ABSTRACT
Only two types of rubber oxygenases, rubber oxygenase (RoxA) and latex clearing protein (Lcp), have been described so far. RoxA proteins (RoxAs) are c-type cytochromes of ≈70 kDa produced by Gram-negative rubber-degrading bacteria, and they cleave polyisoprene into 12-oxo-4,8-dimethyltrideca-4,8-diene-1-al (ODTD), a C15 oligo-isoprenoid, as the major end product. Lcps are common among Gram-positive rubber degraders and do not share amino acid sequence similarities with RoxAs. Furthermore, Lcps have much smaller molecular masses (≈40 kDa), are b-type cytochromes, and cleave polyisoprene to a mixture of C20, C25, C30, and higher oligo-isoprenoids as end products. In this article, we purified a new type of rubber oxygenase, RoxBXsp (RoxB of Xanthomonas sp. strain 35Y). RoxBXsp is distantly related to RoxAs and resembles RoxAs with respect to molecular mass (70.3 kDa for mature protein) and cofactor content (2 c-type hemes). However, RoxBXsp differs from all currently known RoxAs in having a distinctive product spectrum of C20, C25, C30, and higher oligo-isoprenoids that has been observed only for Lcps so far. Purified RoxBXsp revealed the highest specific activity of 4.5 U/mg (at 23°C) of all currently known rubber oxygenases and exerts a synergistic effect on the efficiency of polyisoprene cleavage by RoxAXsp. RoxB homologs were identified in several other Gram-negative rubber-degrading species, pointing to a prominent function of RoxB for the biodegradation of rubber in Gram-negative bacteria.
IMPORTANCE The enzymatic cleavage of rubber (polyisoprene) is of high environmental importance given that enormous amounts of rubber waste materials are permanently released (e.g., by abrasion of tires). Research from the last decade has discovered rubber oxygenase A, RoxA, and latex clearing protein (Lcp) as being responsible for the primary enzymatic attack on the hydrophobic and water-insoluble biopolymer poly(cis-1,4-isoprene) in Gram-negative and Gram-positive rubber-degrading bacteria, respectively. Here, we provide evidence that a third type of rubber oxygenase is present in Gram-negative rubber-degrading species. Due to its characteristics, we suggest the designation RoxB for the new type of rubber oxygenase. Bioinformatic analysis of genome sequences indicates the presence of roxB homologs in other Gram-negative rubber degraders.
KEYWORDS: rubber oxygenase, latex clearing protein, polyisoprene, biodegradation, heme dioxygenase, dioxygenases
INTRODUCTION
Materials that contain or consist completely of rubber [poly(cis-1,4-isoprene)] have been in use by mankind for more than 100 years at an industrial scale. Most of the rubber materials are released into the environment after use. In particular, small rubber particles liberated from car tires by abrasion contribute to a constant supply of rubber to many ecosystems on earth. Therefore, it is not astonishing that rubber-degrading microorganisms are widely distributed, and several research reports have described the isolation of rubber-degrading bacteria and fungi from different sources. For overviews and recent reports, see references 1 to 4. Since rubber is a high-molecular-weight polymer, it cannot be taken up by cells directly; foremost, the polymer has to be cleaved extracellularly into low-molecular-weight compounds that can be transported across the cell membrane and used for metabolism. Therefore, previous research on rubber degradation has concentrated on the investigation of the primary attack on the polyisoprene molecule by extracellular rubber oxygenases. Remarkably, Gram-negative and Gram-positive rubber degraders use two unrelated rubber oxygenases for the primary extracellular cleavage of the polyisoprene molecule: the first purified rubber cleavage enzyme is the rubber oxygenase RoxAXsp (RoxA of Xanthomonas sp. strain 35Y) (5, 6). RoxAXsp was identified as a c-type diheme dioxygenase (7–9). Recently, the three-dimensional RoxAXsp structure and essential amino acids of the active site were determined (10, 11). RoxA homologs were identified in other Gram-negative bacteria such as Haliangium ochraceum and some myxobacteria (12) but not in any Gram-positive rubber degrader. 2-Oxo-4,8-dimethyltrideca-4,8-diene-1-al (ODTD) is the major rubber degradation product of all biochemically characterized RoxAs. Investigation of rubber degradation by Gram-positive species in A. Steinbüchel′s group led to the identification of a second type of rubber oxygenase (named latex clearing protein [Lcp]) (13–18). Meanwhile the Lcps from species of four different genera (Streptomyces, Gordonia, Rhodococcus, and Nocardia) have been purified and biochemically characterized (19–23). Lcps from different species are related in amino acid sequence and share a domain of unknown function, DUF2236 (19, 24), but differ from RoxAs in several properties: the molecular masses of Lcps are only approximately half (≈40 kDa) of that of RoxAs (≈75 kDa), and Lcps don't share amino acid similarities to RoxAs. Moreover, Lcps differ from RoxAs in having only one noncovalently bound heme (b-type cytochromes) and produce a mixture of C20, C25, C30, and higher oligo-isoprenoids as end products. In summary, it seems as if the ability to degrade rubber has evolved twice: Gram-negative rubber degraders use c-type diheme RoxAs to produce a C15 oligo-isoprenoid, and the Gram-positive counterparts use b-type mono-heme Lcps that cleave rubber to a mixture of C20 and higher oligo-isoprenoids via random (endo) cleavage of polyisoprene. Low-molecular-weight degradation products most likely are taken up by the bacteria and further catabolized via β-oxidation (25–27).
Most of our work on RoxAs was done with Xanthomonas sp. strain 35Y, the first isolated Gram-negative rubber degrader (5). Recently, we determined the draft genome sequence of Xanthomonas sp. strain 35Y (V. Sharma, G. Siedenburg, J. Birke, F. Mobeen, D. Jendrossek, and T. Prakash, unpublished data). Interestingly, we identified a gene of 2,046 bp (roxB; accession no. KY498024) in the Xanthomonas sp. strain 35Y genome that coded for a 73.8-kDa protein with 38% amino acid sequence identity to RoxAXsp. Moreover, the gene product harbored two sequence motifs for covalent attachment of two heme cofactors to cysteins (CHACH and CASCH) at the same position in a sequence alignment as in RoxAXsp. This prompted us to investigate the gene product for a potential function in rubber cleavage and was the basis for this study.
RESULTS AND DISCUSSION
Identification of roxBXsp in Xanthomonas sp. strain 35Y.
Screening of the Xanthomonas sp. strain 35Y genome sequence confirmed the presence of one copy of the previously cloned roxAXsp gene (7). Additionally, inspection of the genome sequence for homologs of roxAXsp or lcp genes revealed one open reading frame of 2,046 bp that coded for a protein of 73.8 kDa. The deduced amino acid sequence of this gene product showed a medium degree of identity (38%) to the RoxAXsp sequence and no significant similarity to Lcps. In addition, the amino acid sequence harbored a putative Sec-dependent signal peptide sequence (predicted with the SignalP 4.1 server) of 33 residues, suggesting a putative extracellular localization of the gene product similar to RoxAXsp. Furthermore, the amino acid sequence had two c-type heme binding motifs (C192HAC195H and C391ASC394H) for covalent attachment of heme groups at almost identical positions in the primary amino acid sequence as it is known for RoxA of Xanthomonas sp. strain 35Y (C191HAC194H and C390ASC393H) (7) and RoxAs of other Gram-negative bacteria (12). A MauG motif with a conserved histidine (PYMH517NGSVP), which is present in all currently available RoxA sequences (7, 9), and other residues that are conserved in RoxAs were also present in the newly identified gene product. Table 1 presents an overview of the properties of the rubber oxygenases investigated in this study. These findings suggest that the gene product is a c-type diheme protein like RoxAXsp, and we named it RoxBXsp.
