ABSTRACT
The aim of this study was to demonstrate the capacity of probiotic lactobacilli to hydrolyze immunogenic gluten peptides. Eighteen commercial strains of probiotic lactobacilli with highly variable peptidase activity (i.e., aminopeptidase N, iminopeptidase, prolyl endopeptidyl peptidase, tripeptidase, prolidase, prolinase, and dipeptidase), including toward Pro-rich peptides, were tested in this study. Ten probiotic strains were selected on the basis of their specific enzyme activity. When pooled, these 10 strains provided the peptidase portfolio that is required to completely degrade the immunogenic gluten peptides involved in celiac disease (CD). The selected probiotic mixture was able to completely hydrolyze well-known immunogenic epitopes, including the gliadin 33-mer peptide, the peptide spanning residues 57 to 68 of the α9-gliadin (α9-gliadin peptide 57-68), A-gliadin peptide 62-75, and γ-gliadin peptide 62-75. During digestion under simulated gastrointestinal conditions, the pool of 10 selected probiotic lactobacilli strongly hydrolyzed the wheat bread gluten (ca. 18,000 ppm) to less than 10 ppm after 360 min of treatment. As determined by multidimensional chromatography (MDLC) coupled to nanoelectrospray ionization (nano-ESI)-tandem mass spectrometry (MS/MS), no known immunogenic peptides were detected in wheat bread that was digested in the presence of the probiotics. Accordingly, the level of cytokines (interleukin 2 [IL-2], IL-10, and interferon gamma [IFN-γ]) produced by duodenal biopsy specimens from CD patients who consumed wheat bread digested by probiotics was similar to the baseline value (negative control). Probiotics that specifically hydrolyze gluten polypeptides could also be used to hydrolyze immunogenic peptides that contaminate gluten-free products. This could provide a new and safe adjunctive therapy alternative to the gluten-free diet (GFD).
IMPORTANCE This study confirmed that probiotic Lactobacillus strains have different enzymatic abilities for hydrolyzing polypeptides, including the Pro-rich epitopes involved in the pathology of CD. Ten lactobacilli with complementary peptidase activities that hydrolyze gluten peptides during simulated gastrointestinal digestion were selected and tested. The results collected showed the potential of probiotic formulas as novel dietary treatments for CD patients.
KEYWORDS: celiac disease, probiotic lactobacilli, peptidases, gluten immunogenic peptides, cytokines
INTRODUCTION
Celiac disease (CD) is an immune-mediated enteropathy that is triggered by the ingestion of gluten (in genetically susceptible individuals) and that results from the interaction between gluten and immune, genetic, and environmental factors (1). The prevalence of CD is as high as 1% in European countries (2), and its clinical presentation can include a wide spectrum of insidious symptoms, including abdominal pain, although it is often asymptomatic. The ingestion of gluten is necessary for the development of disease, and the only effective therapy for CD patients is a gluten-free diet (GFD). Additional environmental factors (such as interactions among intestinal microbiota and immunity and dietary factors) seem to be involved in the development of CD (3–5). The intestinal microbiota drives mucosal cell differentiation, intestinal permeability, and the immune response to environmental antigens (3). The salivary and intestinal microbiotas of CD patients differ from those of healthy controls both at the time of diagnosis and after remission following a GFD (6–8). A GFD itself could be involved in the microbial imbalance of CD patients (9, 10). A GFD decreases the relative amount of several beneficial strains of gut bacteria (e.g., Bifidobacterium and Lactobacillus) and reduces the ability of fecal samples to stimulate a host's immunity (10). Recently, novel treatments for CD patients have been proposed (11, 12). Among the novel therapies under investigation is an enzyme strategy that uses microbial proteases and peptidases for gluten detoxification (13, 14). Current research is focused on the oral administration of microbial endopeptidases with various degrees of tolerance to the gut environment, the transamidation of gliadin, and the use of trans-glutaminase inhibitors (15, 16). Indeed, several microbial proteases and peptidases are often inhibited in the stomach by pepsin and pH, which reduce their activity on gluten molecules and enable immunogenic peptides to be generated in the small intestine (17). A biotechnological strategy that hydrolyzes gluten (residual concentration, ≤8 ppm) during food processing and that uses select sourdough lactobacilli and food-grade fungal proteases was recently developed (18, 19). The combined activity of general aminopeptidase type N (PepN; EC 3.4.11.11), endopeptidase (PepO; EC 3.4.23), and prolyl endopeptidyl peptidase (PEP; EC 3.4.21.26) promoted the hydrolysis of the CD immunogenic 33-mer peptide into five small peptides (20). PepN and X-prolyl dipeptidyl aminopeptidase (PepX; EC 3.4.14.5) produced dipeptides from the 33-mer, which were mainly degraded into free amino acids (FAA) via prolidase (PepQ; EC 3.4.13.9) and PepX. The remaining dipeptides were hydrolyzed through PepQ. Overall, 5 peptidases are required to completely degrade the 33-mer and other synthetic immunogenic peptides (20).
