ABSTRACT
The yeast Saccharomyces cerevisiae is widely used to produce biopharmaceutical proteins. However, the limited capacity of the secretory pathway may reduce its productivity. Here, we increased the secretion of a heterologous α-amylase, a model protein used for studying the protein secretory pathway in yeast, by moderately overexpressing SEC16, which is involved in protein translocation from the endoplasmic reticulum to the Golgi apparatus. The moderate overexpression of SEC16 increased α-amylase secretion by generating more endoplasmic reticulum exit sites. The production of reactive oxygen species resulting from the heterologous α-amylase production was reduced. A genome-wide expression analysis indicated decreased endoplasmic reticulum stress in the strain that moderately overexpressed SEC16, which was consistent with a decreased volume of the endoplasmic reticulum. Additionally, fewer mitochondria were observed. Finally, the moderate overexpression of SEC16 was shown to improve the secretion of two other recombinant proteins, Trichoderma reesei endoglucanase I and Rhizopus oryzae glucan-1,4-α-glucosidase, indicating that this mechanism is of general relevance.
IMPORTANCE There is an increasing demand for recombinant proteins to be used as enzymes and pharmaceuticals. The yeast Saccharomyces cerevisiae is a cell factory that is widely used to produce recombinant proteins. Our study revealed that moderate overexpression of SEC16 increased recombinant protein secretion in S. cerevisiae. This new strategy can be combined with other targets to engineer cell factories to efficiently produce protein in the future.
KEYWORDS: protein secretion, mitochondria, ERES, ROS, SEC16
INTRODUCTION
The yeast Saccharomyces cerevisiae is a widely used cell factory for the production of recombinant proteins (1). The advantages of S. cerevisiae are fast growth, easy cultivation, and ability to perform posttranslational modifications and secrete proteins to the extracellular medium, which facilitates the purification of the protein (2, 3). However, S. cerevisiae naturally secretes only a few proteins, such as aspartyl protease, invertase, and α-factor pheromone (4, 5); therefore, it has evolved a secretory pathway with a relatively low capacity. Many different strategies have been used to engineer this pathway with the objective of improving heterologous protein production (6–8). When expressing a protein that has a secretory signal peptide, the protein is cotranslationally translocated into the endoplasmic reticulum (ER) lumen, where disulfide bond formation occurs with the initial glycosylation (9). The secretion of recombinant proteins is affected by their signal peptide sequences (10). A synthetic leader sequence increases human insulin precursor production, whereas the use of the α-factor leader sequence increased α-amylase production (7). Excessive mannose glycosylation of recombinant proteins can reduce the efficiency of protein secretion, and it was recently found that the deletion of mannosyltransferases improves recombinant protein secretion in S. cerevisiae (11, 12). Another approach that has been used to engineer the secretory pathway has been to activate the unfolded protein response (UPR). The UPR is activated when misfolded proteins accumulate in the ER and results in the alternative splicing of the HAC1 gene product, and then the splicing of the HAC1 gene product activates UPR-regulated genes. The secretion of α-amylase was enhanced by 70% through the overexpression of HAC1 (13). The importance of protein folding in the ER lumen was further supported by the overexpression of Pdi1p, a disulfide isomerase that is involved in protein folding; its overexpression has increased the secretion of a wide range of recombinant proteins (14). However, overexpressing KAR2, an ER chaperone that responds to protein misfolding, has varied effects on the secretion of proteins (15).
Correctly folded proteins are transported through the secretory pathway: first, they travel from the ER to the cis side of the Golgi apparatus, then through the Golgi network, and finally from the trans-Golgi network to the plasma membrane (PM). This process starts with the formation of coat protein complex II (COPII) vesicles at the ER exit sites (ERESs) (16). The formation of the COPII vesicles is triggered by Sar1p-GDP binding to the ER membrane. Sec12p, a guanine exchange factor, activates Sar1p by exchanging GDP with GTP. This activation results in the recruitment of the Sec23p-Sec24p COPII inner coat protein complex onto the activated Sar1p-GTP and the binding of the Sec24p subunit to the cargo protein that is to be translocated through the secretory pathway (17). The complex of Sar1p-GTP, Sec23p-Sec24p, and the cargo is called a prebudding complex. The prebudding complex accumulates at the ERESs, and then the outer coat protein complex Sec13p-Sec31p binds to the prebudding complex. The assembly of the intact COPII proteins finally results in the fission of a vesicle from the ER membrane (18). The maturation of the COPII vesicle is accomplished by the disassembly of the coated protein from the liposome, which is driven by the hydrolysis of Sar1p-GTP. The coat proteins used in this process can be reused in a new round of COPII assembly. Sec16p plays an important role in COPII-coated vesicle formation. Sec16p accumulates at ERESs, serves as a scaffold for COPII vesicle assembly, interacts with multiple COPII proteins, and negatively regulates the Sar1p-GTPase (19). However, in S. cerevisiae, the level of Sec16p expression is lower than the level of expression of the other COPII coat proteins and may therefore limit COPII vesicle formation (20).
The trafficking process from the Golgi apparatus to the PM was previously elucidated (21). Sec2p, Sec4p, Sec15p, and Ypt32p are involved in this process. Sec2p is a guanine exchange factor and locates at the Golgi membrane by binding to Ypt32p-GTP and phosphatidyl inositol 4-phosphate [PI(4)P]. The Golgi-derived vesicle then detaches from the Golgi membrane after Sec2p activation of the Rab GTPase Sec4p. The effector Sec15p next is recruited to the complex. During the delivery, the conformation of Sec2p changes with a decrease in the PI(4)P level, which enables the effector Sec15p to substitute for Ypt32p-GTP on Sec2p. Sec2p then is phosphorylated, which drives the reaction forward by strengthening the interaction between Sec2p and Sec15p and preventing the binding of Sec2p to Ypt32p-GTP and PI(4)P (21).
Here, these genes (SAR1, SEC16, SEC2, SEC4, SEC15, and YPT32), which are involved in vesicle formation and vesicle trafficking, were overexpressed to increase protein secretion. Among these genes, only the moderate overexpression of SEC16 was found to significantly increase α-amylase secretion. Transcriptome profiling and the physiological changes in the SEC16 overexpression strain were studied, and lower ER stress and a greater number of ERESs were found in this strain. Furthermore, the moderate overexpression of SEC16 increased the production of two other recombinant proteins, the endoglucanase I from Trichoderma reesei and the glucan-1,4-α-glucosidase from Rhizopus oryzae. These results suggest that the moderate overexpression of SEC16 is a general strategy for improving recombinant protein production in S. cerevisiae.
RESULTS
Moderate expression of SEC16 enhances protein secretion.
In this study, we define high expression as using a high-copy-number plasmid with strong promoters, moderate expression as using a low-copy-number plasmid or a single-copy integration vector with strong promoters, and low expression as using a low-copy-number plasmid or a single-copy integration vector with weak promoters. Both PTEF and PGPD are reported to be strong promoters (22). With the objective being to enhance the capacity of recombinant protein trafficking, several genes that code for proteins that are involved in transport from the ER to the Golgi apparatus or from the Golgi apparatus to the PM were overexpressed using the centromeric plasmid p416TEF. SEC16 overexpression resulted in a 2-fold increase in the yield of secreted α-amylase (defined as the amount of secreted α-amylase per unit of dry cell weight [DCW]) over that of the control strain; however, overexpression of the other trafficking-related genes did not result in a significant increase in the α-amylase yield (Fig. 1; see also Table S1 in the supplemental material). The SEC16 expression level was further increased to enhance COPII formation by using the 2μ high-copy-number plasmid p426TEF. However, the α-amylase yield of the strain with 2μ-SEC16 was reduced to the same level as that of the reference strain (Fig. S1 and Table S1). To obtain a stable strain for further analysis, the native SEC16 promoter in the chromosome was replaced with a strong constitutive promoter, PGPD, resulting in the strain YIGS16, which moderately overexpresses SEC16. Strain YIGS16 produced 2-fold greater secreted α-amylase yield than the wild-type strain AACK did (Fig. S2). Another strong promoter, PTEF1, was used to drive the overexpression of SEC16, but the α-amylase yield was approximately 30% lower than when the PGPD promoter was used. The strain YIGS16 therefore was used for further analysis.
FIG 1.
