ABSTRACT
Vibrio cholerae is an important human pathogen and environmental microflora species that can both propagate in the human intestine and proliferate in zooplankton and aquatic organisms. Cholera is transmitted through food and water. In recent years, outbreaks caused by V. cholerae-contaminated soft-shelled turtles, contaminated mainly with toxigenic serogroup O139, have been frequently reported, posing a new foodborne disease public health problem. In this study, the colonization by toxigenic V. cholerae on the body surfaces and intestines of soft-shelled turtles was explored. Preferred colonization sites on the turtle body surfaces, mainly the carapace and calipash of the dorsal side, were observed for the O139 and O1 strains. Intestinal colonization was also found. The colonization factors of V. cholerae played different roles in the colonization of the soft-shelled turtle's body surface and intestine. Mannose-sensitive hemagglutinin (MSHA) of V. cholerae was necessary for body surface colonization, but no roles were found for toxin-coregulated pili (TCP) or N-acetylglucosamine-binding protein A (GBPA). Both TCP and GBPA play important roles for colonization in the intestine, whereas the deletion of MSHA revealed only a minor colonization-promoting role for this factor. Our study demonstrated that V. cholerae can colonize the surfaces and the intestines of soft-shelled turtles and indicated that the soft-shelled turtles played a role in the transmission of cholera. In addition, this study showed that the soft-shelled turtle has potential value as an animal model in studies of the colonization and environmental adaption mechanisms of V. cholerae in aquatic organisms.
IMPORTANCE Cholera is transmitted through water and food. Soft-shelled turtles contaminated with Vibrio cholerae (commonly the serogroup O139 strains) have caused many foodborne infections and outbreaks in recent years, and they have become a foodborne disease problem. Except for epidemiological investigations, no experimental studies have demonstrated the colonization by V. cholerae on soft-shelled turtles. The present studies will benefit our understanding of the interaction between V. cholerae and the soft-shelled turtle. We demonstrated the colonization by V. cholerae on the soft-shelled turtle's body surface and in the intestine and revealed the different roles of major V. cholerae factors for colonization on the body surface and in the intestine. Our work provides experimental evidence for the role of soft-shelled turtles in cholera transmission. In addition, this study also shows the possibility for the soft-shelled turtle to serve as a new animal model for studying the interaction between V. cholerae and aquatic hosts.
KEYWORDS: Vibrio cholerae, soft-shelled turtle, colonization, mannose-sensitive hemagglutinin, toxin-coregulated pili, N-acetylglucosamine-binding protein A
INTRODUCTION
Cholera is a life-threatening infectious disease that affects millions of people and kills thousands annually (1). Over 200 serogroups have been identified in Vibrio cholerae, but only the toxigenic O1 and O139 serogroups have been responsible for cholera epidemics to date. Seven cholera pandemics have been recorded historically. According to etiological evidence, the sixth pandemic was caused by the classical biotype of serogroup O1, and the seventh pandemic, which began in 1961 and is ongoing, was caused by the O1 El Tor biotype of serogroup O1 (2). In 1992, cholera caused by serogroup O139 emerged in India and spread to many countries in Asia (3, 4). In 1993, cholera from serogroup O139 was found in China, and since then, sporadic O139 cholera cases and local outbreaks have appeared in some provinces.
In addition to human hosts, V. cholerae naturally inhabits aquatic environments, particularly estuarine water, by colonizing zooplankton (5, 6). It can use chitin as a carbon and nitrogen source, which may also benefit the carbon and nitrogen cycles in aquatic environments (7). Water and seafood contaminated with V. cholerae are common sources of infections and outbreaks. In addition, foodborne infections and outbreaks of cholera, mainly of serogroup O139, mediated by soft-shell turtles have been frequently reported in China. Some investigations have reported that the rates of detection of V. cholerae on soft-shelled turtles were higher than for other aquatic products (8, 9). In some areas, the detection rate has reached 8.9% (9). Epidemiological and laboratory investigations of some cholera outbreaks showed that V. cholerae strains isolated from turtle samples had molecular subtyping patterns indistinguishable from those of patient strains, which suggested that the turtles served as the vehicle (10–13). V. cholerae was also found in soft-shelled turtles imported from Malaysia to China and from Bangladesh to Japan (14, 15). V. cholerae strains have been isolated from turtles' surfaces, cloacae, intestines, and eggs (9, 10, 13, 16–18). Taken together, these results provide strong evidence that V. cholerae can adhere to and colonize soft-shelled turtles. Thus, since no appearance or weight changes have been observed in turtles carrying V. cholerae, this colonization may result in the wide spread of the strains through markets.
Soft-shelled turtles are reptiles that inhabit standing or slow-flowing bodies of water, such as reservoirs, ponds, and rivers, and they are known as a nutrient-rich food in Asian countries, particularly, China (19). The large-scale breeding of soft-shelled turtles has been rapidly expanding, particularly, in eastern and southeastern China. Soft-shelled turtle consumption has reached approximately 100 to 150 million kilograms annually (20). Pelodiscus sinensis (commonly known as Chinese soft-shelled turtles), Palea steindachneri, and Pelochelys bibroni are three species of soft-shelled turtles in China. Among them, P. steindachneri and P. bibroni are rare animals protected by Chinese law, and only Chinese soft-shelled turtles can be sold in markets for consumption. The relationship of cholera cases or outbreaks to soft-shelled turtles has mainly been identified through epidemiological investigations and the isolation of strains. Direct observation of V. cholerae colonizing soft-shelled turtles is rare, except for an experiment that showed that V. cholerae could adhere to soft-shelled turtle eggshells and invade eggs (21). Such studies will provide new insight into the colonization of soft-shelled turtles by V. cholerae and will have profound significance for human health and foodborne infectious disease control and prevention.
Mannose-sensitive hemagglutinin (MSHA), toxin-coregulated pili (TCP), and N-acetylglucosamine-binding protein A (GBP) are three important factors of V. cholerae colonization that have been identified in animal models. MSHA, which is a member of the family of type IV pili and is associated with hemagglutinating activity, plays a significant role in biofilm formation on abiotic or biotic surfaces (22) and promotes adherence to zooplankton (23). No apparent role for MSHA in V. cholerae colonization of human or mouse intestines has been reported (24, 25). TCP is another type IV pilus that is necessary for colonization of the mammalian intestine and for pathogenesis (24, 26, 27). GBP is a chitin-binding protein that is involved in the attachment to chitin and mammalian intestine cells (28, 29). Although these three colonization factors have been widely studied, most studies are based on the mammalian intestine, zooplankton, and chitin surfaces. Their roles on the surfaces and in the intestines of soft-shelled turtles have not been investigated.
In this study, we estimated the colonization of the soft-shelled turtle by toxigenic V. cholerae. The colonization ability was certified on the body surface and in the intestine, and the roles of various colonization factors were identified. This study suggests that, aside from its epidemiological significance in the spread of cholera, the soft-shelled turtle can also be developed as a model for V. cholerae colonization.
RESULTS
Colonization of V. cholerae strains on the surface of the soft-shelled turtle.
To observe V. cholerae colonization on the soft-shelled turtle's surface and identify colonization sites, we constructed a bioluminescent strain, VC4251, of the V. cholerae serogroup O139 using the lux reporter plasmid pXEN-pmdh-luxCDABE. Soft-shelled turtles were infected via immersion in phosphate-buffered saline (PBS) containing approximately 107 CFU/ml bioluminescent cells. Because V. cholerae strains grow slowly in aquatic environments and too many rounds of anesthesia will lead to turtle deaths, bioluminescence was measured 24 h postinfection and observed every 24 h. At 96 h, the entire dorsal sides of the turtles emitted photons. Hence, the soft-shelled turtles were not analyzed for bacterial colonization after 96 h postinfection. After the bioluminescence imaging analysis, the surfaces of the soft-shelled turtles were sampled with sterile cotton swabs, and the experimental strains were obtained.
