ABSTRACT
Ergot alkaloids are specialized fungal metabolites that are important as the bases of several pharmaceuticals. Many ergot alkaloids are derivatives of lysergic acid (LA) and have vasoconstrictive activity, whereas several dihydrolysergic acid (DHLA) derivatives are vasorelaxant. The pathway to LA is established, with the P450 monooxygenase CloA playing a key role in oxidizing its substrate agroclavine to LA. We analyzed the activities of products of cloA alleles from different fungi relative to DHLA biosynthesis by expressing them in a mutant of the fungus Neosartorya fumigata that accumulates festuclavine, the precursor to DHLA. Transformants expressing CloA from Epichloë typhina × Epichloë festucae, which oxidizes agroclavine to LA, failed to oxidize festuclavine to DHLA. In substrate feeding experiments, these same transformants oxidized exogenously supplied agroclavine to LA, indicating that a functional CloA was produced. A genomic clone of cloA from Claviceps africana, a sorghum ergot fungus that produces a DHLA derivative, was cloned and expressed in the festuclavine-accumulating mutant of N. fumigata, but several introns in this genomic clone were not processed properly. Expression of a synthetic intron-free version of C. africana cloA resulted in the accumulation of DHLA as assessed by fluorescence high-pressure liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS). In substrate feeding experiments, the C. africana CloA also accepted agroclavine as the substrate, oxidizing it to LA. The data indicate that a specialized allele of cloA is required for DHLA biosynthesis and that the pharmaceutically important compound DHLA can be produced in engineered N. fumigata.
IMPORTANCE Ergot alkaloids are fungal metabolites that have impacted humankind historically as poisons and more recently as pharmaceuticals used to treat dementia, migraines, and other disorders. Much is known about the biosynthesis of ergot alkaloids that are derived from lysergic acid (LA), but important questions remain about a parallel pathway to ergot alkaloids derived from dihydrolysergic acid (DHLA). DHLA-derived alkaloids have minor structural differences compared to LA-derived alkaloids but can have very different activities. To understand how DHLA is made, we analyzed activities of a key enzyme in the DHLA pathway and found that it differed from its counterpart in the LA pathway. Our data indicate a critical difference between the two pathways and provide a strategy for producing DHLA by modifying a model fungus. The ability to produce DHLA in a model fungus may facilitate synthesis of DHLA-derived pharmaceuticals.
KEYWORDS: ergot alkaloids, lysergic acid, dihydrolysergic acid, P450 monooxygenase
INTRODUCTION
Ergot alkaloids are specialized fungal metabolites that have impacted humankind historically as toxins and more recently as pharmaceuticals used to treat dementia, migraines, hyperprolactinemia, and Parkinson's disease (1–4). Many natural ergot alkaloids are derivatives of lysergic acid (LA) and have vasoconstrictive activity. Derivatives of dihydrolysergic acid (DHLA), which are rarely encountered naturally, are the basis for many of the commonly prescribed vasorelaxant ergot-alkaloid-derived pharmaceuticals (5–7). LA and its derivatives differ from dihydroergot alkaloids by the presence of a double bond in the fourth (last assembled) or “D” ring of the ergoline nucleus (Fig. 1) and can be converted to their dihydro forms by reduction of that double bond. Only one known naturally occurring fungus produces a DHLA derivative: Claviceps africana synthesizes dihydroergosine, a dihydroergopeptine containing DHLA (8). Claviceps africana is one of three ergot fungi that parasitize sorghum (Sorghum bicolor) (9, 10); the others, Claviceps sorghicola and Claviceps sorghi, are not known to produce ergot alkaloids. Dihydroergosine is the most complex and abundant dihydroergot alkaloid produced by C. africana, but pathway intermediates festuclavine and dihydroelymoclavine (synonym dihydrolysergol) also accumulate in sorghum infected by this fungus (11, 12). Festuclavine and dihydroelymoclavine must be oxidized to DHLA before incorporation into dihydroergosine (11).
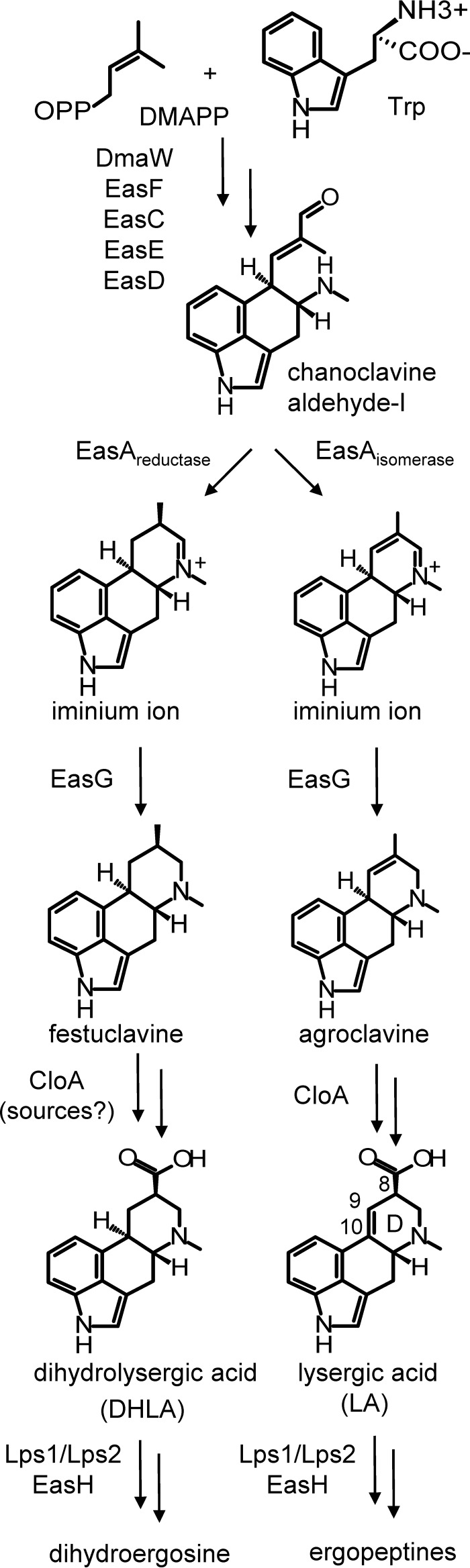
FIG 1.
