ABSTRACT
Functional starter cultures demonstrating superior technological and food safety properties are advantageous to the food fermentation industry. We evaluated the efficacies of single- and double-bacteriocin-producing starters of Lactococcus lactis capable of producing the class I bacteriocins nisin A and/or lacticin 3147 in terms of starter performance. Single producers were generated by mobilizing the conjugative bacteriophage resistance plasmid pMRC01, carrying lacticin genetic determinants, or the conjugative transposon Tn5276, carrying nisin genetic determinants, to the commercial starter L. lactis CSK2775. The effect of bacteriocin coproduction was examined by superimposing pMRC01 into the newly constructed nisin transconjugant. Transconjugants were improved with regard to antimicrobial activity and bacteriophage insensitivity compared to the recipient strain, and the double producer was immune to both bacteriocins. Bacteriocin production in the starter was stable, although the recipient strain proved to be a more efficient acidifier than transconjugant derivatives. Overall, combinations of class I bacteriocins (the double producer or a combination of single producers) proved to be as effective as individual bacteriocins for controlling Listeria innocua growth in laboratory-scale cheeses. However, using the double producer in combination with the class II bacteriocin producer Lactobacillus plantarum or using the lacticin producer with the class II producer proved to be most effective for reducing bacterial load. As emergence of bacteriocin tolerance was reduced 10-fold in the presence of nisin and lacticin, we suggest that the double producer in conjunction with the class II producer could serve as a protective culture providing a food-grade, multihurdle approach to control pathogenic growth in a variety of industrial applications.
IMPORTANCE We generated a suite of single- and double-bacteriocin-producing starter cultures capable of generating the class I bacteriocin lacticin 3147 or nisin or both bacteriocins simultaneously via conjugation. The transconjugants exhibited improved bacteriophage resistance and antimicrobial activity. The single producers proved to be as effective as the double-bacteriocin producer at reducing Listeria numbers in laboratory-scale cheese. However, combining the double producer or the lacticin-producing starter with a class II bacteriocin producer, Lactobacillus plantarum LMG P-26358, proved to be most effective at reducing Listeria numbers and was significantly better than a combination of the three bacteriocin-producing strains, as the double producer is not inhibited by either of the class I bacteriocins. Since the simultaneous use of lacticin and nisin should reduce the emergence of bacteriocin-tolerant derivatives, this study suggests that a protective starter system produced by bacteriocin stacking is a worthwhile multihurdle approach for food safety applications.
KEYWORDS: food safety, protective culture, bacteriocins
INTRODUCTION
The development and characterization of starter cultures that demonstrate superior technological properties, such as improved proteolytic activity and flavor production, exopolysaccharide production, or bacteriophage resistance, are considered highly advantageous within the food fermentation industry (1). The ability of starter strains to produce bacteriocins is also considered an important technological trait for controlling undesirable and/or pathogenic growth in situ and for improving sensory characteristics (1–3). Bacteriocins are ribosomally synthesized, heat-stable antimicrobial peptides that generally act by depolarizing the target cell membrane and/or by inhibiting cell wall synthesis where the producing strain is immune to the antimicrobial effect (4). They comprise a highly heterogeneous group that has recently been divided into three distinct classes (5).
The exploitation of bacteriocin-producing cultures is a particularly attractive option for the food industry owing to the generally recognized as safe (GRAS) status of the cultures, immediately fulfilling the consumers' demand for minimally processed foods lacking artificial food additives. The bacteriocin producer can serve as the starter culture or be added as an additional protective culture. Several studies have highlighted the efficacy of such approaches, where bacteriocin-producing cultures have proven to be effective for inhibiting the growth and proliferation of pathogenic and food spoilage microorganisms (6–9). Despite this, the use of bacteriocins in the food industry remains limited, possibly owing to the fact that a bacteriocin alone may not be capable of providing sufficient protection against contamination (10). The use of bacteriocin combinations or bacteriocin stacking may represent an alternative approach. Indeed, improved antimicrobial activity of bacteriocin combinations has been reported previously (11–13). However, when using multiple bacteriocins, it is essential that other important cultures are not inhibited. This can be overcome to some degree by developing a multibacteriocinogenic culture which is immune to the bacteriocins it produces.
In the present study, we generated single- and double-bacteriocin-producing cultures of Lactococcus lactis CSK2775 with the capacity to produce the class I bacteriocins lacticin 3147 (lacticin), nisin A (nisin), or lacticin and nisin. Both bacteriocins target lipid II to generate pores in the cell membrane, causing proton motive force dissipation and subsequent cell death (14–17). The resulting transconjugants were assessed for bacteriocin production, bacteriophage resistance properties, acidification efficiency, and antimicrobial activity against a spectrum of indicator strains, including food pathogens and other lactic acid bacteria (LAB). The ability of the transconjugants (single and double) to produce bacteriocins in laboratory-scale cheese was assessed, and we also evaluated the antilisterial potential of the class I producers alone and in combination with the class IIa bacteriocin producer Lactobacillus plantarum LMG P-26358 (18). Class IIa bacteriocins cause pore formation by binding to and irreversibly opening the sugar transporter mannose phosphotransferase (Man-PTS) system in the target cell (19). In this study, Listeria innocua served as a surrogate for Listeria monocytogenes for reasons of safety and efficiency (as in many other studies) and because L. innocua has been successfully used in previous studies investigating the antilisterial potential of nisin (20–24), lacticin (25–28), and plantaricin (18) in food systems.
RESULTS
Transconjugant validation.
The presence of the plasmid pMRC01 and the nisin transposon Tn5276 in L. lactis CSK2775 transconjugants was validated by PCR using plasmid- and transposon-specific primers, respectively (Fig. 1). PCR analysis confirmed that the genetic determinants responsible for lacticin production were present in CSK2775(pMRC01), creating the lacticin transconjugant L. lactis CSK3594, and confirmed the presence of the nisin genetic determinants in CSK2775(Tn5276), creating the nisin transconjugant CSK3281. The presence of the lacticin and nisin genetic determinants was confirmed in the double producer, resulting in the nisin-lacticin transconjugant L. lactis CSK3533. To confirm the identity of each transconjugant, genomic fingerprints were generated by pulsed-field gel electrophoresis (PFGE) with the restriction endonuclease SmaI. All transconjugants analyzed generated the same restriction pattern as the recipient strain, CSK2775 (results not shown). Well diffusion assays confirmed that CSK3594 was sensitive to nisin and that CSK3281 was sensitive to lacticin but that the double producer was immune to both bacteriocins.
FIG 1.
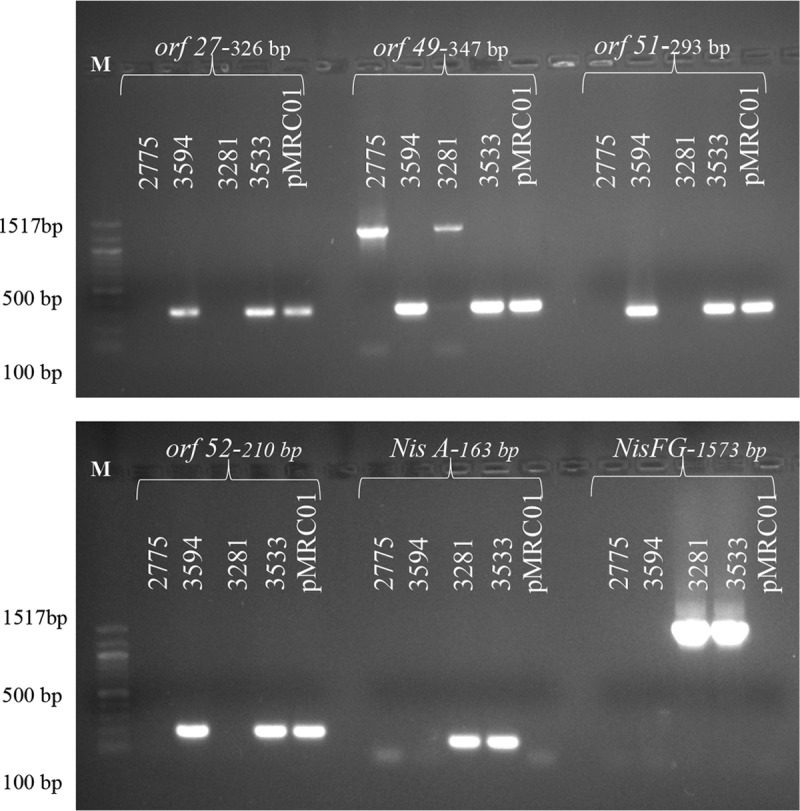
PCR amplification using pMRC01-specific primers (orf27, orf49, orf51, and orf52) to detect the presence of pMRC01 in L. lactis CSK3594 and CSK3533 and using primers designed to regions of the nisin operon (nisA and nisFEG) to confirm the presence of nisin genetic determinants in L. lactis CSK3281 and L. lactis CSK3533. Lane M, 100-bp DNA ladder (New England BioLabs).
