Abstract
Hepatitis C virus (HCV) is a major human pathogen that infects 170 million people. A hallmark of HCV is its ability to establish persistent infections reflecting the evasion of host immunity and interference with α/β-IFN innate immune defenses. We demonstrate that disruption of retinoic acid-inducible gene I (RIG-I) signaling by the viral NS3/4A protease contributes to the ability of HCV to control innate antiviral defenses. RIG-I was essential for virus or HCV RNA-induced signaling to the IFN-β promoter in human hepatoma cells. This signaling was disrupted by the protease activity of NS3/4A, which ablates RIG-I signaling of downstream IFN regulatory factor 3 and NF-κB activation, attenuating expression of host antiviral defense genes and interrupting an IFN amplification loop that otherwise suppresses HCV replication. Treatment of cells with an active site inhibitor of the NS3/4A protease relieved this suppression and restored intracellular antiviral defenses. Thus, NS3/4A control of RIG-I supports HCV persistence by preventing IFN regulatory factor 3 and NF-κB activation. Our results demonstrate that these processes are amenable to restoration through pharmacologic inhibition of viral protease function.
Keywords: interferon, protease, host response, infection, NF-κB
Retinoic acid-inducible gene-I (RIG-I) is a cytosolic DExD/H box RNA helicase (Fig. 1A) that was recently shown to be a dsRNA binding protein that functions independently of Toll-like receptor (TLR) 3 to signal IFN-β production in response to a variety of RNA viruses, including hepatitis C virus (HCV) (1, 2). RIG-I is basally expressed in most tissues, is induced by type 1 IFNs, and is essential for triggering antiviral defenses that regulate HCV RNA replication (1, 2). Its carboxyl-terminal helicase domain specifically binds to a pathogen-associated molecular pattern (PAMP) embedded within the structured 5′ or 3′ nontranslated region of the HCV genome (1). PAMP binding initiates signaling by the RIG-I amino-terminal caspase recruitment domain (CARD) homologues that direct downstream phosphorylation and activation of IFN regulatory factor-3 (IRF-3) and the parallel activation of NF-κB (1, 2).
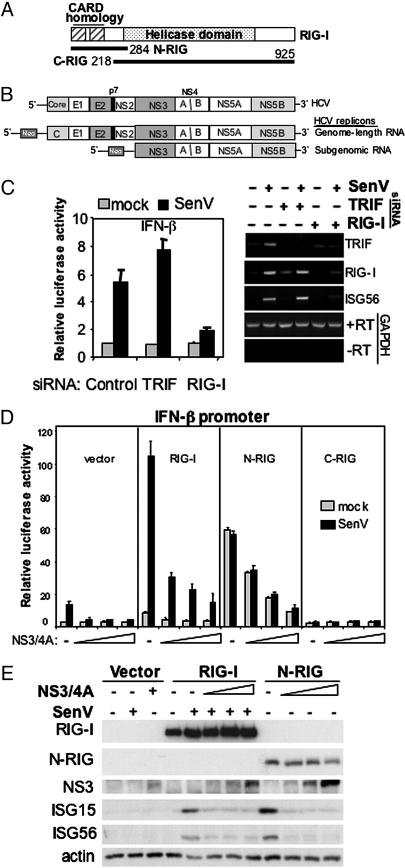
Fig. 1.
RIG-I and NS3/4A regulate innate intracellular defenses. (A) Structural representation of RIG-I, N-RIG, and C-RIG expression constructs. The positions of the caspase recruitment domain, helicase domain, and terminal amino acids are indicated. (B) Genome structure of HCV (Top). Also shown are the genome-length (Middle), and subgenomic HCV RNA replicons (Bottom) used in this study. These autonomously replicating, bicistronic viral RNAs express neomycin phosphotransferase (neo) and are thus selectable. They have been described in detail (6, 10). (C) Huh7 cells transfected with control siRNA or siRNA directed against TRIF or RIG-I were subsequently transfected with plasmids encoding the IFN-β luciferase reporter construct and a constitutively expressed β-galactosidase control construct, mock-infected or infected with SenV and harvested 20 h later for determination of relative luciferase values and RT-PCR analysis of mRNA expression. Bars show average and SD relative luciferase activity from three experiments. Panels at right show agarose gel analysis of the indicated RT-PCR product from cells treated as shown above each lane. The + and - RT designate control RT-PCR reactions to amplify glyceraldehyde dehydrogenase (GAPDH) mRNA in the presence or absence of reverse transcriptase. (D) Huh7 cells were cotransfected with plasmids encoding the IFN-β luciferase reporter construct, Renilla luciferase, and 50 ng of the indicated vector or RIG-I expression construct with increasing amounts (0, 50, 100, and 200 ng) of plasmid DNA encoding NS3/4A. Cells were mock-infected or infected with SenV and harvested for luciferase assay. Bars show the average relative luciferase activity and SD from three experiments. (E) Huh7.5 cells transfected with plasmid constructs encoding vector alone, RIG-I, or N-RIG in the presence of increasing amounts of an NS3/4A expression construct were infected with SenV as indicated and then were harvested for immunoblot analysis.