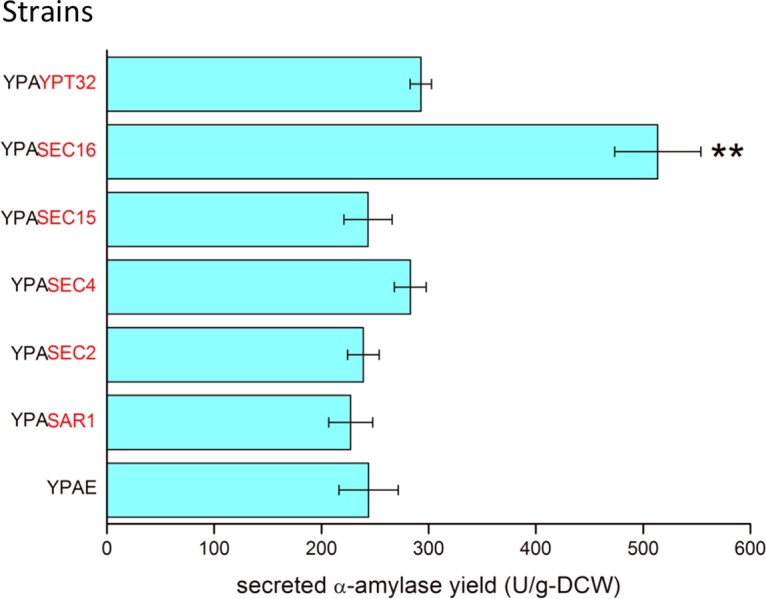
Secreted α-amylase yield of strains that overexpress genes that are involved in ER-to-Golgi or Golgi-to-membrane transport. The overexpressed genes are marked in red. **, P < 0.01. Measurements are reported as the average values ± standard deviations from independent triplicates.
Physiological characterization of YIGS16 in batch fermentation.
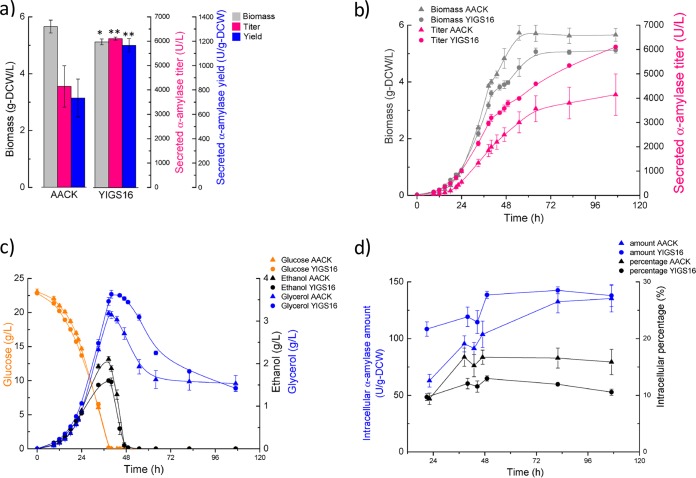
The strains YIGS16 and AACK were characterized in batch fermentations. Both strains produced more α-amylase in bioreactors than in tubes due to the better control of the environmental conditions (pH, oxygen, temperature, etc.). The final biomass yield of YIGS16 was approximately 10% lower than that of AACK, and the α-amylase titer of YIGS16 was higher than that of AACK during the entire culture (Fig. 2a and b). The glucose consumption rate of both strains was almost identical. At the end of the glucose phase (when glucose is depleted), the ethanol titer for AACK was 2.1 g/liter, while it was 1.6 g/liter for YIGS16. In contrast, glycerol production was higher in strain YIGS16 (Fig. 2c). Table 1 summarizes the physiological parameters of both strains. The maximum specific growth rate of YIGS16 was slightly lower than that of AACK. The moderate overexpression of SEC16 resulted in a 54% increase in the specific α-amylase production rate and a 65% increase in the yield of secreted α-amylase from glucose.
FIG 2.
Batch fermentation of strain YIGS16 and the control strain AACK. (a) The final biomass, secreted amylase titer, and secreted amylase yield in batch fermentation. (b) The time course of the biomass and secreted amylase titer. (c) The time course of glucose consumption and ethanol and glycerol production. (d) The intracellular α-amylase amount and the intracellular percentages (calculated by dividing the intracellular α-amylase amount by the total amount of α-amylase) at six different time points. *, P < 0.05; **, P < 0.01. Measurements are reported as the average values ± standard deviations from independent quadruplicates.
TABLE 1.
Physiological parameters of AACK and YIGS16
| Strain | Result fora: |
|||||
|---|---|---|---|---|---|---|
| μmax | rS | rP | Ysα | rE | rG | |
| AACK | 0.249 ± 0.004 | 1.35 ± 0.04 | 103.6 ± 14.5 | 80.6 ± 9.4 | 0.163 ± 0.004 | 0.205 ± 0.008 |
| YIGS16 | 0.233 ± 0.008 | 1.26 ± 0.04 | 159.9 ± 12.0 | 132.2 ± 6.4 | 0.122 ± 0.011 | 0.200 ± 0.006 |
μmax, maximum specific growth rate (h−1) on glucose; rS, specific glucose uptake rate [g/(g-DCW/h)]; rP, specific secreted α-amylase production rate [U/(g-DCW)/h] on glucose; Ysα, yield of secreted α-amylase from glucose (U/g); rE, specific ethanol production rate [g/(g-DCW)/h]; rG, specific glycerol production rate [g/(g-DCW)/h]. Measurements are reported as the average value ± standard deviation from independent quadruplicates.
Lower percentage of intracellular α-amylase in YIGS16.
To evaluate whether the moderate overexpression of SEC16 affects the intracellular accumulation of α-amylase, cell samples were analyzed at six different time points, including the early exponential phase (optical density at 600 nm [OD600] of ≈1), the end of the glucose phase, the ethanol growth phase (when glucose is exhausted and ethanol is used as a carbon source), the end of the ethanol phase (when ethanol is exhausted), the stationary phase, and the end of the fermentation. The intracellular α-amylase level in AACK increased from 60 U/g-DCW in the exponential phase to 140 U/g-DCW at the end of the fermentation. The fraction of α-amylase that remained in AACK cells increased sharply during the glucose phase and was then maintained at a stable level throughout the rest of the fermentation (Fig. 2d). In contrast, the intracellular α-amylase content of YIGS16 remained at a relatively stable level during the whole process (from 110 U/g-DCW in the glucose phase to 140 U/g-DCW at the end of the fermentation). Thus, the intracellular α-amylase levels in YIGS16 increased only slightly during growth on glucose, and throughout fermentation, the fraction of the α-amylase that remained associated with the cells was smaller (Fig. 2d).
Intracellular α-amylase degradation is the same in YIGS16 and AACK.
The secretion and degradation of the retained intracellular α-amylase in AACK and YIGS16 were determined to further quantify the protein secretory capacity of both strains. The cells were incubated in SD-2xSCAA medium until the OD600 reached 1, at which point they were transferred to a fresh carbon-source-free medium, S-2xSCAA, for 48 h. During this time, the synthesis of α-amylase was limited due to the lack of a carbon source; however, the residual intracellular α-amylase would continue to be secreted out of the cells. The secretion of the residual intracellular α-amylase from YIGS16 was more than 2-fold higher than that from AACK. Furthermore, there was no significant difference in the degradation of the residual intracellular α-amylase between the AACK and YIGS16 strains (Table S2).
Moderate expression of SEC16 reduces ROS accumulation from heterologous protein production.
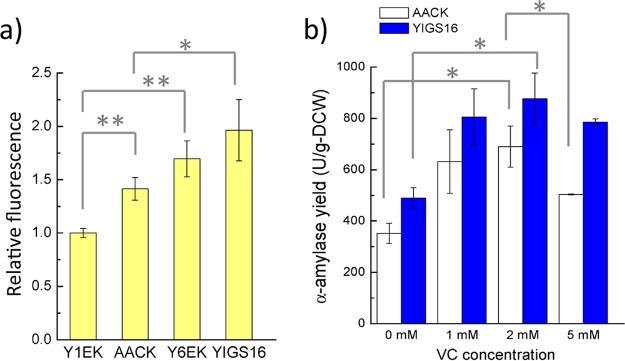
Heterologous protein production often results in the accumulation of reactive oxygen species (ROS) due to an increased demand for protein folding in the ER (23). The ROS accumulation in Y1EK was set as 1 relative fluorescence unit (RFU) (Fig. 3a). When producing α-amylase in the wild-type background 530-1CK, the ROS accumulation increased by 0.41 RFU (AACK versus Y1EK). However, the ROS accumulation resulting from the production of α-amylase did not increase significantly in a strain that moderately overexpressed SEC16 (YIGS16 versus Y6EK), even though the α-amylase yield was 2-fold higher in YIGS16 than in AACK. Interestingly, the moderate overexpression of SEC16 itself increased the intracellular ROS accumulation. The mechanisms behind this behavior are unknown, but this behavior may partly explain why the overexpression of SEC16 with a high-copy-number plasmid did not increase α-amylase production; a higher accumulation of ROS from the high-level expression of SEC16 could be harmful for the cell and for recombinant protein production.