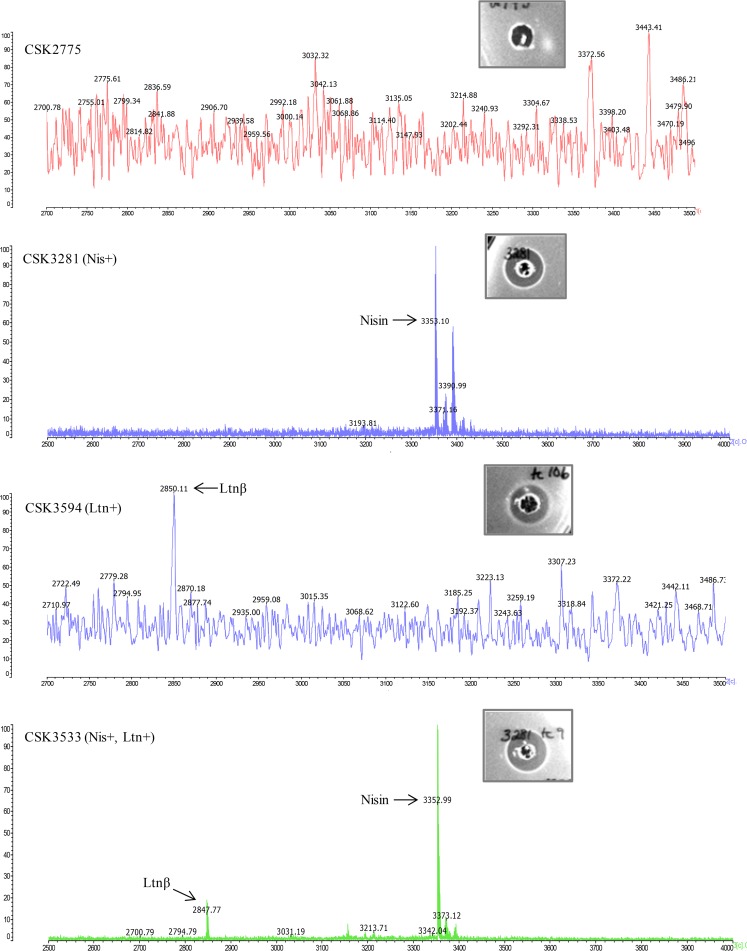
Colony mass spectrometry (CMS) confirmed that CSK3594 and CSK3533 each produced a peptide with a mass of approximately 2,847 ± 4 Da, corresponding to the lacticin peptide Ltnβ (Fig. 2). However, lacticin peptides (Ltnα or Ltnβ) could not be detected in the recipient strain, CSK2775 (Fig. 2). CMS also detected a peptide with a mass of 3,352 ± 3 Da, corresponding to nisin, in strains CSK3281 and CSK3533; this peptide was absent in the recipient strain. These data confirm that lacticin is produced by CSK3594, nisin is produced by CSK3281, and both nisin and lacticin are produced by CSK3533 (Fig. 2).
FIG 2.
Colony mass spectrometry analysis of L. lactis CSK2775, L. lactis CSK3281 (nisin transconjugant, Nis+), L. lactis CSK3594 (lacticin transconjugant, Ltn+), and L. lactis CSK3533 (nisin and lacticin double producer, Nis+, Ltn+). Masses corresponding to the bacteriocins are indicated. Inset photos show inhibition zones produced by each strain against the indicator strain L. lactis HP.
The level of inhibitory activity in the culture supernatant of the double producer, CSK3533, against L. lactis HP was determined to be 1,000 arbitrary units (AU)/ml when measured by agar well diffusion assays, corresponding to a zone size of 4.5 mm, which is equivalent to the zone size produced by the nisin transconjugant L. lactis CSK3281. The lacticin transconjugant, CSK3594, produced a 2.5-mm zone against L. lactis HP. To our knowledge, this is the first report of the successful construction of a food-grade commercial L. lactis starter strain capable of producing both nisin and lacticin 3147, two potent class I bacteriocins.
Strain performance and stability.
The stability of the bacteriocin/lactose-positive phenotype in each transconjugant was confirmed via repeated “passaging” in GM17 followed by bacteriocin activity assays against the indicator, L. lactis HP. Bacteriocin production and immunity in CSK3281, CSK3594, and CSK3533 proved to be stable over time. However, upon passaging of the double producer, CSK3533, in GM17, a mixed culture containing lactose-fermenting and nonfermenting colonies could be observed when plated on lactose indicator agar (LIA). This mixed culture was subsequently attributed to the loss of a large plasmid (>50 kb) present in CSK3533 (which was confirmed by plasmid profile analysis; results not shown) and is presumed to be involved in lactose metabolism. The lactose-fermenting phenotype could be preserved through the supplementation of lactose to the growth medium.
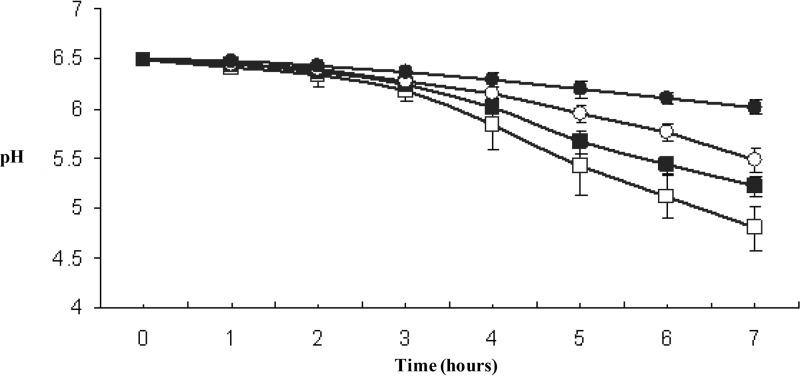
Comparative analyses of the acidification profiles of each transconjugant and the recipient strain revealed that the bacteriocin-free recipient, CSK2775, was the most efficient acidifier (Fig. 3). Although the lacticin single producer (CSK3594) was more efficient than the nisin single producer (CSK3281), both proved to be more efficient than the double producer, CSK3533 (Fig. 3). The addition of 0.1% yeast extract improved lactic acid production in the transconjugants.
FIG 3.
Acidification profiles of L. lactis CSK2775 (□), L. lactis CSK3594 (lacticin) (■), L. lactis CSK3281 (nisin) (○), and L. lactis CSK3533 (nisin and lacticin) (●) grown in 10% reconstituted skim milk.
Spectrum of inhibition and bacteriophage resistance.
The activities of the single-bacteriocin producers CSK3594 and CSK3281 as well as the double producer, CSK3533, were assayed against a range of indicator strains, including food spoilage bacteria, pathogenic bacteria, LAB, and nonstarter LAB (NSLAB) (Table 1). The single lacticin producer was found to inhibit primarily lactococci, lactobacilli, and clostridia, while a wider spectrum of inhibition was observed for both the nisin producer and the double producer. The double producer proved to be more effective than either the lacticin or nisin single producer with regard to Clostridium tyrobutyricum inhibition, producing a 6-mm zone while the lacticin producer, CSK3594, and the nisin producer, CSK3281, produced zones of 4 mm and 3 mm, respectively. Interestingly, the recipient strain, CSK2775, also produced a 1-mm zone against C. tyrobutyricum, suggesting that some other antimicrobial effect is potentially working in conjunction with the bacteriocins in the transconjugants. In addition, the double producer generated a 4-mm zone against CSK3281 in comparison to a 2.5-mm zone produced by CSK3594. This increase in zone size is surprising given that CSK3281 harbors the genetic machinery for nisin immunity and indeed was proven to be immune to nisin in the antimicrobial assays. However, the increased susceptibility of the nisin transconjugant to lacticin may be due to a lower cell density in the seeded plate as a consequence of a lower growth rate, although this has not been confirmed.