Recent studies have shown that the processes of RIG-I signaling and the actions of IRF-3 and NF-κB initiate host defenses that limit HCV RNA replication (1, 3, 4). The single-stranded RNA genome of HCV encodes a 3,010-aa polyprotein that is posttranslationally processed into the mature structural proteins (core, E1 and E2), p7, and the nonstructural (NS) proteins NS2-NS5B (5), of which the NS3-NS5B proteins are sufficient to support viral RNA replication (Fig. 1B). HCV controls induction of the antiviral defense response through the activity of its NS3/4A protease complex, which disrupts intracellular signaling processes that confer IRF-3 activation (3). Viral control of IRF-3 limits IFN expression to thereby support viral persistence and impart fitness to HCV (3, 6). However, the intracellular signaling pathway(s) targeted by NS3/4A to govern IRF-3 function and HCV replication are incompletely defined.
Materials and Methods
Cell Culture. Huh7 are human hepatoma cells (7). Huh7.5 cells (a gift from C. Rice, The Rockefeller University, New York) are a subline of Huh7 and are highly permissive for HCV RNA replication (8). Huh7-HP, Huh7-A7, and Huh7-K2040 are Huh7 cells that harbor culture-adapted variants of the Con1 HCV (genotype 1b) subgenomic replicon RNA and have been described in detail (3, 6, 9). Huh7 2-3 and Huh7 2-3c are Huh7-derived cell lines that harbor the full-length self-replicating HCV genome (derived from the HCV-N strain, genotype 1b) and their IFN-cured counterparts, respectively (3, 10, 11). UNS3/4A (clone 24) are osteosarcoma cells that conditionally express an HCV 1a NS3/4A construct under control of a tetracycline-regulated (tet-off) promoter (a gift from D. Moradpour, University of Lausanne, Lausanne, Switzerland). NS3/4A expression was controlled by culturing cells in the presence (to repress NS3/4A expression) or absence of tetracycline (to induce NS3/4A expression) as presented elsewhere (12). Culture methods have been described (3, 6). Cell treatments were conducted by replacing culture medium with medium containing 10 ng of IL-1 (R & D Systems), the indicated units/ml IFN-α2a (PBL, Piscataway, NJ) or 10 μM SCH6 (a gift from Schering-Plough) (3). For immunofluorescent staining of cells, cultures were seeded into chamber slides and processed for immunostaining as described (3). For virus infection, 2 × 105 or 1 × 105 cells were respectively seeded into wells of a 6-well or 12-well dish. Where indicated, cells were transfected with various expression constructs. Twenty-four hours after culture seeding or transfection, the cells were mock-infected or infected with Sendai virus (SenV) (Charles River Laboratories) or New Castle disease virus (NDV) (2) by using 100 hemagglutinin units/ml of culture exactly as described (3).
Plasmids, Transfection, and Protein Analysis. The RIG-I expression plasmids pEF-flagRIG-Ifull, pEF-flagN-RIG, and pEF-flagC-RIG have been described and encode the indicated RIG-I construct with an amino-terminal FLAG epitope (2). pIFN-β-luc, pCMV-Renilla-luc, pCDNA3.1-NS3/4A, and pPRDII-luc have been described elsewhere (3, 4). pEF-BOS was a gift from K. Fitzgerald (University of Massachusetts Medical School, Worcester) (13). pcDNA1-TBK1 was a gift from T. Maniatis (University of Massachusetts Medical School, Worcester) (13). pIRF-3-5D and pIRF3-ΔN were gifts from J. Hiscott (McGill University, Montreal) (14). pNS5AB was constructed by cloning a PCR amplicon (primer sequences are available upon request) containing the NS5A-NS5B coding region from pHCV 1b pt (3) into pCDNA3.1 (Invitrogen). Plasmid transfection was conducted by using the FuGENE 6 transfection reagent (Roche Applied Science, Indianapolis) and manufacturer's protocol. For promoter-luciferase (luc) assay, 1 × 104 cells per well of a 48-well dish were cotransfected with 100 ng and 25 ng, respectively, of pIFN-β-luc and pCMV-Renilla-luc along with the stated amount of the indicated expression constructs. Cells were further cultured alone or subjected to virus infection as described above and were harvested 18 h later for luciferase assay (3).
For immunoblot analysis, cultures of 2 × 105 cells were harvested, protein extracts were prepared, and immunoblot analysis was conducted as described (3) by using antiserum specific to IRF-3 (from M. David, University of California at San Diego, La Jolla), RIG-I (2), IFN-stimulated gene (ISG) 56 (from G. Sen, Cleveland Clinic Foundation, Cleveland), ISG15 (from A. Haas, Louisiana State University, New Orleans), IRF-3 phosphoserine 396 (from J. Hiscott), IRF-3 phosphoserine 386 (15), or IκB-α (Santa Cruz Biotechnology). Monoclonal anti-PKR and anti-NS3 antibodies were from A. Hovanessian (Pasteur Institute, Paris) and NovoCastra (Newcastle, U.K.), respectively. Monoclonal anti-FLAG M2 antibody was from Sigma and was used to detect RIG-I, N-RIG, and C-RIG proteins. Anti-β-actin serum and secondary antibody reagents were from The Jackson Laboratory. In some experiments, HCV proteins were detected with hyperimmune patient serum [obtained from W. Lee (University of Texas Southwestern Medical Center, Dallas) with informed consent]. Specific methods for analysis of IRF-3 dimerization using nondenaturing gel electrophoresis were followed as described elsewhere (15). Immunostaining procedures used the indicated primary antibodies in combination with Alexa Fluor 488 or Rhodamine-conjugated specific secondary antibodies (Molecular Probes). Analysis of stained cells was conducted in the University of Texas Southwestern Medical Center Pathogen Imaging Facility by using a Zeiss Axiovert or Zeiss Pascal LSM confocal microscope and axiovision software. Electrophoretic mobility-shift analysis of NF-κB DNA binding activity was conducted on nuclear extracts prepared from cultures of 4 × 105 cells as described (4).
