding elements within the crystal structure (25, 26) of its highly related paralog, β-Arr1, are shown in yellow in Fig. 1B. Interestingly, a construct containing full-length β-Arr2 showed minimal binding of [35S]NHE5-C (data not shown).
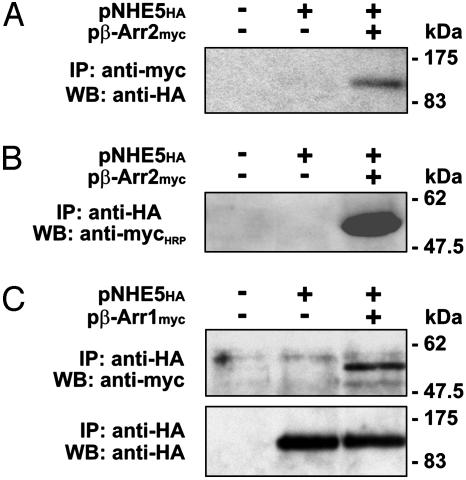
NHE5 and β-Arrestins Coassociate in Transfected Cells. To verify the interaction of full-length NHE5 and β-Arr2 in mammalian cells, an external HA-tagged construct of NHE5 (NHE5HA) was transiently expressed either singly or in combination with myc-tagged β-Arr2 (β-Arr2myc) in a subline of CHO cells (AP-1 cells) that lack endogenous plasmalemmal NHEs. Cell lysates were incubated with an anti-myc antibody, and the resultant immunoprecipitates were resolved by SDS/PAGE and Western blot. As shown in Fig. 2A, probing the blot with an anti-HA antibody revealed a single immunoreactive band corresponding to NHE5HA (≈102 kDa) in immunoprecipitates from cotransfected cells, but not from lysates of cells expressing only NHE5HA or untransfected cells. The reciprocal experiment demonstrated that exogenous β-Arr2myc (≈50 kDa) (Fig. 2B) also associates with NHE5HA when the latter was immunoprecipitated by using an anti-HA antibody. Likewise, the β-Arr1myc isoform also coimmunoprecipitated with NHE5HA when coexpressed in the same cells (Fig. 2C).
Fig. 2.
β-Arrestins form a complex with NHE5 in transfected cells. CHO AP-1 cells were transfected with expression plasmids containing HA-tagged NHE5 alone or with myc-tagged β-Arr2 or β-Arr1 as indicated. After transfection (24 h), cells were lysed and immunoprecipitated (IP) by using anti-myc or anti-HA antibodies. Western blot (WB) analyses were performed as described in Materials and Methods. Data shown are representative of three independent experiments.
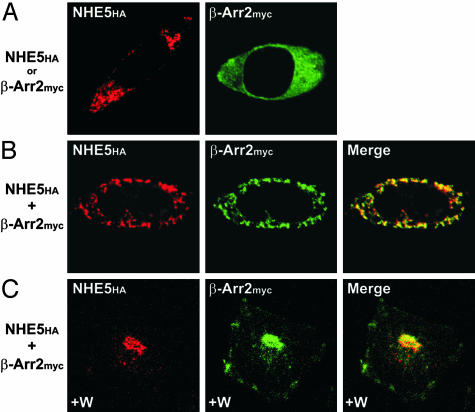
To further demonstrate the association of NHE5HA with β-Arr2myc, their subcellular distribution was compared in AP-1 cells by immunofluorescence confocal microscopy. Previous studies (9) showed that a minor fraction (< 15%) of NHE5 resides on the plasma membrane, whereas the bulk accumulates in recycling endosomes when expressed in AP-1 cells. Consistent with this report, low levels of NHE5HA were detected visually along the plasma membrane, but the majority accumulated in tubulovesicular structures that concentrated in juxtanuclear regions of the cell (Fig. 3A). By contrast, β-Arr2myc was dispersed throughout the cytoplasm when expressed in the absence of NHE5HA (Fig. 3A), a distribution mirroring that found in other cell types in the absence of GPCR agonists (27, 28). However, when expressed simultaneously, the immunofluorescence signal for β-Arr2 redistributed to vesicles that closely coincided with those of NHE5HA (Fig. 3B). It is noteworthy that the overlapping signals are slightly offset, which is consistent with the luminal orientation of the HA-tag of NHE5 and the exofacial membrane location of β-Arr2myc. Similar results were also obtained for NHE5HA and β-Arr1myc (data not shown).