TABLE 1.
Properties of the rubber oxygenases used in this study
| Protein attribute | Result fora: |
||
|---|---|---|---|
| RoxBXsp | RoxAXsp | LcpK30 | |
| Gene length, bp | 2,046 | 2,037 | 1,191 |
| Signal peptideb | Sec dependent | Sec dependent | Tat dependent |
| Mol mass, kDa | |||
| Preprotein | 73.8 | 74.7 | 44.0 |
| Mature protein | 70.3 | 71.5 | 41.0 |
| Pi (theoretical)c | 7.21 | 8.45 | 6.10 |
| Aromatic residues | |||
| % in mature protein | 9.9 | 11.4 | 7.7 |
| Total no. of F, Y, W | 23, 24, 17 | 24, 30, 20 | 7, 10, 12 |
| Heme attachment, N/C terminal | CHACH196/CASCH395 | CSACH195/CASCH394 | None (b-type heme) |
| Axial heme ligands, N/C terminal | H196/H395 and H627 | H195/H394 and H641 | H198 (only N terminal) |
| Fe state “as isolated,” N/C terminal | Fe3+/Fe3+ | Fe2+—O2/Fe3+ | Fe3+/− |
| MauG motif | PYMH517NGSVP | PYFH517NGSVP | − |
| F317 equivalent | F309 | F317 | − |
| W302 equivalent | V327 | W302 | − |
| Soret band maximum, nm | |||
| Oxidized | 404 | 407 | 412 |
| Reduced | 419 | 418 | 430 |
| β-Band (reduced) | 548, 556 | 549, 553 | 562 |
| α-Band (reduced) | 522, 529 | 521 | 532 |
| Molar extinction coefficient, 104 M−1 cm−1 | 21.3 (404 nm) | 20.6 (407 nm) | 8 (412 nm) |
| UV-vis effect upon addition of CO | No | Yes | No |
| Cleavage product(s) | Mixture of ≥C20 | ODTD | Mixture of ≥C20 |
| Sp act (U/mg) at 23, 30, 37°C | 4.5, 5.7, 6.4 | ND, 1.9, 2.6 | 1.5, ND, 4.7 |
ND, not determined; −, feature not present.
Deduced from SignalP4.1 server for RoxAXsp and RoxBXsp and from reference 14 for LcpK30.
Estimated via ExPASy compute pI/Mw tool.
Homologous expression of RoxBXsp in a ΔroxA background of Xanthomonas sp. strain 35Y revealed rubber oxygenase activity of RoxBXsp.
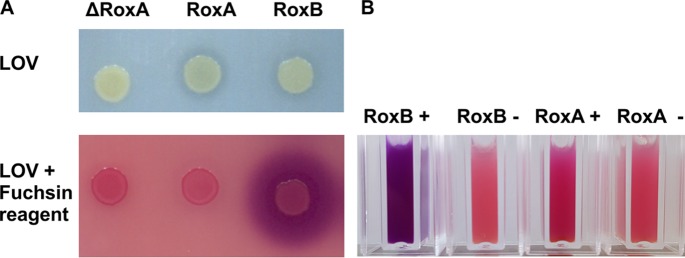
The similarities of the RoxBXsp amino acid sequence to RoxAs suggested that RoxBXsp could have similar biochemical properties to RoxAXsp. To find experimental evidence for this hypothesis, we amplified the roxBXsp gene by PCR and cloned it into several plasmids as described in Materials and Methods. Attempts to express roxBXsp in Escherichia coli were not successful (not shown). The growth-inhibitory effect of plasmid-derived expression of roxA in E. coli has been described previously (11, 28). We therefore integrated roxBXsp under the control of an l-rhamnose-inducible promoter into the genome of a ΔroxA background of Xanthomonas sp. strain 35Y by application of a PhiC31-dependent attB/attP-assisted genome integration. The approach to integrate an inducible roxAXsp gene into a ΔroxA background of Xanthomonas sp. strain 35Y had been previously used successfully for RoxAXsp expression (11). When we cultivated a clone of the constructed ΔroxA Xanthomonas sp. strain 35Y with the genome-integrated roxB gene on opaque polyisoprene latex overlay (LOV) agar that had been supplemented with 0.1% l-rhamnose, the arising colonies developed a very weak, hardly visible clearing zone surrounding the colonies (Fig. 1A). In comparison to a roxAXsp-expressing control, the clearing zones were smaller in diameter and clearing intensity. However, when the agar plates were stained with fuchsin reagent, large and intensively stained pink-blue zones appeared around RoxBXsp-expressing colonies (Fig. 1A). We assume that the bacteria expressed and secreted RoxBXsp, which in turn featured the ability to cleave polyisoprene to aldehyde-containing products that can be stained with fuchsin, as shown in Fig. 1B. Interestingly, the RoxAXsp-expressing control strain did not show a fuchsin-stained halo, although it produced a weak clearing zone. The reason for this presumably lies in the ability of Xanthomonas sp. strain 35Y to take up and utilize the RoxAXsp-derived rubber cleavage product ODTD very efficiently. The finding that the RoxBXsp-overexpressing colonies (in a ΔroxA background) produced such large halos upon staining with fuchsin reagent suggested that the products generated by overexpressed RoxBXsp could not or could only partially be taken up and metabolized by the bacteria. When fuchsin solution was added to cell-free culture supernatants of roxAXsp- or roxBXsp-overexpressing strains that had been allowed to react with added polyisoprene latex for 2 h, a positive reaction was obtained (Fig. 1B). This indicated that both enzymes were active and cleaved polyisoprene latex to aldehyde-containing products.
FIG 1.
Rubber oxygenase activities of RoxAXsp- and RoxBXsp-harboring Xanthomonas sp. (A) Cells of ΔroxA Xanthomonas sp. strain 35Y harboring a roxAXsp gene or a roxBXsp gene under the control of a rhamnose-dependent promoter were spotted onto a polyisoprene latex overlay (LOV) agar plate supplemented with 0.1% rhamnose and were incubated at room temperature for 10 days (top row). Note the formation of weak clearing zones around the colonies of RoxAXsp- and RoxBXsp-expressing clones on LOV plates and its absence in the control strain (ΔroxA Xanthomonas sp. strain 35Y). The bottom row shows the same colonies after staining with fuchsin solution. Note the formation of large pink halos around the RoxBXsp-expressing colony, which indicates the formation of degradation products with aldehyde groups. The almost complete absence of pink halos around the RoxAXsp colony can be explained by the efficient uptake and consumption of RoxAXsp-produced ODTD. (B) Polyisoprene latex (0.1% final concentration) was added to cell-free culture fluid of roxAXsp- or roxBXsp-expressing clones that had been grown on NB medium in the presence (+) or absence (−) of 0.1% rhamnose for 3 days at room temperature. After incubation for 2 h at room temperature, 100 μl fuchsin solution was added to stain aldehyde-containing compounds.
Expression of the roxAXsp and roxBXsp genes is upregulated during growth on polyisoprene.