Pathways to lysergic acid and dihydrolysergic acid and their derivatives. Enzymes responsible for catalysis are indicated between intermediates and products. Multiple arrows indicate the omission of intermediates to simplify the figure. Relevant customary ring and carbon labeling is indicated in the lysergic acid structure. DMAPP, dimethylallylpyrophosphate.
All fungi that make 4-ring ergot alkaloids share five pathway genes (dmaW, easF, easC, easE, and easD) (Fig. 1) required for assembly of chanoclavine-I aldehyde, the final intermediate prior to formation of the fourth or D ring (7, 13–15). The formation of the D ring may proceed to yield a saturated form, festuclavine, or an unsaturated form, agroclavine, depending on the allele of the gene easA present (16–18) (Fig. 1). For example, the allele of easA from the fungus Neosartorya fumigata (Aspergillus fumigatus) reduces the C-8–C-9 double bond in chanoclavine-I aldehyde, allowing rotation of the aldehyde functional group such that it can react with the secondary amine to form an iminium ion that is subsequently reduced by the enzyme EasG (also called FgaFS) to festuclavine (Fig. 1) (19). Epichloë festucae (formerly called Neotyphodium lolii) has an allele of easA that encodes a functionally different enzyme (17). EasA from E. festucae appears to only temporarily reduce the C-8–C-9 double bond of chanoclavine-I aldehyde to allow the isomerization that facilitates the formation of the iminium ion before oxidizing the product to restore the C-8–C-9 double bond as found in agroclavine. LA is produced from agroclavine via primary alcohol (elymoclavine) and aldehyde oxidation intermediates by the activity of the P450 monooxygenase named CloA (20, 21). Early labeling studies (11) indicated that DHLA was produced from festuclavine via an analogous set of reactions, but the genetic and enzymatic bases of DHLA production have not been analyzed.
The difference between LA and DHLA is the double bond in the D ring, with associated differences in numbers of hydrogens on C-9 and C-10 (Fig. 1). One possibility is that the only difference between LA and DHLA pathways is formation of agroclavine, in the LA pathway, versus festuclavine, in the DHLA pathway, as intermediates resulting from the differences in activities between the products of the two alleles of easA. This scenario would require the four enzymes or enzyme complexes downstream of EasA (EasG, CloA, the Lps1/Lps2 complex, and EasH) in the pathway to ergopeptines or dihydroergopeptines to act on intermediates with either a saturated or unsaturated D ring. This hypothesis is supported by the observations that three of the four enzymes downstream of EasA have been documented to accept saturated and unsaturated forms of their substrates. EasG of N. fumigata, which normally acts on the fully reduced iminium ion precursor to festuclavine (Fig. 1), also reduced the unsaturated iminium ion precursor to agroclavine (17–19). Moreover, Lps2 of Claviceps purpurea (together with Lps1, as part of the lysergyl peptide synthetase complex) acted on DHLA with approximately the same Km with which it accepted its natural substrate LA (22). Finally, EasH, the last enzyme in the pathway to LA-derived ergopeptines in C. purpurea, accepted DHLA-derived dihydroergopeptam substrates as well as its typical LA-derived ergopeptams (23). The alternative hypothesis is that a specialized festuclavine-oxidizing version of CloA will be required to produce DHLA from festuclavine. In this present study, we address these two alternative hypotheses by expressing alleles of cloA from an LA producer and a DHLA producer in a strain of N. fumigata that had been modified previously to accumulate festuclavine as its end product by knocking out the gene easM (24). In N. fumigata (which does not have an ortholog of cloA in its genome), festuclavine is typically modified by a unique series of enzymatic reactions (initiated by the hydroxylation of festuclavine at C-9 by EasM) to produce a family of ergot alkaloids called fumigaclavines that differ structurally from DHLA and its derivatives (7, 14, 15, 25). The easM step, thus, serves as the branch point between fumigaclavine and DHLA pathways, and its mutation provides a tool for analysis of DHLA biosynthesis.