Fig. 3.
Subcellular distribution of NHE5 and β-Arr2 in transfected cells. (A and B) Immunofluorescence confocal microscopy images of AP-1 cells transfected (24 h) with NHE5HA (red) or β-Arr2myc (green) alone (A) or in combination (B). (C) AP-1 cells were cotransfected (24 h) with NHE5HA and β-Arr2myc and then treated with 100 nM wortmannin (W) for 60 min at 37°C to block PI3-K activity. Overlapping signals in merged images in B and C are yellow. Data are representative of at least three independent experiments.
Previous studies (9) have shown that trafficking of NHE5-containing vesicles along the recycling endocytic pathway is regulated by phosphatidylinositol 3′-kinase (PI3-K). Selective inhibition of PI3-K with wortmannin causes vesicles containing NHE5 to tightly coalesce in a juxtanuclear region of the cell while concurrently diminishing peripheral staining due to sustained endocytosis coupled with decreased exocytosis (9). To evaluate the distribution of β-Arr2myc as a function of PI3-K activity when coexpressed with NHE5HA, transfected cells were treated with 100 nM wortmannin for 60 min. As expected, inhibition of PI3-K not only caused the mergence of NHE5HA-containing vesicles to a juxtanuclear region but also caused the bulk of β-Arr2myc to concentrate in an identical intracellular compartment (Fig. 3C). Comparable results were also obtained with LY294002, a chemically unrelated antagonist of PI3-K (29) (data not shown).
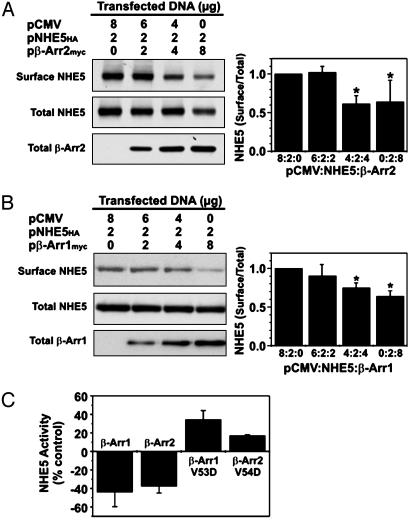
β-Arrestins Decrease Cell-Surface Abundance of NHE5. To determine whether the interaction of β-arrestins with NHE5HA has any functional consequences, AP-1 cells were transiently cotransfected with a fixed amount of the NHE5HA cDNA in the presence of increasing amounts of β-Arr2myc or β-Arr1myc, and the abundance of plasmalemmal NHE5 was measured by using a cell-surface biotinylation assay (23). As illustrated in Fig. 4A, the amount of plasma membrane NHE5HA relative to total cellular NHE5HA decreased significantly as a function of the abundance of β-Arr2myc. Quantitation of the band intensities of surface to total NHE5HA showed a maximal decrease of ≈40% (Fig. 4A), and correlated with a similar decrease in plasmalemmal NHE5 activity (Fig. 4C). Comparable results were also observed with β-Arr1 (Fig. 4 B and C). By contrast, coexpression of NHE5HA and dominant-negative β-arrestins, β-Arr1/V53D or β-Arr2/V54D, resulted in significant increases in plasmalemmal NHE5 activity (Fig. 4C), demonstrating that endogenous β-arrestins are involved in regulation of transfected NHE5HA.
Fig. 4.