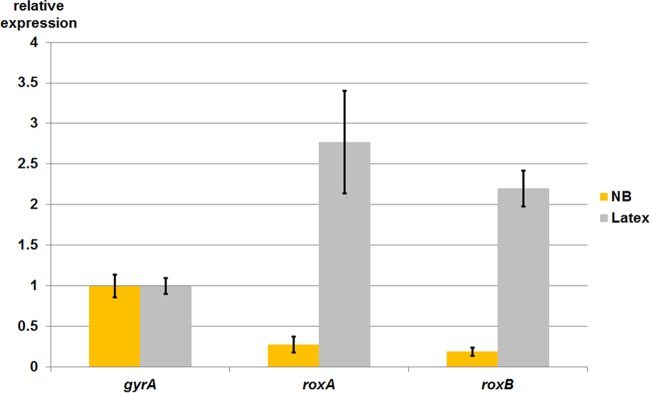
To analyze if and to what extent the roxAXsp and roxBXsp genes were expressed in Xanthomonas sp. strain 35Y, we determined transcription of roxAXsp and roxBXsp in the wild-type strain that had been grown on polyisoprene latex or on nutrient broth (NB). Quantitative reverse transcription-PCR (qRT-PCR) of isolated RNA samples was performed as described in Materials and Methods. Only basal transcription levels were determined for roxAXsp and roxBXsp in NB-grown cells (Fig. 2), and this corresponds to the absence of detectable rubber oxygenase activity of NB-grown cells. In contrast, about 10-fold-higher levels of roxAXsp and roxBXsp transcripts were detected in polyisoprene latex-grown cells. These data suggest that roxAXsp and roxBXsp are specifically expressed in polyisoprene-containing media. This finding is in line with a function of both gene products in the cleavage of polyisoprene.
FIG 2.
qRT-PCR of RNA isolated from the Xanthomonas sp. strain 35Y wild type. Cells were grown on nutrient broth (yellow columns) or Tuschii and Takeda medium with polyisoprene latex (gray columns) for 4 days, respectively, and RNA was isolated and quantified as described in Materials and Methods. RNA levels were normalized to gyrA RNA levels. Three technical replicates were performed. Error bars show standard deviations.
Purification and biophysical characterization of untagged wild-type RoxBXsp.
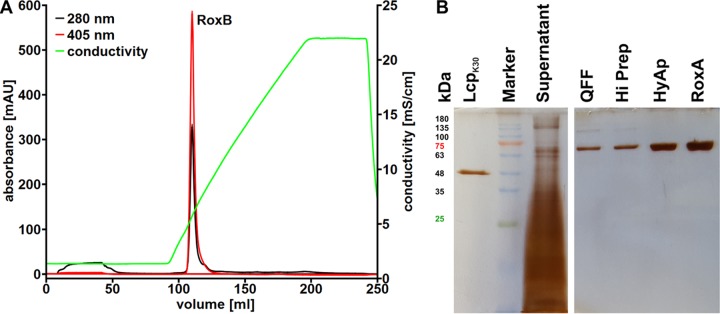
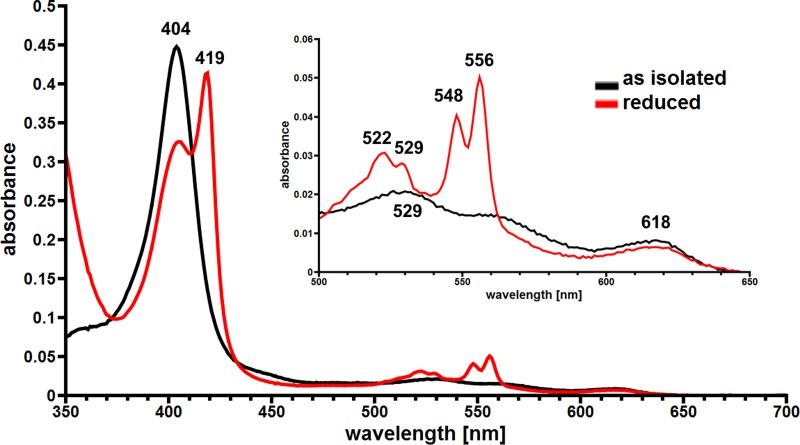
We cultivated the ΔroxA Xanthomonas sp. strain 35Y mutant with genome-integrated roxBXsp on LB medium that had been supplemented with 0.1% of l-rhamnose as inducer. The roxBXsp-harboring strain produced and secreted large amounts of an ≈70-kDa protein into the culture medium in comparison to a control without overexpression of the roxBXsp gene. A weak reddish color of the culture fluid already indicated the presence of a heme-containing protein. The combined culture fluid of 7.2 liters was concentrated via ultrafiltration and diafiltrated against Tris-HCl buffer. RoxBXsp was purified by two subsequent chromatographic steps on Q Sepharose and hydroxyapatite as described in Materials and Methods. Figure 3A shows the elution profile of the second chromatographic step on hydroxyapatite. A “homogeneous” peak with absorption at 280 and at 404 nm was eluted by a potassium phosphate buffer gradient. The combined fraction had a volume of 14 ml, and a protein concentration of 0.44 mg/ml was determined. This corresponded to a yield of 6.2 mg pure, untagged protein from 7.2 liters of culture fluid. SDS-PAGE (Fig. 3B) revealed that a protein with an apparent molecular mass of ≈70 kDa had been purified to homogeneity, corresponding to the theoretical molecular mass of 70.3 kDa calculated for the mature form of RoxBXsp. The supposed presence of covalently attached heme groups (c-type cytochrome) in purified RoxBXsp was confirmed by (i) a red-brownish color of concentrated purified RoxBXsp solutions, (ii) a positive staining reaction for pseudoperoxidase activity after SDS-PAGE, (iii) the determination of an absorption maximum of 550 nm in a hemochrome pyridine assay with the purified protein that is typical for c-type cytochromes (29), and (iv) the inability to solvent extract the heme group from purified RoxBXsp (details not shown). To characterize the heme cofactors of RoxBXsp a spectral analysis (UV-visible light [UV-vis] spectrum) of a concentrated RoxBXsp solution was performed. As shown in Fig. 4, RoxBXsp in the form as isolated has a strong absorption at 404 nm that is characteristic of the Soret band of heme-containing proteins. Extinction coefficients for purified RoxBXsp at 404 and 280 nm of 213,000 and 129,000 M−1 cm−1, respectively, were calculated after experimental determination of the protein concentration using the bicinchoninic acid (BCA) assay. Additionally, weak absorption at 529 (α-band), 562 (β-band), and 618 nm was also detected. When RoxBXsp was chemically reduced by the addition of dithionite, the Soret band partially shifted to 419 nm and the α- and β-bands increased. Remarkably, both the α- and β-bands were split into two separate signals (α-band, 522 and 529 nm; β-band, 548 and 556 nm). This indicates two distinguishable heme groups in RoxBXsp in the reduced state.
FIG 3.
Purification of RoxBXsp. (A) Elution profile of RoxBXsp during the second chromatographic step on hydroxyapatite. (B) SDS-PAGE analysis of RoxBXsp preparations at various stages of purification. Purified LcpK30 and RoxAXsp are shown for comparison. QFF, Q Sepharose Fast Flow; HiPrep, gel filtration for buffer exchange; HyAp, hydroxyapatite.
FIG 4.
UV-vis spectral analysis of purified RoxBXsp. The figure shows the absorption of purified RoxBXsp in the form as isolated (black line) and after reduction with dithionite (red line) between 350 and 700 nm. Note that dithionite led only to a partial shift of the Soret band from 404 to 419 nm. The inset shows an enlargement of the spectrum around 550 nm in the region of the Q-bands.
RoxB has high rubber oxygenase activity.