RESULTS
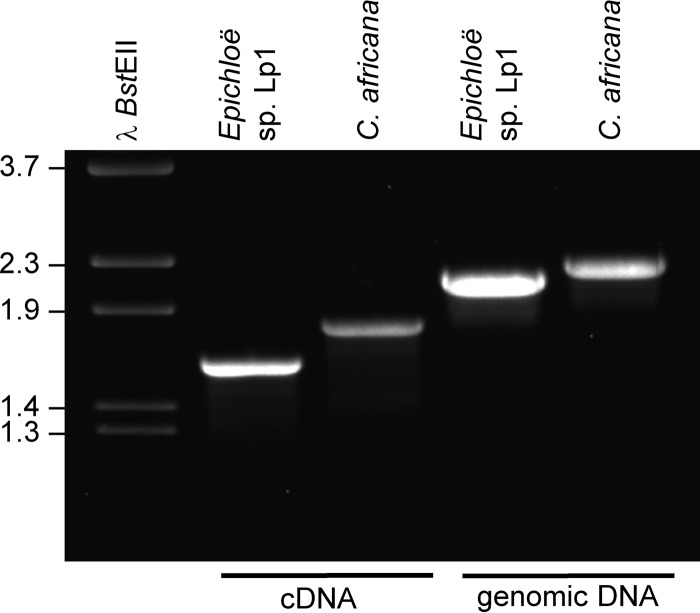
The ability of the Epichloë typhina × E. festucae isolate Lp1 CloA to oxidize festuclavine to DHLA was evaluated by heterologously expressing its encoding gene in a festuclavine-accumulating mutant of N. fumigata. Expression of E. typhina × E. festucae Lp1 cloA under the control of the N. fumigata easA promoter did not result in accumulation of DHLA (Fig. 2), and no other novel metabolites were evident in high-pressure liquid chromatography (HPLC) chromatograms. PCR assays indicated that the desired fragments were in the genomes of recipient strains (see Fig. S1 in the supplemental material), and reverse transcriptase PCR (RT-PCR) assays indicated accumulation of cloA mRNA (Fig. 3). Sequencing of the major band of cloA cDNA from strains containing E. typhina × E. festucae Lp1 cloA demonstrated that cloA transcripts were processed correctly (see Fig. S2 in the supplemental material). Therefore, substrate feeding studies were conducted to test whether functional CloA was present in transformants. Agroclavine supplied exogenously to E. typhina × E. festucae Lp1 cloA transformants was oxidized to LA (Table 1), indicating that functional CloA was present. Controls indicated that accumulation of LA was dependent on the presence of cloA in the fungus and exogenously supplied agroclavine. Collectively, the data indicate that expression of E. typhina × E. festucae Lp1 cloA was successful, but festuclavine was not recognized as the substrate by CloA encoded by that particular allele.
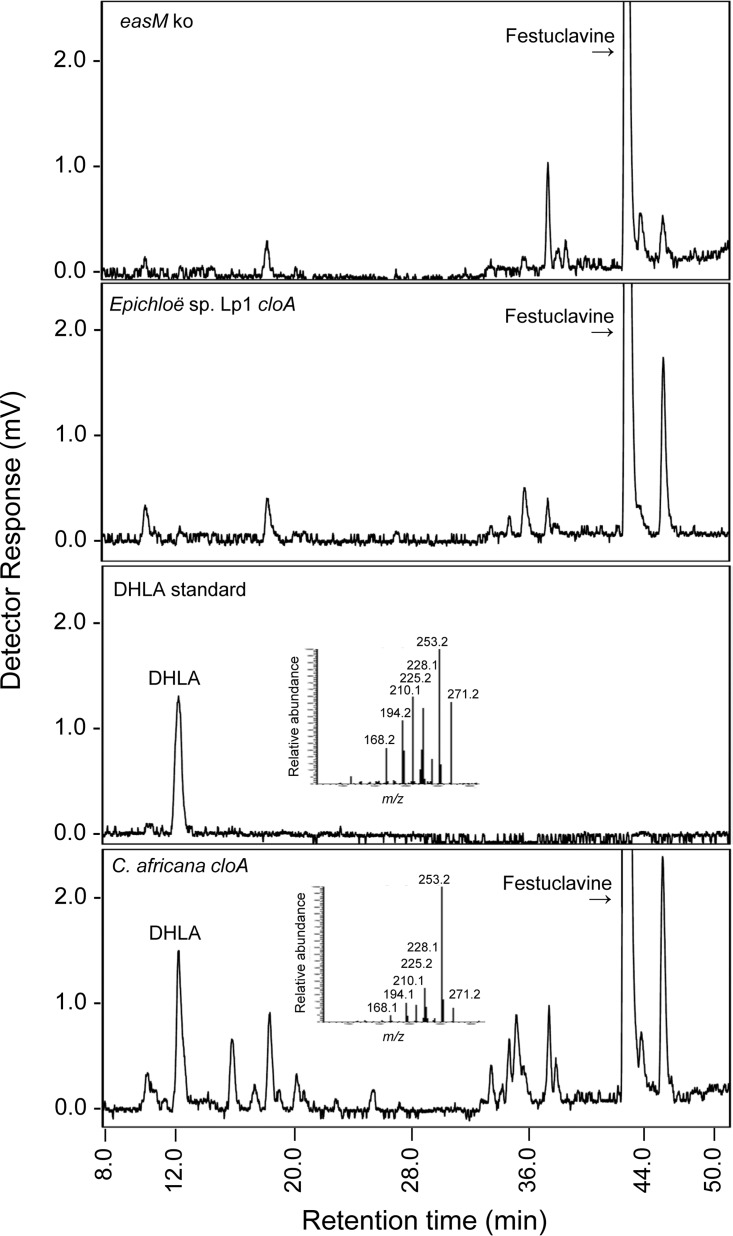
FIG 2.
Analysis of ergot alkaloids from transformed strains. HPLC chromatograms of recipient strain (N. fumigata easM knockout), transformed strains containing E. typhina × E. festucae isolate Lp1 cloA or synthetic C. africana cloA, and DHLA standard. Fluorescence was detected at 372 nm after exciting at 272 nm. Mass spectra of fragmented parent ion of m/z 271.2 from DHLA standard and N. fumigata expressing synthetic C. africana cloA are inserted in relevant chromatograms.
FIG 3.
Products derived from cDNA or genomic DNA isolated from N. fumigata expressing E. typhina × E. festucae isolate Lp1 or C. africana genomic alleles of cloA. Sizes of relevant fragments from BstEII-digested bacteriophage lambda DNA are indicated to the left. Gel was stained with ethidium bromide.
TABLE 1.