TABLE 1.
Antimicrobial spectrum of the L. lactis isogenic family of nisin and lacticin transconjugants
| Indicator strain | Growth medium | Zone size (mm) for strain: |
Source | |||
|---|---|---|---|---|---|---|
| CSK2275 | CSK3594 (Ltn) | CSK3281 (Nis) | CSK3533 (Ltn, Nis) | |||
| Bacillus cereus DPC6085/6086 | BHIb | No zone | No zone | 1 | 1 | TFRCe |
| Bacillus subtilis DPC6511 | BHIb | No zone | No zone | No zone | No zone | TFRC |
| Enterococcus faecalis DPC5055/LMG 7973 | BHIb | No zone | No zone | 3 | 3 | TFRC |
| Enterococcus faecium DPC5056a | BHIb | No zone | No zone | 6 | 6 | TFRC |
| Escherichia coli P1432-DPC6054 | BHIb | No zone | No zone | No zone | No zone | TFRC |
| Clostridium sporogenes DPC6341a | RCMb | No zone | 1.5 | 1.5 | 1 | TFRC |
| Clostridium tyrobutyricum DPC6342a | RCMb | 1 | 4 | 3 | 6 | TFRC |
| Lactobacillus casei DPC6125 | MRSd | No zone | No zone | 7 | 7 | TFRC |
| Lactobacillus acidophilus DPC5378 | MRSb | No zone | 1 | 3 | 3 | TFRC |
| Lactobacillus delbrueckii subsp. delbrueckii DPC5385 | MRSb | No zone | No zone | 6 | 6 | TFRC |
| Lactobacillus delbrueckii subsp. lactis DPC5387 | MRSd | No zone | 1.5 | 6 | 6 | TFRC |
| Lactobacillus delbrueckii subsp. bulgaricus DPC5383 | MRSd | No zone | 2 | 9.5 | 9.5 | TFRC |
| Lactobacillus helveticus DPC4571 | MRSb | No zone | 1.5 | 8 | 8 | TFRC |
| L. lactis subsp. lactis biovar diacetylactis CSK1411 | LM17c | No zone | 1.5 | 4 | 4 | CSK, The Netherlands |
| L. lactis subsp. cremoris HP DPC5718 | LM17d | No zone | 2.5 | 4.5 | 4.5 | TFRC |
| L. lactis subsp. lactis DPC4268/303 | LM17d | No zone | No zone | 0.5 | 0.25 | TFRC |
| L. lactis subsp. lactis CSK2775 | LM17d | No zone | 2.5 mm | 4.5 mm | 4.5 mm | CSK, The Netherlands |
| L. lactis subsp. lactis CSK3594 | LM17d | No zone | No zone | 4.5 mm | 4.5 mm | CSK, The Netherlands |
| L. lactis subsp. lactis CSK3281 | LM17d | No zone | 2.5 mm | No zone | 4 mm | CSK, The Netherlands |
| L. lactis subsp. lactis CSK3533 | LM17d | No zone | No zone | No zone | No zone | CSK, The Netherlands |
| Leuconostoc lactis DPC3838 | MRSd | No zone | No zone | 1.5 | 1.5 | TFRC |
| L. innocua DPC6578 | GM17b | No zone | No zone | No zone | No zone | TFRC |
Cultures were grown anaerobically for up to 48 h.
Cultures were grown at 37°C for up to 48 h.
Cultures were grown at 35°C for up to 48 h.
Cultures were grown at 30°C for up to 48 h.
TFRC, Teagasc Food Research Centre, Moorepark, Fermoy, County Cork, Ireland.
Bacteriophage sensitivity assays confirmed that CSK2775, the nonbacteriocinogenic recipient, was sensitive to all bacteriophages analyzed (Table 2). The nisin producer, CSK3281 was resistant to 50% of the bacteriophages analyzed, while the lacticin single producer and the double producer were each resistant to 80% of bacteriophages analyzed (Table 2).
TABLE 2.
L. lactis transconjugants surveyed for bacteriophage sensitivity
| Bacteriophage | Sensitivitya of L. lactis strain: |
|||
|---|---|---|---|---|
| CSK2775 | CSK3281 (Nis) | CSK3594 (Ltn) | CSK3533 (Nis, Ltn) | |
| 5410F | + | + | − | − |
| 5163F | + | − | − | − |
| 5210 F | + | + | + | + |
| 5167F | + | − | − | − |
| 5385F (Bacteriophage cocktail) | + | − | − | − |
| 5386F (Bacteriophage cocktail) | + | + | − | − |
+, bacteriophage sensitivity observed by a clearing of the bacterial population; −, bacteriophage insensitivity observed as growth (turbidity) of the bacterial population.
Laboratory-scale cheese production.
To analyze the in situ inhibitory activity of the bacteriocin producers (single, double, and in combination with the plantaricin producer Lb. plantarum LMG P-26358), laboratory-scale cheeses were manufactured with the fast acidifier L. lactis DPC4268 and the bacteriocin producers served as protective cultures. The cheeses were spiked with 104 CFU/ml of L. innocua. Each cheese was ripened for 4 weeks at 7°C; Listeria was enumerated weekly during the ripening period. Figure 4 shows Listeria viable cell counts over the 4-week period where bacteriocin-containing vats were compared with vat 1 (no bacteriocin) at each week. At week 0, Listeria numbers were significantly different between vat 1 (5.6 log CFU/g) and all other vats, with the lowest Listeria numbers recorded for vat 7 (CSK3533 and Lb. plantarum) at 3.5 log CFU/g (P < 0.001), followed by vat 2 (CSK3281) at 3.8 log CFU/g (P < 0.001). Listeria numbers in the remaining vats were reduced by 0.9 to 1.5 logs compared to those in vat 1. By week 1, Listeria numbers in vat 7 continued to decrease significantly compared to those in vat 1, with a 3-log reduction recorded (P < 0.001). Listeria numbers in vat 6 (CSK3594 and Lb. plantarum) were also significantly different from those in vat 1, with a 2.7-log reduction (P < 0.001). Significant reductions were also observed for vat 9 (CSK3281, CSK3594, and Lb. plantarum) (1.6-log reduction; P < 0.01) and vat 5 (CSK3281 and Lb. plantarum) (0.8-log reduction; P < 0.05). At week 2, the lowest Listeria numbers were recorded for vat 6 (0.4 log CFU/g) and vat 7 (0.7 log CFU/g), representing 2.6- and 2.3-log reductions compared to numbers in vat 1 (3 log CFU/g) (P < 0.01). Vat 5 (CSK3281 and Lb. plantarum) was also significantly different from vat 1 (1.8-log reduction; P < 0.01). By week 3, vat 7 exhibited the lowest Listeria numbers (0.4 log CFU/g), followed by vat 9 (0.8 log CFU/g), which were both significantly different from vat 1 (2.5 log CFU/g) (P < 0.05). By week 4, Listeria numbers for vat 1 (2.9 log CFU/g) were significantly different from those for most other vats, with no Listeria detected in vat 6 (P < 0.01) and numbers reduced to 0.3 log CFU/g for vat 7 (P < 0.01). Listeria numbers in the remaining vats (vats 2, 3, 4, 8, and 9) ranged from 0.8 to 1 log CFU/g, representing significant differences compared to vat 1 (P < 0.05). While Listeria numbers in vat 5 were not deemed significantly different from those in vat 1, they approached a significant reduction (P = 0.07).
FIG 4.
Counts of viable L. innocua cells in laboratory-scale cheeses. Bacteriocin-containing vats were compared to vat 1 (no bacteriocin) at each week. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In terms of the lacticin and nisin transconjugants (without Lb. plantarum), no significant differences were observed between the double producer (vat 4) and either of the single producers (vats 2 and 3) with regard to Listeria numbers at any week. We then compared vat 8, which consists of the two single producers (CSK3281 and CSK3594) (4.4 log CFU/g at week 0) with vats 2, 3, and 4. While vat 8 was found to be significantly different from vat 2 (3.7 log CFU/g) at week 0 (P < 0.05) whereby the nisin producer generated greater Listeria reductions than the combination of nisin and lacticin single producers, no significant differences were observed for weeks 1 to 4. Likewise, no significant differences were observed between vat 8 and vat 3 or 4 over the ripening period.