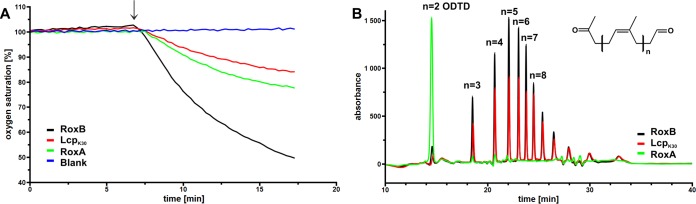
As pointed out above, the RoxBXsp amino acid sequence has similarities to RoxAXsp of Xanthomonas sp. strain 35Y and other RoxAs. To test whether RoxBXsp is able to oxidatively cleave rubber, we incubated purified RoxBXsp in a phosphate-buffered emulsion of polyisoprene latex and monitored the consumption of dissolved oxygen. To compare the expected activity of RoxBXsp with those of other previously described rubber oxygenases, we performed the same experiment in parallel cuvettes using purified RoxAXsp as well as Lcp of Streptomyces sp. strain K30 (LcpK30). In all three experiments, the same concentrations of purified enzyme (4 μg/ml) were used. As shown in Fig. 5A, the presence of RoxBXsp provoked a strong decrease of the dissolved oxygen concentration at room temperature (23°C). This decrease was not observed if latex was absent or if heat-inactivated RoxBXsp (10 min, 90°C) was used (not shown). Apparently, RoxBXsp is able to react with polyisoprene under consumption of oxygen. Interestingly, the slope of the decrease of the oxygen concentration indicated the highest specific activity (4.5 U/mg) of all investigated rubber oxygenases at 23°C (room temperature).
FIG 5.
Activity assay of purified rubber oxygenases. Purified preparations of RoxBXsp, LcpK30, or RoxAXsp (4 μg of each protein) were added to polyisoprene latex (arrow) in an OXY-4 miniapparatus at 30°C. The concentration of dissolved oxygen was monitored over time (A). The blank corresponds to assay buffer without enzyme. The produced products were solvent extracted and analyzed by HPLC (B). Note formation of identical products for LcpK30 and RoxBXsp, while RoxAXsp produced mainly ODTD.
RoxB cleaves polyisoprene to a mixture of C20 and higher oligo-isoprenoids.
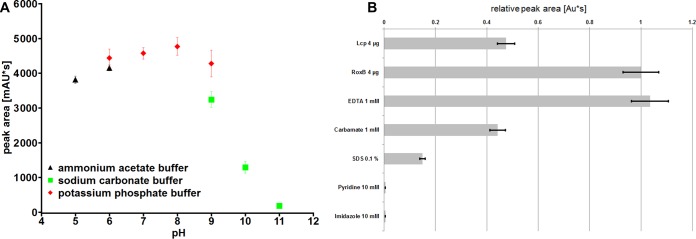
Purified RoxBXsp, RoxAXsp, and LcpK30 were allowed to react with purified polyisoprene latex for 1 h at 23°C. The products were extracted with ethyl-acetate, dissolved in methanol, and separated by high-performance liquid chromatography (HPLC). Figure 5B shows the product profiles obtained for RoxBXsp, RoxAXsp, and LcpK30. To our surprise, the identified products of the RoxBXsp-catalyzed reaction were the same as those previously determined for LcpK30 (21) or for LcpRr from Rhodococcus rhodochrous (22). In all cases, a mixture of C20, C25, and higher oligo-isoprenoids with terminal aldehyde and keto groups were identified. This result was in contrast to RoxAXsp, which cleaved polyisoprene to only one main product, namely, the C15 oligo-isoprenoid ODTD. A peak that corresponded to ODTD was only present in trace amounts in cleavage reactions with LcpK30 or with RoxBXsp. In conclusion, RoxBXsp—although related to RoxAXsp in the primary amino acid sequence and in other properties—resembles Lcps with respect to the produced cleavage products. We therefore propose that RoxBXsp is the first member of a third class of rubber oxygenases (RoxB type) that combines properties of both previously characterized classes of rubber oxygenases (RoxAs and Lcps). The identification of multiple oligo-isoprenoids of different lengths suggests that RoxBXsp is similar to Lcps and in contrast to RoxAXsp cleaves polyisoprene in an endo-type manner. The pH optimum of RoxBXsp-catalyzed polyisoprene cleavage was between pH 6 and 8 (Fig. 6A). Purified RoxBXsp was highly sensitive to imidazole and pyridine (Fig. 6B), suggesting that binding of these compounds to the active heme site inhibited the enzyme. A partial inhibition of RoxBXsp was determined for SDS and for diethyl-dithio-carbamate. The use of 1 or 10 mM EDTA had no effect on the rubber-cleaving activity of RoxBXsp.
FIG 6.
pH profile and inhibition assay of RoxBXsp. The activity of purified RoxBXsp was assayed at different pH values, as indicated (A). The activities of purified LcpK30 (top bar) and of RoxBXsp in the absence or presence of potential inhibitors are shown in panel B. The values show the relative peak areas of the 23.7-min product peak (C35 oligo-isoprenoid). Each experiment was performed in duplicate.
RoxBXsp differs from RoxAXsp in its binding affinity for carbon monoxide.
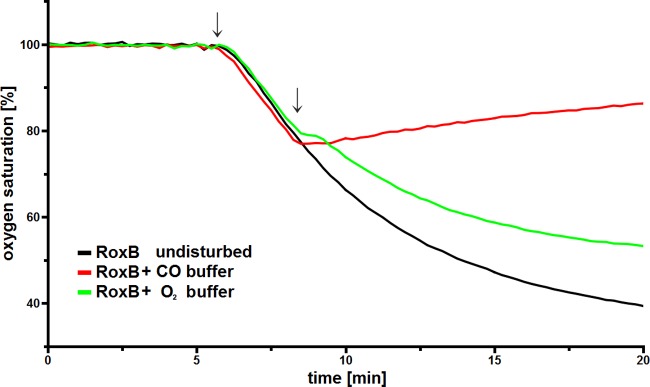
Carbon monoxide is able to bind to RoxAXsp in the form as isolated, as revealed by the CO-dependent appearance of an absorption peak at 415 nm in the difference spectrum (21). This binding can be explained by the CO affinity of the reduced, oxygen-bearing heme center (10). Carbon monoxide strongly inhibits the polyisoprene cleavage reaction of RoxAXsp (21). When we incubated purified RoxBXsp as isolated with carbon monoxide, no change in the UV-vis spectrum was detected. This indicated that both heme sites of RoxBXsp—in contrast to RoxAXsp—are present in an oxidized Fe3+ form that cannot bind dioxygen or carbon monoxide. Nevertheless, oxygen consumption by RoxBXsp was inhibited when carbon monoxide was added to an ongoing polyisoprene cleavage reaction (Fig. 7). This indicated that a reduced heme species occurs during the reaction cycle that is able to bind and to activate dioxygen.
FIG 7.
Effect of carbon monoxide (CO) on RoxBXsp activity. Purified RoxBXsp was added to assay solution (500 μl) at ≈6 min (black graph, left arrow), and the consumption of dissolved oxygen was monitored. In a second cuvette, 200 μl carbon monoxide saturated buffer was added (red graph, right arrow), and in the third cuvette (green graph), 200 μl oxygenated buffer was added. Note the immediate stopping of oxygen consumption in the presence of carbon monoxide.
Synergistic effect of RoxBXsp on RoxAXsp activity.