Overexpression of β-arrestins decreases cell-surface NHE5 levels. AP-1 cells in 10-cm dishes were transiently cotransfected (24 h) with 10 μg of cDNA containing a fixed amount of NHE5HA (2 μg) and increasing amounts (0–8 μg) of β-Arr2myc (A) or β-Arr1myc (B). Cell-surface NHE5 protein was isolated by using a cell-surface biotinylation assay (see Materials and Methods) and compared with total cellular levels of NHE5 (10% of lysate). The fractions were resolved by SDS/PAGE and Western blotting, using antibodies to the respective epitope tags and the intensities of the bands were quantified by densitometry. For comparative purposes, the ratio of surface/total NHE5 for cells expressing NHE5HA alone (control) was normalized to a value of 1. (C) AP-1 cells were transiently cotransfected (24 h) with NHE5HA and β-Arr1myc (1:4 DNA ratio), β-Arr2myc (1:2 DNA ratio), β-Arr1/V53D (1:3 DNA ratio), or β-Arr1/V53D (1:3 DNA ratio), and plasmalemmal NHE5 activity was measured. Values are the means ± SD of at least three to four independent experiments (*, P < 0.05).
Discussion
Members of the NHE family are differentially sorted to the plasma membrane and distinct organellar compartments by mechanisms that are poorly understood. In this report, we have identified β-arrestins as interacting partners of the neuronal NHE5 isoform through yeast two-hybrid screening of a human brain cDNA library. This interaction was confirmed in vitro by protein-binding pull-down assays, and in transfected cells by coimmunoprecipitation experiments and dual-immunolabeling fluorescence microscopy. The data indicate that both β-Arr1 and β-Arr2 interact directly with the cytoplasmic C terminus of NHE5. Functional studies demonstrated that overexpression of either β-arrestin decreased NHE5 activity by reducing its abundance at the cell surface. Conversely, overexpression of dominant-negative β-Arr1/V53D and β-Arr2/V54D increased plasma membrane NHE5 activity, indicative of a role for endogenous β-arrestins in regulating NHE5 activity in CHO cells. Previous studies (30) have also inferred the presence of endogenous β-Arr1 and β-Arr2 in CHO cells by using an identical dominant-negative approach. However, our attempts to detect endogenous hamster β-arrestins (in cell lysates or in a complex with NHE5HA) with several commercially available β-arrestin antibodies were unsuccessful, despite the ability of at least one of them to recognize overexpressed human β-Arr1myc. This finding can be explained by low abundance of the endogenous β-arrestins, poor species crossreactivity, and/or low affinity of the antibodies. The negative effect of β-arrestin on plasmalemmal NHE5 levels can be achieved either by enhancing its rate of internalization into recycling endosomes, and/or by decreasing its rate of recycling back to the cell surface. Additional experimentation will be required to distinguish between these possibilities, although they are not necessarily mutually exclusive. Notwithstanding, the results collectively indicate a role for β-arrestins in regulating, at least in part, the trafficking of NHE5 along the endocytic recycling pathway.
These observations are intriguing in light of recent observations that β-arrestins can enhance the internalization and signaling of another class of receptors other than GPCRs; specifically single-membrane-spanning receptors for insulin-like growth factor I (31), transforming growth factor-β (TGFβR-III) (32) and low-density lipoprotein (LDLR) (33). Interestingly, in the cases of TGFβR-III and LDLR, association with β-Arr2 was ligand-independent and supported constitutive endocytosis, a scenario closely mimicking that of NHE5. A distinguishing aspect of the present study is the revelation that β-arrestins can associate with integral plasma membrane proteins that are not receptors, in this case, a 12-membrane-spanning cation transporter.
The fate of newly formed receptor–β-arrestin complexes is heterogeneous and varies, depending on the receptor. For certain GPCRs such as the β2-adrenergic receptor (termed class A receptor), ligand activation and phosphorylation of the receptor results in preferential binding of β-Arr2 relative to β-Arr1, but the complex dissociates quickly after formation of clathrin-coated pits; thereby allowing for rapid recycling of receptors (28). By contrast, class B receptors such as the angiotensin II type 1A receptor bind both β-arrestins with comparable affinity, forming stable complexes that are retained with internalized vesicles and dissociate more slowly before recycling, or are targeted for degradation in lysosomes (28). The trafficking profile of the NHE5/β-Arr(1 or 2) complex in AP-1 cells best resembles class B receptors, in as much as the association appears relatively persistent after vesicle formation and internalization.