RNA Analysis. Procedures for RNA extraction, Northern blot analysis, probe preparation, semiquantitative PCR, and real-time quantitative PCR have been described (1, 3). For short interfering RNA (siRNA) silencing of gene expression, Huh7 cells were transfected with 80 nM siRNA (Dharmacon, Lafayette, CO) targeting TRIF (Toll-IL-1 receptor domain-containing adapter-inducing IFN-β), RIG-I, or a scrambled negative-control (Ambion, Austin, TX) by using Oligofectamine (Invitrogen) according to the manufacturer's recommendation. Cells were cotransfected with pIFN-β-luc and pCMVβgal 24 h later. After an additional 24 h, cells were mock-infected or infected with 100 hemagglutinin units/ml SenV and harvested 20 h later for luciferase and β-galactosidase assay (Promega). In these experiments, luciferase activity was normalized to the β-galactosidase activity. siRNA and PCR primer sequences are available upon request.
Microarray analyses were carried out within the Molecular Genomics Core Facility of the University of Texas Medical Branch. Huh7 2-3 and Huh7 2-3c cells were grown in 10-cm dishes and were either mock-infected or infected with 100 hemagglutinin units/ml SenV and harvested 20 h later for total RNA extraction. Twentyfive micrograms of total RNA was used as template for synthesis of first-strand cDNA, which was used subsequently for second-strand synthesis and production of biotinylated cRNA probes that were hybridized to an Affymetrix human GeneChip (Hu133A) containing 22,283 oligonucleotide probe sets, according to the manufacturer's protocol (Affymetrix, Santa Clara, CA). GeneChips were scanned by using an Affymetrix confocal scanner (Agilent, Palo Alto, CA), and data were analyzed with microarray genesuite 5.0 software.
Results
HCV NS3/4A Disrupts RIG-I Signaling. We conducted RNA silencing studies to define the nature of the virus responsive pathways that signal to IRF-3 and IFN-β in Huh7 cells. These cells support HCV RNA replication (16). They do not basally express TLR3 and therefore do not signal through the TLR3 pathway (17), allowing us to evaluate virus signaling and regulation by HCV independently of TLR3. Transfection of siRNA directed against the TLR adaptor protein, TRIF (18), efficiently ablated its expression but did not affect signaling to the IFN-β promoter induced by infection with SenV, a potent activator of IRF-3 (14). Similar results were obtained when TRIF-null mouse embryonic fibroblasts (MEFs) were infected with SenV, which efficiently triggered IFN-β promoter induction, but this response was abrogated by expression of NS3/4A (data not shown). However, siRNA directed against RIG-I abolished virus signaling to the IFN-β promoter in Huh7 cells and prevented the expression of ISG56, an IRF-3 target gene (Fig. 1C) (19). These results indicate that NS3/4A disrupts an intracellular virus-responsive pathway that signals the IFN-β promoter independent of TLR3 or TRIF but involves RIG-I. We therefore evaluated the affect of NS3/4A on signaling by WT or mutant RIG-I proteins. Ectopic expression of RIG-I conferred superstimulation of virus signaling to the IFN-β promoter, but this signaling was suppressed in Huh7 cells upon coexpression of increasing amounts of NS3/4A (Fig. 1D). NS3/4A also blocked constitutive activation of the IFN-β promoter directed by ectopic expression of an N-RIG mutant representing the amino-terminal caspase recruitment domain homology region, but had no effect on the carboxyl-terminal helicase domain of RIG-I (C-RIG), which confers a dominant-negative block to virus-induced signaling (2). When expressed in a Huh7 variant (Huh7.5 cells) in which RIG-I is defective (1), ectopic RIG-I or N-RIG restored induction of ISG expression. This response was potently suppressed by NS3/4A (Fig. 1E), indicating that NS3/4A antagonizes signaling events that trigger a host response directed by RIG-I.