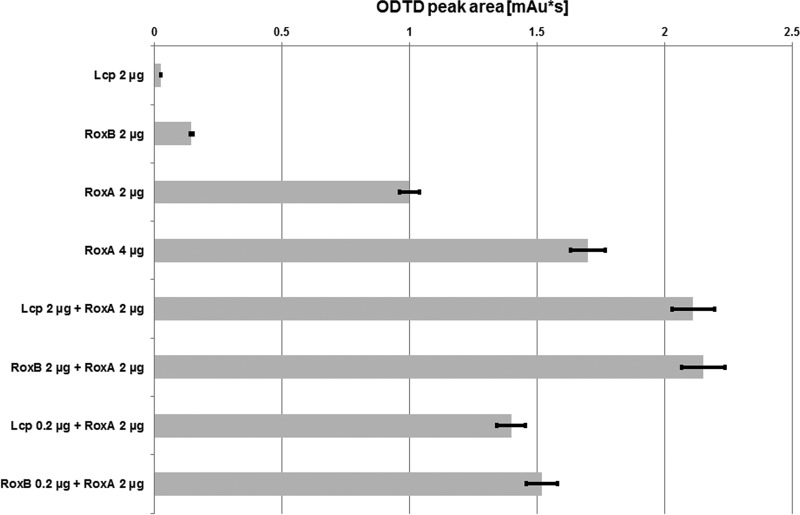
As pointed out above, RoxAXsp and RoxBXsp are both able to efficiently cleave rubber to small and/or medium-sized oligo-isoprenoids. Interestingly, a roxA-deficient Xanthomonas sp. strain 35Y isolate was not able to substantially consume higher oligo-isoprenoids that accumulated during rhamnose-induced expression of roxBXsp. This is evident from Fig. 1A, where the fuchsin halo attests the presence of aldehyde-containing, not-yet-metabolized polyisoprene cleavage products. This raised the question of the putative physiological function of RoxBXsp. Production and excretion of RoxBXsp, containing two costly cofactors, would not seem advantageous for the bacteria if they are not able to use the reaction products for metabolism. One reason for the presence of a roxB gene might be the fact that RoxAXsp is an exo-type cleavage enzyme that needs free polyisoprene ends to cleave the polymer in a processive manner to give the only observed end product, ODTD (10). The presence of RoxBXsp, which cleaves rubber in an endo-type fashion to a mixture of oligo-isoprenoids, would enlarge the number of poly/oligo-isoprenoid chains with free, accessible ends. This could result in a more efficient cleavage of these products by RoxAXsp to ODTD, which can be taken up by Xanthomonas sp. strain 35Y efficiently and used for growth. To find evidence for a postulated synergistic effect of RoxBXsp for the enhanced production of ODTD molecules by RoxAXsp, we performed experiments examining cleavage of polyisoprene latex by RoxAXsp alone, by RoxBXsp alone, and by combinations of RoxAXsp and RoxBXsp. Since LcpK30 produces the same oligo-isoprenoids from polyisoprene latex as RoxBXsp, we also tested combinations of LcpK30 with RoxAXsp for their efficiency to produce ODTD. The results are shown in Fig. 8: LcpK30 alone and RoxBXsp alone (2 μg each of the purified enzymes) produced only trace amounts (<0.05) or rather small amounts of ODTD (<0.15), respectively, in the absence of RoxAXsp. (The relative amount of ODTD produced by RoxAXsp alone was set to 1.0.) Doubling the amount of RoxAXsp from 2 to 4 μg increased the amount of produced ODTD ≈1.7-fold, suggesting that the amount of free polyisoprene ends was already limiting the cleavage reaction. When we mixed 2 μg each of RoxAXsp and RoxBXsp, the amount of ODTD increased more than 2-fold (≈2.2-fold). This result is in agreement with our assumption that the generation of free oligo-isoprenoid ends by RoxBXsp increases the efficiency of ODTD production by RoxAXsp. A similar result was obtained when we used a 10-fold-smaller amount of RoxBXsp (0.2 μg) in combination with 2 μg of RoxAXsp. The amount of liberated ODTD was increased 1.5-fold. Replacement of RoxBXsp by LcpK30 resulted in a similar positive effect of LcpK30 on ODTD production. In conclusion, our data strongly suggest that the presence of only small amounts of RoxBXsp can increase the amount of RoxAXsp-produced ODTD. This finding provides a plausible explanation for a possible physiological function of the secretion of two rubber oxygenases (RoxAXsp plus RoxBXsp) by enlarging the efficiency of ODTD generation that can be used for metabolism.
FIG 8.
Synergistic effect of RoxBXsp and RoxAXsp on polyisoprene cleavage. Polyisoprene latex was cleaved by different amounts and combinations of rubber oxygenase as indicated. The bars correspond to the peak areas of the ODTD product (C15 oligo-isoprenoid). The area obtained for 2 μg of RoxAXsp activity was set as 1.0. Testing of each combination was performed in duplicate.
RoxB is present in other Gram-negative bacteria.
A BLASTp analysis (performed in January 2017) with the RoxBXsp amino acid sequence of Xanthomonas sp. strain 35Y as the query sequence revealed 13 putative RoxB homologs in other bacteria with at least 60% amino acid sequence identity. The protein with highest similarity (83% amino acid identity) was identified as the product of a gene (2,040 bp) from Rhizobacter gummiphilus, a well-known rubber-degrading species (30). Interestingly, this protein had been previously annotated as a putative rubber oxygenase (accession no BAS44780.1). During revision of this article, experimental evidence for the function of the roxB homolog in R. gummiphilus as a rubber oxygenase gene was published (31). We assume that the gene product (here designated RoxBRgu; 74.0 kDa, [LatA in reference 31]) has rubber oxygenase activity similar to RoxB from Xanthomonas sp. strain 35Y. Remarkably, all other Gram-negative species for which a functional RoxA protein has been described (Haliangium ochraceum, Myxococcus fulvus, and Corallococcus coralloides [12]) also harbor a putative roxB gene. Figure 9 shows a phylogenetic tree of biochemically characterized and postulated RoxA and RoxB proteins. All currently known RoxA proteins form a separate cluster clearly distinct from RoxB sequences. The cooccurrence of RoxB proteins with RoxA proteins in Gram-negative rubber-degrading bacteria indicates a prominent function of RoxB for rubber degradation.
FIG 9.
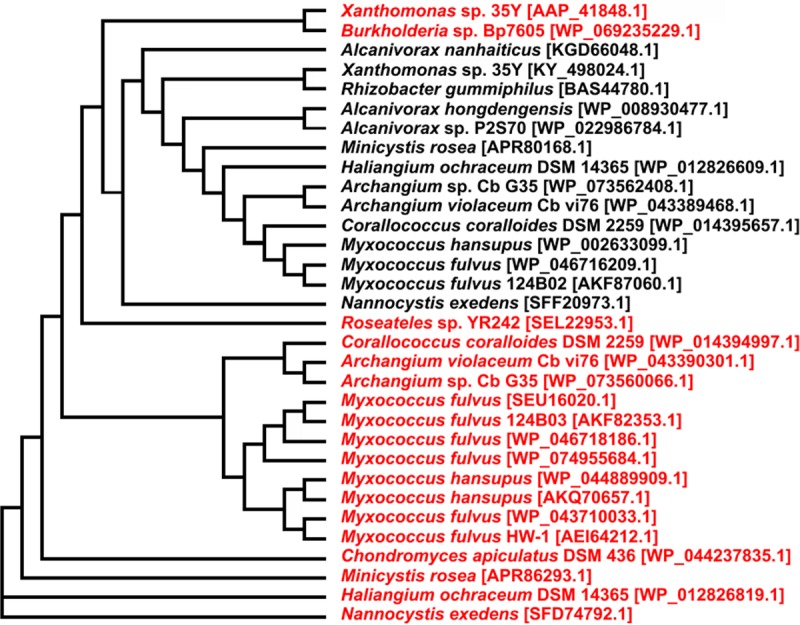
Phylogenetic tree of RoxAs and RoxBs. A multiple-sequence alignment of described and postulated rubber oxygenases was performed using Clustal omega and is shown as a cladogram. The black species designations refer to RoxB proteins, and the red designations indicate RoxAs. The sequences were obtained using a NCBI-provided BLASTp search with the RoxAXsp or RoxBXsp sequences from Xanthomonas sp. strain 35Y as queries. All shown RoxA and RoxB orthologous sequences have a coverage of ≥95% and an identity of >60% to the query sequence, respectively.