The precise molecular mechanism underlying the association of β-arrestin with NHE5 remains to be resolved. It is generally recognized that β-arrestins are recruited to agonist-activated GPCRs after the receptors have been phosphorylated at clusters of serine/threonine residues, rather than at a single site, within their third-intracellular loop and/or C-terminal domains by GPCR kinases (14, 34, 35). Upon recognition of phosphoreceptor sites, β-arrestins undergo major conformational rearrangements that include exposure of additional elements within the two concave antiparallel β-sheet domains (see Fig. 1B) that bind with high affinity to multiple receptor sites, and displacement of the C-terminal region that facilitates binding of clathrin and AP-2 (25, 26, 36–38).
It is unclear whether phosphorylation of NHE5 is a prerequisite for binding β-Arr1 or β-Arr2. However, it is noteworthy that a GST construct containing full-length β-Arr2 showed minimal binding of the in vitro-synthesized, unphosphorylated C terminus of NHE5 (data not shown). This result could arise from misfolding of full-length β-Arr2 when synthesized in bacteria, but a more appealing possibility is that full-length β-Arr2 requires phosphorylation of NHE5 for high-affinity binding. This region of NHE5 contains several clusters of serine/threonine residues, some of which are amenable to phosphorylation by serine/threonine kinases in vitro (E.Z.S., M.N., V.L., and J.O., unpublished data). Additional preliminary 32P-radiolabeling experiments indicate that NHE5 is also phosphorylated in intact cells. Although circumstantial, these findings suggest that phosphorylation may be a potential determinant for NHE5–β-arrestin interactions and warrants further investigation.
To provide some indication of where NHE5 might bind within β-Arr2, we evaluated the ability of various GST–β-Arr2 fusion constructs to bind the in vitro-synthesized C terminus of NHE5 by using conventional protein pull-down assays. The analyses revealed three noncontiguous elements that line the inner face of the N-terminal (residues 52–78 and 133–183, which encompass β-strands S5 plus S6 and S9 plus S10 with adjoining loops, respectively) and C-terminal (residues 318–332 of β-strand S18) concave domains of β-Arr2 (illustrated in Fig. 1B). The first element (residues 52–78) coincides precisely with the conserved β-strand S5 plus S6 segment required for high-affinity receptor binding (38), suggesting that part of the NHE5 C terminus assumes a conformation that resembles part of the β-arrestin-binding domain of GPCRs. By contrast, the other two elements appear more unique to recognition of NHE5. All three elements are conserved in both β-Arr1 and β-Arr2. Significantly, the location of NHE5-binding sites is compatible with simultaneous binding of other proteins involved in β-arrestin-mediated endocytosis, such as clathrin and AP-2. Further mutagenesis and functional measurements using full-length molecules will be required to validate the importance of these NHE5-binding sites.
The physiological relevance of an association between the β-arrestins and NHE5 in neurons remains to be defined. However, it is tempting to speculate that this linkage may facilitate their inclusion in clathrin-coated pits that also contain neuromodulator-activated GPCR–β-arrestin complexes; thereby enhancing vesicle acidification, maturation, and recycling of receptors (39). Consistent with this notion, rapid acidification of early endosomes was found to enhance dephosphorylation and resensitization of the β2-adrenergic receptor by facilitating its physical association with a GPCR-specific phosphatase (40). It will be of interest to determine whether the β-arrestins also associate with NHEs in other tissues, particularly the epithelial NHE3 isoform, which shares high sequence identity with NHE5, and displays similar trafficking behavior. Overall, the findings in this report indicate that β-arrestins play a broader role in promoting endocytosis of integral plasma membrane proteins than previously suspected.
Acknowledgments
We thank Caroline Girard for technical assistance. This work was supported by funding from the Canadian Institutes of Health Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NHE, Na+/H+ exchanger; β-Arr1, β-arrestin1; β-Arr2; β-arrestin2; AP-n, adaptor protein n; HA, hemagglutinin; PI3-K, phosphatidylinositol 3′-kinase; GPCR, G protein-coupled receptor; 35S-NHE5-C, in vitro-synthesized 35S-labeled NHE5 C terminus.