The surface bioluminescence imaging results showed that strong bioluminescent signals could be detected at 24 h and that the bioluminescence increased gradually with time (Fig. 1). Compared with that on the ventral sides, the dorsal sides emitted more photons. Photon emission was strongest and most stable on the calipashes during the experiments, showing that V. cholerae had the strongest ability to colonize the calipash. In addition, relatively strong bioluminescent signals could also be monitored on the dorsal sides of the carapaces, and these signals increased gradually with time (Fig. 1). The entire dorsal sides of the soft-shelled turtles' carapaces emitted photons at 96 h. The bioluminescent signals could also be detected on the dorsal sides of the limbs and necks. Poor photon emission was detected on the ventral sides, and the main bioluminescent sites on this side were on the limbs. Lower photon emission was detected on the plastrons, calipashes, and necks on the ventral sides (Fig. 1). A bioluminescent strain of V. cholerae serogroup O1 (VC1898) was also constructed based on the bioluminescent plasmid pXEN-pmdh-luxCDABE to estimate the colonization by V. cholerae O1 on soft-shelled turtles. Using the same infection and bioluminescence imaging procedures, the colonization by this serogroup O1 strain on the soft-shelled turtle surface was observed. The colonization sites and the expanded bioluminescent areas were similar to those of O139 strain VC4251.
FIG 1.
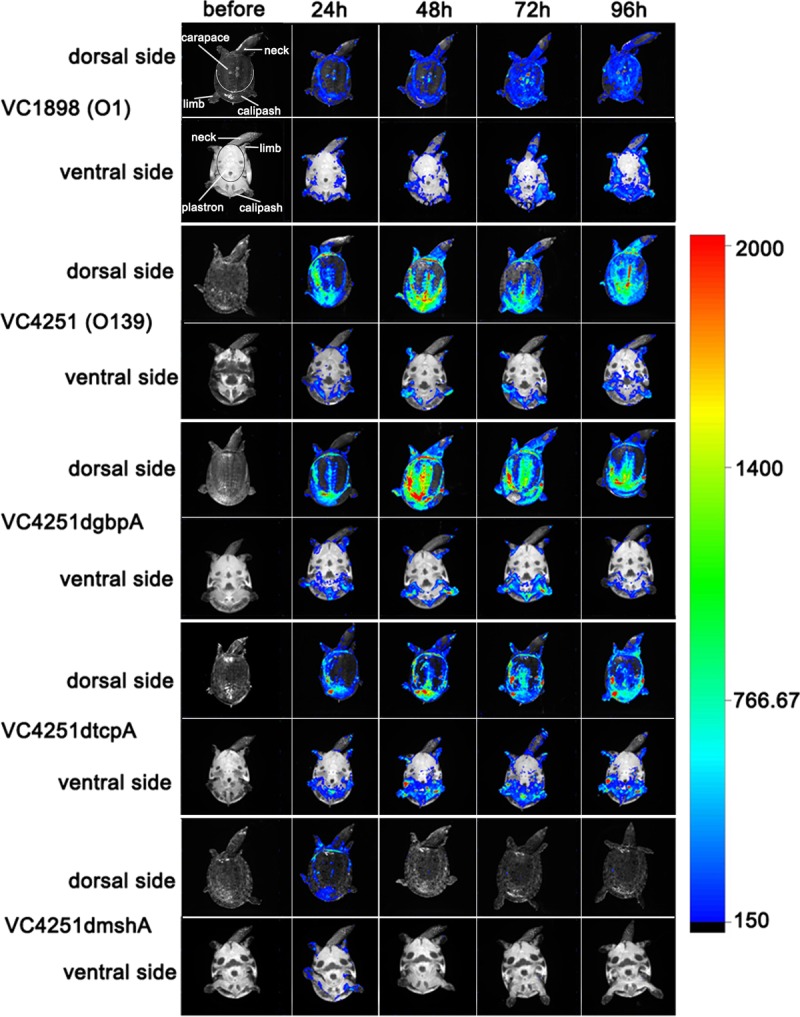
Body surface bioluminescence imaging of soft-shelled turtles infected with V. cholerae strains. The soft-shelled turtles were separately immersed in bacterial suspensions of bioluminescence-labeled O1 strain VC1898, O139 strain VC4251, and three VC4251-derived gene mutant bioluminescent strains. All the dorsal and ventral body surfaces were scanned before infection (as negative controls) and at 24, 48, 72, and 96 h postinfection. The figure shows one representative turtle for each tested strain. The color bar on the right shows the intensity of bioluminescence, representing increasing densities of bacteria. The anatomical structures on the dorsal side are labeled in the upper left corner image.
To determine the possible outer membrane factors of V. cholerae involved in its colonization on the body surfaces of soft-shelled turtles, mutations were made in the genes encoding TCP, MSHA, and GBP. The mutant strains were then tested for their ability to colonize and were compared with their corresponding isogenic wild-type strains. We first determined the colonization functions of these gene mutants with competition tests in the infant mouse intestine, the commonly used colonization model for evaluation of these factors in V. cholerae, and confirmed their effects for strain VC4251 (Fig. 2). In addition, no obvious growth defects were found between the wild-type strain and mutants carrying the pXEN-pmdh-luxCDABE plasmid, and all strains produced similar levels of bioluminescent light at the same time points (see Fig. S1C and D in the supplemental material). Using reverse transcription-PCR (RT-PCR) assays, the transcription of these three genes at 28°C was also confirmed, whereas the knockout mutants did not express mRNAs for the deleted genes (data not shown). These strains were used in the following experiments. Bioluminescence imaging of the turtles' body surfaces showed that in comparison to the wild-type strain, VC4251, the gbpA and tcpA mutant strains had similar photon emission trends, i.e., expanded bioluminescent areas on the dorsal sides and stable areas on the ventral sides of the turtles (Fig. 1). However, the mshA mutant strain had poor photon emission after infection, and after 48 h, photon emission from the strain could barely be detected on the soft-shelled turtles' surfaces (Fig. 1). These data suggested that compared with the wild-type strain, VC4251, the gbpA and tcpA mutant strains had similar colonization abilities on the body surfaces of the soft-shelled turtles, whereas the deletion of mshA led to a colonization defect.
FIG 2.
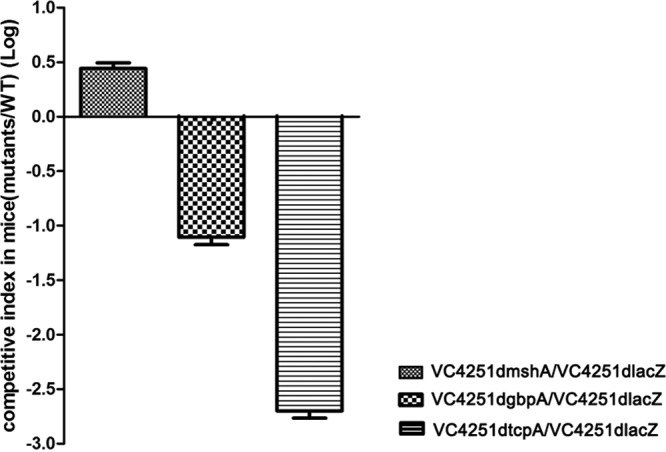
Small intestine colonization competition assays in infant mice with O139 strain VC4251dlacZ and mshA, gbpA, and tcpA deletion mutants from strain VC4251. The competition indexes were measured in the small intestines of infant mice; the y axis represents the log values of the competition index (mutant/VC4251dlacZ). The bars represent competition pairs of strain VC4251dlacZ and the mutants.
Colonization competition assays of V. cholerae on the turtle surface.