Conclusion.
The discovery of a third type of a highly active rubber oxygenase (RoxBXsp) in Xanthomonas sp. strain 35Y and the in vitro evidence for a synergistic effect of the simultaneous presence of RoxAXsp and RoxBXsp on the efficiency of polyisoprene cleavage highlight the prominent function of RoxBXsp. Furthermore, this protein might explain the superior growth of Xanthomonas sp. strain 35Y on polyisoprene in comparison to most other clearing-zone-forming rubber-degrading species. Moreover, our finding might raise the perspective of more efficient biodegradation of rubber materials and/or the biotechnological synthesis of defined oligo-isoprenoids.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Table 2 shows the bacterial strains, plasmids, and oligonucleotides that were used in this study. Plasmid-carrying recombinant E. coli strains were grown with lysogeny broth (LB) medium at 37°C in the presence of the appropriate antibiotic. ΔroxA Xanthomonas sp. strain 35Y mutants carrying a roxBXsp gene at the position of the former roxAXsp gene were grown in nutrient broth (NB) or in modified LB medium (per liter: 5 g NaCl, 0.3 g yeast extract, 10 g tryptone) that had been supplemented with 0.1% (wt/vol) l-rhamnose as an inducer as described in detail elsewhere (11). Polyisoprene latex was kindly provided by Weber and Schaer, Hamburg, Germany, and was used after 3 washing steps in 0.1% (wt/vol) Nonidet P-40 to remove stabilizing compounds. The preparation of latex overlay agar in mineral salts medium (Tsuchii and Takeda medium, [5] supplemented with 0.1% yeast extract) has been described previously (11).
TABLE 2.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, oligonucleotide, or primer | Relevant characteristic(s)a | Reference(s) |
|---|---|---|
| Strains | ||
| E. coli | ||
| S17-1 | Conjugation strain | 34 |
| JM109/pUC9::lcpK30 (SN5339) | Cloning vector for roxBXsp, Apr | 20 |
| JM109/p4782.1::lcpK30 (SN5496) | Expression of lcpK30 under rhamnose promoter control, cloning vector for roxBXsp, Kmr | 21 |
| Xanthomonas sp. | ||
| 35Y (SN5065) | Growth on poly(cis-1,4-isoprene) latex, clearing zone formation | 5; Sharma et al., unpublished |
| 35-CM ΔroxA-attB (SN4114) | Chromosomal deletion of roxAXsp, attB at former roxA site, Cmr | 11 |
| 35-CM ΔroxA-attB/pNH1::roxA (SN4230) | Expression of roxAXsp from rhamnose promoter of genome-integrated roxA, Kmr Cmr | 11 |
| 35-CM ΔroxA-attB/pNH1::roxB (SN6487) | Expression of roxBXsp from rhamnose promoter of genome-integrated roxB, Kmr Cmr | This study |
| Oligonucleotides | ||
| roxB-f | GGAATTCCATATGAGTTCAAAGCAACACCGGGCGCGCGCCAAGG | |
| roxB-r | CCCAAGCTTCTACAATGTCTTCAGATACTCGATGATGG | |
| Int-f | TCTCCTGCAAACTGCTTTTAC | |
| Int-r | GCGAATCTGAACTATCTCATCC | |
| Primers | ||
| Sequencing | ||
| pInt-f | CCCATTTTCCTGTCAGTAAC | |
| pInt-r | CTCCACGGGGAGAGCCTGAG | |
| roxB500-f | CGATGAGCTGGTTGCCGAGC | |
| qRT-PCR | ||
| GyrA-f | AGAGCAATAACGTCTCCTCCCG | |
| GyrA-r | GGTAGCGCATCGAGAAGTTCTG | |
| RoxA-f | TTGTTCATAGGACAGGGAGCCG | |
| RoxA-r | CCGCGTTTGGATTGGAATTCAC | |
| RoxB-f | ACGGATCTCATCAAGAGCAGCC | |
| RoxB-r | ATGCATTGAATGCAACCGCAC |
Kmr, kanamycin resistance; Apr, ampicillin resistance; Cmr, chloramphenicol resistance. The orientation of each of the oligonucleotide and primer sequences shown is 5'→3'.
Cloning of roxB.
The roxB gene of Xanthomonas sp. strain 35Y was amplified from the genomic DNA by PCR (for primer sequences, see Table 2, roxB-f and -r) and cloned into pUC9 via NdeI and HindIII sites. The plasmid pUC9::roxB was cleaved with NdeI/SacI, and the roxBXsp gene-containing DNA fragment was cloned into the expression vector pNH1 using the same restriction sites. The resulting plasmid, pNH1::roxBXsp, was conjugatively transferred from E. coli S17-1 to Xanthomonas sp. strain 35Y ΔroxA-attB and chromosomally integrated via attP/attB recombination, as previously described in detail (11). The correct integration of roxBXsp was confirmed by colony PCR (for primer sequences, see Table 2, Int-f and -r) and subsequent DNA sequencing with primers roxB500-f and pInt-f and -r.
Purification of rubber oxygenases LcpK30, RoxAXsp, and RoxBXsp.
Purifications of LcpK30 and RoxAXsp were performed as described previously (9, 21). RoxBXsp was purified from the supernatant of a Xanthomonas sp. strain 35Y-CM ΔroxA-attB/pNH1::roxB culture that was grown in 12 individual 600-ml cultures of modified LB medium (each in a 3-liter Erlenmeyer flask), supplemented with 0.1% (wt/vol) l-rhamnose for 72 h at 22°C at 120 rpm. Cells were harvested by centrifugation (4°C at 16,000 × g), and the supernatant was concentrated by ultrafiltration (10-kDa cutoff) to a volume of 350 ml and applied to a Q Sepharose fast-flow column (Q-FF 50/11; bed volume, 250 ml) that had been equilibrated with 20 mM Tris-HCl (pH 8.0; flow rate, 8 ml/min). RoxBXsp was detected by monitoring the absorbance at 404 nm and was eluted in a subsequent step gradient at ≈50 mM NaCl in equilibration buffer. Combined RoxBXsp-containing fractions were concentrated to a volume of 50 ml (Amicon 30-kDa cutoff), and Tris-HCl buffer was exchanged with potassium phosphate buffer (10 mM, pH 6.8) by gel filtration with a HiPrep 26/10 desalting column (GE Healthcare, United Kingdom). Subsequently, the RoxBXsp pool was applied to a hydroxyapatite column (CHT5-I; Bio-Rad [bed volume, 20 ml]) that had been equilibrated with the same buffer. RoxBXsp was eluted with a linear gradient of 10 to 200 mM potassium phosphate buffer (pH 6.8) at ≈50 mM. RoxBXsp fractions were pooled and stored on ice or frozen in liquid nitrogen and stored at −70°C. Purity was tested by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and by determination of the absorption quotient 404 nm/280 nm (1.66 for pure RoxBXsp).