FIG 3.
Oxidative stress study. (a) Intracellular ROS level of strains Y1EK (wt), AACK (production of α-amylase in wt strain), Y6EK (SEC16 overexpression), and YIGS16 (production of α-amylase in SEC16 overexpression) in shake flask cultivation. (b) The final yield of α-amylase with different concentrations of vitamin C (VC) supplied to the medium, with tube fermentation. *, P < 0.05; **, P < 0.01. Measurements are reported as the average values ± standard deviations from independent triplicates (for ROS) and duplicates (for VC test).
Vitamin C is an antioxidant that can protect cells against the harmful effects of ROS (24). To evaluate whether the reduction of the ROS level could benefit α-amylase production, different concentrations of vitamin C were added to the medium. The pH of the medium was only slightly affected by the vitamin C since the medium was buffered. The α-amylase yield of both strains increased when low concentrations of vitamin C were added to medium (Fig. 3b). However, when the concentration of vitamin C reached 5 mM, the α-amylase yield of AACK was less than that of the medium supplied with 2 mM vitamin C (Fig. 3b). When the vitamin C concentration increased to 10 mM, the growth of both strains was strongly inhibited, and consequently the α-amylase titer was very low (data not shown). Thus, a moderate amount of antioxidant helped cells resist the damage caused by ROS accumulation and was beneficial for protein production. The α-amylase yield in the AACK strain decreased 27% when the vitamin C was increased from 2 to 5 mM.
More ERESs in YIGS16.
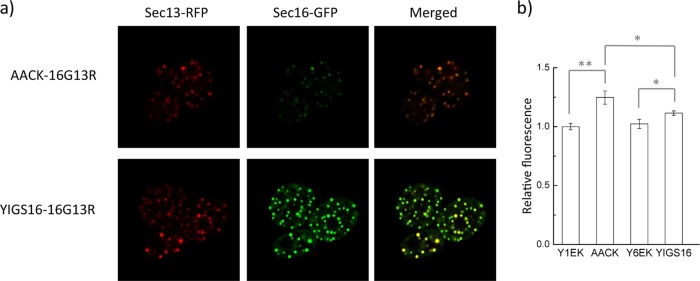
To monitor the ERESs, green fluorescent protein (GFP) and red fluorescent protein (RFP) were fused with the C termini of Sec16p and Sec13p, respectively. Sec13p is an outer coat protein of the COPII vesicles and colocalizes with Sec16p at the ERESs (25). The fluorescence signal intensity of SEC16-GFP in YIGS16-16G13R was approximately 5-fold higher than that in AACK-16G13R (Fig. 4a and Fig. S3), which confirms that the level of Sec16p is increased in YIGS16. The RFP signal intensity in YIGS16-16G13R was also stronger than that in AACK-16G13R, although the mRNA level of SEC13 was about the same in AACK and in YIGS16 (Table S3). This result might have occurred because more Sec13p in YIGS16 participated in COPII vesicle formation and therefore less Sec13p was directed to degradation.
FIG 4.
Strain YIGS16 has more ERESs and a reduced amount of ER membrane compared to strain AACK. (a) Airyscan images showing ERESs by the colocalization of Sec16-GFP and Sec13-RFP. SEC16 was fused with GFP, and SEC13 was fused with RFP in strains AACK and YIGS16. (b) ER staining was performed, and the ER membrane changes were quantified by their fluorescence intensity. *, P < 0.05; **, P < 0.01. The data are reported as the mean values ± standard deviations from independent triplicates.
The YIGS16 strain has a reduced amount of ER membrane.
As Sec16p is a peripheral ER protein that is involved in transport from the ER to the Golgi apparatus, we wondered if the moderate overexpression of SEC16 affected the amount of ER membrane. The ER membranes of Y1EK, AACK, Y6EK, and YIGS16 were detected using an ER membrane dye, the ER-tracker blue-white DPX (26). When the yeast cells produced α-amylase (AACK), the ER membrane expanded by approximately 25% (Fig. 4b). However, when the cells overexpressed only SEC16, the amount of ER membrane that was produced was similar to that of the control strain. Interestingly, when producing α-amylase in the SEC16 moderate expression strain (YIGS16), the amount of ER membrane that was produced was smaller than that of the AACK strain. Additionally, we detected a smaller amount of Kar2p in the extracellular medium of YIGS16 than in that of AACK (Fig. S4). Kar2p is an ER resident protein. The abundance of Kar2p increases when UPR is activated to assist protein folding (27), and the secretion of Kar2p is a result of an impaired secretory pathway (28).
Transcriptome analysis.
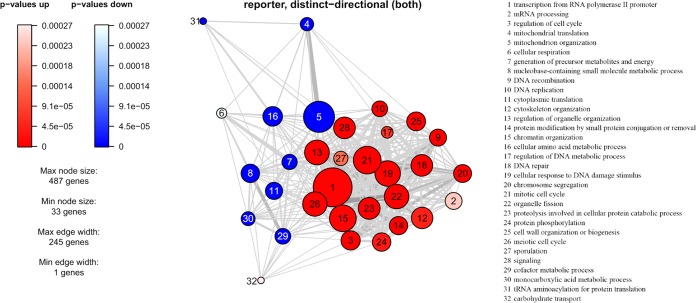
To understand the global gene expression in the engineered strain, a transcriptome analysis was performed on samples from the glucose growth phase and the ethanol growth phase. A principal component analysis showed that the genes of YIGS16 had significantly different expression levels than those of the AACK strain in both growth phases (Fig. 5 and Fig. S5). Even though SEC16 was intentionally moderately overexpressed, the expression levels of 612 (Table S3) and 579 (Table S4) genes changed significantly more than 2-fold in the glucose phase and the ethanol phase, respectively. In total, 203 gene transcript levels changed significantly more than 2-fold [log2(fold change) of ≤−1 or log2(fold change) of ≥1 and an adjusted P value of <0.05] in both phases (Table S5). Many of these 203 genes were involved in three processes: cell wall organization, trafficking, and the cell cycle.
FIG 5.
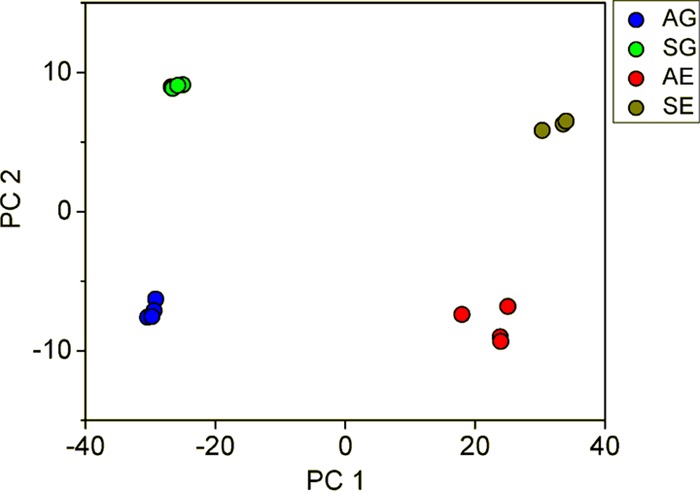
Principal component analysis (PCA) of AG (transcriptome of AACK in the glucose phase), SG (transcriptome of YIGS16 in the glucose phase), AE (transcriptome of AACK in the ethanol phase), and SE (transcriptome of YIGS16 in the ethanol phase).
To better understand the process underlying these results, reporter gene ontology (GO) term and reporter transcription factor (TF) analyses were performed using the transcriptome data for the glucose phase. The reporter transcription factor analysis was used to show transcript level changes of genes known to be regulated by certain transcription factors. TFs were scored by the modulation in the expression levels of the genes that were controlled by the same TF. These GO terms (Fig. 6) and reporter TFs (Fig. S6) reflect the cellular processes that changed significantly when SEC16 is moderately overexpressed. From the reporter TF analysis, genes such as ENO1/2, PGK1, TDH3, and CDC19, which are regulated by the glycolytic transcriptional activators Tye7p and Gcr1p, were found to be downregulated, indicating changes in the carbohydrate metabolism of strain YIGS16 (Fig. S6). This downregulation can be explained by the expression levels of TYE7 and GCR1 themselves, as they were downregulated approximately 2.6-fold and 2-fold, respectively (Table S3).
FIG 6.
Reporter GO term analysis of transcriptome profiling in the glucose phase. The nodes represent GO terms, and the thickness of the edges correlate to the number of shared genes. The size of the nodes represents the number of genes within the GO terms.