Oxidation of exogenously fed agroclavine to lysergic acid by N. fumigata easM knockout cultures expressing different alleles of cloA
| Straina | Treatmentb | Agroclavine (nmol/culture [mean ± SE]) | Lysergic acid (nmol/culture [mean ± SE]) |
|---|---|---|---|
| easM ko | Agroclavine | 12.3 ± 2.3 Ac | NDd |
| easM ko + Epichloë sp. Lp1 cloA | Agroclavine | 11.0 ± 1.3 A | 0.25 ± 0.02 A |
| easM ko + C. africana synthetic cloA | Agroclavine | 12.4 ± 0.7 A | 0.47 ± 0.02 B |
| Medium | Agroclavine | 11.3 ± 2.7 A | ND |
| easM ko + Epichloë sp. Lp1 cloA | Methanol | ND | ND |
| easM ko + C. africana cloA | Methanol | ND | ND |
n = 6.
Agroclavine, 20 nmol.
Means followed by the letter A did not differ significantly in an ANOVA (P > 0.05).
ND, not detected; limit of detection = 0.01 pmol/culture.
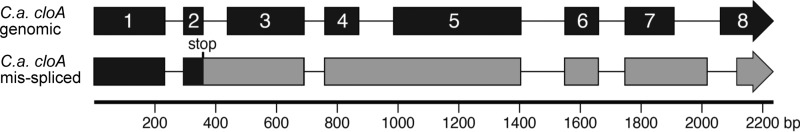
A genomic clone of the allele of cloA from C. africana was tested in a parallel set of experiments. Claviceps africana produces dihydroergosine, the dihydroergopeptine derived from DHLA. Neosartorya fumigata easM knockout (ko) transformed with C. africana cloA under the control of the N. fumigata easA promoter failed to oxidize festuclavine to DHLA (data not shown). The cloA cDNA from this strain was larger than the expected 1,610-bp fragment (Fig. 3), and when sequenced, it was found to be derived from improperly processed mRNA (Fig. 4; see also Fig. S3 in the supplemental material). Introns 2 and 4 were fully retained, and intron 7 was processed from erroneous 5′ splice junction and 3′ splice junction sequences such that the 5′ portion of the intron was retained, and part of the final exon was removed. Due to the retention of intron 2, the protein translated from this mRNA would have a premature termination codon after 99 amino acids (Fig. 4; see also Fig. S4 in the supplemental material), whereas the full-length protein should be 509 amino acids long.
FIG 4.
Graphic representation of C. africana cloA allele to demonstrate issues with intron processing in N. fumigata. Exons in genomic sequence are numbered and colored black. Introns are indicated by thinner lines. In the misspliced transcript, introns 2 and 4 and part of intron 7 are retained in the mRNA as indicated by the thicker, exonic lines. The stop codon resulting from the retention of intron 2 is indicated and exonic sequences downstream of that stop codon are shaded gray.
To circumvent the processing issue, a synthetic, intron-free version of C. africana cloA was prepared that contained the coding sequences of the gene along with 506 nt of 3′-untranslated DNA fused to the N. fumigata gpdA promoter. Codons were optimized for N. fumigata translation (see Fig. S5 in the supplemental material), but the amino acid sequence of the gene product was not altered compared to the wild-type version (Fig. S4). Transformation of N. fumigata easM ko with the synthetic, coding-sequence-only version of C. africana cloA resulted in strains that oxidized festuclavine into DHLA, as evidenced by HPLC with fluorescence detection and mass spectrometry (Fig. 2). In HPLC analyses, a novel peak fluorescing with an excitation wavelength of 272 nm and emission wavelength of 372 coeluted with DHLA standard. In positive mode electrospray ionization liquid chromatography-mass spectrometry (LC-MS), the peak had a parent ion with m/z of 271.2 which corresponds to the mass of [DHLA + H]+. When fragmented, the m/z 271.2 ion gave rise to fragment ions of the same mass as those liberated by fragmenting the DHLA standard (Fig. 2). Additional novel but minor peaks were detected eluting at approximately 16 and 26 min in the chromatogram of strains expressing the synthetic C. africana cloA construct. The identities of these peaks and their potential relationship to DHLA were not investigated further.
Feeding agroclavine, the precursor to LA, to the strain expressing the synthetic C. africana cloA resulted in accumulation of LA, indicating that the C. africana version of CloA accepts agroclavine as well as festuclavine as the substrate (Table 1). This finding contrasts with the results obtained with CloA from E. typhina × E. festucae isolate Lp1, which accepted agroclavine but not festuclavine as the substrate. Controls indicated that LA accumulated only when a fungus expressing cloA was exposed to exogenously supplied agroclavine.
DISCUSSION
The ergot alkaloid pathway branch to DHLA differs from the branch to LA in two significant ways. First, as previously established (17–20), the reductase form of EasA is required to synthesize the intermediate festuclavine (with its fully reduced D ring) in the DHLA branch, as opposed to production of the unsaturated agroclavine intermediate by the isomerase type of EasA in the LA branch of the pathway (Fig. 1). Second, as shown in this present study, a version of CloA with altered or relaxed substrate specificity is required to oxidize festuclavine to DHLA in the DHLA branch. CloA from the LA-producing fungus E. typhina × E. festucae isolate Lp1 oxidized agroclavine but not festuclavine as the substrate, whereas the product of the C. africana allele oxidized both agroclavine and festuclavine to their respective carboxylic acid forms. The results support the hypothesis that a specialized version of CloA is required to produce DHLA from festuclavine.