Assay of rubber oxygenase activity.
To determine the activities of purified rubber oxygenases (RoxAXsp, RoxBXsp, and LcpK30), two different assays were applied: in the first assay, the consumption of dissolved oxygen was determined in an OXY-4 miniapparatus (PreSens, Regensburg, Germany) as described previously (20, 32). This assay allows the online determination of the polyisoprene cleavage reaction by following the consumption of the cosubstrate dioxygen. For the second assay, polyisoprene latex was incubated in the presence of the test enzyme for 1 or 2 h at 23 or 30°C. The cleavage products were extracted with ethyl-acetate and separated by HPLC as described previously (20). For quantification, the peak area of the C35 peak with a retention time of ≈23 min was used.
Fuchsin staining.
The presence of polyisoprene cleavage products was indicated by the developed purple color after addition of 100 μl of a fuchsin solution (2 g of fuchsin dissolved in 50 ml of glacial acetic acid, 10 g Na2S2O5, 100 ml of 0.1 N HCl, and 50 ml H2O) to the assay mixture (0.2% polyisoprene in 100 mM KP buffer with rubber oxygenase or cell-free culture supernatants of roxAXsp- or roxBXsp-expressing strains at a volume of 400 to 1,000 μl). Latex overlay agar plates with Xanthomonas sp. strain 35Y colonies were stained with the same fuchsin solution; after 1 to 5 min, the purple color arising from the aldehyde-containing products became visible.
qRT-PCR.
Cells for qRT-PCR were grown on either NB agar or latex overlay agar plates for 2 to 5 days at 22°C, respectively. RNA was extracted using the RNeasy minikit (Qiagen, Hilden, Germany). To this end, cells were resuspended in 350 μl buffer RLT (containing 10 μl 2-mercaptoethanol per ml of buffer), supplemented with zirconia/silica beads (0.1 μm; BioSpec, Bartlesville, OK) and disrupted on a shaker mill (Silamat S6; Ivoclar Vivadent, Liechtenstein) for 30 s. The lysate was transferred into 250 μl ethanol and used for RNA purification according to the manual supplied by the manufacturer, including an on-column DNase treatment of 30 min. The quality of the purified RNA was determined by agarose gel electrophoresis and determination of the nucleic acid concentration via a NanoDrop 2000 photometer (Thermo Scientific, Wilmington, DE). Reverse transcription was performed using random hexamer primers and RevertAid transcriptase (Thermo Scientific, Wilmington, DE) in a 10-μl reaction mixture with one temperature cycle: 10 min at 25°C and 60 min at 42°C. RNA was digested by RNase H for 30 min at 37°C, and the DNA concentration was determined spectroscopically (NanoDrop). The transcribed cDNA was purified using the DNA Clean & Concentrator-5 kit (Zymo Research, Irvine, CA). Fifty nanograms of cDNA derived from cultures grown on NB or latex overlay agar was used as the template for the quantitative PCR. In a reaction volume of 10 μl, 20 pmol of each primer (Table 2) was supplemented with 5 μl innuMix qPCR mastermix SyGreen (Analytik, Jena, Germany), and the volume was adjusted using PCR-grade water. The reaction was carried out in a qTOWER 2.0 (Analytik, Jena, Germany) controlled by the software qPCRsoft 3.2. The PCR program was as specified as 95°C for 3 min followed by 40 cycles of 5 s at 95°C, 5 s at 52°C, and 20 s at 72°C. Subsequently, the melting curves of the synthesized DNA strands were recorded by monitoring of heating from 60°C to 95°C. Data analysis was performed via cycle threshold (ΔΔCT) quantification relative to the expression level of the single-copy DNA gyrase housekeeping gene.
Other techniques.
The concentration of protein solutions was determined by the bicinchoninic acid (BCA) method using a commercial BCA kit (Pierce). Separation of proteins was performed by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate (SDS-PAGE) under reducing (2-mercaptoethanol) conditions. SDS-PAGE gels were stained with silver (33). Assays for pseudoperoxidase activity and fuchsin straining of agar plates were performed as described previously (9, 22). Heme extraction, the pyridine hemochrome assay (29), and the RoxBXsp inhibitor assays were carried out as previously described (22). A phylogenetic tree of RoxA and RoxB amino acid sequences was generated by a multisequence alignment using the Clustal Omega online tool (www.ebi.ac.uk/Tools/msa/clustalo).
Accession number(s)
The DNA sequences of roxBXsp and gyrAXsp are available under GenBank accession no. KY498024 and MF033387, respectively.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft.
We thank Weber and Schaer Co., Hamburg, Germany, for providing polyisoprene and PreSens, Regensburg, Germany, for sensor spots. We thank K. Gottlieb for assistance during qRT-PCR.
REFERENCES
- 1.Jendrossek D, Tomasi G, Kroppenstedt RM. 1997. Bacterial degradation of natural rubber: a privilege of actinomycetes? FEMS Microbiol Lett 150:179–188. doi: 10.1016/S0378-1097(97)00072-4. [DOI] [PubMed] [Google Scholar]
- 2.Rose K, Steinbüchel A. 2005. Biodegradation of natural rubber and related compounds: recent insights into a hardly understood catabolic capability of microorganisms. Appl Environ Microbiol 71:2803–2812. doi: 10.1128/AEM.71.6.2803-2812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yikmis M, Steinbüchel A. 2012. Historical and recent achievements in the field of microbial degradation of natural and synthetic rubber. Appl Environ Microbiol 78:4543–4551. doi: 10.1128/AEM.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imai S, Ichikawa K, Muramatsu Y, Kasai D, Masai E, Fukuda M. 2011. Isolation and characterization of Streptomyces, Actinoplanes, and Methylibium strains that are involved in degradation of natural rubber and synthetic poly(cis-1,4-isoprene). Enzyme Microb Technol 49:526–531. doi: 10.1016/j.enzmictec.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Tsuchii A, Takeda K. 1990. Rubber-degrading enzyme from a bacterial culture. Appl Environ Microbiol 56:269–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braaz R, Fischer P, Jendrossek D. 2004. Novel type of heme-dependent oxygenase catalyzes oxidative cleavage of rubber (poly-cis-1,4-isoprene). Appl Environ Microbiol 70:7388–7395. doi: 10.1128/AEM.70.12.7388-7395.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jendrossek D, Reinhardt S. 2003. Sequence analysis of a gene product synthesized by Xanthomonas sp. during growth on natural rubber latex. FEMS Microbiol Lett 224:61–65. doi: 10.1016/S0378-1097(03)00424-5. [DOI] [PubMed] [Google Scholar]
- 8.Braaz R, Armbruster W, Jendrossek D. 2005. Heme-dependent rubber oxygenase RoxA of Xanthomonas sp. cleaves the carbon backbone of poly(cis-1,4-isoprene) by a dioxygenase mechanism. Appl Environ Microbiol 71:2473–2478. doi: 10.1128/AEM.71.5.2473-2478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt G, Seiffert G, Kroneck PMH, Braaz R, Jendrossek D. 2010. Spectroscopic properties of rubber oxygenase RoxA from Xanthomonas sp., a new type of dihaem dioxygenase. Microbiology (Reading, Engl) 156:2537–2548. doi: 10.1099/mic.0.038992-0. [DOI] [PubMed] [Google Scholar]