The heat shock response (HSR), which is regulated by the transcription factor HSF1, has been reported to relieve ER stress (29), and the constitutive activation of HSR through the HSF1 mutation HSF1-R206S can improve protein secretion (30). Interestingly, less HSR may be present in YIGS16, because the reporter TF analysis found that genes such as YOP1, DDR2, and HSP104, which are regulated by Hsf1p, were downregulated (Fig. S6).
The UPR is activated by an increase in ER stress when unfolded proteins accumulate there. The transcription factors Hac1p and Gcn4p co-upregulate the expression of hundreds of genes in response to the UPR (31). From our transcriptome analysis, we found that approximately 50% of the genes that are coregulated by Hac1p and Gcn4p were downregulated in YIGS16 during its growth on glucose (Table S6). Moreover, some genes that are required for protein folding were also downregulated, such as CNE1, STI1, SSE2, SSA4, and SIS1 (30). This relationship indicates that the moderate overexpression of SEC16 resulted in less ER stress and provided better conditions for protein folding.
We performed a similar analysis on the transcriptome data from the ethanol growth phase and found similar results, thus showing that the moderate overexpression of SEC16 has the same effect during both fermentative and respiratory metabolism (Fig. S7 and S8).
Moderate expression of SEC16 results in fewer mitochondria.
Based on the transcriptome analysis, we found that several key genes, such as ACC1, FAA1, FAA2, FAA4, and MSS4 (32), which are involved in lipid metabolism, were significantly up- or downregulated due to the moderate overexpression of SEC16 (Table S3). Mitochondrial biogenesis is tightly related to lipid metabolism (33); hence, mitochondrial biogenesis may be affected in the SEC16 moderate expression strain. Furthermore, the higher accumulation of ROS in the SEC16 moderate expression strain may stimulate lipid peroxidation, which can also affect mitochondrial organization and function (34). We therefore examined the mitochondria in the strains and found that there were fewer mitochondria in YIGS16 than in AACK (Fig. S9a and S9b). From the reporter GO term analysis, the transcript levels of the genes involved in mitochondrial translation and mitochondrial organization were significantly downregulated (Fig. 6). This observation confirmed our speculation that the mitochondrial function is reduced in YIGS16, which may be due to a combined effect of high ROS accumulation and dysregulated lipid metabolism. The presence of fewer mitochondria is in agreement with the reduced respiration of YIGS16.
General effect on secretion of other recombinant proteins.
To investigate whether the moderate overexpression of SEC16 can increase the production of other recombinant proteins, two heterologous fungal proteins, endoglucanase I from T. reesei and glucan-1,4-α-glucosidase from R. oryzae, were also evaluated. There were higher yields of both proteins in the SEC16 moderate overexpression strain than in the corresponding reference strain (Fig. 7). This result confirmed the importance of ER-to-Golgi transport in protein secretion and showed that the moderate overexpression of SEC16 may be a general strategy for increasing the secretion of recombinant proteins.
FIG 7.
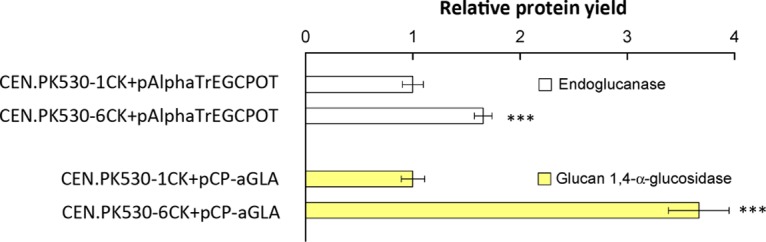
Secretion of two recombinant proteins, endoglucanase and glucan 1,4-α-glucosidase, in the wild-type and SEC16 overexpression strains. ***, P < 0.001. The data are reported as the mean values ± standard deviations from independent quadruplicates.
DISCUSSION
The effects of overexpressing several different secretion-related proteins on the production of α-amylase were determined. We found that only moderate overexpression of SEC16 significantly increased the α-amylase production. It has been reported that overexpression of SAR1, which is also involved in COPII formation in transport from the ER to the Golgi apparatus, can partially complement the loss of function of an SEC16 mutation (sec16-2) (35). However, the overexpression of SAR1 did not increase α-amylase secretion in our study, suggesting a limitation of Sec16p in the recruitment step. Moreover, α-amylase secretion is not significantly increased when target genes that are involved in Golgi-to-plasma membrane transport are overexpressed (Fig. 1), indicating that the transport from the Golgi apparatus to the PM is not the primary limiting step in the reference strain. When SEC16 was overexpressed by a high-copy-number plasmid with a strong promoter, there was no significant increase in heterologous protein secretion. This result is consistent with previous findings that showed that the deletion of SEC16 or the overexpression of SEC16 by a high-copy-number plasmid would block transport from the ER to the Golgi apparatus or even be lethal for the cell (36). Thus, clearly, the expression level of Sec16p must be tuned to a suitable level to ensure efficient transport from the ER to the Golgi apparatus. A similar phenomenon is also observed in mammalian cells, which require an appropriate amount of Sec16p to maintain ER-to-Golgi transport (37).
In our bioreactor experiment, we found that the moderate overexpression of SEC16 lowered the specific growth rate and the final biomass that was reached during culture. Meanwhile, the lower ethanol and higher glycerol production in the SEC16 overexpression strain, compared with the reference strain, implied that the redox balance was maintained differently in YIGS16 than in AACK. Intracellular α-amylase samples of both strains taken at six different time points during the fermentations were analyzed. The increase in the intracellular α-amylase levels (from 110 U/g-DCW to 140 U/g-DCW) in YIGS16 during the fermentation process was much less than the increase (from 60 U/g-DCW to 140 U/g-DCW) in AACK, which suggests that the moderate overexpression of SEC16 provides a higher secretion capacity. The amounts of residual intracellular α-amylase also confirm the improved secretion capacity in the SEC16 moderate overexpression strain. The relatively stable intracellular α-amylase levels during the fermentations, combined with the lack of a significant change in the degradation of residual intracellular α-amylase in YIGS16, indicate that the secretory capacity in YIGS16 matches protein synthesis throughout the whole fermentation process. In contrast, the secretory capacity of AACK may lag behind its protein synthesis, as the concentration of intracellular α-amylase increased during the fermentation.
Since Sec16p mediates COPII-coated vesicle formation, it colocalizes with COPII coat proteins at the ERESs and is related to ERES organization (19, 25, 38). In Pichia pastoris, the abundance of Sec16p is lower than that of Sec13p, which is an essential COPII protein; however, the overexpression of SEC16 does not further increase the number of ERESs in P. pastoris (38). This lack of change is because the number of ERESs is relatively constant in P. pastoris, and each ERES is associated with a Golgi stack. The situation is different in S. cerevisiae, which has small and numerous ERESs (19). Feizi et al. reported that Sec16p in S. cerevisiae is present at lower levels than other COPII proteins (20). Airyscan images showed an increased number of ERESs in the SEC16 moderate overexpression strain. The increase in α-amylase production from overexpressing SEC16 may be due to the increased number of ERESs in S. cerevisiae. When the abundance of Sec16p increases, the number of ERESs increases; this change increases COPII formation and thus facilitates the translocation of α-amylase from the ER to the Golgi apparatus.
ER expansion helps to alleviate the ER stress of the cell (39–41). Schuck et al. found that a hac1Δ yeast strain was unable to properly expand its ER membrane and increase chaperone levels. The yeast strain with the HAC1 deletion was hypersensitive to tunicamycin, which introduces ER stress to the cells (39). However, the deletion of OPI1, a transcriptional regulator that negatively mediates phospholipid biosynthesis by binding to the Ino2p/Ino4p complex, or the overexpression of INO2, a transcriptional activator of lipid biosynthesis, and its Opi1p-insensitive variant ino2(L119A) enhanced the ER stress tolerance of the hac1Δ mutant by expanding the ER (39). In our study, SEC16 moderate overexpression resulted in a less-expanded ER than was found in AACK. The moderate overexpression of SEC16 generated more ROS in yeast cells (Y1EK versus Y6EK), but fewer ROS were produced by recombinant protein production in the SEC16 moderate overexpression strain (AACK-Y1EK versus YIGS16-Y6EK), although the secretion of the α-amylase was higher in YIGS16 than in AACK. The transcriptome analysis showed that half of the genes that are coregulated by Hac1p and Gcn4p were significantly downregulated, and some ER chaperones, such as Kar2p, Pdi1p, and Lhs1p, had similar expression levels in YIGS16 and AACK (Table S7). These results suggest that fewer ROS were generated from recombinant protein production in YIGS16 than in AACK. Another explanation for the less-expanded ER in YIGS16 is that the increased COPII vesicle transport delivered more lipids from the ER to the Golgi apparatus in this strain compared with the reference strain.