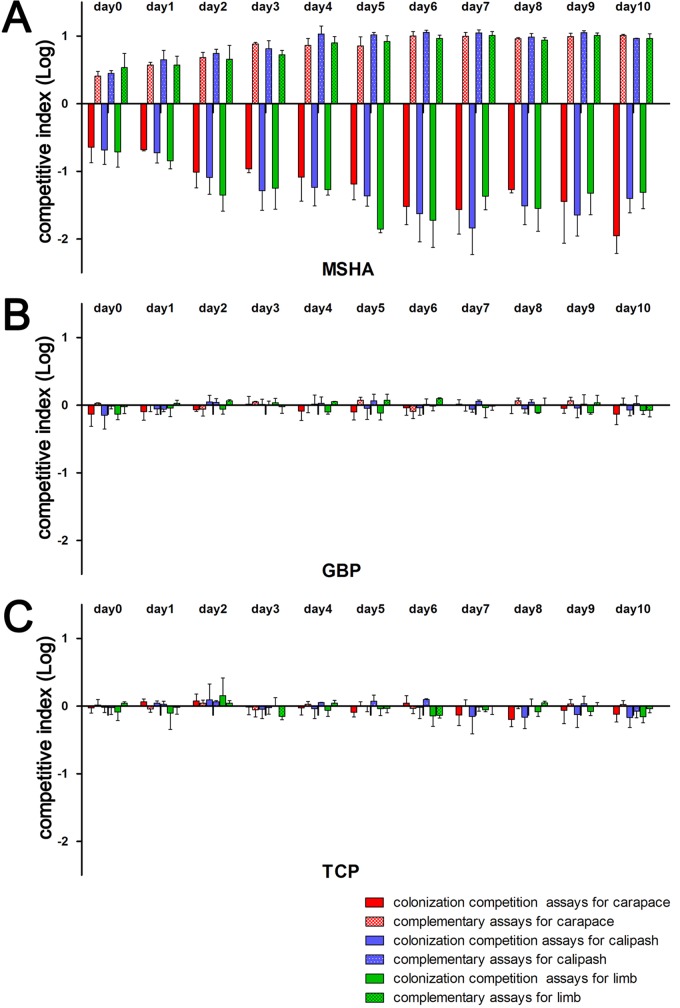
To accurately evaluate the colonization abilities of VC4251dlacZ and mshA, gbpA, and tcpA mutant strains, colonization competition assays were performed. According to the surface bioluminescence imaging results, the main colonization sites for V. cholerae on soft-shelled turtles' surfaces (the dorsal side carapace, calipash, and limbs) were sampled in the colonization competition assays. We measured the optical density at 600 nm (OD600) of these strains in LB medium, and no obvious growth difference was found between the VC4251 wild-type strain and any mutant (Fig. S1C). According to surface bioluminescence imaging, the dorsal side carapace, calipash, and limbs were colonized; t tests were performed to compare the differences between the output ratios and input ratios for each competition strain pair. The results showed that for gbpA and tcpA mutants, from day 0 to day 10 on the carapace, calipash, or limbs, no significant differences between the input and output ratios of the competition pairs (P > 0.05 by t test) were observed. The competitive indexes of the gbpA and tcpA mutants were always close to 1 (Fig. 3, shown with the log10 values). However, for the mshA mutant strain, significant colonization differences were observed on the carapaces, calipashes, and limbs (P < 0.05 by t test). From day 2, the competitive indexes were relatively stable and showed that loss of mshA decreased colonization by 10- to 81-fold compared with that of the wild type (Fig. 3).
FIG 3.
Colonization competition and complementary assays of V. cholerae strains on turtle body surfaces. Competition indexes were measured at different sites on the soft-shelled turtles' body surfaces. Colonization competition and complementary assays for mshA (A), gbpA (B), and tcpA (C) strains. The y axis represents log values of the competitive index [mutants/VC4251dlacZ or complementation strains/VC4251dlacZ(pBAD)]. Bars with different colors indicate competitive indexes for different competition pairs. For the mshA gene competition and complementary assays, the results always showed statistically significant differences (day 0 to day 10, regardless of carapace, calipash, or limbs; all P < 0.05 by t test). For the gbpA and tcpA genes, no competition pairs showed significance (all P > 0.05).
We also tested the colonization abilities of the three colonization factor gene mutants which were restored by introducing the plasmids pBADmshA, pBADgbpA, and pBADtcpA to express mshA, gbpA, and tcpA, respectively, on the carapaces, calipashes, or limbs, from day 0 to day 10. The colonization ability for the mshA-complemented strain was restored and even 2.5- to 11.4-fold higher than for VC4251dlacZ(pBAD), the wild-type strain characterized by the lack of lacZ and carrying the control plasmid pBAD24, on the carapace, calipash, and limbs (Fig. 3). For gbpA and tcpA complementary strains, the colonization competitive indexes were always close to 1 and showed no colonization differences from strain VC4251dlacZ(pBAD) (P > 0.05 by t test).
Colonization of V. cholerae in turtle intestine.
To detect whether V. cholerae can colonize the intestines of soft-shelled turtles, the animals were intragastrically inoculated with the bioluminescent O139 strain, VC4251. It was previously found that the food digestion time of soft-shelled turtles weighing approximately 150 g was between 34.03 and 56.56 h at 28°C (30). Thus, we chose 72 h as the colonization time. Because soft-shelled turtles have thick carapaces, bioluminescent signals could not be directly detected by our in vivo imaging system. Thus, at 72 h postinfection, the soft-shelled turtles were euthanized, and the internal organs, including the hearts, livers, lungs, kidneys, spleens, and digestive tracts, were separated and placed on the observation platform of the imaging system. In vivo bioluminescence imaging assays showed that bioluminescent signals could only be detected in the intestines (Fig. 4), and the V. cholerae strain was isolated only from these segments. No bioluminescent photons were emitted from other organs and no V. cholerae strains were isolated in other organs.
FIG 4.
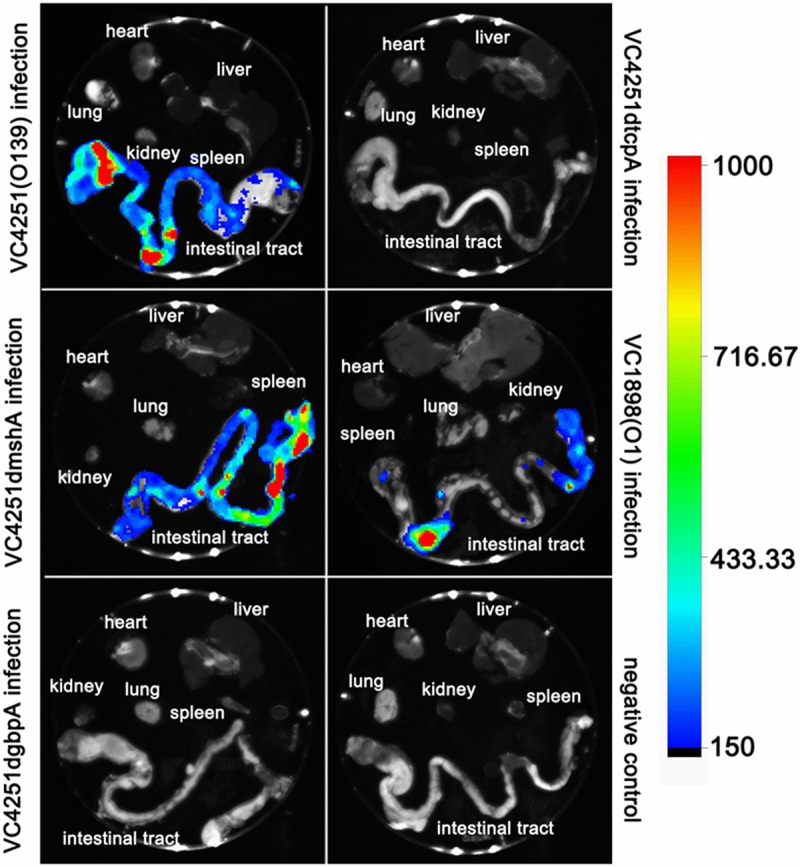
Bioluminescence imaging of organs of soft-shelled turtles intravenously infected with bioluminescence-labeled V. cholerae strains. Six groups of soft-shelled turtles (three turtles per group) were intravenously infected with bioluminescence-labeled serogroup O1 strain VC1898, serogroup O139 strain VC4251, and VC4251-derived gene mutant bioluminescent strains. The negative control was injected with sterile PBS. All soft-shelled turtles were euthanized at 72 h, and internal organs, including the hearts, livers, lungs, spleens, kidneys, and digestive tracts, were separated and scanned. The experiments were repeated three times, and this figure shows one representative test. The color bar on the right shows bioluminescence intensity, representing increasing densities of bacteria. Anatomical structures are labeled in each picture.
To determine the colonization by the O1 serogroup V. cholerae strain and the possible outer membrane factors involved in the in vivo colonization of soft-shelled turtles, the turtles were also intragastrically inoculated with the bioluminescent-labeled serogroup O1 strain, VC1898, and all VC4251-derived gene mutant bioluminescent strains. For the bioluminescence-labeled serogroup O1 strain, VC1898, and VC4251dmshA gene mutant bioluminescent strains, the same colonization sites (i.e., only in the intestinal tracts) were suggested by bioluminescence imaging and bacterial isolation. However, for the VC4251dgbpA and VC4251dtcpA gene mutant bioluminescent strains, no bioluminescent photons were emitted from the intestines, and relatively few VC4251dgbpA or VC4251dtcpA strains were isolated. In other organs, both the bioluminescence imaging detection results and bacterial isolation results were negative.