- 10.Seidel J, Schmitt G, Hoffmann M, Jendrossek D, Einsle O. 2013. Structure of the processive rubber oxygenase RoxA from Xanthomonas sp. Proc Natl Acad Sci U S A 110:13833–13838. doi: 10.1073/pnas.1305560110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Birke J, Hambsch N, Schmitt G, Altenbuchner J, Jendrossek D. 2012. Phe317 is essential for rubber oxygenase RoxA activity. Appl Environ Microbiol 78:7876–7883. doi: 10.1128/AEM.02385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birke J, Röther W, Schmitt G, Jendrossek D. 2013. Functional identification of rubber oxygenase (RoxA) in soil and marine myxobacteria. Appl Environ Microbiol 79:6391–6399. doi: 10.1128/AEM.02194-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rose K, Tenberge KB, Steinbüchel A. 2005. Identification and characterization of genes from Streptomyces sp strain K30 responsible for clear zone formation on natural rubber latex and poly(cis-1,4-isoprene) rubber degradation. Biomacromolecules 6:180–188. doi: 10.1021/bm0496110. [DOI] [PubMed] [Google Scholar]
- 14.Yikmis M, Arenskoetter M, Rose K, Lange N, Wernsmann H, Wiefel L, Steinbüchel A. 2008. Secretion and transcriptional regulation of the latex-clearing protein, Lcp, by the rubber-degrading bacterium Streptomyces sp. strain K30. Appl Environ Microbiol 74:5373–5382. doi: 10.1128/AEM.01001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yikmis M, Steinbüchel A. 2012. Importance of the latex-clearing protein (Lcp) for poly(cis-1,4-isoprene) rubber cleavage in Streptomyces sp K30. Microbiologyopen 1:13–24. doi: 10.1002/mbo3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibrahim E, Arenskötter M, Luftmann H, Steinbüchel A. 2006. Identification of poly(cis-1,4-isoprene) degradation intermediates during growth of moderately thermophilic actinomycetes on rubber and cloning of a functional lcp homologue from Nocardia farcinica strain E1. Appl Environ Microbiol 72:3375–3382. doi: 10.1128/AEM.72.5.3375-3382.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bröker D, Dietz D, Arenskötter M, Steinbüchel A. 2008. The genomes of the non-clearing-zone-forming and natural-rubber-degrading species Gordonia polyisoprenivorans and Gordonia westfalica harbor genes expressing Lcp activity in Streptomyces strains. Appl Environ Microbiol 74:2288–2297. doi: 10.1128/AEM.02145-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiessl S, Schuldes J, Thuermer A, Halbsguth T, Broeker D, Angelov A, Liebl W, Daniel R, Steinbüchel A. 2012. Involvement of two latex-clearing proteins during rubber degradation and insights into the subsequent degradation pathway revealed by the genome sequence of Gordonia polyisoprenivorans strain VH2. Appl Environ Microbiol 78:2874–2887. doi: 10.1128/AEM.07969-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiessl S, Boese D, Oetermann S, Eggers J, Pietruszka J, Steinbüchel A. 2014. Latex clearing protein—an oxygenase cleaving poly(cis-1,4-isoprene) rubber at the cis double bonds. Appl Environ Microbiol 80:5231–5240. doi: 10.1128/AEM.01502-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birke J, Jendrossek D. 2014. Rubber oxygenase and latex clearing protein cleave rubber to different products and use different cleavage mechanisms. Appl Environ Microbiol 80:5012–5020. doi: 10.1128/AEM.01271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birke J, Röther W, Jendrossek D. 2015. Latex clearing protein (Lcp) of Streptomyces sp. strain K30 is a b-type cytochrome and differs from rubber oxygenase A (RoxA) in its biophysical properties. Appl Environ Microbiol 81:3793–3799. doi: 10.1128/AEM.00275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watcharakul S, Röther W, Birke J, Umsakul K, Hodgson B, Jendrossek D. 2016. Biochemical and spectroscopic characterization of purified latex clearing protein (Lcp) from newly isolated rubber degrading Rhodococcus rhodochrous strain RPK1 reveals novel properties of Lcp. BMC Microbiol 16:92. doi: 10.1186/s12866-016-0703-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linh DV, Huong NL, Tabata M, Imai S, Iijima S, Kasai D, Anh TK, Fukuda M. 2017. Characterization and functional expression of a rubber degradation gene of a Nocardia degrader from a rubber-processing factory. J Biosci Bioeng 1123:412–418. doi: 10.1016/j.jbiosc.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Röther W, Austen S, Birke J, Jendrossek D. 2016. Molecular insights in the cleavage of rubber by the latex clearing protein (Lcp) of Streptomyces sp. strain K30. Appl Environ Microbiol 82:6593–6602. doi: 10.1128/AEM.02176-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bode HB, Kerkhoff K, Jendrossek D. 2001. Bacterial degradation of natural and synthetic rubber. Biomacromolecules 2:295–303. doi: 10.1021/bm005638h. [DOI] [PubMed] [Google Scholar]
- 26.Shah AA, Hasan F, Shah Z, Kanwal N, Zeb S. 2013. Biodegradation of natural and synthetic rubbers: a review. Int Biodeterior Biodegrad 83:145–157. doi: 10.1016/j.ibiod.2013.05.004. [DOI] [Google Scholar]
- 27.Luo Q, Hiessl S, Poehlein A, Daniel R, Steinbüchel A. 2014. Insights into the microbial degradation of rubber and gutta-percha by analysis of the complete genome of Nocardia nova SH22a. Appl Environ Microbiol 80:3895–3907. doi: 10.1128/AEM.00473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hambsch N, Schmitt G, Jendrossek D. 2010. Development of a homologous expression system for rubber oxygenase RoxA from Xanthomonas sp. J Appl Microbiol 109:1067–1075. doi: 10.1111/j.1365-2672.2010.04732.x. [DOI] [PubMed] [Google Scholar]
- 29.Berry EA, Trumpower BL. 1987. Simultaneous determination of hemes-A, hemes-B, and hemes-C from pyridine hemochrome spectra. Anal Biochem 161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 30.Imai S, Yoshida R, Endo Y, Fukunaga Y, Yamazoe A, Kasai D, Masai E, Fukuda M. 2013. Rhizobacter gummiphilus sp. nov., a rubber-degrading bacterium isolated from the soil of a botanical garden in Japan. J Gen Appl Microbiol 59:199–205. doi: 10.2323/jgam.59.199. [DOI] [PubMed] [Google Scholar]
- 31.Kasai D, Imai S, Asano S, Tabata M, Iijima S, Kamimura N, Masai E, Fukuda M. 2017. Identification of natural rubber degradation gene in Rhizobacter gummiphilus NS21. Biosci Biotechnol Biochem 81:614–620. doi: 10.1080/09168451.2016.1263147. [DOI] [PubMed] [Google Scholar]
- 32.Röther W, Birke J, Jendrossek D. 2017. Assays for the detection of rubber oxygenase activities. Bio-protocol 7:1–14. doi: 10.21769/BioProtoc.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blum H, Beier H, Gross HJ. 1987. Improved silver staining of plant-proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93–99. doi: 10.1002/elps.1150080203. [DOI] [Google Scholar]
- 34.Simon R, Priefer R, Pühler A. 1983. A broad-host-range mobilization system for in vivo engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]