Conclusions.
In summary, we report that the moderate overexpression of SEC16 increases recombinant protein secretion in S. cerevisiae. An increased number of ERESs and a reduced level of ER stress are the mechanisms that enable this phenomenon. The findings demonstrate a new strategy to increase protein secretion that can be combined with other targets to engineer cell factories for the efficient production of protein in the future.
MATERIALS AND METHODS
Strain and plasmid construction.
The strains and plasmids used in this study are listed in Table 2. The primers used in this study are listed in Table 3. The plasmid p416TEFSAR1 was constructed as follows. (i) The SAR1 gene was amplified from the CEN.PK530-1C genome by using the primers sar1F and sar1R; (ii) the plasmid backbone was amplified using p416TEF-PTEF-F and p416TEF-TCYC1-R; and (iii) the SAR1 gene fragment was ligated with the plasmid backbone by Gibson Assembly (42). The other plasmids, p416TEFSEC16, p416TEFSEC2, p416TEFSEC4, p416TEFSEC15, p416TEFYPT32, and p426TEFSEC16, were constructed in the same manner. The plasmid pAlphaTrEG was constructed as follows. (i) The endoglucanase gene from T. reesei with the yeast alpha factor leader sequence was synthesized by GenScript and amplified using primers KpnI-alpha-leader-F and XhoI-cellulase-R, resulting in fragment alphaTrEG; (ii) the alphaTrEG fragment was digested with KpnI and XhoI and inserted into the corresponding sites of the vector CPOTud. The yeast strains YPAE, YPASAR1, YPASEC2, YPASEC4, YPASEC15, YPASEC16, YPAYPT32, YPA2uE, and YPA2uSEC16 were constructed by cotransforming pAlphaAmyCPOT with the plasmids p416TEF, p416TEFSAR1, p416TEFSEC2, p416TEFSEC2, p416TEFSEC15, p416TEFSEC16, p416TEFYPT32, p426TEF, and p426TEFSEC16, respectively, into CEN.PK 530-1D.
TABLE 2.
Plasmids and strains used in this study
| Name | Description | Origin or reference |
|---|---|---|
| Plasmids | ||
| CPOTud | TPI promoter and terminator from S. cerevisiae, POT marker from Schizosaccharomyces pombe (2μ) | 7 |
| pAlphaAmyCPOT | α-Factor leader with α-amylase gene inserted into CPOTud | 7 |
| p416TEF | TEF1 promoter (centromeric) | 22 |
| p426TEF | TEF1 promoter (2μ) | 22 |
| p416TEF-SAR1 | PTEF1-SAR1 | This study |
| p416TEF-SEC16 | PTEF1-SEC16 | This study |
| p416TEF-SEC2 | PTEF1-SEC2 | This study |
| p416TEF-SEC4 | PTEF1-SEC4 | This study |
| p416TEF-SEC15 | PTEF1-SEC15 | This study |
| p416TEF-YPT32 | PTEF1-YPT32 | This study |
| p426TEF-SEC16 | PTEF1-SEC16 (2μ) | This study |
| pAlphaTrEGCPOT | α-Factor leader with endoglucanase I gene inserted into CPOTud | This study |
| pCP-aGLA | α-Factor leader with glucan 1,4-α-glucosidase gene inserted into CPOTud | 3 |
| Strains | ||
| CEN.PK530-1C | MATa URA3 HIS3 LEU2 TRP1 SUC2 MAL2-8c tpi1(41-707)::loxP-KanMX4-loxP | 7 |
| CEN.PK530-1CK | MATa URA3 HIS3 LEU2 TRP1 SUC2 MAL2-8c tpi1(41-707)::loxP | This study |
| CEN.PK530-1D | MATa ura3-52 HIS3 LEU2 TRP1 SUC2 MAL2-8c tpi1(41-707)::loxP-KanMX4-loxP | 6 |
| CEN.PK530-6CK | MATa URA3 HIS3 LEU2 TRP1 SUC2 MAL2-8c tpi1(41-707)::loxP PGPD-SEC16 | This study |
| YPAE | CEN.PK530-1D with pAlphaAmyCPOT and p416TEF | This study |
| YPASAR1 | CEN.PK530-1D with pAlphaAmyCPOT and p416TEF-SAR1 | This study |
| YPASEC16 | CEN.PK530-1D with pAlphaAmyCPOT and p416TEF-SEC16 | This study |
| YPASEC2 | CEN.PK530-1D with pAlphaAmyCPOT and p416TEF-SEC2 | This study |
| YPASEC4 | CEN.PK530-1D with pAlphaAmyCPOT and p416TEF-SEC4 | This study |
| YPAYPT32 | CEN.PK530-1D with pAlphaAmyCPOT and p416TEF-YPT32 | This study |
| YPA2uE | CEN.PK530-1D with pAlphaAmyCPOT and p426TEF | This study |
| YPA2uSEC16 | CEN.PK530-1D with pAlphaAmyCPOT and p426TEF-SEC16 | This study |
| AACK | CEN.PK530-1CK with pAlphaAmyCPOT | This study |
| YIGS16 | CEN.PK530-6CK with pAlphaAmyCPOT | This study |
| AACK-16G13R | AACK SEC16-GFP; SEC13-RFP | This study |
| YIGS16-16G13R | YIGS16 SEC16-GFP; SEC13-RFP | This study |
| Y1EK | CEN.PK530-1CK with CPOTud | This study |
| Y6EK | CEN.PK530-6CK with CPOTud | This study |
TABLE 3.
Primers used in this study
| Name | Sequence (5′ to 3′) |
|---|---|
| sar1F | GCAATCTAATCTAAGTTTTCTAGAACTAGTGGATCCCCCGGGAACAAAATGGCTGGTTGGGATATTTTTGGTTGGTTCAGAGATGTGTTGGCTTCCCTTGGTCTGTGGA |
| sar1R | CATAACTAATTACATGACTCGAGGTCGACGGTATCGATAAGCTTGTTAAATATATTGAGATAACCATTGGAACGCCT |
| p416TEF-PTEF-F | CCGGGGGATCCACTAGTTC |
| p416TEF-TCYC1-R | CAAGCTTATCGATACCGTCG |
| sec2F | GCATAGCAATCTAATCTAAGTTTTCTAGAACTAGTGGATCCAACAAAATGGATGCTTCTGAGGAAGCAAA |
| sec2R | CATAACTAATTACATGACTCGAGGTCGACGGTATCGATAAGCTTGTTATTGCTGTTCCTGGGCATC |
| sec4F | GCATAGCAATCTAATCTAAGTTTTCTAGAACTAGTGGATCCAACAAAATGTCAGGCTTGAGAACTGTTTC |
| sec4R | CGTGACATAACTAATTACATGACTCGAGGTCGACGGTATCGATTCAACAGCAATTTGATTTAGAAC |
| sec15F | GCATAGCAATCTAATCTAAGTTTTCTAGAACTAGTGGATCCAACAAAATGGACCAAGAAGGCCAGCCAGTT |
| sec15R | GCGTGACATAACTAATTACATGACTCGAGGTCGACGGTATCGATTTAACGTCTATTAAAAAATTTGGC |
| sec16F | GCATAGCAATCTAATCTAAGTTTTCTAGAACTAGTGGATCCAACAAAATGACACCTGAAGCCAAGAA |
| sec16R | CGTGACATAACTAATTACATGACTCGAGGTCGACGGTATCGATTTATTGTATGTTATCCATTAC |
| ypt32F | GCATAGCAATCTAATCTAAGTTTTCTAGAACTAGTGGATCCAACAAAATGAGCAACGAAGATTACGG |
| ypt32R | GCGTGACATAACTAATTACATGACTCGAGGTCGACGGTATCGATTTAACAACAGTTGCTGGAT |
| Psec16-GPD-R | ATTATTTACGTATTCTTTGAAATGGCGAGTATTGATAATGATAAACTGAGAGAAGGAGAAAGCACTTAAAAG |
| Psec16-GPD-F | CTTCTTTTAAGTGCTTTCTCCTTCTCTCAGTTTATCATTATCAATACTCGCCATTTC |
| SEC16pro-L-F | GTCAGGGTGTTCTTCGATTTTCC |
| SEC16pro-L-R | GTCAAGGAGGGTATTCTGGGCCTCCATGTCATGGCTCGTCATGGAAAAGGCC |
| amdSYM-F | GACATGGAGGCCCAGAATAC |
| amdSYM-R | CAGTATAGCG ACCAGCATTC |
| SEC16pro-rec-F | GTATGTGAATGCTGGTCGCTATACTGTCGGTTAACCTTGGTTGTTGC |
| SEC16pro-rec-R | ATGGCTCGTCATGGAAAAGGCC |