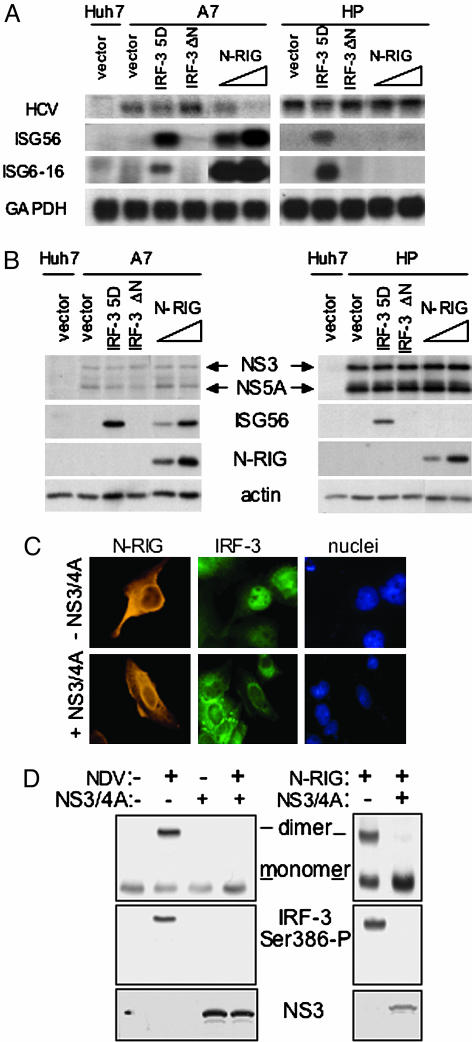
The RIG-I Pathway Can Modulate HCV Replication and Is Regulated by NS3/4A. Disruption of RIG-I signaling may allow HCV to block IRF-3 activation and IFN defenses that could limit viral replication. In support of this finding, overexpression of the constitutively active IRF-3-5D or N-RIG proteins (1, 14) induced a response that suppressed the replication of an HCV RNA replicon (A7) that is poorly adapted to Huh7 cells, whereas only expression of IRF-3-5D induced a host response restricting replication of a highly adapted replicon (HP, Fig. 2 A and B). This result could reflect the reduced abundance of NS3/4A expressed by A7, or differences in the cell culture adaptive mutations present within the NS3/4A coding regions of these isogenic replicons that specifically affect downstream IRF-3 regulation (3). Nonetheless, these results define RIG-I as an essential transducer of a virus-responsive cellular pathway that has the capacity to control HCV RNA replication.
Fig. 2.
RIG-I signals IRF-3 to control HCV replication and is regulated by NS3/4A. (A and B) Control Huh7 cells were transfected with vector only. Huh7-A7 cells harboring the A7 HCV replicon (A7) or Huh7-HP cells harboring the HP HCV replicon (HP) were transfected with 1 μg of plasmid DNA encoding vector alone, IRF-3-5D or IRF-3-ΔNor1 μgor2 μg of N-RIG expression plasmid. Cells were harvested 48 h posttransfection, and extracts were subjected to Northern blot analysis (A) and immunoblot analysis (B) using specific DNA or antibody probes, respectively. (C) UNS3/4A cells, cultured to suppress (-NS3/4A) or induce (+NS3/4A) NS3/4A expression, were transfected with the N-RIG expression construct, and 24 h later were subjected to dual immunostaining for ectopic N-RIG and endogenous IRF-3. Panels show N-RIG, IRF-3, and DAPI-stained nuclei. (D) UNS3/4A cells cultured to suppress or induce NS3/4A expression were infected with NDV as shown (Left) or were transfected with N-RIG expression plasmid (Right). After 24 h, cells were harvested, and protein extracts were separated on nondenaturing gels and subjected to immunoblot analysis to define the dimer, monomer, and phosphoserine 386 (Ser-386-P) isoforms of IRF-3. (Bottom) NS3 levels derived by standard denaturing gel immunoblot analysis.
We examined the impact of NS3/4A regulation of RIG-I signaling on IRF-3 activation. NS3/4A prevented the nuclear accumulation of IRF-3 in response to SenV infection or transfection of cells with HCV RNA (Fig. 5, which is published as supporting information on the PNAS web site). In osteosarcoma cells conditionally expressing NS3/4A, ectopic N-RIG induced the cytosol to nuclear redistribution of IRF-3, but this response was blocked upon NS3/4A expression (Fig. 2C). IRF-3 nuclear retention depends upon its carboxyl-terminal phosphorylation at sites that include serine 386 (S386) and serine 396 (S396), which results in the stable formation of IRF-3 dimers (15, 20). We therefore evaluated the regulation of IRF-3 phosphorylation at these sites by NS3/4A. In control experiments, infection of cells with SenV (data not shown) or NDV (a paramyxovirus related to SenV) induced S386 phosphorylation and dimerization of IRF-3, but both were blocked by NS3/4A. Ectopic expression of N-RIG similarly conferred IRF-3 S386 phosphorylation and dimerization, but these events were also blocked by NS3/4A (Fig. 2D). Similarly, SenV infection induced the accumulation of the phosphoserine 396 isoform of IRF-3, but this response was blocked by NS3/4A (see Fig. 3A). Thus, RIG-I signals IRF-3 activation by directing IRF-3 carboxyl-terminal phosphorylation, and this process is disrupted by NS3/4A.
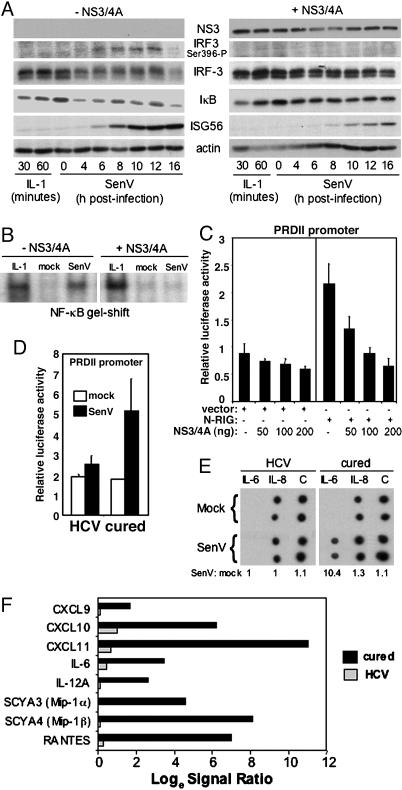
Fig. 3.