Bacteriocin detection in vats 4, 5, 6, and 7.
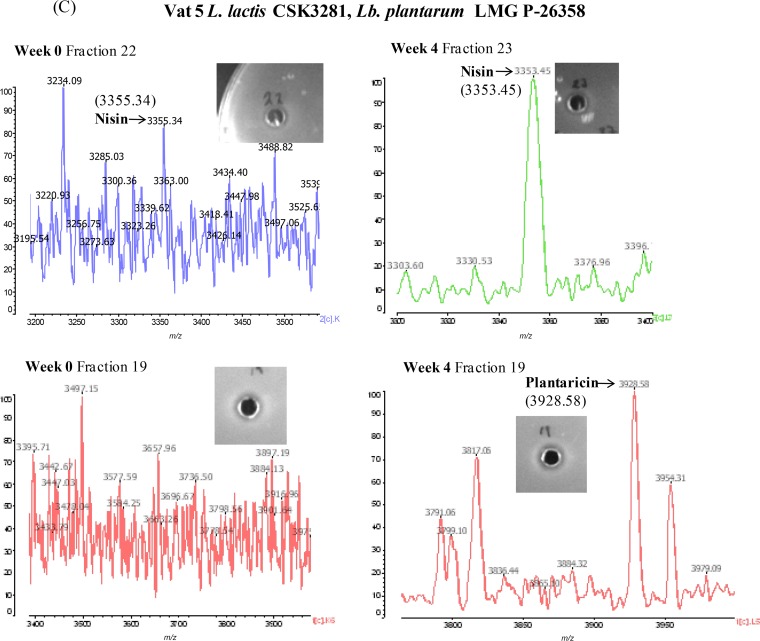
The correct masses for nisin (fraction 21), Ltnβ (fraction 37), and plantaricin (fraction 19) were detected by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) in vat 7 (double producer and plantaricin) at week 0 (Fig. 5A). Antimicrobial assays also revealed that these fractions contained activity (lacticin activity was restored by combining fractions 23 [Ltnα] and 37).
FIG 5.
MALDI-TOF MS analysis of vat 7 (lacticin, nisin, and plantaricin) (A), vat 4 (lacticin and nisin) (B), vat 5 (nisin and plantaricin) (C), and vat 6 (lacticin and plantaricin) (D). Masses corresponding to the bacteriocins are indicated. Inset photos show inhibition zones produced by correct-mass-containing fractions against the indicator strain L. lactis HP (nisin and lacticin) or L. innocua (plantaricin). F, fraction.
At week 4, a mass corresponding to plantaricin was detected in fraction 19, but there was no antimicrobial activity. Nisin was detected in fraction 21 at week 4, and activity was also confirmed. The correct mass for Ltnβ could not be detected at week 4. Despite this, combining fractions 23 and 37 did yield a zone of inhibition against the indicator strain, suggesting that the bacteriocin is present.
The correct mass for nisin was not detected in vat 4 (double producer) at week 0 or 4; however, fraction 21, which is generally expected to contain nisin, yielded a zone of inhibition against the indicator strain on both weeks, suggesting that the bacteriocin is present (Fig. 5B). The correct mass for Ltnβ was detected in vat 4 (double producer) at weeks 0 and 4 (Fig. 5B). Lacticin activity was confirmed when fractions 23 and 37 were positioned beside each other in the agar well diffusion assays.
Vat 5 (nisin and plantaricin) was found to contain the nisin mass at weeks 0 and 4, although the correct mass was found in fraction 22 at week 0 and in fraction 23 at week 4 (Fig. 5C). This is presumably due to slight variations in the times that the peptide eluted from the high-performance liquid chromatography (HPLC) column. These fractions exhibited antimicrobial activity against the indicator strain, but the zones were smaller than previously observed. The plantaricin mass was not detected in vat 5 at week 0, but a zone of inhibition against Listeria was observed for fraction 19. However, the plantaricin mass was detected in vat 5 at week 4, and antimicrobial activity was confirmed.
Vat 6 (lacticin and plantaricin) was shown to contain the correct Ltnβ and plantaricin masses at weeks 0 and 4 in fractions 37 and 19, respectively (Fig. 5D). Antimicrobial activity was confirmed for fraction 19, although the zone was smaller at week 0 than at week 4. In the case of lacticin, combining fractions 24 (Ltnα) and 37 confirmed antimicrobial activity.
Development of bacteriocin tolerance.
The frequency of tolerance/resistance development in L. innocua was assessed using 1,000 AU/ml of each bacteriocin. The frequency of resistance against 1,000 AU/ml of nisin was calculated to be 6.56 × 10−4. Resistance development could not be observed when Listeria was exposed to 1,000 AU/ml lacticin or 1,000 AU/ml nisin and lacticin. On the other hand, the frequencies of resistance development against 320 AU/ml nisin or 320 AU/ml lacticin (representing arbitrary concentrations in cheese) were much lower, at 4.9 × 10−1 and 3.02 × 10−1, respectively. Simultaneous exposure to lacticin and nisin at 320 AU/ml decreased the frequency of resistance to 3.18 × 10−2. However, bacteriocin-resistant colonies remained sensitive to 1,000 AU/ml, indicating that Listeria cells were tolerant rather than completely resistant.
DISCUSSION
In this study, an isogenic family of nisin and lacticin transconjugants was developed with a view to better understand the impact of multiple bacteriocin production and genetic load on starter culture functionality. Genotypic and phenotypic analyses, including PCR, well diffusion assays, and CMS, confirmed the acquisition of lacticin and/or nisin in each transconjugant.
In agreement with previous findings (29) which revealed that pMRC01 imposes a burden on lactococcal metabolism affecting growth and acidification rates, the presence of pMRC01 was shown to influence lactococcal acidification in the lacticin transconjugant, as did the presence of the nisin transposon in the nisin transconjugant. However, bacteriocin stacking resulted in the slowest acidification rates, but this can be overcome with the addition of 0.1% yeast extract to the double producer. Yeast extract presumably lessens the burden of plasmid and transposon acquisition, as it provides amino acids along with purine and pyrimidine bases and inorganic constituents which have been shown to stimulate lactococcal growth (30). We therefore suggest that the double producer has potential to serve as a protective culture when used in conjunction with a suitable acidifier. Despite this, the lactose utilization phenotype in the double producer, CSK3533, was found to be unstable in GM17, apparently due to the loss of a large (>50-kb) plasmid likely involved in lactose utilization. Both the plasmid instability and slower acidification profiles observed in the double producer (CSK3533) may be attributed to the metabolic burden imposed by the presence of pMRC01 and the nisin transposon. In an effort to ease the metabolic load, it is possible that as the energy demand of the cell increases and metabolites are exhausted, a reduction in growth rate and perhaps the loss of nonessential plasmids may occur (29, 31). This is supported by the fact that the addition of lactose to M17 broth maintains the lactose utilization phenotype in the double producer.
Antimicrobial activity assays confirmed that coproduction of nisin and lacticin by CSK3533 was as effective as its single-producing counterparts against most of the indicator strains tested. This indicates that bacteriocin production is not affected by the lower growth rate observed in the double producer. Furthermore, with the exception of C. tyrobutyricum, the coproduction of the two potent bacteriocins did not result in an increase in antimicrobial activity. Studies suggest that bacteriocin production and subsequent inhibition may be influenced by the growth medium (32, 33). The multibacteriocinogenic strain L. lactis INIA 415 was capable of producing the class I bacteriocin lacticin 481 in M17 broth only and produced nisin in milk only (32). Therefore, it is possible that growth of CSK3533 under different conditions could result in higher levels of nisin or lacticin being produced.
In terms of bacteriophage resistance, transconjugants were shown to be more resistant to bacteriophage attack than the recipient strain, with superior resistance properties observed in derivatives of pMRC01, which is known to harbor an abortive infection mechanism (34).