Colonization competition assays of V. cholerae in the turtle intestine.
To accurately evaluate the roles of MSHA, TCP, and GBP in the intestinal colonization by V. cholerae, colonization competition assays were performed by coinfection of the strain VC4251dlacZ and tcpA, mshA, and gbpA deletion mutants. After 72 h, the entire intestines were isolated for plate counting of the competition strain pairs. Similar competition effects were observed with the gbpA and tcpA groups, and the comparisons between the ratios of the inputs and outputs were all statistically significant (P < 0.05 by t tests). Competition index calculations showed that mutations of gbpA and tcpA decreased the colonization abilities of the mutants by 44- and 225-fold, respectively (Fig. 5, shown with the log10 values). The colonization abilities of the gbpA and tcpA mutants were restored by the complemented plasmids pBADgbpA and pBADtcpA, respectively, and were 2.69- and 3.57-fold higher than that for VC4251dlacZ(pBAD), respectively, as deduced by the overexpression of these two complementary genes resulting from the strong promoter induced by arabinose. For mshA, comparisons of the ratios between the inputs and outputs of the VC4251dmshA-VC4251dlacZ pairs were statistically significant (P < 0.05 by t test). The competition index analysis showed that colonization by the mshA mutant was 3.71-fold higher than that by the wild-type strain and 1.96-fold lower when mshA was complemented (Fig. 5).
FIG 5.
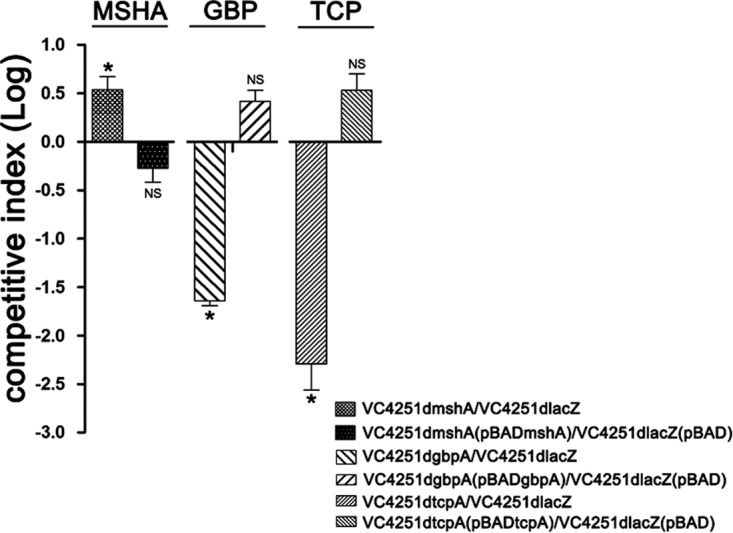
Colonization competition assays and complementary assays of V. cholerae strains in turtle intestines. Competition indexes were calculated for isolated intestines of soft-shelled turtles. The y axis represents the log values of the competitive index [mutants/VC4251dlacZ or complementation strains/VC4251dlacZ(pBAD)]. The columns indicate competitive indexes for different competition pairs. *, P < 0.05 by t test). NS, not significant.
DISCUSSION
V. cholerae has two typical living environments, namely, an aquatic habitat and the human intestine (31, 32). The organism possesses different colonization and proliferation mechanisms in each environment (33). Soft-shelled turtles have already caused several foodborne cholera outbreaks (10–13) and have become a notable spreading factor for cholera. In this study, we demonstrated that the soft-shelled turtle could be colonized by V. cholerae on the surface and in the intestine, therefore playing a role in V. cholerae transmission as a medium.
Using bioluminescence-labeled V. cholerae strains and bioluminescence imaging techniques, the colonization sites on the soft-shelled turtle surface and the proliferation of V. cholerae were observed directly and successively. Combined with competition assays, these data were used to estimate the roles of the colonization factors of V. cholerae on the soft-shelled turtle surface and in the digestive tract. Both methods showed good repeatability. In V. cholerae, the animal models commonly used for colonization and pathogenesis studies are infant mice and rabbits, as well as the ileal loop of the adult rabbit (34–37). Compared with those using mammalian animal models, studies with soft-shelled turtles provide a new method for evaluating V. cholerae colonization on aquatic animal surfaces; animals can be continuously observed without euthanization. In addition, zooplankton, fish, shellfish, chironomid egg masses, waterfowl, and crustaceans are environmental hosts of V. cholerae (6, 38–43), and fish and shellfish are proven vectors of V. cholerae (44). In studies of colonization in aquatic organisms, soft-shelled turtles can be anesthetized and sampled out of water for relatively long periods without the risk of hydropenia and death, due to their amphibious nature, making them a better model than zebrafish and other fish or shrimp models. Therefore, it should be possible to develop specific-pathogen-free soft-shelled turtles as a model for studying V. cholerae survival mechanisms in aquatic environments.
From bioluminescence imaging assays, we found the colonizations by V. cholerae on soft-shelled turtles surfaces were uneven and that the main colonization sites were the carapace, calipash, limbs, and the dorsal side of the neck. Soft-shelled turtle muscle and skin (45), particularly the calipash (46), are highly abundant in collagen. V. cholerae produces collagenase (47), and collagen may be a nutrient source for the growth of V. cholerae. The carapace and plastron have different anatomical and physiological structures, which possibly lead to the different colonization abilities of V. cholerae at these two sites.
MSHA, TCP, and GBP are the major colonization factors of V. cholerae and are recognized as such in mouse models (24, 26–29) and in zooplankton (23). In the soft-shelled turtle's intestine and on the body surface, we found different effects of these factors. In the soft-shelled turtle's intestine, effects similar to those seen in the mouse intestine were observed for these colonization factors. TCP was important in the colonization by V. cholerae in the turtle intestine, similar to that in the mouse model (24–26). MSHA pili showed an extremely important role in colonization on the turtle body surface. In the bioluminescence imaging assays, the weak bioluminescence of the mshA mutant strain was found in the first 24 h but hardly observed later, showing the colonization loss caused by MSHA. We suspected it may represent nonspecific binding in the first 24 h because of the higher bacterial concentration, and the mutant with the defective colonization was washed away, since every 24 h the turtles were washed and the sterile PBS was replaced during the experiments. The complement of intact mshA to the mshA mutant restored the colonization ability and produced a slightly increased colonization. MshA is the major pilin subunit of the MSHA pili, and we suspect that the overexpressed MshA from the complementary plasmid could promote the assembly of MSHA pili and result in the slightly increased colonization compared with that of the wild-type strain. However, in the intestine, this appeared to depress colonization. A similar effect was also found in the mouse model (25). In the suckling mouse intestine, MSHA pili bound secretory immunoglobulin A (sIgA) and reduced the colonization ability of V. cholerae by preventing the bacteria from penetrating the mucus barrier and adhering to epithelial cells (48). As reptiles, soft-shelled turtles can also produce sIgA (49), and reptilian sIgA is evolutionarily related to mammalian sIgA (50). V. cholerae has a low level of MSHA gene expression in the intestine (48). Therefore, we hypothesize that the slightly lower colonization of the wild type compared with that of the mshA deletion mutant might result from the interaction between sIgA and weakly expressed MSHA in the wild-type strain; cells could bind to sIgA to some degree, whereas the mshA mutant should lack MSHA pili.
Compared with that of the wild-type strain, the colonization ability of the gbpA deletion mutant obviously decreased in the soft-shelled turtle intestine, similar to that in the mouse intestine colonization model (51). However, on the body surfaces of soft-shelled turtles, inactivation of gbpA has no significant effect on colonization. Previous results have shown that deletion of the gbpA gene of V. cholerae led to a significant decrease in colonization on the shells of crustaceans, such as crab, shrimp, and shellfish (28, 29). Chitin is the main element of crustacean shells (52, 53), and its β-1,4-linked N-acetylglucosamine (GlcNAc) residues are receptors for GBP-mediated binding (54). Thus, GBP interacts with the chitinous exoskeleton and promotes the colonization ability of V. cholerae. Soft-shelled turtles are reptiles with endoskeletons and are distinct from crustaceans with exoskeletons (55). Epidermis, dermis, and subcutaneous tissues cover the outside of the carapace skeleton, and these tissues are rich in collagen (56). GBP had no effect on the V. cholerae colonization of soft-shelled turtle shells, possibly because of this difference in structure and material composition.