NS3/4A controls RIG-I signaling to NF-κB. (A and B) UNS3/4A cells, cultured to suppress (-NS3/4A) or induce (+NS3/4A) NS3/4A expression, were treated with IL-1, mock-infected or infected with SenV. (A) Cells were harvested at the times indicated, and extracts were subjected to immunoblot analysis to detect NS3, the phosphoserine 396 isoform of IRF-3 (IRF-3 Ser-396-P), total IRF-3, IκB-α, ISG56, and actin. (B) Cells were harvested 30 min post-IL-1 treatment or 16 h postinfection. Nuclear extracts were prepared and subjected to EMSA by using a DNA probe encoding the PRDII element of the IFN-β promoter to detect NF-κB DNA-binding activity. (C) Huh7 cells were cotransfected with plasmids encoding the PRDII-luciferase promoter construct and Renilla luciferase along with 50 ng of vector only or N-RIG expression plasmid and the indicated amount of NS3/4A expression plasmid. Cells were mock-infected or infected with SenV and processed for luciferase assay 16 h postinfection. Bars show the average relative luciferase and SD values from three experiments. (D) Huh7 2-3 cells harboring replicating genome-length RNA (HCV) or their IFN-cured Huh7 2-3c counterparts were cotransfected with plasmids encoding the PRDII-luciferase promoter construct and Renilla luciferase. Cells were mock-infected or infected with SenV and processed for luciferase assay. Bars show the average relative luciferase and SD values from three experiments. (E) Medium from cultures of mock or SenV-infected Huh7 2-3 (HCV) and Huh 2-3c cells (cured) was subjected to cytokine blot analysis by using the Cytokine Array III kit and the manufacturer's protocol (Ray Biotech, Norcross, GA). Numbers show the ratio of mock to SenV signal derived from averaged densitometric values of each spot. C denotes the reference control. (F) Huh7 2-3 (HCV) and Huh7 2-3c cells (cured) were mock-infected or infected with SenV for 16 h. Total RNA was extracted and subjected to microarray analysis by using Affymetrix U133A GeneChips. Bars show the quantified signal ratio of average hybridization levels for the indicated cytokine and chemokine mRNAs.
Previous work has demonstrated that the RIG-I pathway bifurcates to signal IRF-3 and NF-κB activation (2). We further evaluated the role of RIG-I in NF-κB activation and its potential regulation by NS3/4A. In the absence of NS3/4A, SenV infection of osteosarcoma cells induced a rapid decay in the abundance of the NF-κB inhibitor, IκB-α, accompanied by accumulation of the IRF-3 phosphoserine 396 isoform (Fig. 3A Left). This response was followed by induction of ISG56 expression. NS3/4A blocked virusmediated IκB-α decay and IRF-3 S396 phosphorylation but did not prevent IκB-α degradation induced by treatment of cells with IL-1 (Fig. 3A Right). NS3/4A disrupted virus-induced NF-κB binding to the cognate PRDII DNA element (Fig. 3B) and prevented induction of an NF-κB-dependent PRDII promoter-luciferase construct signaled by ectopically expressed N-RIG in Huh7 cells (Fig. 3C). Huh7 2-3 cells containing replicating genome-length HCV RNA (10) similarly exhibited a block in NF-κB induction of the PRDII promoter construct (Fig. 3D). However, virus-responsive promoter activity was restored in their cured Huh7 2-3c counterparts lacking HCV RNA. Protein blot and microarray analyses (Fig. 3 E and F, respectively) demonstrated that HCV replication was associated with a blockade in virus-induced accumulation of secreted IL-6, an NF-κB target gene (21). This blockade associated with a general attenuation of NF-κB-dependent chemokine expression (21) in Huh7 2-3 cells, but virus-responsiveness of each was restored in the cured Huh7 2-3c cells. NS3/4A therefore ablates RIG-I signaling to NF-κB, likely by controlling events that signal IκB degradation. This regulation of RIG-I signaling thus attenuates both IRF-3 (3) and NF-κB target gene expression during HCV RNA replication.