Phylogenetic data indicate that the dihydroergot alkaloid pathway in C. africana evolved in a lineage of fungi that produced LA-derived ergot alkaloids (13). Two novel traits were required in the evolution of the DHLA branch of the ergot alkaloid pathway, alternative alleles of easA and cloA. The change in alleles of easA leading to production of festuclavine as opposed to agroclavine is clearly a prerequisite to DHLA synthesis, but the necessity for a novel allele of cloA for DHLA biosynthesis was not evident a priori. Reasons for initially hypothesizing that enzymes downstream of festuclavine and agroclavine would accept either of those intermediates as the substrates included previous studies indicating that the three downstream enzymes (or enzyme complexes) other than CloA that interact with the relevant intermediates (EasG, Lps2 as part of the lysergyl peptide synthetase complex, and EasH) accepted saturated (festuclavine-derived) and unsaturated (agroclavine-derived) substrates (17, 18, 22, 23). Hypothetically, the failure of the E. typhina × E. festucae isolate Lp1 version of CloA to recognize festuclavine as the substrate may result from steric hindrances due to additional hydrogens on both carbon 9 and carbon 10. The changes in the C. africana version of CloA that allowed festuclavine to serve as the substrate still allowed it to accept agroclavine.
One can envision the novel alleles of easA and cloA arising in either of two possible sequences. If evolution of the version of CloA with relaxed or altered specificity occurred first, the fungus probably would have been able to continue to produce LA-derived ergot alkaloids from the agroclavine substrate, since in this current study, C. africana CloA oxidized agroclavine to LA. When in this lineage easA mutated to the reductase allele, the fungus would then have been preadapted to accept festuclavine as the substrate and make DHLA and DHLA-derivatives. Alternatively, if the version of easA changed first, festuclavine would have accumulated and there would have then been pressure on the fungus to evolve an allele of cloA that encoded a product capable of accepting festuclavine as the substrate. The events in either scenario fit well with the screening hypothesis of Firn and Jones (26), since the events increase chemical diversity and form a new pathway branch.
The inability of N. fumigata to process the C. africana cloA introns properly was unexpected. Retained intron 2 had a 3′ splice site of AAG, which differs from the more common YAG sequence typically found in fungi (27); however, intron 1 also ended with AAG and was processed properly. The introns that were retained had a predominance of adenosine homopolymers. Retained intron 2 contained a run of 25 consecutive A residues immediately before and into the 3′ splice site sequence. Retained intron 4 had a series of 9 consecutive A residues between the predicted branch site and the 3′ splice site. Retained intron 7 had a string of 69 nucleotides in which 60 A residues occurred between the 5′ splice site and the presumed branch site. It should be noted, however, that intron 5, which was processed properly, contained a string of 54 nucleotides, of which 42 were A residues between its 5′ splice site and the presumed branch site.
In addition to removal of introns, the synthetic C. africana cloA allele differed from the native allele in two other ways, (i) a gpdA promoter from N. fumigata was used on the synthetic allele, whereas an N. fumigata easA promoter drove expression of the native allele, and (ii) codon usage was altered in the synthetic allele to conform with preferences observed in N. fumigata (28) without altering the protein sequence. These changes might affect the quantity of DHLA produced; however; the qualitative presence/absence of DHLA observed almost certainly resulted from the production of a full-length protein. RT-PCR analyses indicated that abundant mRNA was produced from the original C. africana cloA genomic construct, although that mRNA was not properly processed. Another factor that potentially affected relative expression of the different constructs in this study was that their integration in the genome was likely random. Fifteen to 20 transformants for each construct were thus analyzed to control any position effects on expression via randomization.
Whereas our results show accumulation of DHLA in N. fumigata transformants expressing the intron-free allele of C. africana cloA, relatively large quantities of festuclavine were retained, as evidenced by relative peak height in HPLC chromatograms. Determination of the reason for this relatively poor turnover was not the purpose of this current study, but potential reasons include poor expression or stability of the introduced CloA or differences in localization of CloA and its substrate in the heterologous host, N. fumigata. Substrate feeding studies with agroclavine produced conversions comparable to those observed in a previous study in which native N. fumigata enzymes prenylated when exogenously fed agroclavine substrate (21). This observation indicates that neither protein expression nor stability was a major factor. Issues related to localization or other potential reasons for the modest turnover will need to be addressed in order for a DHLA-producing strain to have practical value. An ability to produce abundant DHLA in a manipulable fungus like N. fumigata might provide opportunities for improved synthesis of DHLA-derived compounds.
MATERIALS AND METHODS
Modification of fungi.
All cloA constructs were expressed in a mutant of N. fumigata (Aspergillus fumigatus) Af293 in which the easM gene had been knocked out (N. fumigata easM ko) to cause the strain to accumulate festuclavine (24), a precursor to DHLA (11). Since the N. fumigata easM ko strain is resistant to hygromycin, all cloA expression constructs were prepared in pBCphleo (29; Fungal Genetics Stock Center, Manhattan, KS), which confers resistance to phleomycin.