In summary, we studied the colonization ability, colonization sites, and roles of major colonization-related factors for V. cholerae on soft-shelled turtles. Soft-shelled turtles' surfaces and intestines carried V. cholerae and allowed it to proliferate. These outer membrane accessory structures play different but important and even aquatic animal-specific roles in the colonization by V. cholerae on the body surfaces and in the intestines of soft-shelled turtles. The results from this study strongly implicate soft-shelled turtles in cholera transmission in areas with high soft-shelled turtle consumption. These soft-shelled turtles are thus an important risk factor for foodborne cholera outbreaks. In addition, soft-shelled turtles could be developed as a valuable animal model to study the interaction between V. cholerae and aquatic hosts.
MATERIALS AND METHODS
Bacterial strains.
V. cholerae serogroup O139 strain VC4251 and serogroup O1 strain VC1898 were isolated from soft-shelled turtles in markets in Jiangxi and Guizhou provinces in China, respectively. Both strains have natural resistances to streptomycin (100 μg/ml) and polymyxin B (5 μg/ml). The mshA, gbpA, tcpA, and lacZ mutants were constructed from strain VC4251. A list of all strains used in this study is presented in Table 1.
TABLE 1.
Bacterial strains used in this work
| Strain | Relevant propertiesa | Source |
|---|---|---|
| VC1898 | O1 serogroup, Smr, PBr | Lab strain |
| VC4251 | O139 serogroup, Smr, PBr | Lab strain |
| VC4251dmshA | Strain VC4251 with mshA deleted | This work |
| VC4251dgbpA | Strain VC4251 with gbpA deleted | This work |
| VC4251dtcpA | Strain VC4251 with tcpA deleted | This work |
| VC4251dlacZ | Strain VC4251 with lacZ deleted | This work |
| VC4251dlacZ(pBAD) | VC4251dlacZ containing pBAD24 | This work |
| VC4251dmshA(pBADmshA) | VC4251dmshA containing pBADmshA | This work |
| VC4251dgbpA(pBADgbpA) | VC4251dgbpA containing pBADgbpA | This work |
| VC4251dtcpA(pBADtcpA) | VC4251dtcpA containing pBADtcpA | This work |
| VC4251(pXEN-pmdh-luxCDABE) | VC4251 containing pXEN-pmdh-luxCDABE | This work |
| VC4251dmshA(pXEN-pmdh-luxCDABE) | VC4251dmshA containing pXEN-pmdh-luxCDABE | This work |
| VC4251dgbpA(pXEN-pmdh-luxCDABE) | VC4251dgbpA containing pXEN-pmdh-luxCDABE | This work |
| VC4251dtcpA(pXEN-pmdh-luxCDABE) | VC4251dtcpA containing pXEN-pmdh-luxCDABE | This work |
| SM10 λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir | Lab strain |
Sm, streptomycin; PB, polymyxin B.
These strains were obtained from swab samples of the turtles and identified according to the Manual for Prevention and Control of Cholera, 6th edition (57). The serogroups were determined with diagnosis typing sera (Wantai Biotech, Zhengzhou, China). The Escherichia coli strain SM10 λpir was constructed from SM10 (58) by lysogenizing with λpir to allow the replication of an R6K suicide plasmid (http://www.openwetware.org/wiki/E._coli_genotypes) used to construct a mutant allele in allelic exchange to create a gene mutation, which was obtained from the strain collection in our lab.
Soft-shelled turtles.
Soft-shelled turtles were obtained from an aquaculture company in Shanghai. Turtles weighing 150 g and 9 cm in length (diameter of calipash) were selected for use in these studies. To ensure that the turtles used in this study did not carry V. cholerae before the experiments, we screened the turtles with PCR and with isolation for V. cholerae. Surface and anus swabs were collected from the turtles. PCR for V. cholerae was performed as previously described (ompW and ctxA were detected as the targets of V. cholerae [59]). The surface and anus swabs were also placed in alkaline peptone water for V. cholerae enrichment, and V. cholerae was isolated and identified according to the Manual for Prevention and Control of Cholera, 6th edition (57). Turtles positive for any gene in the PCR and/or isolation were excluded, and turtles negative for both detections were used in subsequent experiments.
Each selected soft-shelled turtle was carefully washed with sterile distilled water and kept in sterile distilled water for 48 h to remove mud from the surface and intestine. The soft-shelled turtles were kept in sterile PBS for another 48 h to maintain osmotic pressure for the subsequent experiments. For anesthesia, the soft-shelled turtles were placed in a sealed beaker with five ether cotton balls (containing 10 ml of ether) for 10 min. For euthanasia, the dose of ether was doubled, and the soft-shelled turtles were kept in the sealed beaker for 60 min.
This study was performed in strict accordance with animal protocols approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention, China CDC, in accordance with the medical research regulations of the Ministry of Health, China.
Construction of bioluminescent V. cholerae strains.
To enhance the expression of luxCDABE and bioluminescence intensity, the promoter of the housekeeping gene mdh (malate dehydrogenase gene), a highly transcribed gene (by transcriptome analysis, data not shown), from strain VC4251 was amplified (primers are listed in Table 2). The amplicons were digested with KpnI and BamHI and cloned into pXEN-luxCDABE (PerkinElmer) through the multiple cloning site, to generate the recombinant plasmid pXEN-pmdh-luxCDABE. O1 strain VC1898, O139 strain VC4251, and all mutants generated from strain VC4251 were transformed with the plasmid pXEN-pmdh-luxCDABE. These strains were plated on LB (Luria-Bertani) agar plates containing ampicillin (100 μg/ml), and lux-expressing clones were selected with the In-vivo FX Pro imaging system (Bruker) and were verified by sequencing.
TABLE 2.
Primers for bioluminescent plasmid and mutant strain construction
| Primer | Sequencea | Restriction enzyme |
|---|---|---|
| gbpA-1 | 5′-CCGCTCGAGAGCGCATGGTGCTCCTTA-3′ | XhoI |
| gbpA-2 | 5′-AGGATAACTTCACAGACTCTTCTTTGTTAGCTG-3′ | |
| gbpA-3 | 5′-AGAGTCTGTGAAGTTATCCTCCCTCTTACA-3′ | |
| gbpA-4 | 5′-ATTTGCGGCCGCGCCTTGGGATGTTCTACG-3′ | NotI |
| mshA-1 | 5′-CCGCTCGAGGGTGGAACTGGTCATCGT-3′ | XhoI |
| mshA-2 | 5′-AGCCTATGTCCTCTCTTTCATGTGAATACGCA-3′ | |
| mshA-3 | 5′-TGAAAGAGAGGACATAGGCTTCAATGGTTA-3′ | |
| mshA-4 | 5′-ATTTGCGGCCGCGCCTAAACTATCGAAATCAA-3′ | NotI |
| tcpA-1 | 5′-CCCTCGAGTCCACAGTCAAAGTGAC-3′ | XhoI |
| tcpA-2 | 5′-GTGATATTAGATTTATATAACTCCACCATTTGT-3′ | |
| tcpA-3 | 5′-TTATATAAATCTAATATCACGCATGTTGAG-3′ | |
| tcpA-4 | 5′-ATTTGCGGCCGCTAATCAGCATCACAAGCA-3′ | NotI |
| lacZ-1 | 5′-CCGCTCGAGCCACCACGATGATAACCAAT-3′ | XhoI |
| lacZ-2 | 5′-GAGTGAGCAACCCTCAAGCCGAGGAGTAAA-3′ | |
| lacZ-3 | 5′-GGCTTGAGGGTTGCTCACTCAGCCGCACTA-3′ | |
| lacZ-4 | 5′-ATTTGCGGCCGCGCGAACAGGCGATGACTAAC-3′ | NotI |
| prom-mdh-F | 5′-CGGGGTACCAGCCAAAGCGTTCTTCTTTTAGTAA-3′ | KpnI |
| prom-mdh-R | 5′-CGCGGATCCCGTAAATCTCCTTGAGAGTAATCTC-3′ | BamHI |
| gbpC-F | 5′-CCGGAATTCATGAAAAAACAACCTAAAATGACCG-3′ | EcoRI |
| gbpC-R | 5′-CTAGTCTAGATTAACGTTTATCCCACGCCATTTCC-3′ | XbaI |
| mshC-F | 5′-CCGGAATTCATGGTAATAATGAAAAGACAAGGTG-3′ | EcoRI |
| mshC-R | 5′-CTAGTCTAGATTATTGCGCTGGTTTACCACAAGCA-3′ | XbaI |
| tcpC-F | 5′-CCGGAATTCATGCAATTATTAAAACAGCTTTTTA-3′ | EcoRI |
| tcpC-R | 5′-CTAGTCTAGATTAACTGTTACCAAAAGCTACTGTG-3′ | XbaI |
Restriction endonuclease sites are underlined.