| GPDpro-F | GTTAGTTTTTGGGCCTTTTCCATGACGAGCCATAGTTTATCATTATCAATACTCGCCATTTCAAAGAATACG |
| GPDpro-R | GGTTTTTCCTTTTCTTGGCTTCAGGTGTCATTTTGTTATCCGTCGAAACTAAGTTCTGGTGTTTTAAAACTAAAAAAAAGAC |
| SEC16pro-R-F | ATGACACCTGAAGCCAAGAAAAGGAAAAACCAAAAGAAG |
| SEC16pro-R-R | CTCTTCGTATTCTTGAGACATGGC |
| sec16-end-w/o TAA-F | GGTTGGTTGAAGAAAGATACTGGC |
| sec16-end-w/o TAA-R | TTGTATGTTATCCATTACATTAACATAGCC |
| sec16-G4S-GFP-F | GAAGAAAACAGCGAGAGGCTATGTTAATGTAATGGATAACATACAAGGTGGAGGTGGATCTATGCGAATCCCCGGGTTAATTAAC |
| GFP-R | CTATTTGTATAGTTCATCCATGCCATGTG |
| GFP-tSEC16-F | GCTGCTGGGATTACACATGGCATGGATGAACTATACAAATAGTTATAGTAATACGTTTTTCTTTCATGGTACCC |
| tSEC16-R | GGTATGATAGAATAATATTAGTATAAGCAAAGTTTCAG |
| tSEC16-amds-f | GATGAATGCGATCTGAAACTTTGCTTATACTAATATTATTCTATCATACCGACATGGAGGCCCAGAATACCCTCC |
| amds-R | CAGTATAGCGACCAGCATTCACATACG |
| amds-tSEC16-overlap-f | GAAAGTAATATCATGCGTCAATCGTATGTGAATGCTGGTCGCTATACTGTTATAGTAATACGTTTTTCTTTCATGGTACCC |
| tSEC16-overlap-R | AGTACAAGACTTTTCAAATGCTAACG |
| sec13-end-w/o TAA-F | CCATTGGCGTTAACTCTGCTTC |
| sec13-end-w/o TAA-R | CTGATGAACTTCACCAGCGGGTTC |
| sec13-G4S-RFP-F | GGAAAATCTTGAGGGTAAATGGGAACCCGCTGGTGAAGTTCATCAGGGAGGAGGTGGTTCTATGGCCTCCTCCGAGGACGTCATC |
| RFP-R | TTAGGCGCCGGTGGAGTGGCGGCCC |
| RFP-tSEC13-F | GTGGAACAGTACGAGCGCGCCGAGGGCCGCCACTCCACCGGCGCCTAAAGAGTATCAAGAATTTAAAATGAAACATCTCAAAAGAAAAAAG |
| tSEC13-R | GTTCGCTTGTTTTTGTATAAGAAATATCTATGAC |
| tSEC13-amds-f | CTTCTTATATTAATATTGTCATAGATATTTCTTATACAAAAACAAGCGAACGACATGGAGGCCCAGAATACCCTCC |
| amds-R | CAGTATAGCGACCAGCATTCACATACG |
| amds-tSEC13-overlap-f | GAAAGTAATATCATGCGTCAATCGTATGTGAATGCTGGTCGCTATACTGAGAGTATCAAGAATTTAAAATGAAACATCTCAAAAGAAAAAAG |
| tSEC13-overlap-R | GGATGTTGGGAGGAGATGCTGTCG |
| KpnI-alpha-leader-F | AAGGTACCAACAAAATGAGATTTCCATCTATTTTTACTGC |
| XhoI-cellulase-R | GAAAAGAAGATAATATTTTTATATAATTATATTAATCTTAGTTTCTAG |
| qpcr-ACT1-F | GCCTTCTACGTTTCCATCCA |
| qpcr-ACT1-R | GGCCAAATCGATTCTCAAAA |
| qpcr-sar1-f | AGCTCGTCGTTTATGGAAGGA |
| qpcr-sar1-r | GCGTTTGGAGCATCGATCTT |
| qpcr-sec2-f | GTGTTTGCTAAAGTTGCGGC |
| qpcr-sec2-r | ACCCTGCTGTCTCTTTTGGT |
| qpcr-sec4-f | TGGTGATTCTGGTGTTGGGA |
| qpcr-sec4-r | CCAGCGGTATCCCAAAGTTG |
| qpcr-sec15-f | TGCATGACACACCAAGTACA |
| qpcr-sec15-r | ACACCGTTGGCATTTGGAAA |
| qpcr-ypt32-f | CAGTACCAACCGATGAAGCG |
| qpcr-ypt32-r | CCGACCCGCTTAAGTCTACT |
| qpcr-sec16-f | ACGTTCAGGCCCCATCAATA |
| qpcr-sec16-r | GGGAGAGGATGGTGATGGAG |
The promoter replacement cassette SL-amdSYM-GPD-SR was constructed as follows. The left arm, SL, which is homologous to the 5′ upstream region of the SEC16 native promoter, was amplified by the primer pair SEC16pro-L-F and SEC16pro-L-R based on the yeast genome; the right arm, SR, which is homologous to the 3′ downstream region of the SEC16 native promoter, was amplified by the primer pair SEC16pro-R-F and SEC16pro-R-R based on the yeast genome; the selection marker amdSYM was amplified from plasmid pamdSYM by the primers amdSYM-F and amdSYM-R; the GPD promoter was amplified from the yeast genome by GPDpro-F and GPDpro-R; all four of these parts were fused together by fusion PCR, resulting in the fragment SL-amdSYM-GPD-SR. The replacement of the SEC16 native promoter by the GPD promoter was completed by transforming the replacement cassette SL-amdSYM-GPD-SR into the cell. The amdSYM marker was removed from the genome by transformation of a marker-free fragment (SL-GPD), and transformants were selected by counterselecting on a fluoroacetamide plate.
The yeast CEN.PK 530-6CK was generated by eliminating the plasmid pAlphaAmyCPOT from YIGS16 in yeast extract-peptone-ethanol (YPE). The yeast AACK-16G13R was constructed by fusing enhanced green fluorescent protein (EGFP) to the C′ terminus of Sec16p and RFP to the C′ terminus of Sec13p. The yeast YIGS16-16G13R was constructed in the same way as AACK-16G13R.
Media.
The yeast extract-peptone-dextrose (YPD) medium contained 10 g/liter yeast extract, 20 g/liter peptone, and 20 g/liter glucose. The YPE medium contained 10 g/liter yeast extract, 20 g/liter peptone, 10 g/liter ethanol, and 0.5 g/liter glucose. The SD-URA medium contained 6.9 g/liter yeast nitrogen base without amino acids (YNB w/o AA), 0.77 g/liter CSM-URA, and 20 g/liter glucose. The α-amylase production experiment was performed with SD-2xSCAA medium (35), which contained 20 g/liter glucose, 6.9 g/liter YNB w/o AA, 190 mg/liter Arg, 400 mg/liter Asp, 1,260 mg/liter Glu, 130 mg/liter Gly, 140 mg/liter His, 290 mg/liter Ile, 400 mg/liter Leu, 440 mg/liter Lys, 108 mg/liter Met, 200 mg/liter Phe, 220 mg/liter Thr, 40 mg/liter Trp, 52 mg/liter Tyr, 380 mg/liter Val, 1 g/liter bovine serum albumin (BSA), 5.4 g/liter Na2HPO4, and 8.56 g/liter NaH2PO4 · H2O (pH 6.0 by NaOH). In the bioreactor batch fermentations, SD-2xSCAA medium was used. Strains were inoculated into 600 ml of SD-2xSCAA medium in a 1-liter bioreactor (DasGip, Jülich, Germany) with an initial OD of 0.02 to 0.032. The bioreactor system was run at 30°C, 600 rpm, and 36 liters/h airflow, and the pH value was maintained at 6 by the addition of NaOH. In the intracellular α-amylase degradation test, the components of S-2xSCAA medium are the same as those of SD-2xSCAA, except for the lack of glucose. In the vitamin C test, a vitamin C stock (500 mM) was sterilized by filtration and added to the SD-2xSCAA medium to reach the desired experimental concentrations.
Physiological characterization.
The dry cell weight and the concentrations of glucose, ethanol, and glycerol were measured as described previously (6). A summit high-pressure liquid chromatograph (Dionex, Sunnyvale, CA, USA) was used with an Aminex HPX-87H column (Bio-Rad, Hercules, CA, USA) at 65°C, using 5 mM H2SO4 as the mobile phase at a flow rate of 0.6 ml/min.