Two cloA expression constructs were prepared by PCR from genomic DNA in a series of reactions catalyzed by Phusion DNA polymerase (Thermo Fisher, Waltham, MA). In general, PCR analysis was conducted as follows: an initial denaturation at 98°C for 30 s, followed by 35 cycles of denaturation at 98°C for 15 s, primer annealing at 62°C for 15 s, and primer extension at 72°C for 60 s. Variations in annealing temperature or extension time are noted below for particular primer combinations. The E. typhina × E. festucae isolate Lp1 (30) cloA construct consisted of the easA promoter of N. fumigata (operationally defined as the 787 bp immediately upstream of the initiation codon) fused to the genomic E. typhina × E. festucae Lp1 cloA coding sequences along with 622 nt of 3′-untranslated sequences. The promoter/coding sequence fragment was PCR amplified from the E. typhina × E. festucae isolate Lp1 easA-cloA strain described in Robinson and Panaccione (21) in a reaction with primer combination 1 (Table 2). PCR conditions were as described above, but the annealing temperature was 67°C. The PCR product was cleaned with the DNA Clean and Concentrator-5 kit (Zymo Research, Irvine, CA), digested with EcoRI and NotI (New England BioLabs, Ipswich, MA) (sites for which were included in primers), and ligated into EcoRI/NotI-digested pBCphleo. To prepare the C. africana isolate Cla9 (10) genomic cloA construct, coding sequences of cloA and 300 bp of 3′ flanking sequences were PCR amplified from primer combination 2 (Table 2). Primers were designed based on the sequence of a contig obtained from a draft genome sequence of C. africana isolate Cla 9, which has been deposited in GenBank as accession number KY677717. The N. fumigata easA promoter (amplified with primer combination 3; Table 2) was attached to coding sequence fragments in a fusion PCR with approximately equimolar concentrations of C. africana cloA coding sequences and N. fumigata easA promoter fragments as the template and primer combination 4 (Table 2). Reaction conditions were as described above but with an extension time of 75 s. The fusion PCR product was cleaned as described above, digested with EcoRI and NotI (sites included in primers for fusion PCR), and ligated into EcoRI/NotI-digested pBCphleo. Sequence integrity of constructs was confirmed by Sanger methodology at Eurofins Genomics (Louisville, KY).
TABLE 2.
PCR primers and products
| Primer combination | Primer sequences (5′→3′) | Product | Amplicon length(s) (bp) |
|---|---|---|---|
| 1 | CATCGAATTCGTTGACATTGCTTCTAATCCACC, GACAGCGGCCGCAACAAGCGATAAGCGTTAG | Epichloë sp. Lp1 cloA + N. fumigata promoter | 3,526 |
| 2 | GTACTTGGTGGATTAGAAGCAATGTCGCAACTATGGCTATACAAGGC, GACAGCGGCCGCGTGTTCACGCCACGTCACC | C. africana cloA with promoter extensiona | 2,569 |
| 3 | CATCGAATTCTGCCTACTCTATAGAAGATGGATC, GCCTTGTATAGCCATAGTTGCGACATTGCTTCTAATCCACCAAGTAC | Promoter with C. africana cloA extension | 811 |
| 4 | CATCGAATTCTGCCTACTCTATAGAAGATGGATC, GACAGCGGCCGCGTGTTCACGCCACGTCACC | Promoter/C. africana cloA fusion construct | 3,334 |
| 5 | CTATAGAGTAGGCACTCCGCAC, CTCCAAGTCAAGATACATCCCTC | Epichloë sp. Lp1 cloA | 1,610 (cDNA), 2,178 (gDNAb) |
| 6 | CTATAGAGTAGGCACTCCGCAC, GCGAGATAAATGAGAAGAACCATTG | C. africana cloA | 1,861 (cDNA), 2,318 (gDNA) |
Extension refers to incorporation of an additional 21 or 23 nucleotide (nt) at the 5′ end of a primer to add sequences that will facilitate a later fusion PCR.
Genomic DNA.
The coding sequence of synthetic cloA construct (GenBank accession no. KY681814) was derived from the C. africana allele by deleting introns (as marked in Fig. S3 in the supplemental material) and changing nucleotides to favor codons considered optimal for N. fumigata in the analyses of Iriarte et al. (28) without altering amino acid sequence of the product (see Fig. S4). The synthetic coding sequences were placed under the control of the promoter of the N. fumigata glyceraldehyde-3-phosphate dehydrogenase gene, gpdA (GenBank accession no. AM999768), operationally defined as the 962 bp immediately preceding the initiation codon. The synthetic construct also contained 506 bp of 3′-untranslated sequence as found immediately downstream of the stop codon of the native C. africana cloA allele. The construct was prepared and sequenced by GenScript (Piscataway, NJ). The expression construct was removed from its original pUC19 vector by digestion with EcoRI and ligated into EcoRI-digested pBCphleo for transformation into N. fumigata easM ko.
The N. fumigata easM ko strain was transformed essentially as described previously (18, 24). Spheroplasts were prepared with cell wall-degrading enzymes Vinotaste (Gusmer Enterprises, Mountainside, NJ) and lysing enzyme from Trichoderma harzianum (Sigma Chemical, St. Louis, MO), and DNA was introduced via the polyethylene glycol–CaCl2 method. Transformants were selected on complete regeneration medium containing phleomycin (InvivoGen, San Diego, CA) at 400 μg/ml and purified to nuclear homogeneity by culturing from single conidia. Fifteen to 20 transformants obtained with each construct were analyzed by HPLC (described below) to account for potential variation associated with random integration of the cloA expression constructs.
Analysis of transformants.
DNA was isolated from transformants with a GeneClean kit (MP Biomedical, Santa Ana, CA) and analyzed for the presence of introduced fragments by PCR as described above with primer combination 1 (Table 2). RNA was isolated from 100 mg of 2-day-old surface cultures of native and transformed strains of N. fumigata grown on malt extract broth by application of the Qiagen RNeasy Plant kit. Polyadenylated mRNA was primed with oligo(dT) and reverse transcribed into cDNA in a reaction with Superscript IV reverse transcriptase (Thermo Fisher). Resulting cDNA was amplified with primer combination 5 for E. typhina × E. festucae Lp1 or primer combination 6 for C. africana (Table 2). PCR conditions were as described above but with an annealing temperature of 58°C and an extension time of 45 s. PCR products were sequenced by Sanger methodology at Eurofins Genomics (Louisville, KY).