Laboratory-scale cheese inoculated with Listeria was used to assess the efficacy of single- and double-bacteriocin producers alone and in combination with the plantaricin producer Lb. plantarum LMG P-26358 in situ. The latter strain was previously shown to have a narrow spectrum of inhibition, inhibiting Listeria and enterococcal strains but not clostridia, Escherichia coli, Bacillus species, Salmonella, or members of the LAB (18). The strain proved to be an effective adjunct for controlling Listeria growth in a cheese model (18). In the present study, CSK3594, CSK3281, and CSK3533 failed to inhibit L. innocua by agar well diffusion assay; however, by the end of the ripening period, Listeria numbers from cheeses prepared with these starters were significantly reduced compared to those for the control, vat 1 (no bacteriocin). Interestingly, the double producer combined with the plantaricin producer exhibited the greatest reduction in Listeria numbers at week 0, a trend which continued to week 1, suggesting the effectiveness of this combination for reducing the initial bacterial load. The inhibitory effect of this combination was on the whole significantly better than using both single producers with the plantaricin producer. This can be explained by the fact that the nisin-producing transconjugant inhibits the lacticin transconjugant and vice versa, whereas the double producer is immune to both bacteriocins. The combination of the lacticin producer with Lb. plantarum LMG P-26358 also significantly reduced Listeria numbers by week 1, and by week 4, Listeria could not be detected in this vat. Overall, the double producer combined with the plantaricin producer, followed by the lacticin producer combined with plantaricin, exhibited the most significant reductions in Listeria numbers over the ripening period.
In general, a similar inhibitory trend was observed among the single class I producers, the double producer, or the combined single producers, which were significantly different from vat 1 at week 0 and week 4. While the double producer did not alter Listeria numbers significantly compared to the single producers alone, a 10-fold reduction in the emergence of bacteriocin tolerance was observed when Listeria was exposed to both nisin and lacticin, suggesting that bacteriocin stacking could be an effective method to prevent pathogen growth in food applications. However, combining bacteriocins from different classes or subclasses is considered most effective for reducing the emergence of resistance (35), which explains the increased antimicrobial efficacy for vats containing the class I and class II bacteriocins in this study.
Interestingly, cheese prepared with nonbacteriocinogenic CSK2775 also resulted in reduced Listeria numbers over the 4-week period, although to a lesser extent than the bacteriocin-containing cheeses. Therefore, it is probable that bacterial competition coupled with unfavorable conditions relating to cheese manufacture, including lactic acid and high salt concentrations, have provided a hurdle effect to cause the observed reductions. Indeed, several intrinsic factors, including moisture content, acidity, and competitive flora, are known to dictate pathogen survival in cheese (36, 37).
MALDI-TOF MS of cheeses from vats 4, 5, 6, and 7 indicated that nisin and lacticin were present in the appropriate cheeses, implying that bacteriocin integrity was not compromised in the cheese environment. The presence of plantaricin could not be confirmed in vat 7 cheese (double producer and plantaricin) at week 4, but it was present in vats 5 and 6 at both times, as expected. MALDI-TOF MS is not quantitative and is also subject to preferential ionization in that some peptides ionize better than others. The peptide content in a cheese increases during ripening due to the breakdown of casein, so a number of bacteriocin purification steps were performed to increase the chances of detecting bacteriocin masses. Cheeses were passed through C18 solid-phase extraction, columns and peptides were further separated using reverse-phase HPLC (RP-HPLC). Each HPLC fraction potentially contains numerous peptides, making it difficult to detect the bacteriocin masses which are present at low concentrations. Usually the bacteriocin mass and a concomitant zone of inhibition are taken as proof of the presence of bacteriocin, but in the case of a cheese fraction, the presence of a zone of inhibition alone may be taken as indicative of bacteriocin presence.
Natural isolates capable of producing multiple bacteriocins have been reported in the literature (38–42). Most recently, L. lactis LMG2081 was shown to produce two different classes of bacteriocins, a novel lantibiotic and the class IIb bacteriocin lactococcin G (43). However, the ability to generate a multibacteriocin producer from an already-established culture through food-grade enabling technologies has several benefits. First, the technological properties of the culture are known. The number of cultures required to produce multiple bacteriocins is reduced. The risk of bacteriocin inhibition is removed, since the multibacteriocin-producing starter will also harbor the genetic machinery for bacteriocin immunity. As conjugation is a natural process, the resulting transconjugants do not fall under current European regulations governing the use of genetically modified microorganisms (44, 45). Therefore, transconjugants can be used in food applications in a manner similar to that for the recipient strain (46). While the double bacteriocin producer generated in this study proved to be a slower acidifier than the recipient strain, it has potential to serve as a protective culture. However, studies generating multiple bacteriocin producers have been rare (13). This can most likely be attributed to the complex biosynthetic process required for bacteriocin production and secretion. Indeed, previous attempts to construct nisin-lacticin transconjugants were unsuccessful, which was often attributed to the incompatibility of bacteriocin modification machinery or bacteriocin sensitivity (13, 34). Traditionally, the discovery of technologically valuable industrial strains has focused on large-scale screening strategies from a variety of sources (47). However, the successful transfer of lacticin and nisin to commercial starter cultures as reported in this study may provide additional avenues for the development of multihurdle protective cultures using food-grade methods.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in this study are listed in Tables 1 and 3. L. lactis strains were routinely propagated at 30°C in M17 medium (Difco Laboratories, Detroit, MI, USA) supplemented with 0.5% (wt/vol) lactose (LM17) or glucose (GM17). Lb. plantarum was grown in MRS medium (48) (Difco Laboratories) at 30°C. L. innocua was routinely propagated in GM17 broth at 37°C containing 500 μg/ml streptomycin (Sigma-Aldrich, Ireland). Other media used in this study include brain heart infusion (BHI) broth (Oxoid Ltd., Basingstoke, Hampshire, England) and reinforced clostridial medium (RCM) (Merck, Darmstadt, Germany). All strains were stored in 50% glycerol at −20°C.
TABLE 3.
Bacterial strains used in this study
| Bacterial strain | Relevant detail(s) | Relevant phenotypea | Source or reference |
|---|---|---|---|
| L. lactis HP | Bacteriocin-sensitive indicator strain | Ltn− Nis− | TFRCb |
| L. lactis MG1363 (pMRC01) | Donor strain harboring pMRC01, lacticin producer | Lac− Ltn+ Nis− | TFRC |
| L. lactis CSK2583 | Donor strain harboring Tn5276, nisin producer | Lac− Nis+ Ltn− | CSK, The Netherlands |
| L. lactis CSK2775 | Recipient strain | Lac+ Nis− Ltn− | CSK, The Netherlands |
| L. lactis CSK3281 | CSK2775 transconjugant, nisin producer | Lac+ Nis+ | This study |
| L. lactis CSK3594 | CSK2775 transconjugant harboring pMRC01, lacticin producer | Lac+ Ltn+ | This study |
| L. lactis CSK3533 | CSK3281 transconjugant harboring pMRC01, nisin-lacticin double producer | Lac+ Nis+ Ltn+ | This study |
| Lb. plantarum LMG P-26358 | Plantaricin 423 producer | Pln+ | 18 |
| L. lactis DPC4268 | Starter culture for cheese manufacture | Lac+ | TFRC |
Lac, lactose utilization; Ltn, lacticin genetic determinants; Nis, nisin genetic determinants; Pln, plantaricin genetic determinants.
TFRC, Teagasc Food Research Centre, Moorepark, Fermoy, County Cork, Ireland.
Strain construction and analytical tests. (i) Strain construction.
The conjugation method of Coakley et al. (34) was used with slight modifications to generate lacticin transconjugants. Inocula (2%) of both donor and recipient were grown for 4 h in GM17 broth at 30°C. After the growth period, 1 ml of recipient and 1 ml of donor were harvested by centrifugation (16,000 × g for 1 min) and rinsed twice with GM17 broth. After the final rinse, each strain was resuspended in 50 μl of GM17 broth. The concentrated recipient and donor (20×) were then mixed with each other at ratios of 1:1, 2:1, and 20:1. Each mixture was spotted onto the center of a GM17 agar plate and incubated for 18 h at 30°C. The following day, spots were harvested in 1 ml of maximum recovery diluent (MRD) (Oxoid) and serially diluted before plating on LIA containing lacticin (400 AU/ml) as described previously (34). Following 48 h of incubation at 30°C, the lacticin-containing LIA plates were examined for lactose-positive colonies (yellow) against a background of lactose-negative colonies (white), and lactose-positive colonies were selected and grown in LM17 broth for further analysis.