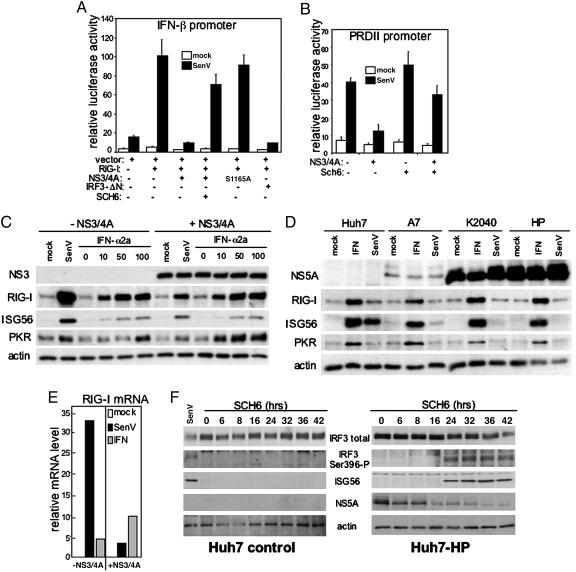
NS3/4A Protease Activity Imparts Regulation of RIG-I Signaling. We next determined whether regulation of RIG-I signaling depends on the protease activity of NS3/4A. In the context of ectopic RIG-I expression, SenV infection of Huh7 cells resulted in IFN-β promoter superstimulation that was blocked by NS3/4A (Fig. 4A). This blockade was relieved in cells treated with SCH6, a peptidomimetic active site inhibitor of the NS3/4A protease (3). Mutation of the active site Ser to Ala at HCV codon 1165 ablates NS3/4A protease activity (22), and also abrogated NS3/4A control of RIG-I signaling to the IFN-β promoter. Expression of the dominant-negative IRF-3-ΔN mutant (14) similarly ablated RIG-I signaling, confirming that IRF-3 is a downstream effector of the RIG-I pathway in hepatoma cells. We also assessed whether or not protease function was required for NS3/4A regulation of the NF-κB-responsive PRDII promoter element (Fig. 4B). The NS3/4A-mediated blockade to SenV-induced NF-κB-dependent promoter activity was relieved in osteosarcoma cells treated with SCH6. Thus, NS3/4A disrupts the host response by mediating protease-dependent control of virus-induced RIG-I signaling to IRF-3 and NF-κB.
Fig. 4.
NS3/4A protease-dependent regulation of RIG-I disrupts an IFN amplification loop that controls HCV replication. (A and B) Transfected cells were cultured alone or in the presence of 10 μM SCH6 for 24 h before mock-infection or infection with SenV and luciferase assay. Bars show the average relative luciferase and SD values from three experiments. (A) Huh7 cells were cotransfected with plasmids encoding the IFN-β-luciferase promoter construct and Renilla luciferase along with 50 ng of plasmid DNA expressing vector only or RIG-I and 200 ng of plasmid DNA encoding WT NS3/4A, S1165A mutant NS3/4A, or IRF-3-ΔN as indicated. (B) UNS3/4A cells, cultured to suppress or induce NS3/4A expression as indicated, were cotransfected with plasmids encoding the PRDII-luciferase promoter construct and Renilla luciferase. (C) UNS3/4A cells, cultured to suppress or induce NS3/4A expression as indicated, were mock-infected, infected with SenV, or treated with 0, 10, 50, or 100 units/ml IFN-α2a as shown above each lane. Twenty hours later, cells were harvested for immunoblot analysis of endogenous protein levels. (D) Huh7 control or Huh7-A7, Huh7-K2040, and Huh-HP cells harboring the respective HCV replicon were mock-infected, infected with SenV, or treated with 50 units/ml IFN-α2a. After 20 h, the cells were harvested, and extracts were subjected to immunoblot analysis. (E) UNS3/4A cells, cultured to suppress or induce NS3/4A expression, were mock-infected, infected with SenV, or treated with 50 units/ml IFN-α2a. Twenty hours later, cells were harvested. Total RNA was isolated and reverse transcribed by using an oligo(dT) primer. Equal amounts of cDNA from each sample were subjected to quantitative real-time PCR analysis of RIG-I and GAPDH mRNA. Bars show the RIG-I mRNA level relative to the GAPDH internal control. (F) Huh7 cells (Left) or Huh7-HP cells harboring the HP HCV replicon (Right) were cultured in the presence of 10 μM SCH6 for the time indicated. Cells were harvested, and extracts were subjected to immunoblot analysis for determination of total IRF-3, the phosphoserine 396 isoform of IRF-3 (Ser-396-P), ISG56, NS5A, and actin levels. A parallel culture of Huh7 cells was infected with SenV and harvested 20 h later for use as an internal control (far left lane).
RIG-I Directs an IFN Amplification Loop That Is Blocked by NS3/4A. These results prompted us to evaluate the effect of NS3/4A on the abundance and stability of RIG-I. RIG-I is an ISG (2) and, in the absence of NS3/4A, SenV infection or IFN treatment of UNS3/4A cells induced a marked increase in the abundance of RIG-I and other ISGs, including ISG56 and PKR. Expression of NS3/4A prevented the virus-induced accumulation of RIG-I and ISGs but failed to prevent their increase after treatment of cultures with exogenous IFN (Fig. 4C). Consistent with this finding, NS3/4A expression blocked virus-induced RIG-I mRNA accumulation but did not prevent the increase in RIG-I mRNA abundance conferred by IFN treatment (Fig. 4E). Similar regulation of RIG-I expression occurred in the context of HCV RNA replication. In these experiments, control Huh7 cells responded to both IFN and SenV infection with induction of RIG-I and ISG expression, but cells harboring genetically distinct HCV replicon variants all exhibited a blockade to SenV-induced RIG-I and ISG accumulation while responding normally to exogenous IFN (Fig. 4D). These results indicate that RIG-I is not a substrate for the NS3/4A protease, but that NS3/4A disrupts the activation of an IFN amplification loop that is first signaled by RIG-I and then supported subsequently through an NS3/4A-resistant, IFN-mediated increase in RIG-I mRNA and protein accumulation.