Ergot alkaloids were extracted from conidiating cultures of N. fumigata in HPLC-grade methanol and analyzed by reverse-phase HPLC as described previously (31). Briefly, the column was a 150 × 4.6 mm Prodigy ODS3 with 5-μm particle size (Phenomenex, Torrance, CA), and the mobile phase was a binary, multilinear gradient from 5% acetonitrile (Fisher, Pittsburgh, PA) + 95% 50 mM ammonium acetate to 75% acetonitrile + 25% 50 mM aqueous ammonium acetate. Analytes were detected by fluorescence at two wavelengths: excitation at 272 nm with emission at 372 nm for ergot alkaloids lacking a C-9–C-10 unsaturation in the D ring (such as festuclavine and DHLA) (Fig. 1), and 310-nm excitation with 410-nm emission for ergot alkaloids containing a C-9–C-10 double bond in the D ring (such as LA). DHLA standard was prepared by hydrolyzing the peptide bonds in dihydroergocristine (Sigma) in 1.2 M NaOH at 75°C for 6 h. The solution was then brought to neutral pH with the addition of 0.5 volume of 2.4 M acetic acid. Standard prepared in this manner was analyzed for fluorescence properties characteristic of DHLA (excitation at 272 nm with emission at 372 nm) and by high-resolution mass spectrometry (calculated m/z, 271.1441; measured m/z, 271.1440; deviation, 0.37 ppm). Electrospray ionization liquid chromatography–mass spectrometry (LC-MS) was performed in positive mode on a Thermo LCQ Deca XP Plus equipped with a Surveyor HPLC as described by Ryan et al. (32). DHLA standard was analyzed on a Thermo Q Exactive mass spectrometer as described by Robinson and Panaccione (21).
Production of LA through a substrate feeding approach.
Substrate feeding studies were conducted as described by Robinson and Panaccione (21) with minor modifications. Briefly, six replicate cultures of each treatment or control group were grown from 60,000 conidia in 200 μl malt extract broth supplemented with 20 nmol agroclavine in 1 μl of methanol or, in controls, with 1 μl of methanol. After 1 week, ergot alkaloids were extracted by the addition of 300 μl of methanol and analyzed by HPLC with fluorescence detection as described above. Agroclavine was quantified by comparing peak areas to standard curve prepared from known quantities of agroclavine (Fisher), whereas LA was quantified relative to a standard curve prepared from ergonovine (Sigma). Quantities of alkaloids across treatments and controls were compared by analysis of variance (ANOVA). All statistical analyses were performed with JMP software (SAS, Cary, NC).
Accession number(s).
The sequences described were deposited in GenBank under the accession numbers KY677717 and KY681814.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Tooley for generously supplying genomic DNA of C. africana, Christopher Schardl for advice on sequencing, Sarah Robinson for helpful suggestions on the manuscript, and Matthew Maust and Protea Biosciences for assistance with high-resolution mass spectrometry.
Research with lysergic acid was conducted with licenses from the West Virginia Board of Pharmacy (TI0555042) and the U.S. Drug Enforcement Agency (RP0463353).
This work was supported by grant R15GM114774 from the National Institutes of Health, National Institute of General Medical Sciences, and United States Department of Agriculture Hatch funds and published with permission of the West Virginia Agriculture and Forestry Experiment Station as scientific article number 3309.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00805-17.
REFERENCES
- 1.Baskys A, Hou AC. 2007. Vascular dementia: pharmacological treatment approaches and perspectives. Clin Interv Aging 2:327–335. [PMC free article] [PubMed] [Google Scholar]
- 2.Winblad B, Fioravanti M, Dolezal T, Logina I, Milanov IG, Popescu DC, Solomon A. 2008. Therapeutic use of nicergoline. Clin Drug Investig 28:533–552. doi: 10.2165/00044011-200828090-00001. [DOI] [PubMed] [Google Scholar]
- 3.Morren JA, Galvez-Jimenez N. 2010. Where is dihydroergotamine mesylate in the changing landscape of migraine therapy? Expert Opin Pharmacother 11:3085–3093. doi: 10.1517/14656566.2010.533839. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Lloret S, Rascol O. 2010. Dopamine receptor agonists for the treatment of early or advanced Parkinson's disease. CNS Drugs 24:941–968. doi: 10.2165/11537810-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 5.Jähnichen S, Horowski R, Pertz HH. 2005. Agonism at 5-HT2B receptors is not a class effect of the ergolines. Eur J Pharmacol 513:225–228. doi: 10.1016/j.ejphar.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 6.Iliff LD, Du Boulay GH, Marshall J, Russell RWR, Symon L. 1977. Effect of nicergoline on cerebral blood flow. J Neurol Neurosurg Psychiatry 40:746–747. doi: 10.1136/jnnp.40.8.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallwey C, Li S-M. 2011. Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat Prod Rep 28:496–510. doi: 10.1039/C0NP00060D. [DOI] [PubMed] [Google Scholar]
- 8.Mantle PG, Waight ES. 1968. Dihydroergosine: a new naturally occurring alkaloid from the sclerotia of Sphacelia sorghi (McRae). Nature 218:581–582. doi: 10.1038/218581a0. [DOI] [Google Scholar]
- 9.Pažoutová S, Bandyopadhyay R, Frederickson DE, Mantle PG, Frederiksen RA. 2000. Relations among sorghum ergot isolates from the Americas, Africa, India, and Australia. Plant Dis 84:437–442. doi: 10.1094/PDIS.2000.84.4.437. [DOI] [PubMed] [Google Scholar]