Enzyme quantification.
The enzyme activity of α-amylase in the medium was measured using an α-amylase assay kit (K-CERA; Megazyme, Wicklow, Ireland), and α-amylase from Aspergillus oryzae (Sigma, St. Louis, MO, USA) was used as a standard. The enzyme activity of the endoglucanase was measured using a cellulase assay kit (K-CELLG3; Megazyme, Wicklow, Ireland) at 50°C for 10 min. The enzyme activity of the glucan 1,4-α-glucosidase was measured using an amyloglucosidase assay kit (R-AMGR3; Megazyme, Wicklow, Ireland).
Degradation and secretion of intracellular α-amylase.
The seed cultures of the AACK and YIGS16 strains were individually transferred to 100 ml of SD-2xSCAA medium, and the secondary cultures were incubated at 30°C. When the OD600 of the secondary cultures reached 1, yeast cells were collected from 20 ml of culture by centrifugation. Cell pellets were washed twice with S-2xSCAA medium and then resuspended in 20 ml of S-2xSCAA medium. The cells were incubated at 30°C for another 48 h at a speed of 200 rpm. The amount of degraded α-amylase was the difference between the amount of intracellular α-amylase at the beginning and the sum of the amounts of intracellular and extracellular α-amylase after 48 h. The amount of secreted α-amylase was equal to the amount of extracellular α-amylase after 48 h. The amount of residual α-amylase was equal to the amount of intracellular α-amylase after 48 h.
Intracellular α-amylase extraction.
Yeast cells from 1 ml of culture were harvested, washed with 1× phosphate-buffered solution (pH 7.4) (PBS) twice, and then resuspended in 1 ml of PBS with 10 μl of Halt protease inhibitor cocktail (Thermo Fisher, Waltham, MA, USA). A 500-μl cell resuspension aliquot was transferred to 1.0-mm silica spheres in a lysing matrix tube (MP Biomedicals, Santa Ana, CA, USA). The cells were lysed using a FastPrep-24 tissue and cell homogenizer (MP Biomedicals, Santa Ana, CA, USA) for 2 min at 6.5 m/s. The cells were put on ice during the 5-min interval between the two runs. The supernatant was collected for α-amylase quantification by centrifugation at 16,000 × g for 5 min. The supernatant includes cell wall-associated and intracellular α-amylase.
Transcriptome analysis.
AACK and YIGS16 cells were collected at an OD600 of ≈1 (22.2 h for AACK and 21 h for YIGS16) in the glucose phase during batch fermentation for RNA extraction and the subsequent transcriptome profiling. For the transcriptome profiling of cells harvested in the ethanol phase, the cells were collected at 44.5 h (for AACK) and 47 h (for YIGS16). The total RNA was extracted by using an RNeasy kit (Qiagen, Valencia, CA, USA). The RNA samples were prepared using the TruSeq mRNA sample preparation kit (Illumina, San Diego, CA, USA) and sequenced with a mid-output flow cell using the NextSeq run 2 × 75 method (Illumina, San Diego, CA, USA). The raw RNA sequencing (RNA-seq) data were processed by Bowtie2 (43), SAMTools (44, 45), and BEDTools (46) for aligning raw counts. The differentially expressed transcripts were analyzed using the DESeq R package (47). The reporter GO terms and reporter transcription factor analyses were performed using the PIANO R package (48).
ROS detection.
The cells for the ROS detection experiments were cultivated in shake flasks and harvested at an OD600 of ≈1. The cells then were washed twice with PBS and once with 50 mM SCB (sodium citrate buffer, pH 5). The cell pellets were resuspended in 1 ml of SCB containing 1 μl of 50 mM dihydrorhodamine 123 (Thermo Fisher, Waltham, MA, USA) and were then incubated in the dark for 30 min at room temperature. After incubation, the cells were washed twice with SCB and resuspended in 1 ml of SCB. Two-hundred-microliter aliquots of the cell suspensions were loaded into wells of a 96-well black plate for fluorescence measurements with a fluorescence microplate reader (FLUOstar Omega; BMG Labtech, Germany) equipped with a 485-nm excitation filter and a 520-nm emission filter.
ER staining.
The cells for the ER staining experiments were cultivated in shake flasks and harvested at an OD600 of ≈1. The cells then were washed twice with PBS and once with Hanks' balanced salt solution without phenol red (HBSS). The cell pellets were resuspended in 1 ml of HBSS that contained 5 μl of ER-tracker blue-white and incubated for 30 min at 30°C. After their incubation, the cells were washed twice with HBSS and resuspended in 1 ml of HBSS. Two-hundred-microliter aliquots of each sample were loaded into the wells of a 96-well black plate for fluorescence measurements with a fluorescence microplate reader equipped with a 355-nm excitation filter and a 590-nm emission filter.
Kar2p detection.
The AACK and YIGS16 cells were cultivated in SD-2xSCAA medium for 96 h using tube fermentation. The supernatant then was harvested by centrifugation at 16,000 × g for 5 min, and the extracellular Kar2p in the supernatant was detected by Western blotting. The rabbit polyclonal antibody Kar2 (y-115) (Santa-Cruz, CA, USA) was used as the primary antibody, and goat anti-rabbit IgG-horseradish peroxidase (Santa-Cruz, CA, USA) was used as the secondary antibody for the Western blot.
Mitochondrial staining.
The mitochondrial staining method was described in a previous publication (49). Briefly, the cells were cultivated in SD-2xSCAA medium using tube fermentation and harvested when the OD600 was ≈1. The cells then were resuspended in 10 mM HEPES buffer (pH 7.4, 5% glucose with 100 nM MitoTracker Green FM; Thermo Fisher, Waltham, MA, USA) and incubated in the dark for 30 min at room temperature. After their incubation, the cells were resuspended in 10 mM HEPES without MitoTracker Green FM and were observed with a Leica DMI4000 B fluorescence microscope (Leica, Wetzlar, Germany) with differential interference contrast and GFP filters. Aliquots of the cell suspensions (200 μl) were loaded into the wells of a 96-well black plate for fluorescence measurement by a fluorescence microplate reader with a 485-nm excitation filter and a 520-nm emission filter.
Airyscan imaging.
The AACK and YIGS16 cells were incubated in SD-2xSCAA medium and harvested when the OD600 reached 1. The cells then were washed twice with PBS. The ERES colocalization was observed with an LSM 880 with an Airyscan (Carl Zeiss, Jena, Germany) system at room temperature with a Plan-Apochromat 63×/1.45 oil DIC M27 objective. Light with wavelengths of 561 nm and 488 nm were used to excite the RFP and GFP, respectively. The emission wavelengths used for RFP and GFP detection were 579 nm and 523 nm, respectively.
Fluorescence signal detection of ERESs.
The samples were incubated in tubes in the SD-2xSCAA medium. The samples were collected when the OD600 reached 1. The samples then were washed twice with PBS, and the OD of samples was adjusted to exactly 1. An aliquot of each sample (200 μl) was loaded into each well. The experiment was performed with a 96-well black flat-bottom plate. The samples were excited at 485 and 544 nm (for the GFP and RFP, respectively), and their emissions were collected at 520 and 590 nm.
qPCR.
The cells for the quantitative PCR (qPCR) experiment were incubated in SD-2xSCAA medium using shake flask fermentation. The cells were collected when the OD was ≈1. The total RNA was extracted by using the RNeasy kit. The quality of total RNA was controlled by a NanoDrop 2000 (Thermo Fisher, Waltham, MA, USA). The cDNA was reverse transcribed from the total mRNA using the QuantiTect reverse transcription kit (Qiagen, Valencia, CA, USA). The qPCR analysis was performed using the DyNAmo flash SYBR green qPCR kit (Thermo Fisher, Waltham, MA, USA). ACT1 was used as the normalization standard. All data were presented as the averages of results from three trials.
Accession number(s).
The RNA-seq data were submitted to the ENA database and have been assigned accession no. PRJEB15327.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by the Novo Nordisk Foundation, the Knut and Alice Wallenberg Foundation, and the European Research Council (grant no. 247013).
We thank Verena Siewers, José L. Martínez, Guokun Wang, Xin Chen, Michael Gossing, and Markus Bisschops for useful comments and discussions. We thank the National Microscopy Infrastructure, NMI (VR-RFI 2016-00968), for helping with the Airyscan imaging. We thank David Bergenholm for helping with the RNA-seq analysis.
J.B., M.H., and J.N. conceived the study. J.B. and M.H. designed and performed the experiments. D.P. and J.N. supervised the project. J.B., M.H., D.P., and J.N. analyzed the data and wrote the manuscript.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03400-16.