Nisin transconjugants were generated according to the method of Gireesh et al. (49) with modifications. Inocula of the donor (1.5%) and recipient (2%) were grown for 4 h in GM17 broth at 30°C. The donor and recipient were then mixed at ratios of 1:10 and 1:100 in the presence of 400 μg/ml α-chymotrypsin (Sigma-Aldrich). The cells were collected onto membrane filters (0.45-μm pore size; Merck, Millipore, Darmstadt, Germany), after which the filters were placed on GM17 agar plates (cell side down). Following 18 h of incubation at 30°C, cells were harvested from the filter and added to 10% reconstituted skim milk (RSM) containing 400 AU/ml nisin (Sigma-Aldrich) and incubated at 30°C for 24 to 48 h. Clotted samples were serially diluted and plated on LIA, and following 48 h of incubation at 30°C, yellow colonies were selected for further analysis.
(ii) Bacteriocin production and immunity.
Bacteriocin production and immunity was assessed by performing the agar well diffusion assay as described by Ryan et al. (50). Indicator organisms are listed in Table 1. Bacteriocin sensitivity was scored according to the diameter of the zone of inhibition surrounding the well which contained cell-free supernatant from the bacteriocin producer. The concentration of bacteriocin produced by the double producer was measured by agar well diffusion assay using a serial 2-fold dilution of the filtered culture supernatant, and bacteriocin activity was calculated as the inverse of the last dilution that gave a definite zone of clearance after overnight incubation, where AU were expressed per ml.
(iii) CMS.
CMS was performed according to the method described by Field et al. (51).
(iv) PCR scan.
Genomic DNA was extracted from 1.5 ml of 18-h cultures according to the method of Hoffman and Winston (52) slightly modified as described previously (53). Primer pairs used to scan strains for the presence of pMRC01 as well as the genes associated with nisin production are listed in Table 4. PCR was performed in a Hybaid PCR express unit (Hybaid Ltd., Middlesex, UK) using MyTaq Red Mix polymerase (Bioline Ltd., London, UK) according to the manufacturers' specifications combined with an annealing temperature of 55°C.
TABLE 4.
Primer pairs used in this study
| Primer | Sequence | Target gene(s) | Size (bp) |
|---|---|---|---|
| 27-F | 5′-GGGGAACAATCTTACCTA | orf27 | 326 |
| 27-R | 5′-ATTATTTTTATTGCATTCTACTA | ||
| 49-F | 5′-CCAATACCCGCCAAAATAAAGT | orf49 | 347 |
| 49-R | 5′-CTAAGCGCAGAGGAAATACAACC | ||
| 51-F | 5′-TTCTCAAAATCATCAAAATCAAGT | orf51 | 293 |
| 51-R | 5′-GTACGAACAGGAGCGAAAAA | ||
| 52-F | 5′-CCTAAGTTGTCTATTCGTGTCCA | orf52 | 210 |
| 52-R | 5′-ATTAGGTGAGTGCTCTGATTTTTC | ||
| nisA-F | 5′-CAAAAGATTTTAACTTGGATTTG | nisA | 163 |
| nisA-R | 5′-ACGTGAATACTACAATGACAAG | ||
| nisFG-F | 5′-GGTTTAATTTCTGCAGATACTG | nisFEG | 1,573 |
| nisFG-R | 5′-GTAATTATCCAGATCATTGCTG |
(v) PFGE.
PFGE was performed as described by Mills et al. (53) using the restriction enzyme SmaI (New England BioLabs, Hertfordshire, UK). DNA fragments were run on a CHEF-DR III pulsed-field system (Bio-Rad Laboratories, Hercules, CA, USA) at 6 V/cm for 22 h with a 1- to 30-s linear ramp pulse time. Molecular size markers (N0340S and N0350S) were purchased from New England BioLabs.
(vi) Bacteriophage assays.
Bacteriophages were propagated according to a method outlined previously (54). Sensitivity to bacteriophage infection was performed by the double agar layer plaque assay as described previously (34).
(vii) Characterization of acid production.
Acid production was monitored in 10% RSM in the presence and absence of 0.1% yeast extract according to the method of Harrington and Hill (55).
Laboratory-scale cheese manufacture.
Cultures were grown from frozen stocks in their respective media for 18 h (Table 1). The cultures were then inoculated at 1% (vol/vol) into 10% (wt/vol) RSM and incubated for a further 18 h at 30°C. In the case of Lb. plantarum LMG P-26358, the culture was grown in 10% RSM containing 0.1% (vol/vol) yeast extract and 0.2 g/liter MnSO44H2O as previously reported (18).
One-liter vats of whole milk heated to 31°C were inoculated with the 18-h RSM cultures as follows: vat 1, 0.75% (vol/vol) L. lactis DPC4268 and 0.75% (vol/vol) L. lactis CSK2775 (no bacteriocin); vat 2, 0.75% (vol/vol) L. lactis DPC4268 and 0.75% (vol/vol) L. lactis CSK3281 (nisin producer); vat 3, 0.75% (vol/vol) L. lactis DPC4268 and 0.75% (vol/vol) L. lactis CSK3594 (lacticin producer); vat 4, 0.75% (vol/vol) L. lactis DPC4268 and 0.75% (vol/vol) L. lactis CSK3533 (nisin-lacticin double producer); vat 5, 0.75% (vol/vol) L. lactis DPC4268, 0.5% (vol/vol) L. lactis CSK3281 (nisin producer), and 0.5% (vol/vol) Lb. plantarum LMG P-26358 (plantaricin producer); vat 6, 0.75% (vol/vol) L. lactis DPC4268, 0.5% (vol/vol) L. lactis CSK3594 (lacticin producer), and 0.5% (vol/vol) Lb. plantarum LMG P-26358 (plantaricin producer); vat 7, 0.75% (vol/vol) L. lactis DPC4268, 0.5% (vol/vol) L. lactis CSK3533 (nisin-lacticin double producer), and 0.5% (vol/vol) Lb. plantarum LMG P-26358 (plantaricin producer); vat 8, 0.75% (vol/vol) L. lactis DPC4268, 0.5% (vol/vol) CSK3594 (lacticin producer), and 0.5% (vol/vol) CSK3281 (nisin producer); and vat 9, 0.75% (vol/vol) L. lactis DPC4268, 0.5% (vol/vol) CSK3594 (lacticin producer), 0.5% (vol/vol) CSK3281 (nisin producer), and 0.5% (vol/vol) Lb. plantarum LMG P-26358 (plantaricin producer).
A streptomycin-resistant derivative of L. innocua (DPC6578) grown for 18 h was added to each vat at a level of 104 CFU/ml. Thirty minutes after inoculation, 150 international milk clotting units/ml Kalase rennet (CSK Food Enrichment, The Netherlands) was added according to the manufacturer's specifications and after a further 15 min, the curd was cut into cubes. Following a 10-min stirring step, approximately 35% of the whey was removed, and the curd was stirred for a further 5 min. The temperature was then elevated to 36°C over a 5-min period, and the curd was stirred for a further 20 min. The curd was further drained and lightly pressed into molds for 20 min before pressing overnight. After 24 h, the cheeses were submerged in a brine bath (23% [wt/vol] NaCl, 0.22% [vol/vol] phosphoric acid, 0.1% [wt/vol] NaOH, 0.6% [wt/vol] CaCl2) at 10 to 12°C for 5 h, after which they were vacuum packed and ripened at 7°C for 4 weeks. L. innocua DPC6578 was enumerated in each cheese on a weekly basis by homogenizing 1 g of cheese in 2% sterile trisodium citrate and plating serial dilutions on selective medium (GM17 agar with 500 μg/ml of streptomycin). The cheese trial was performed in triplicate, and sampling for each trial was performed in duplicate.
Nisin (3,352 ± 3 Da), lacticin (Ltnβ) (2,847 ± 4 Da), and plantaricin (3,928 ± 3 Da) present within cheese samples from vats 4, 5, 6, and 7 were verified by MALDI-TOF MS as described previously (18). In the case of lacticin, the presence of the correct mass for Ltnβ was indicative of lacticin, since Ltnα can be difficult to detect in a complex fraction. All fractions were tested for antimicrobial activity by agar well diffusion assays against the appropriate indicator strains (lacticin and nisin against L. lactis HP and plantaricin against L. innocua), where mass and concomitant activity were indicative of bacteriocin presence. Fractions expected to contain the lacticin peptides (fractions 23 and 24, Ltnα; fraction 37, Ltnβ) were combined or wells were positioned near each other to assess lacticin activity.
Frequency of bacteriocin resistance/tolerance.