- 10.Tooley P, Goley E, Carras M, Frederick R, Weber E. 2001. Characterization of Claviceps species pathogenic on sorghum by sequence analysis of the β-tubulin gene intron 3 region and EF-1α gene intron 4. Mycologia 93:541–551. doi: 10.2307/3761739. [DOI] [Google Scholar]
- 11.Barrow KD, Mantle PG, Quigley FR. 1974. Biosynthesis of dihydroergot alkaloids. Tet Lett 15:1557–1560. doi: 10.1016/S0040-4039(01)93135-1. [DOI] [Google Scholar]
- 12.Blaney BJ, Maryam R, Murray S-A, Ryley MJ. 2003. Alkaloids of the sorghum ergot pathogen (Claviceps africana): assay methods for grain and feed and variation between sclerotia/sphacelia. Crop Pasture Sci 54:167–175. doi: 10.1071/AR02095. [DOI] [Google Scholar]
- 13.Florea S, Panaccione DG, Schardl CL. 2017. Ergot alkaloids of the family Clavicipitaceae. Phytopathology 107:504–518. doi: 10.1094/PHYTO-12-16-0435-RVW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson SL, Panaccione DG. 2015. Diversification of ergot alkaloids in natural and modified fungi. Toxins 7:201–218. doi: 10.3390/toxins7010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerhards N, Neubauer L, Tudzynski P, Li SM. 2014. Biosynthetic pathways of ergot alkaloids. Toxins 6:3281–3295. doi: 10.3390/toxins6123281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng JZ, Coyle CM, Panaccione DG, O'Connor SE. 2010. A role for old yellow enzyme in ergot alkaloid biosynthesis. J Am Chem Soc 132:1776–1777. doi: 10.1021/ja910193p. [DOI] [PubMed] [Google Scholar]
- 17.Cheng JZ, Coyle CM, Panaccione DG, O'Connor SE. 2010. Controlling a structural branch point in ergot alkaloid biosynthesis. J Am Chem Soc 132:12835–12837. doi: 10.1021/ja105785p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coyle CM, Cheng JZ, O'Connor SE, Panaccione DG. 2010. An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways. Appl Environ Microbiol 76:3898–3903. doi: 10.1128/AEM.02914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallwey C, Matuschek M, Xie XL, Li SM. 2010. Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I aldehyde to festuclavine by the festuclavine synthase FgaFS in the presence of the old yellow enzyme FgaOx3. Org Biomol Chem 8:3500–3508. doi: 10.1039/c003823g. [DOI] [PubMed] [Google Scholar]
- 20.Haarmann T, Ortel I, Tudzynski P, Keller U. 2006. Identification of the cytochrome P450 monooxygenase that bridges the clavine and ergoline alkaloid pathways. Chembiochem 7:645–652. doi: 10.1002/cbic.200500487. [DOI] [PubMed] [Google Scholar]
- 21.Robinson SL, Panaccione DG. 2014. Heterologous expression of lysergic acid and novel ergot alkaloids in Aspergillus fumigatus. Appl Environ Microbiol 80:6465–6472. doi: 10.1128/AEM.02137-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Riederer B, Han M, Keller U. 1996. d-Lysergyl peptide synthetase from the ergot fungus Claviceps purpurea. J Biol Chem 271:27524–27530. doi: 10.1074/jbc.271.44.27524. [DOI] [PubMed] [Google Scholar]
- 23.Havemann J, Vogel D, Loll B, Keller U. 2014. Cyclolization of d-lysergic acid alkaloid peptides. Chem Biol 21:146–155. doi: 10.1016/j.chembiol.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Bilovol Y, Panaccione DG. 2016. Functional analysis of the gene controlling hydroxylation of festuclavine in the ergot alkaloid pathway of Neosartorya fumigata. Curr Genet 62:853–860. doi: 10.1007/s00294-016-0591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panaccione DG, Coyle CM. 2005. Abundant respirable ergot alkaloids from the common airborne fungus Aspergillus fumigatus. Appl Environ Microbiol 71:3106–3111. doi: 10.1128/AEM.71.6.3106-3111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firn RD, Jones CG. 2003. Natural products—a simple model to explain chemical diversity. Nat Prod Rep 20:382–391. doi: 10.1039/b208815k. [DOI] [PubMed] [Google Scholar]
- 27.Kupfer DM, Drabenstot SD, Buchanan KL, Lai H, Zhu H, Dyer DW, Roe BA, Murphy JW. 2004. Introns and splicing elements of five diverse fungi. Eukaryot Cell 3:1088–1100. doi: 10.1128/EC.3.5.1088-1100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iriarte A, Sanguinetti M, Fernández-Calero T, Naya H, Ramón A, Musto H. 2012. Translational selection on codon usage in the genus Aspergillus. Gene 506:98–105. doi: 10.1016/j.gene.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 29.Silar P. 1995. Two new easy to use vectors for transformations. Fungal Genet Rep 42:73. doi: 10.4148/1941-4765.1353. [DOI] [Google Scholar]
- 30.Schardl CL, Leuchtmann A, Tsai H-F, Collett MA, Watt DM, Scott DB. 1994. Origin of a fungal symbiont of perennial ryegrass by interspecific hybridization of a mutualist with the ryegrass choke pathogen, Epichloë typhina. Genetics 136:1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panaccione DG, Ryan KL, Schardl CL, Florea S. 2012. Analysis and modification of ergot alkaloid profiles in fungi. Methods Enzymol 515:267–290. doi: 10.1016/B978-0-12-394290-6.00012-4. [DOI] [PubMed] [Google Scholar]
- 32.Ryan KL, Moore CT, Panaccione DG. 2013. Partial reconstruction of the ergot alkaloid pathway by heterologous gene expression in Aspergillus nidulans. Toxins 5:445–455. doi: 10.3390/toxins5020445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.