Construction of mutants.
The mshA mutant strain was constructed from strain VC4251 using a homologous recombination method. We designed primers for a region upstream of the mshA gene (mshA-1 and mshA-2) and introduced a XhoI endonuclease recognition site into the primer mshA-1. Downstream primers (mshA-3 and mshA-4) were also designed, and a NotI endonuclease recognition site was introduced into the primer mshA-4. The upstream and downstream DNA fragments flanking the target sequences within mshA were amplified from VC4251 genomic DNA using the primer pairs mshA-1/mshA-2 and mshA-3/mshA-4, respectively. Because portions of the primers mshA-2 and mshA-3 were complementary to one another, the upstream and downstream amplicons were mixed at equimolar concentrations and used as the template to amplify the chromosomal fragment, excluding the target-deleted fragment, using the primer pair mshA-1/mshA-4. After digestion with XhoI and NotI, the resulting fragment was cloned into the suicide plasmid, pWM91, containing a sacB counterselectable marker (60) to generate the plasmid pWM91-ΔmshA. The resulting suicide plasmid was maintained in E. coli SM10 λpir and transformed to the strain VC4251 by conjugation. The transconjugants were selected on LB agar plates containing streptomycin (100 μg/ml) and ampicillin (100 μg/ml), and the clones were streaked on LB agar plates with 8% sucrose and without NaCl at 22°C. After 48 h, double-crossover recombination mutants were selected and verified by sequencing to generate the mutant strain, VC4251dmshA. The gbpA, tcpA, and lacZ mutants were constructed using the same method described above. The primers used in the mutant construction of these genes are listed in Table 2.
Construction of complementary strains.
The complementation plasmids carrying mshA, gbpA, and tcpA were constructed with plasmid pBAD24 (61). The open reading frames (ORFs) of mshA, gbpA, and tcpA were amplified from VC4251 chromosomal DNA using the primer pairs mshC-F/mshC-R, gbpC-F/gbpC-R, and tcpC-F/tcpC-R, respectively. After digestion with EcoRI and XbaI, the resulting fragments were cloned downstream of the pBAD promoter in the plasmid pBAD24 (as a result, all transcriptions of these genes were driven by the pBAD promoter), to generate the complementation plasmids pBADmshA, pBADgbpA, and pBADtcpA, respectively, which were verified further by sequencing. Then, the mutant strains VC4251dmshA, VC4251dgbpA, and VC4251dtcpA were transformed with the complementation plasmids pBADmshA, pBADgbpA, and pBADtcpA, respectively, to generate the complementation strains VC4251dmshA(pBADmshA), VC4251dgbpA(pBADgbpA), and VC4251dtcpA(VC4251dtcpA). Strain VC4251dlacZ was transformed with the control plasmid pBAD24 to generate strain VC4251dlacZ(pBAD).
Small intestine colonization competition assays in infant mice.
VC4251dlacZ and mshA, gbpA, and tcpA mutant strains were cultured overnight on LB agar plates at 37°C. The colonies were scraped and resuspended in 3 ml of PBS to an OD600 of 1.0 (the amount of each strain was measured by plate counting), which was approximately equal to 109 CFU/ml. Approximately 105 cells of mshA, gbpA, and tcpA mutants were mixed with VC4251dlacZ at a 1:1 ratio and were inoculated intragastrically into 5-day-old CD-1 infant mice. The mixtures were diluted for enumeration to calculate the input ratios (mutant to wild type). After 24 h, the mice were euthanized and the small intestines were removed and homogenized. Serial dilutions were made and plated on LB agar plates supplemented with polymyxin B (5 μg/ml), streptomycin (100 μg/ml), and X-Gal ([5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] 50 μg/ml) for enumeration and calculation of the output ratios (mutant to wild type). Referring to the colony counting method from the previous study (62), the lacZ mutant strain was also constructed from strain VC4251. Because lacZ-positive V. cholerae strains (all colonization gene mutants, including VC4251dmshA, VC4251dgbpA, and VC4251dtcpA) show blue colonies on LB agar plates in the presence of X-Gal and lacZ-negative V. cholerae strain colonies are white, the colony numbers of different strains for colonization competition assays can be calculated directly. The competitive index was calculated from the output ratio and normalized against the input ratio. Three mice were used for each group of competition assays.
Turtle body surface bioluminescence imaging assays.
The bioluminescence-labeled serogroup O1 strain VC1898, serogroup O139 strain VC4251, and VC4251-derived mshA, gbpA, and tcpA mutants were cultured overnight on LB agar plates containing 100 μg/ml ampicillin at 37°C. The colonies were scraped and resuspended in 30 ml of PBS containing 100 μg/ml ampicillin to obtain an OD600 value of 1.0 (the amount of each strain was measured by plate counting), which was approximately 109 CFU/ml. The suspensions were diluted in 3 liters of PBS containing 100 μg/ml ampicillin. Soft-shelled turtles were immersed in the bacterial suspension for 2 h for surface colonization. Each soft-shelled turtle was washed twice with 2 liters of sterile PBS to remove cells that were not attached. The soft-shelled turtles were kept in 3 liters of sterile PBS containing 100 μg/ml ampicillin at room temperature (28°C). At 24, 48, 72, and 96 h postinfection, the soft-shelled turtles were washed and placed in fresh sterile PBS containing 100 μg/ml ampicillin. After administration of anesthesia, the soft-shelled turtles were analyzed for bioluminescent bacteria with an In-vivo FX Pro imaging system. Bioluminescent signals were also detected before infection as negative controls. After bioluminescence imaging, the surfaces of the soft-shelled turtles were sampled with sterile cotton swabs for V. cholerae isolation. In these assays, five turtles were immersed in the bacterial suspensions of the above five strains, and the tests were repeated three times (15 turtles in total were used).
Body surface colonization competition assays.
The mshA, gbpA, and tcpA mutants and VC4251dlacZ were cultured overnight on LB agar plates at 37°C, and the colonies were scraped and resuspended in PBS to an OD600 of 1.0. VC4251dlacZ was mixed with the mshA, gbpA, and tcpA mutants at a 1:1 ratio. The mixtures were diluted for enumeration to calculate the input ratios. Then, 30 ml of the mixtures was diluted with 3 liters of PBS. The soft-shelled turtles were immersed in the bacterial suspension for 2 h for surface infection; the turtles were separated into three groups to detect the three gene mutants. Each soft-shelled turtle was washed twice with 2 liters of sterile PBS to remove cells that were not attached. The dorsal carapace, calipash, and limbs of each turtle were sampled with sterile cotton swabs. The swabs were washed with 300 μl of sterile PBS. Serial dilutions were made and plated on LB agar plates supplemented with polymyxin B (5 μg/ml), streptomycin (100 μg/ml), and X-Gal (100 μg/ml) for enumeration and calculation of the output ratios (this detection time point was assigned as day-0 time point). The soft-shelled turtles were then kept in 3 liters of sterile PBS at 28°C. Every 24 h postinfection, the soft-shelled turtles were washed, sampled, and placed in fresh sterile PBS. Bacterial enumerations were performed as described above to calculate the output ratios. The detections were continuously performed for 10 days (day-1 time point to day-10 time point). The competitive index was calculated from the output ratio and normalized against the input ratio. Three soft-shelled turtles were used for each group in the competition assays, and the assays were repeated three times.
In the competition assays using strain VC4251dlacZ(pBAD) and the complementation strains VC4251dmshA(pBADmshA), VC4251dgbpA(pBADgbpA), and VC4251dtcpA(pBADtcpA), the LB agar plates and PBS were supplemented with ampicillin and arabinose to final concentrations of 100 μg/ml and 0.5%, respectively, to culture these strains. LB agar plates for the selective isolation of these strains were supplemented with the following components: ampicillin (final concentration of 100 μg/ml), polymyxin B (5 μg/ml), streptomycin (100 μg/ml), and X-Gal (50 μg/ml). Other procedures of the competition assay with these strains were performed as described above.