To determine the frequency of bacteriocin resistance/tolerance development in L. innocua, freshly prepared 18-h cultures were serially diluted in MRD and spread plated on to GM17 or GM17 containing either 1,000 AU/ml or 320 AU/ml of the appropriate bacteriocin or bacteriocin combination; the latter concentration represents the arbitrary in situ concentration of the bacteriocins in the cheeses. Plates were incubated aerobically at 37°C for up to 48 h, at which time the frequency of bacteriocin resistance/tolerance was calculated as described previously (56). All experiments were performed in triplicate.
Statistical analysis.
Listeria counts in laboratory-scale cheeses were statistically analyzed using one-way analysis of variance (ANOVA). Post hoc multiple comparisons were done by Tukey's test, and differences were considered to be statistically significant at a P value of <0.05. Statistical tests were performed using XLSTAT statistical software.
ACKNOWLEDGMENT
This project was funded by CSK Food Enrichment B.V., The Netherlands.
REFERENCES
- 1.Mills S, O'Sullivan O, Hill C, Fitzgerald GF, Ross RP. 2010. The changing face of dairy starter culture research: from genomics to economics. Int J Dairy Technol 63:149–170. doi: 10.1111/j.1471-0307.2010.00563.x. [DOI] [Google Scholar]
- 2.O'Sullivan L, Morgan SM, Ross RP. 2007. Bacteriocins: changes in cheese flora and flavour, p 326–348. In Weimer BC. (ed), Improving the flavour of cheese. Woodhead Publishing, Cambridge, England. [Google Scholar]
- 3.O'Connor PM, Ross RP, Hill C, Cotter PD. 2015. Antimicrobial antagonists against food pathogens: a bacteriocin perspective. Curr Opin Food Sci 2:51–57. doi: 10.1016/j.cofs.2015.01.004. [DOI] [Google Scholar]
- 4.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 5.Alvarez-Sieiro P, Montalbán-López M, Mu D, Kuipers OP. 2016. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. doi: 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez RC, Staliano CD, Vieira AD, Villarreal ML, Todorov SD, Saad SM, Franco BD. 2015. Bacteriocin production and inhibition of Listeria monocytogenes by Lactobacillus sakei subsp. sakei 2a in a potentially synbiotic cheese spread. Food Microbiol 48:143–152. doi: 10.1016/j.fm.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Diaz-Ruiz G, Omar NB, Abriouel H, Canamero MM, Galvez A. 2012. Inhibition of Listeria monocytogenes and Escherichia coli by bacteriocin-producing Lactobacillus plantarum EC52 in a meat sausage model system. Afr J Microbiol Res 6:1103–1108. doi: 10.5897/AJMR10.175. [DOI] [Google Scholar]
- 8.Garde S, Avila M, Arias R, Gaya P, Nunez M. 2011. Outgrowth inhibition of Clostridium beijerinckii spores by a bacteriocin-producing lactic culture in ovine milk cheese. Int J Food Microbiol 150:59–65. doi: 10.1016/j.ijfoodmicro.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Gómez-Sala B, Herranz C, Díaz-Freitas B, Hernández PE, Sala A, Cintas LM. 2016. Strategies to increase the hygienic and economic value of fresh fish: biopreservation using lactic acid bacteria of marine origin. Int J Food Microbiol 223:41–49. doi: 10.1016/j.ijfoodmicro.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Mills S, Stanton C, Hill C, Ross RP. 2011. New developments and applications of bacteriocins and peptides in foods. Annu Rev Food Sci Technol 2:299–329. doi: 10.1146/annurev-food-022510-133721. [DOI] [PubMed] [Google Scholar]
- 11.Horn N, Martinez MI, Martinez JM, Hernandez PE, Gasson MJ, Rodriguez JM, Dodd HM. 1999. Enhanced production of pediocin PA-1 and coproduction of nisin and pediocin PA-1 by Lactococcus lactis. Appl Environ Microbiol 65:4443–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reviriego C, Fernandez L, Rodriguez JM. 2007. A food-grade system for production of pediocin PA-1 in nisin-producing and non-nisin-producing Lactococcus lactis strains: application to inhibit Listeria growth in a cheese model system. J Food Prot 70:2512–2517. doi: 10.4315/0362-028X-70.11.2512. [DOI] [PubMed] [Google Scholar]
- 13.O'Sullivan L, Ryan MP, Ross RP, Hill C. 2003. Generation of food-grade lactococcal starters which produce the lantibiotics lacticin 3147 and lacticin 481. Appl Environ Microbiol 69:3681–3685. doi: 10.1128/AEM.69.6.3681-3685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl HG, de Kruijff B. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 15.Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. 2008. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci 65:455–476. doi: 10.1007/s00018-007-7171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiedemann I, Bottiger T, Bonelli RR, Wiese A, Hagge SO, Gutsmann T, Seydel U, Deegan L, Hill C, Ross P, Sahl HG. 2006. The mode of action of the lantibiotic lacticin 3147—a complex mechanism involving specific interaction of two peptides and the cell wall precursor lipid II. Mol Microbiol 61:285–296. doi: 10.1111/j.1365-2958.2006.05223.x. [DOI] [PubMed] [Google Scholar]
- 17.Suda S, Cotter PD, Hill C, Ross RP. 2012. Lacticin 3147—biosynthesis, molecular analysis, immunity, bioengineering and applications. Curr Protein Pept Sci 13:193–204. doi: 10.2174/138920312800785021. [DOI] [PubMed] [Google Scholar]
- 18.Mills S, Serrano LM, Griffin C, O'Connor PM, Schaad G, Bruining C, Hill C, Ross RP, Meijer WC. 2011. Inhibitory activity of Lactobacillus plantarum LMG P-26358 against Listeria innocua when used as an adjunct starter in the manufacture of cheese. Microb Cell Fact 10(Suppl 1):S7. doi: 10.1186/1475-2859-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui Y, Zhang C, Wang Y, Shi J, Zhang L, Ding Z, Qu X, Cui H. 2012. Class IIa bacteriocins: diversity and new developments. Int J Mol Sci 13:16668–16707. doi: 10.3390/ijms131216668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Tassell ML, Ibarra-Sánchez LA, Takhar SR, Amaya-Llano SL, Miller MJ. 2015. Use of a miniature laboratory fresh cheese model for investigating antimicrobial activities. J Dairy Sci 98:8515–8524. doi: 10.3168/jds.2015-9967. [DOI] [PubMed] [Google Scholar]
- 21.Schelegueda LI, Delcarlo SB, Gliemmo MF, Campos CA. 2016. Effect of antimicrobial mixtures and modified atmosphere packaging on the quality of Argentine hake (Merluccius hubbsi) burgers. LWT Food Sci Technol 68:258–264. doi: 10.1016/j.lwt.2015.12.012. [DOI] [Google Scholar]
- 22.Olle Resa CP, Gerschenson LN, Jagus RJ. 2014. Natamycin and nisin supported on starch edible films for controlling mixed culture growth on model systems and Port Salut cheese. Food Control 44:146–151. doi: 10.1016/j.foodcont.2014.03.054. [DOI] [Google Scholar]
- 23.Fernandez MV, Jagus RJ, Mugliaroli SL. 2014. Effect of combined natural antimicrobials on spoilage microorganisms and Listeria innocua in a whey cheese “Ricotta.” Food Bioprocess Technol 7:2528–2537. doi: 10.1007/s11947-013-1243-0. [DOI] [Google Scholar]
- 24.Mingming G, Jin TZ, Wang L, Scullen J, Sommers CH. 2014. Antimicrobial films and coatings for inactivation of Listeria innocua on ready-to-eat deli turkey meat. Food Control 40:64–70. doi: 10.1016/j.foodcont.2013.11.018. [DOI] [Google Scholar]
- 25.Morgan SM, Ross RP, Beresford T, Hill C. 2000. Combination of hydrostatic pressure and lacticin 3147 causes increased killing of Staphylococcus and Listeria. J Appl Microbiol 88:414–420. doi: 10.1046/j.1365-2672.2000.00975.x. [DOI] [PubMed] [Google Scholar]
- 26.Scannell AG, Ross RP, Hill C, Arendt K. 2000. An effective lacticin biopreservative in fresh pork sausage. J Food Prot 63:370–375. doi: 10.4315/0362-028X-63.3.370. [DOI] [PubMed] [Google Scholar]