REFERENCES
- 1.Huang M, Bao J, Nielsen J. 2014. Biopharmaceutical protein production by Saccharomyces cerevisiae: current state and future prospects. Pharma Bioprocess 2:167–182. doi: 10.4155/pbp.14.8. [DOI] [Google Scholar]
- 2.Benjaphokee S, Hasegawa D, Yokota D, Asvarak T, Auesukaree C, Sugiyama M, Kaneko Y, Boonchird C, Harashima S. 2012. Highly efficient bioethanol production by a Saccharomyces cerevisiae strain with multiple stress tolerance to high temperature, acid and ethanol. New Biotechnol 29:379–386. doi: 10.1016/j.nbt.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Huang M, Bai Y, Sjostrom SL, Hallstrom BM, Liu Z, Petranovic D, Uhlen M, Joensson HN, Andersson-Svahn H, Nielsen J. 2015. Microfluidic screening and whole-genome sequencing identifies mutations associated with improved protein secretion by yeast. Proc Natl Acad Sci U S A 112:E4689–E4696. doi: 10.1073/pnas.1506460112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komano H, Rockwell N, Wang GT, Krafft GA, Fuller RS. 1999. Purification and characterization of the yeast glycosylphosphatidylinositol-anchored, monobasic-specific aspartyl protease yapsin 2 (Mkc7p). J Biol Chem 274:24431–24437. doi: 10.1074/jbc.274.34.24431. [DOI] [PubMed] [Google Scholar]
- 5.Carlson M, Taussig R, Kustu S, Botstein D. 1983. The secreted form of invertase in Saccharomyces cerevisiae is synthesized from mRNA encoding a signal sequence. Mol Cell Biol 3:439–447. doi: 10.1128/MCB.3.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou J, Tyo K, Liu Z, Petranovic D, Nielsen J. 2012. Engineering of vesicle trafficking improves heterologous protein secretion in Saccharomyces cerevisiae. Metab Eng 14:120–127. doi: 10.1016/j.ymben.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Tyo KE, Martinez JL, Petranovic D, Nielsen J. 2012. Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae. Biotechnol Bioeng 109:1259–1268. doi: 10.1002/bit.24409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solow SP, Sengbusch J, Laird MW. 2005. Heterologous protein production from the inducible MET25 promoter in Saccharomyces cerevisiae. Biotechnol Prog 21:617–620. doi: 10.1021/bp049916q. [DOI] [PubMed] [Google Scholar]
- 9.Hou J, Tyo KE, Liu Z, Petranovic D, Nielsen J. 2012. Metabolic engineering of recombinant protein secretion by Saccharomyces cerevisiae. FEMS Yeast Res 12:491–510. doi: 10.1111/j.1567-1364.2012.00810.x. [DOI] [PubMed] [Google Scholar]
- 10.Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. 2009. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol Bioeng 103:1192–1201. doi: 10.1002/bit.22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoshida H, Fujita T, Cha-aim K, Akada R. 2013. N-Glycosylation deficiency enhanced heterologous production of a Bacillus licheniformis thermostable alpha-amylase in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97:5473–5482. doi: 10.1007/s00253-012-4582-2. [DOI] [PubMed] [Google Scholar]
- 12.Tang H, Wang S, Wang J, Song M, Xu M, Zhang M, Shen Y, Hou J, Bao X. 2016. N-hypermannose glycosylation disruption enhances recombinant protein production by regulating secretory pathway and cell wall integrity in Saccharomyces cerevisiae. Sci Rep 6:25654. doi: 10.1038/srep25654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valkonen M, Penttila M, Saloheimo M. 2003. Effects of inactivation and constitutive expression of the unfolded-protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 69:2065–2072. doi: 10.1128/AEM.69.4.2065-2072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schröder M. 2008. Engineering eukaryotic protein factories. Biotechnol Lett 30:187–196. doi: 10.1007/s10529-007-9524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroder M. 2007. The cellular response to protein unfolding stress, p 117–139. In Robson GD. (ed), Exploitation of fungi. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 16.Budnik A, Stephens DJ. 2009. ER exit sites–localization and control of COPII vesicle formation. FEBS Lett 583:3796–3803. doi: 10.1016/j.febslet.2009.10.038. [DOI] [PubMed] [Google Scholar]
- 17.Wendeler MW, Paccaud JP, Hauri HP. 2007. Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep 8:258–264. doi: 10.1038/sj.embor.7400893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen D, Schekman R. 2011. COPII-mediated vesicle formation at a glance. J Cell Sci 124:1–4. doi: 10.1242/jcs.069773. [DOI] [PubMed] [Google Scholar]
- 19.Bharucha N, Liu Y, Papanikou E, McMahon C, Esaki M, Jeffrey PD, Hughson FM, Glick BS. 2013. Sec16 influences transitional ER sites by regulating rather than organizing COPII. Mol Biol Cell 24:3406–3419. doi: 10.1091/mbc.E13-04-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feizi A, Österlund T, Petranovic D, Bordel S, Nielsen J. 2013. Genome-scale modeling of the protein secretory machinery in yeast. PLoS One 8:e63284. doi: 10.1371/journal.pone.0063284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stalder D, Mizuno-Yamasaki E, Ghassemian M, Novick PJ. 2013. Phosphorylation of the Rab exchange factor Sec2p directs a switch in regulatory binding partners. Proc Natl Acad Sci U S A 110:19995–20002. doi: 10.1073/pnas.1320029110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mumberg D, Müller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:4. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 23.Chakravarthi S, Jessop CE, Bulleid NJ. 2006. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep 7:271–275. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branduardi P, Fossati T, Sauer M, Pagani R, Mattanovich D, Porro D. 2007. Biosynthesis of vitamin C by yeast leads to increased stress resistance. PLoS One 2:e1092. doi: 10.1371/journal.pone.0001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yorimitsu T, Sato K. 2012. Insights into structural and regulatory roles of Sec16 in COPII vesicle formation at ER exit sites. Mol Biol Cell 23:2930–2942. doi: 10.1091/mbc.E12-05-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Echevarria W, Leite MF, Guerra MT, Zipfel WR, Nathanson MH. 2003. Regulation of calcium signals in the nucleus by a nucleoplasmic reticulum. Nat Cell Biol 5:440–446. doi: 10.1038/ncb980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil C, Walter P. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol 13:349–355. doi: 10.1016/S0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- 28.Semenza JC, Hardwick KG, Dean N, Pelham HR. 1990. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61:1349–1357. doi: 10.1016/0092-8674(90)90698-E. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Chang A. 2008. Heat shock response relieves ER stress. EMBO J 27:1049–1059. doi: 10.1038/emboj.2008.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou J, Osterlund T, Liu ZH, Petranovic D, Nielsen J. 2013. Heat shock response improves heterologous protein secretion in Saccharomyces cerevisiae. Appl Microbiol Biotechnol 97:3559–3568. doi: 10.1007/s00253-012-4596-9. [DOI] [PubMed] [Google Scholar]
- 31.Patil CK, Li H, Walter P. 2004. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol 2:E246. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henry SA, Kohlwein SD, Carman GM. 2012. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics 190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voelker DR. 1991. Organelle biogenesis and intracellular lipid transport in eukaryotes. Microbiol Rev 55:543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott M, Gogvadze V, Orrenius S, Zhivotovsky B. 2007. Mitochondria, oxidative stress and cell death. Apoptosis 12:913–922. doi: 10.1007/s10495-007-0756-2. [DOI] [PubMed] [Google Scholar]
- 35.Nakano A, Muramatsu M. 1989. A novel Gtp-binding protein, Sar1p, is involved in transport from the endoplasmic-reticulum to the Golgi-apparatus. J Cell Biol 109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Espenshade P, Gimeno RE, Holzmacher E, Teung P, Kaiser CA. 1995. Yeast Sec16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J Cell Biol 131:311–324. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ. 2006. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic 7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Connerly PL, Esaki M, Montegna EA, Strongin DE, Levi S, Soderholm J, Glick BS. 2005. Sec16 is a determinant of transitional ER organization. Curr Biol 15:1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- 39.Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. 2009. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol 187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J, Kaufman RJ. 2006. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ 13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 41.Hetz C. 2012. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13:89–102. [DOI] [PubMed] [Google Scholar]
- 42.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Varemo L, Nielsen J, Nookaew I. 2013. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res 41:4378–4391. doi: 10.1093/nar/gkt111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martínez JL, Meza E, Petranovic D, Nielsen J. 2016. The impact of respiration and oxidative stress response on recombinant α-amylase production by Saccharomyces cerevisiae. Metab Eng Commun 3:205–210. doi: 10.1016/j.meteno.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.