Bioluminescence imaging assays of the internal organs.
The bioluminescence-labeled serogroup O1 strain VC1898, serogroup O139 strain VC4251, and VC4251-derived mshA, gbpA, and tcpA mutant bioluminescent strains were cultured overnight on LB agar plates containing 100 μg/ml ampicillin at 37°C. The next day, the strains were scraped and resuspended in 3 ml of PBS to obtain an OD600 of 1.0. Soft-shelled turtles were anesthetized as described above. Next, 200-μl bacterial suspensions were inoculated intragastrically into the soft-shelled turtles' stomachs. To ensure the entry of all bacterial suspensions into the stomachs, polyethylene tubes that were at least 15 cm long were inserted into the turtles' mouths. In the assays, six turtles were inoculated intragastrically with the above six strains, and the assays were repeated three times (18 turtles in total were used).
The turtles were cultured in 3 liters of sterile PBS containing 100 μg/ml ampicillin at 28°C. Every 24 h postinfection, the soft-shelled turtles were placed in new sterile PBS containing 100 μg/ml ampicillin. At 72 h, the soft-shelled turtles were euthanized, and the internal organs (including the hearts, livers, lungs, spleens, kidneys, and digestive tracts) were analyzed for bioluminescent bacteria. Bioluminescent signals were also detected for the soft-shelled turtles inoculated with sterile PBS as negative controls. The internal organs were homogenized with 5 ml of sterile PBS. Serial dilutions were made and plated on selective LB agar plates supplemented with polymyxin B (5 μg/ml), streptomycin (100 μg/ml), and ampicillin (100 μg/ml) for V. cholerae isolation.
Intestinal colonization competition assays.
In the competition assays with the gene mutant strains/VC4251dlacZ and complementary strains/VC4251dlacZ(pBAD), the strains were cultured overnight on LB agar plates (100 μg/ml ampicillin was used when strains carrying plasmids were cultured) at 37°C. The next day, the colonies were scraped and resuspended in 3 ml of PBS to obtain an OD600 of 1.0. The paired strains used for the competition assays were mixed at approximately a 1:1 ratio. The mixtures were diluted for enumeration to calculate the input ratios. Soft-shelled turtles were inoculated with 200 μl of the bacterial suspensions and kept as described above. At 72 h, the soft-shelled turtles were euthanized, the intestines were aseptically removed, and the intestinal tubes were washed with sterile PBS three times to remove cells that were not attached and homogenized with 5 ml of sterile PBS. Serial dilutions were made and plated on LB agar plates supplemented with the corresponding antibiotics as above and X-Gal for enumeration of the corresponding strains and the calculation of output ratios. The competitive index was calculated from the output ratio and normalized against the input ratio. Three soft-shelled turtles were used for each competition strain pair in these colonization competition assays, and the assays were repeated three times.
Accession number(s).
The gene sequences of strain VC4251 which were sequenced in this study were deposited in GenBank with the accession numbers MF002592 (gbpA), MF002593 (tcpA), MF002594 (mshA), and MF002595 (lacZ).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation (grant no. 81401715) and State Key Laboratory of Infectious Disease Prevention and Control (grant no. 2014SKLID101).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00713-17.
REFERENCES
- 1.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. 2012. Cholera. Lancet 379:2466–2476. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaper JB, Morris JG Jr, Levine MM. 1995. Cholera. Clin Microbiol Rev 8:48–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert MJ, Siddique AK, Islam MS, Faruque AS, Ansaruzzaman M, Faruque SM, Sack RB. 1993. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet 341:704. doi: 10.1016/0140-6736(93)90481-U. [DOI] [PubMed] [Google Scholar]
- 4.Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Nair GB, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhury MA, Huq A, Xu B, Madeira FJ, Colwell RR. 1997. Effect of alum on free-living and copepod-associated Vibrio cholerae O1 and O139. Appl Environ Microbiol 63:3323–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol 45:275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. 2004. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci U S A 101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang ZR, Zhang J, Wang DC, Zhong HJ, Xu J, Ran L, Wang MW, Wang ZJ, Kan B. 2007. Identification and molecular study on vibrio cholerae in sea products. Zhonghua Yu Fang Yi Xue Za Zhi 41:304–306. (In Chinese.) [PubMed] [Google Scholar]
- 9.Lu HK, Chen EF, Xie SY, Chai CL, Wei YD, Mo ST, Ye JL, Luo Y. 2006. Investigation on vibrio cholera carried in aquatic products of littoral areas, Zhejiang Province. Zhonghua Yu Fang Yi Xue Za Zhi 40:336–338. (In Chinese.) [PubMed] [Google Scholar]
- 10.Jiang Z, Zhou L, Wang X. 2007. Pathogenic research on cholera epidemic caused by infected soft-shell turtle. Xian dai Yu fang Yi xue 22:069 (In Chinese.) [Google Scholar]
- 11.Long Z, Hu S, Deng Z. 2010. Epidemic factors and rapid assessment of Vibrio cholerae O139 in Hunan province. Pract Prev Med 17:846–851. [Google Scholar]
- 12.Li H, Wang X, Cui Z, Zhou H, Xia S, Kan B. 2008. Analysis of the source of a cholera outbreak caused by O139 Vibrio cholerae. Dis Surveill 23:218–220. [Google Scholar]
- 13.Wang C, Yang X, Tao W, Yang X. 2008. Epidemiological investigation on an outbreak of Cholera O139. Parasit Infect Dis 6:15–17. [Google Scholar]
- 14.Tokunaga A, Yamaguchi H, Morita M, Arakawa E, Izumiya H, Watanabe H, Osawa R. 2010. Novel PCR-based genotyping method, using genomic variability between repetitive sequences of toxigenic Vibrio cholerae O1 El Tor and O139. Mol Cell Probes 24:99–103. doi: 10.1016/j.mcp.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Huang S, Ye X. 2001. Vibrio cholerae ogawa 9K was isolated from imported live soft-shelled turtles. Zhongguo Wei Sheng Jian Yan 11:74 (In Chinese.) [Google Scholar]
- 16.Ding J, Zhang H, Zhao N, Li D. 2003. Vibrio choleae O139 was isolated from the surface of soft-shelled turtles. Zhongguo Wei Sheng Jian Yan 13:484 (In Chinese.) [Google Scholar]
- 17.Qiu X, Wu J, Gao Y. 2013. Investigation on a foodborne cholera O139 outbreak in a rural banquet and the turtle markets. Pract Prev Med 2:171–174. [Google Scholar]
- 18.Wu J, Zeng Y, Luo X. 2005. Sampling survey of Vibrio cholerae among soft-shelled turtles sold in Hengyang city markets. Pract Prev Med 12:1102–1103. [Google Scholar]
- 19.Ip YK, Lee SM, Wong WP, Chew SF. 2013. The Chinese soft-shelled turtle, Pelodiscus sinensis, decreases nitrogenous excretion, reduces urea synthesis and suppresses ammonia production during emersion. J Exp Biol 216:1650–1657. doi: 10.1242/jeb.078972. [DOI] [PubMed] [Google Scholar]
- 20.Li XL, Zhang CL, Fang WH, Lin FC. 2008. White-spot disease of Chinese soft-shelled turtles (Trionyx sinens) caused by Paecilomyces lilacinus. J Zhejiang Univ Sci B 9:578–581. doi: 10.1631/jzus.B0720009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L, Chen G, Chen R, Lin X, Chen K. 2005. Investigation on carrying bacteria in terrapin eggs and infected artificially by Vibrio cholerae (VC) III. Experiment observation on terrapin eggs contaminated by O139 VC. Strait J Prev Med 11:18–20. [Google Scholar]
- 22.Watnick PI, Fullner KJ, Kolter R. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol 181:3606–3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiavelli DA, Marsh JW, Taylor RK. 2001. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl Environ Microbiol 67:3220–3225. doi: 10.1128/AEM.67.7.3220-3225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thelin KH, Taylor RK. 1996. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun 64:2853–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Attridge SR, Manning PA, Holmgren J, Jonson G. 1996. Relative significance of mannose-sensitive hemagglutinin and toxin-coregulated pili in colonization of infant mice by Vibrio cholerae El Tor. Infect Immun 64:3369–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhine JA, Taylor RK. 1994. TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol Microbiol 13:1013–1020. doi: 10.1111/j.1365-2958.1994.tb00492.x. [DOI] [PubMed] [Google Scholar]