- 27.Soriano A, Ulmer HM, Scannell AGM, Ross RP, Hill C, Garcia-Ruiz A, Arendt EK. 2004. Control of food spoiling bacteria in cooked meat products with nisin, lacticin 3147, and a lacticin 3147-producing starter culture. Eur Food Res Technol 219:6–13. doi: 10.1007/s00217-004-0910-9. [DOI] [Google Scholar]
- 28.Scannell AG, Hill C, Ross RP, Marx S, Hartmeier W, Elke Arendt K. 2000. Development of bioactive food packaging materials using immobilised bacteriocins lacticin 3147 and nisaplin. Int J Food Microbiol 60:241–249. doi: 10.1016/S0168-1605(00)00314-7. [DOI] [PubMed] [Google Scholar]
- 29.Fallico V, McAuliffe O, Fitzgerald GF, Hill C, Ross RP. 2009. The presence of pMRC01 promotes greater cell permeability and autolysis in lactococcal starter cultures. Int J Food Microbiol 133:217–224. doi: 10.1016/j.ijfoodmicro.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 30.Smith JS, Hillier AJ, Lees GJ. 1975. The nature of the stimulation of the growth of Streptococcus lactis by yeast extract. J Dairy Res 42:123–138. doi: 10.1017/S0022029900015156. [DOI] [PubMed] [Google Scholar]
- 31.Friehs K. 2004. Plasmid copy number and plasmid stability. Adv Biochem Eng Biotechnol 86:47–82. [DOI] [PubMed] [Google Scholar]
- 32.Bravo D, Rodriguez E, Medina M. 2009. Nisin and lacticin 481 coproduction by Lactococcus lactis strains isolated from raw ewes' milk. J Dairy Sci 92:4805–4811. doi: 10.3168/jds.2009-2237. [DOI] [PubMed] [Google Scholar]
- 33.Avonts L, E Van Uytven, De Vuyst L. 2004. Cell growth and bacteriocin production of probiotic Lactobacillus strains in different media. Int Dairy J 14:947–955. doi: 10.1016/j.idairyj.2004.04.003. [DOI] [Google Scholar]
- 34.Coakley M, Fitzgerald G, Ross RP. 1997. Application and evaluation of the bacteriophage resistance- and bacteriocin-encoding plasmid pMRC01 for the improvement of dairy starter cultures. Appl Environ Microbiol 63:1434–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bastos Mdo C, Coelho ML, Santos OC. 2015. Resistance to bacteriocins produced by Gram-positive bacteria. Microbiology 161:683–700. doi: 10.1099/mic.0.082289-0. [DOI] [PubMed] [Google Scholar]
- 36.Donnelly CW. 2004. Growth and survival of microbial pathogens in cheese, p 541–560. In Fox PF, McSweeney PLH, Cogan TM, Guinee TP (ed), Cheese chemistry, physics and microbiology, 3rd edition, vol 1 Elsevier Academic Press, London, England. [Google Scholar]
- 37.Johnson EA, Nelson JH, Johnson M. 1990. Microbiological safety of cheese made from heat-treated milk, part II. Microbiology J Food Prot 53:519–540. doi: 10.4315/0362-028X-53.6.519. [DOI] [PubMed] [Google Scholar]
- 38.O'Shea EF, O'Connor PM, Raftis EJ, O'Toole PW, Stanton C, Cotter PD, Ross RP, Hill C. 2011. Production of multiple bacteriocins from a single locus by gastrointestinal strains of Lactobacillus salivarius. J Bacteriol 193:6973–6982. doi: 10.1128/JB.06221-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quadri LE, Sailer M, Roy KL, Vederas JC, Stiles ME. 1994. Chemical and genetic characterization of bacteriocins produced by Carnobacterium piscicola LV17B. J Biol Chem 269:12204–12211. [PubMed] [Google Scholar]
- 40.Rodríguez E, González B, Gaya P, Nuñez M, Medina M. 2000. Diversity of bacteriocins produced by lactic acid bacteria isolated from raw milk. Int Dairy J 10:7–15. doi: 10.1016/S0958-6946(00)00017-0. [DOI] [Google Scholar]
- 41.Kojic M, Strahinic I, Fira D, Jovcic B, Topisirovic L. 2006. Plasmid content and bacteriocin production by five strains of Lactococcus lactis isolated from semi-hard homemade cheese. Can J Microbiol 52:1110–1120. doi: 10.1139/w06-072. [DOI] [PubMed] [Google Scholar]
- 42.Himeno K, Fujita K, Zendo T, Wilaipun P, Ishibashi N, Masuda Y, Yoneyama F, Leelawatcharamas V, Nakayama J, Sonomoto K. 2012. Identification of enterocin NKR-5-3C, a novel Class IIa bacteriocin produced by a multiple bacteriocin producer, Enterococcus faecium NKR-5-3. Biosci Biotechnol Biochem 76:1245–1247. doi: 10.1271/bbb.120089. [DOI] [PubMed] [Google Scholar]
- 43.Mirkovic N, Polovic N, Vukotic G, Jovcic B, Miljkovic M, Radulovic Z, Diep DB, Kojic M. 2016. Lactococcus lactis LMG2081 produces two bacteriocins, a nonlantibiotic and a novel lantibiotic. Appl Environ Microbiol 82:2555–2562. doi: 10.1128/AEM.03988-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derkx PMF, Janzen T, Sørenson KI, Christensen JE, Stuer-Lauridsen B, Johansen E. 2014. The art of strain improvement of industrial lactic acid bacteria without the use of recombinant DNA technology. Microb Cell Fact 13(Suppl 1):S5. doi: 10.1186/1475-2859-13-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedersen MB, Iversen SL, Sørensen KI, Johansen E. 2005. The long and winding road from the research laboratory to industrial applications of lactic acid bacteria. FEMS Microbiol Rev 29:611–624. doi: 10.1016/j.fmrre.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 46.Hill C, Ross RP. 1998. Starter cultures for the dairy industry, p 174–192. In Roller S, Harlander S (ed), Genetic modification in the food Industry: a strategy for food quality improvement. Springer Science, Dordrecht, The Netherlands. [Google Scholar]
- 47.Hansen EB. 2002. Commercial bacterial starter cultures for fermented foods of the future. Int J Food Microbiol 78:119–131. doi: 10.1016/S0168-1605(02)00238-6. [DOI] [PubMed] [Google Scholar]
- 48.De Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J Appl Microbiol 23:130–135. [Google Scholar]
- 49.Gireesh T, Davidson BE, Hillier AJ. 1992. Conjugal transfer in Lactococcus lactis of a 68-kilobase-pair chromosomal fragment containing the structural gene for the peptide bacteriocin nisin. Appl Environ Microbiol 58:1670–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryan MP, Rea MC, Hill C, Ross RP. 1996. An application in Cheddar cheese manufacture for a strain of Lactococcus lactis producing a novel broad-spectrum bacteriocin, lacticin 3147. Appl Environ Microbiol 62:612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Field D, Begley M, O'Connor PM, Daly KM, Hugenholtz F, Cotter PD, Hill C, Ross RP. 2012. Bioengineered nisin A derivatives with enhanced activity against both Gram positive and Gram negative pathogens. PLoS One 7:e46884. doi: 10.1371/journal.pone.0046884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffman CS, Winston F. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 53.Mills S, Griffin C, Coffey A, Meijer WC, Hafkamp B, Ross RP. 2010. CRISPR analysis of bacteriophage insenstitive mutants (BIMs) of Streptococcus thermophilus—implications for starter design. J Appl Microbiol 108:945–955. doi: 10.1111/j.1365-2672.2009.04486.x. [DOI] [PubMed] [Google Scholar]
- 54.Mills S, Coffey A, McAuliffe OE, Meijer WC, Hafkamp B, Ross RP. 2007. Efficient method for generation of bacteriophage insensitive mutants of Streptococcus thermophilus yoghurt and mozzarella strains. J Microbiol Methods 70:159–164. doi: 10.1016/j.mimet.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Harrington A, Hill C. 1991. Construction of a bacteriophage-resistant derivative of Lactococcus lactis subsp. lactis 425A by using the conjugal plasmid pNP40. Appl Environ Microbiol 57:3405–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gravesen A, Jydegaard Axelsen AM, Mendes da Silva J, Hansen TB, Knochel S. 2002. Frequency of bacteriocin resistance development and associated fitness costs in Listeria monocytogenes. Appl Environ Microbiol 68:756–764. doi: 10.1128/AEM.68.2.756-764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]