References
- 1.Orlowski, J. & Grinstein, S. (2004) Pflügers Arch. Eur. J. Physiol. 447, 549–565. [DOI] [PubMed] [Google Scholar]
- 2.de Silva, M. G., Elliott, K., Dahl, H. H., Fitzpatrick, E., Wilcox, S., Delatycki, M., Williamson, R., Efron, D., Lynch, M. & Forrest, S. (2003) J. Med. Genet. 40, 733–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schultheis, P. J., Clarke, L. L., Meneton, P., Miller, M. L., Soleimani, M., Gawenis, L. R., Riddle, T. M., Duffy, J. J., Doetschman, T., Wang, T., et al. (1998) Nat. Genet. 19, 282–285. [DOI] [PubMed] [Google Scholar]
- 4.D'Souza, S., Garcia-Cabado, A., Yu, F., Teter, K., Lukacs, G. L., Skorecki, K., Moore, H. P., Orlowski, J. & Grinstein, S. (1998) J. Biol. Chem. 273, 2035–2043. [DOI] [PubMed] [Google Scholar]
- 5.Kurashima, K., Szabó, E. Z., Lukacs, G. L., Orlowski, J. & Grinstein, S. (1998) J. Biol. Chem. 273, 20828–20836. [DOI] [PubMed] [Google Scholar]
- 6.Chow, C. W., Khurana, S., Woodside, M., Grinstein, S. & Orlowski, J. (1999) J. Biol. Chem. 274, 37551–37558. [DOI] [PubMed] [Google Scholar]
- 7.Akhter, S., Kovbasnjuk, O., Li, X., Cavet, M., Noël, J., Arpin, M., Hubbard, A. L. & Donowitz, M. (2002) Am. J. Physiol. 283, C927–C940. [DOI] [PubMed] [Google Scholar]
- 8.Baird, N. R., Orlowski, J., Szabó, E. Z., Zaun, H. C., Schultheis, P. J., Menon, A. G. & Shull, G. E. (1999) J. Biol. Chem. 274, 4377–4382. [DOI] [PubMed] [Google Scholar]
- 9.Szászi, K., Paulsen, A., Szabó, E. Z., Numata, M., Grinstein, S. & Orlowski, J. (2002) J. Biol. Chem. 277, 42623–42632. [DOI] [PubMed] [Google Scholar]
- 10.Gekle, M., Drumm, K., Mildenberger, S., Freudinger, R., Gassner, B. & Silbernagl, S. (1999) J. Physiol. (London) 520, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gekle, M., Freudinger, R. & Mildenberger, S. (2001) J. Physiol. (London) 531, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slepnev, V. I. & De Camilli, P. (2000) Nat. Rev. Neurosci. 1, 161–172. [DOI] [PubMed] [Google Scholar]
- 13.Maxfield, F. R. & McGraw, T. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 121–132. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson, S. S. (2001) Pharmacol. Rev. 53, 1–24. [PubMed] [Google Scholar]
- 15.Lefkowitz, R. J. & Whalen, E. J. (2004) Curr. Opin. Cell Biol. 16, 162–168. [DOI] [PubMed] [Google Scholar]
- 16.Krupnick, J. G., Goodman, O. B., Jr., Keen, J. H. & Benovic, J. L. (1997) J. Biol. Chem. 272, 15011–15016. [DOI] [PubMed] [Google Scholar]
- 17.Laporte, S. A., Oakley, R. H., Holt, J. A., Barak, L. S. & Caron, M. G. (2000) J. Biol. Chem. 275, 23120–23126. [DOI] [PubMed] [Google Scholar]
- 18.McDonald, P. H., Cote, N. L., Lin, F. T., Premont, R. T., Pitcher, J. A. & Lefkowitz, R. J. (1999) J. Biol. Chem. 274, 10677–10680. [DOI] [PubMed] [Google Scholar]
- 19.Claing, A., Chen, W., Miller, W. E., Vitale, N., Moss, J., Premont, R. T. & Lefkowitz, R. J. (2001) J. Biol. Chem. 276, 42509–42513. [DOI] [PubMed] [Google Scholar]
- 20.Gietz, R. D., Schiestl, R. H., Willems, A. R. & Woods, R. A. (1995) Yeast 11, 355–360. [DOI] [PubMed] [Google Scholar]
- 21.Rotin, D. & Grinstein, S. (1989) Am. J. Physiol. 257, C1158–C1165. [DOI] [PubMed] [Google Scholar]
- 22.Smith, D. B. & Corcoran, L. M. (1994) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York), pp. 16.7.1–16.7.7.