Inhibition of NS3/4A Protease Activity Enables Host Recognition of the HCV Pathogen-Associated Molecular Pattern (PAMP). To determine whether HCV RNA replication results in expression of PAMPs that are capable of activating the RIG-I signaling pathway, we attempted to unmask the NS3/4A-mediated blockade of RIG-I signaling in Huh7-HP cells harboring a subgenomic HCV replicon by treatment of the cells with the SCH6 NS3/4A protease inhibitor. SenV infection of control Huh7 cells induced a host response marked by IRF-3 S396 phosphorylation and ISG56 expression (Fig. 4F Left). Within 24 h of the addition of SCH6 to the culture medium, a similar host response was induced in the Huh7-HP cells in the absence of SenV infection (Fig. 4F Right). Identical SCH6 treatment did not evoke these responses in control Huh7 cells (Fig. 4F Left). Thus, HCV RNA replication has an inherent capacity to signal a RIG-I-dependent host response, thereby triggering an IFN amplification loop that would otherwise limit viral replication in the absence of an effective NS3/4A blockade (see Figs. 2A and 4C).
Discussion
Our results define RIG-I as a critical component of innate intracellular defense against HCV and show that the RIG-I pathway is targeted by NS3/4A, thereby allowing HCV to avoid triggering the host antiviral response. We provide further evidence that this pathway branches to signal both IRF-3 and NF-κB activation during virus infection. NS3/4A control of RIG-I signaling depends on its protease activity, implying that NS3/4A may target and cleave component(s) of the RIG-I pathway with actions essential for IRF-3 phosphorylation and IκB degradation. Immunostaining of cells harboring the HP HCV replicon demonstrated colocalization of endogenous RIG-I and NS3 (Fig. 6, which is published as supporting information on the PNAS web site), suggesting that NS3/4A may regulate virus signaling through proteolytic inactivation of components of a RIG-I signaling complex. The Tankbinding kinase (TBK1) and IκB kinase-ε (IKKε) protein kinases were originally identified as upstream agonists of NF-κB activation (23, 24). Further studies have shown that TBK1 is essential for IRF-3 activation in nonlymphoid tissues (13, 25–28). This finding indicates an essential role for TBK1 in RIG-I signaling to IRF-3, although the redundant actions of IKKε may compensate to signal IRF-3 in the absence of TBK1 expression (2). TBK1 and IKKε provide an attractive link between IRF-3 and NF-κB signaling by RIG-I. However, in vitro studies have shown that neither serves as an NS3/4A proteolytic substrate (data not shown). Moreover, cell-based protein expression and stability analyses demonstrated that NS3/4A expression does not alter the abundance or stability of TBK1 or RIG-I (Fig. 7, which is published as supporting information on the PNAS web site) nor does it affect the stability of the TBK1 signaling adaptor protein, TANK (23) (data not shown). Thus, the disruption of RIG-I signaling is most likely due to proteolysis of undefined protein partner(s) involved in the RIG-I pathway.
Related studies have shown that HCV can antagonize IFN defenses through multiple mechanisms (29). Moreover, recent work has identified TRIF as a proteolytic substrate of NS3/4A whose proteolysis may ablate TLR3 signaling events (30). The current results show that TRIF is not a component of the RIG-I pathway, and thus indicate that the HCV protease has evolved to effectively target multiple pathways of host defense through proteolysis of more than one cellular protein. In the case of RIG-I signaling, our results support a model in which proteolytic cleavage of a RIG-I pathway component attenuates cellular antiviral defense gene expression, including ISGs and immunomodulatory cytokines, thereby providing a local cellular environment conducive to HCV persistence. In the context of HCV infection, the regulation of these pathways could control viral “priming” of the host response and activation of an IFN amplification loop signaled by RIG-I that provides cellular control of HCV replication, and possibly also influences viral sensitivity to IFN-based therapy. NS3/4A protease inhibitors thus offer a particularly attractive approach to therapeutic intervention, in that their use may lead to restoration of innate intracellular immune defenses that control HCV replication while also providing a direct antiviral effect.
Supplementary Material
Acknowledgments
We thank the indicated contributors for antibody and cell line reagents, and B. Malcolm and the Schering-Plough Research Institute for SCH6. This work was supported by National Institutes of Health Grants AI48235 and AI060389 (to M.G.), U19-AI40035 (to S.M.L.), R21-DA018054 (to K.L.), T32-GM08203 (to R.S.), and T32 AI 07520 (to E.F.), by a Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease Award, by Ellison Medical Foundation New Scholars in Global Infectious Disease Research Program ID-NS-0032, and by a gift from Mr. and Mrs. R. Batcheldor (to M.G.). K.L. is the John Mitchell Hemophilia of Georgia Liver Scholar of the American Liver Foundation. M.G. is the Nancy C. and Jeffrey A. Marcus Scholar in Medical Research in Honor of Dr. Bill S. Vowell.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RIG-I, retinoic acid-inducible gene I; TLR, Toll-like receptor; HCV, hepatitis C virus; IRF-3, IFN regulatory factor-3; NS, nonstructural; Huh, human hepatoma; SenV, Sendai virus; siRNA, short interfering RNA; TRIF, Toll-IL-1 receptor domain-containing adapter-inducing IFN-β; ISG, IFN-stimulated gene; TBK1, Tank-binding kinase 1; NDV, New Castle disease virus.