- 27.Kirn TJ, Bose N, Taylor RK. 2003. Secretion of a soluble colonization factor by the TCP type 4 pilus biogenesis pathway in Vibrio cholerae. Mol Microbiol 49:81–92. doi: 10.1046/j.1365-2958.2003.03546.x. [DOI] [PubMed] [Google Scholar]
- 28.Kirn TJ, Jude BA, Taylor RK. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863–866. doi: 10.1038/nature04249. [DOI] [PubMed] [Google Scholar]
- 29.Wong E, Vaaje-Kolstad G, Ghosh A, Hurtado-Guerrero R, Konarev PV, Ibrahim AF, Svergun DI, Eijsink VG, Chatterjee NS, van Aalten DM. 2012. The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog 8:e1002373. doi: 10.1371/journal.ppat.1002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Z, Hu J, Yan C. 2000. Impact of water temperature to digestion time and growth of Pelodiscus sinensis under artificial feeding condition. Sichuan J Zool 19:48. [Google Scholar]
- 31.Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, Levine MM. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med 168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colwell RR, Spira WM. 1992. The ecology of Vibrio cholerae, p 107–127. In Barua D, Greenough WB III (ed), Cholera. Springer US, New York, NY. [Google Scholar]
- 33.Reidl J, Klose KE. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev 26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 34.Klose KE. 2000. The suckling mouse model of cholera. Trends Microbiol 8:189–191. doi: 10.1016/S0966-842X(00)01721-2. [DOI] [PubMed] [Google Scholar]
- 35.Spira WM, Sack RB, Froehlich JL. 1981. Simple adult rabbit model for Vibrio cholerae and enterotoxigenic Escherichia coli diarrhea. Infect Immun 32:739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie JM, Rui H, Bronson RT, Waldor MK. 2010. Back to the future: studying cholera pathogenesis using infant rabbits. mBio 1:e00047-10. doi: 10.1128/mBio.00047-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma DP, Thomas C, Hall RH, Levine MM, Attridge SR. 1989. Significance of toxin-coregulated pili as protective antigens of Vibrio cholerae in the infant mouse model. Vaccine 7:451–456. doi: 10.1016/0264-410X(89)90161-8. [DOI] [PubMed] [Google Scholar]
- 38.Islam MS. 1990. Increased toxin production by Vibrio cholerae O1 during survival with a green alga, Rhizoclonium fontanum, in an artificial aquatic environment. Microbiol Immunol 34:557–563. doi: 10.1111/j.1348-0421.1990.tb01032.x. [DOI] [PubMed] [Google Scholar]
- 39.Senderovich Y, Izhaki I, Halpern M. 2010. Fish as reservoirs and vectors of Vibrio cholerae. PLoS One 5:e8607. doi: 10.1371/journal.pone.0008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morris JG., Jr 2003. Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell. Clin Infect Dis 37:272–280. doi: 10.1086/375600. [DOI] [PubMed] [Google Scholar]
- 41.Nahar S, Sultana M, Naser MN, Nair GB, Watanabe H, Ohnishi M, Yamamoto S, Endtz H, Cravioto A, Sack RB, Hasan NA, Sadique A, Huq A, Colwell RR, Alam M. 2011. Role of shrimp chitin in the ecology of toxigenic Vibrio cholerae and cholera transmission. Front Microbiol 2:260. doi: 10.3389/fmicb.2011.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halpern M, Broza YB, Mittler S, Arakawa E, Broza M. 2004. Chironomid egg masses as a natural reservoir of Vibrio cholerae non-O1 and non-O139 in freshwater habitats. Microb Ecol 47:341–349. doi: 10.1007/s00248-003-2007-6. [DOI] [PubMed] [Google Scholar]
- 43.Halpern M, Senderovich Y, Izhaki I. 2008. Waterfowl: the missing link in epidemic and pandemic cholera dissemination? PLoS Pathog 4:e1000173. doi: 10.1371/journal.ppat.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Runft DL, Mitchell KC, Abuaita BH, Allen JP, Bajer S, Ginsburg K, Neely MN, Withey JH. 2014. Zebrafish as a natural host model for Vibrio cholerae colonization and transmission. Appl Environ Microbiol 80:1710–1717. doi: 10.1128/AEM.03580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagai N, Kobayashi H, Katayama S, Munekata M. 2009. Preparation and characterization of collagen from soft-shelled turtle (Pelodiscus sinensis) skin for biomaterial applications. J Biomater Sci Polym Ed 20:567–576. doi: 10.1163/156856209X426394. [DOI] [PubMed] [Google Scholar]
- 46.Lu J, Wan Q, Yin Z, Lin L, Weng S, Ye Y, Jiang S. 2010. Extraction and characterization of collagen from calipash of Chinese soft-shelled turtle (Pelodiscus sinensis). J Fish China 34:801–808. doi: 10.3724/SP.J.1231.2010.06800. [DOI] [Google Scholar]
- 47.Park BR, Zielke RA, Wierzbicki IH, Mitchell KC, Withey JH, Sikora AE. 2015. A metalloprotease secreted by the type II secretion system links Vibrio cholerae with collagen. J Bacteriol 197:1051–1064. doi: 10.1128/JB.02329-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsiao A, Liu Z, Joelsson A, Zhu J. 2006. Vibrio cholerae virulence regulator-coordinated evasion of host immunity. Proc Natl Acad Sci U S A 103:14542–14547. doi: 10.1073/pnas.0604650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Z, Wang GL, Nie P. 2009. IgM, IgD and IgY and their expression pattern in the Chinese soft-shelled turtle Pelodiscus sinensis. Mol Immunol 46:2124–2132. doi: 10.1016/j.molimm.2009.03.028. [DOI] [PubMed] [Google Scholar]
- 50.Deza FG, Espinel CS, Beneitez JV. 2007. A novel IgA-like immunoglobulin in the reptile Eublepharis macularius. Dev Comp Immunol 31:596–605. doi: 10.1016/j.dci.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Bhowmick R, Ghosal A, Das B, Koley H, Saha DR, Ganguly S, Nandy RK, Bhadra RK, Chatterjee NS. 2008. Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect Immun 76:4968–4977. doi: 10.1128/IAI.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaur S, Dhillon GS. 2015. Recent trends in biological extraction of chitin from marine shell wastes: a review. Crit Rev Biotechnol 35:44–61. doi: 10.3109/07388551.2013.798256. [DOI] [PubMed] [Google Scholar]
- 53.Younes I, Hajji S, Frachet V, Rinaudo M, Jellouli K, Nasri M. 2014. Chitin extraction from shrimp shell using enzymatic treatment. Antitumor, antioxidant and antimicrobial activities of chitosan. Int J Biol Macromol 69:489–498. [DOI] [PubMed] [Google Scholar]
- 54.Stauder M, Huq A, Pezzati E, Grim CJ, Ramoino P, Pane L, Colwell RR, Pruzzo C, Vezzulli L. 2012. Role of GbpA protein, an important virulence-related colonization factor, for Vibrio cholerae's survival in the aquatic environment. Environ Microbiol Rep 4:439–445. doi: 10.1111/j.1758-2229.2012.00356.x. [DOI] [PubMed] [Google Scholar]
- 55.Hirasawa T, Nagashima H, Kuratani S. 2013. The endoskeletal origin of the turtle carapace. Nat Commun 4:2107. doi: 10.1038/ncomms3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alibardi L, Toni M. 2006. Skin structure and cornification proteins in the soft-shelled turtle Trionyx spiniferus. Zoology (Jena) 109:182–195. doi: 10.1016/j.zool.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 57.Disease Control Bureau of the Ministry of Health of China. 2013. Manual for prevention and control of cholera, 6th ed People's Medical Publishing House, Beijing, China. [Google Scholar]
- 58.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nature Biotech 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 59.Nandi B, Nandy RK, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC. 2000. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J Clin Microbiol 38:4145–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 61.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheng Y, Fan F, Jensen O, Zhong Z, Kan B, Wang H, Zhu J. 2015. Dual zinc transporter systems in Vibrio cholerae promote competitive advantages over gut microbiome. Infect Immun 83:3902–3908. doi: 10.1128/IAI.00447-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.