- 23.Le Bivic, A., Real, F. X. & Rodriguez-Boulan, E. (1989) Proc. Natl. Acad. Sci. USA 86, 9313–9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chien, C.-T., Bartel, P. L., Sternglanz, R. & Fields, S. (1991) Proc. Natl. Acad. Sci. USA 88, 9578–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han, M., Gurevich, V. V., Vishnivetskiy, S. A., Sigler, P. B. & Schubert, C. (2001) Structure (London) 9, 869–880. [DOI] [PubMed] [Google Scholar]
- 26.Milano, S. K., Pace, H. C., Kim, Y. M., Brenner, C. & Benovic, J. L. (2002) Biochemistry 41, 3321–3328. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, J., Barak, L. S., Anborgh, P. H., Laporte, S. A., Caron, M. G. & Ferguson, S. S. (1999) J. Biol. Chem. 274, 10999–11006. [DOI] [PubMed] [Google Scholar]
- 28.Oakley, R. H., Laporte, S. A., Holt, J. A., Caron, M. G. & Barak, L. S. (2000) J. Biol. Chem. 275, 17201–17210. [DOI] [PubMed] [Google Scholar]
- 29.Vlahos, C. J., Matter, W. F., Hui, K. Y. & Brown, R. F. (1994) J. Biol. Chem. 269, 5241–5248. [PubMed] [Google Scholar]
- 30.Bremnes, T., Paasche, J. D., Mehlum, A., Sandberg, C., Bremnes, B. & Attramadal, H. (2000) J. Biol. Chem. 275, 17596–17604. [DOI] [PubMed] [Google Scholar]
- 31.Lin, F. T., Daaka, Y. & Lefkowitz, R. J. (1998) J. Biol. Chem. 273, 31640–31643. [DOI] [PubMed] [Google Scholar]
- 32.Chen, W., Kirkbride, K. C., How, T., Nelson, C. D., Mo, J., Frederick, J. P., Wang, X. F., Lefkowitz, R. J. & Blobe, G. C. (2003) Science 301, 1394–1397. [DOI] [PubMed] [Google Scholar]
- 33.Wu, J. H., Peppel, K., Nelson, C. D., Lin, F. T., Kohout, T. A., Miller, W. E., Exum, S. T. & Freedman, N. J. (2003) J. Biol. Chem. 278, 44238–44245. [DOI] [PubMed] [Google Scholar]
- 34.Orsini, M. J., Parent, J. L., Mundell, S. J., Benovic, J. L. & Marchese, A. (1999) J. Biol. Chem. 274, 31076–31086. [DOI] [PubMed] [Google Scholar]
- 35.Oakley, R. H., Laporte, S. A., Holt, J. A., Barak, L. S. & Caron, M. G. (2001) J. Biol. Chem. 276, 19452–19460. [DOI] [PubMed] [Google Scholar]
- 36.Gurevich, V. V., Dion, S. B., Onorato, J. J., Ptasienski, J., Kim, C. M., Sterne-Marr, R., Hosey, M. M. & Benovic, J. L. (1995) J. Biol. Chem. 270, 720–731. [DOI] [PubMed] [Google Scholar]
- 37.Vishnivetskiy, S. A., Paz, C. L., Schubert, C., Hirsch, J. A., Sigler, P. B. & Gurevich, V. V. (1999) J. Biol. Chem. 274, 11451–11454. [DOI] [PubMed] [Google Scholar]
- 38.Vishnivetskiy, S. A., Hosey, M. M., Benovic, J. L. & Gurevich, V. V. (2004) J. Biol. Chem. 279, 1262–1268. [DOI] [PubMed] [Google Scholar]
- 39.Mellman, I., Fuchs, R. & Helenius, A. (1986) Annu. Rev. Biochem. 55, 663–700. [DOI] [PubMed] [Google Scholar]
- 40.Krueger, K. M., Daaka, Y., Pitcher, J. A. & Lefkowitz, R. J. (1997) J. Biol. Chem. 272, 5–8. [DOI] [PubMed] [Google Scholar]