References
- 1.Sumpter, R., Loo, Y.-M., Foy, E., Li, K., Yoneyama, M., Fujita, T., Lemon, S. M. & Gale, M. J. (2005) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 2.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S. & Fujita, T. (2004) Nat. Immunol. 5, 730-737. [DOI] [PubMed] [Google Scholar]
- 3.Foy, E., Li, K., Wang, C., Sumpter, R., Ikeda, M., Lemon, S. M. & Gale, M., Jr. (2003) Science 300, 1145-1148. [DOI] [PubMed] [Google Scholar]
- 4.Fredericksen, B., Akkaraju, G., Foy, E., Wang, C., Pflugheber, J., Chen, Z. J. & Gale, M., Jr. (2001) Viral Immunol. 15, 29-40. [DOI] [PubMed] [Google Scholar]
- 5.Reed, K. E. & Rice, C. M. (1998) in Hepatitis C Virus, ed. Reesink, H. W. (Karger, Basel), pp. 1-37.
- 6.Sumpter, R., Wang, C., Foy, E., Loo, Y.-M. & Gale, M. J. (2004) J. Virol. 78, 11591-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakabayashi, H., Taketa, K., Miyano, K., Yamane, T. & Sato, J. (1982) Cancer Res. 42, 3858-3863. [PubMed] [Google Scholar]
- 8.Blight, K. J., McKeating, J. A. & Rice, C. M. (2002) J. Virol. 76, 13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, C., Pflugheber, J., Sumpter, R., Sodora, D., Hui, D., Sen, G. C. & Gale, M., Jr. (2002) J. Virol. 77, 3898-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda, M., Yi, M., Li, K. & Lemon, S. M. (2002) J. Virol. 76, 2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scholle, F., Li, K., Bodola, F., Ikeda, M., Luxon, B. A. & Lemon, S. M. (2004) J. Virol. 78, 1513-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolk, B., Sansonno, D., Krausslich, H., Dammacco, F., Rice, C., Blum, H. & Moradpour, D. (2000) J. Virol. 74, 2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., Coyle, A. J., Liao, S. M. & Maniatis, T. (2003) Nat. Immunol. 4, 491-496. [DOI] [PubMed] [Google Scholar]
- 14.Lin, R., Mamane, Y. & Hiscott, J. (1999) Mol. Cell. Biol. 19, 2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mori, M., Yoneyama, M., Ito, T., Takahashi, K., Inagaki, F. & Fujita, T. (2004) J. Biol. Chem. 279, 9698-9702. [DOI] [PubMed] [Google Scholar]
- 16.Lohmann, V., Korner, F., Kock, J.-O., Theilmann, L. & Bartenschlager, R. (1999) Science 285, 110-113. [DOI] [PubMed] [Google Scholar]
- 17.Lanford, R. E., Guerra, B., Lee, H., Averett, D. R., Pfeiffer, B., Chavez, D., Notvall, L. & Bigger, C. (2003) J. Virol. 77, 1092-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamoto, M., Sato, S., Mori, K., Hoshino, K., Takeuchi, O., Takeda, K. & Akira, S. (2002) J. Immunol. 169, 6668-6672. [DOI] [PubMed] [Google Scholar]
- 19.Grandvaux, N., Servant, M. J., tenOever, B., Sen, G. C., Balachandran, S., Barber, G. N., Lin, R. & Hiscott, J. (2002) J. Virol. 76, 5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Servant, M. J., Grandvaux, N., tenOever, B. R., Duguay, D., Lin, R. & Hiscott, J. (2003) J. Biol. Chem. 278, 9441-9447. [DOI] [PubMed] [Google Scholar]
- 21.Richmond, A. (2002) Nat. Rev. Immunol. 2, 664-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Francesco, R. & Steinkuhler, C. (2000) Curr. Top. Microbiol. Immunol. 242, 149-169. [DOI] [PubMed] [Google Scholar]
- 23.Pomerantz, J. L. & Baltimore, D. (1999) EMBO J. 18, 6694-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters, R. T., Liao, S. M. & Maniatis, T. (2000) Mol. Cell 5, 513-522. [DOI] [PubMed] [Google Scholar]
- 25.Hemmi, H., Takeuchi, O., Sato, S., Yamamoto, M., Kaisho, T., Sanjo, H., Kawai, T., Hoshino, K., Takeda, K. & Akira, S. (2004) J. Exp. Med. 199, 1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry, A. K., Chow, E. K., Goodnough, J. B., Yeh, W. C. & Cheng, G. (2004) J. Exp. Med. 199, 1651-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma, S., Benjamin, R., tenOever, B., Grandvaux, N., Zhou, G.-P., Lin, R. & Hiscott, J. (2003) Science 300, 1148-1151. [DOI] [PubMed] [Google Scholar]
- 28.McWhirter, S. M., Fitzgerald, K. A., Rosains, J., Rowe, D. C., Golenbock, D. T. & Maniatis, T. (2004) Proc. Natl. Acad. Sci. USA 101, 233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katze, M. G., He, Y. & Gale, M., Jr. (2002) Nat. Rev. Immunol. 2, 675-667. [DOI] [PubMed] [Google Scholar]
- 30.Li, K., Foy, E., Ferreon, J. C., Nakamura, M., Ferreon, A. C. M., Ikeda, M., Ray, S. C., Gale, M., Jr., & Lemon, S. M. (2005) Proc. Natl. Acad. Sci. USA 102, 2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.