In vitro studies (3) have demonstrated that the proteinase and peptidase activities of the gut microbiota degrade gliadin peptides, which affects their toxicity. Indeed, gluten-degrading bacteria have been isolated from the gastrointestinal tracts of humans and pigs (21–25). Previously, it was shown that Lactobacillus rhamnosus GG improves the intestinal permeability of Caco-2 cells when exposed to gliadin peptides (26). Strains belonging to the Bifidobacterium genus (e.g., Bifidobacterium longum CECT 7347, Bifidobacterium bifidum CECT 7365) decreased the cytotoxic and inflammatory effects of gluten peptides (25, 27). On the basis of this scientific evidence, the use of select probiotic strains for their ability to hydrolyze gluten epitopes under human gastrointestinal conditions could be a new strategy to improve/maintain the health of CD patients.
The ability of probiotic lactobacilli to hydrolyze gluten epitopes under simulated gastrointestinal conditions was evaluated in this study. A combined approach of multidimensional chromatography (MDLC) and nanoelectrospray ionization (nano-ESI)-tandem mass spectrometry (MS/MS) with immunological analysis was performed to determine the gluten-detoxifying activity of the probiotic strains.
RESULTS
Peptidase activities of probiotic lactobacilli.
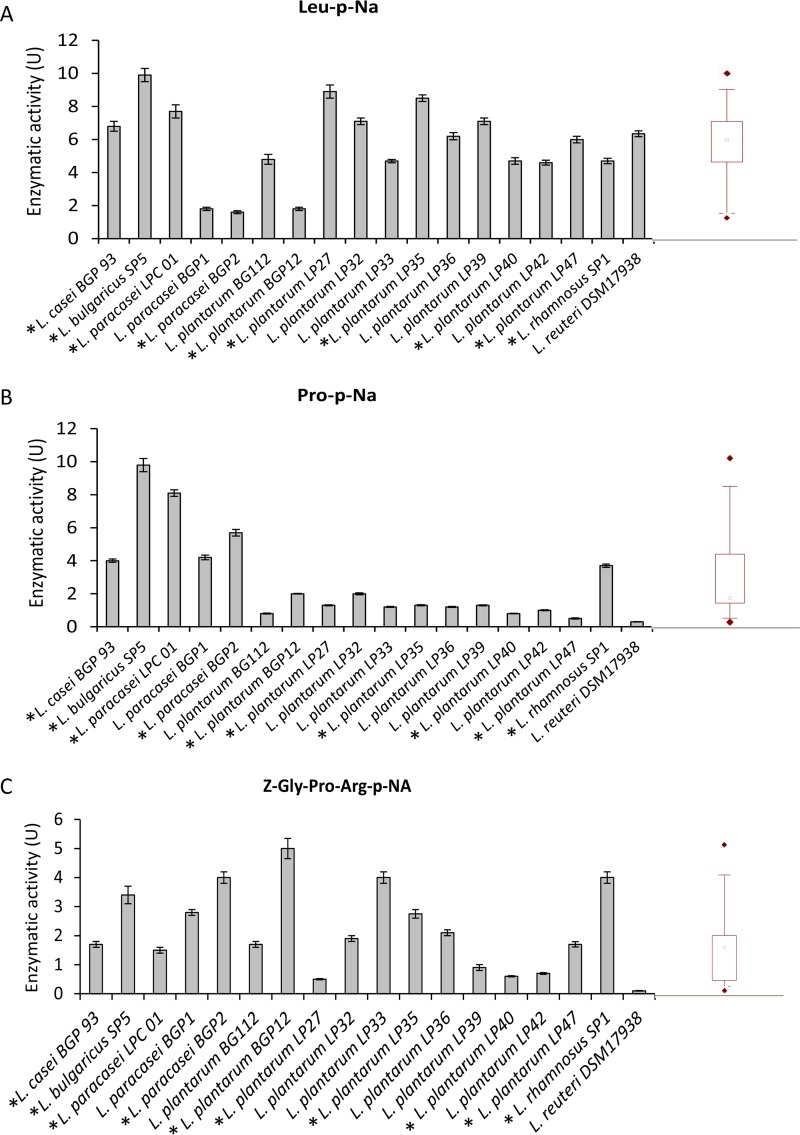
All Lactobacillus strains showed general PepN activities for Leu-p-nitroanilides (Leu-p-NA) which ranged from 1.79 to 9.96 units (U) (median value, 6.08 U) (Fig. 1A). The strains Lactobacillus delbrueckii subsp. bulgaricus SP5 and L. plantarum LP27 and LP35 showed the highest activities (9.96 ± 0.44, 9.94 ± 0.37, and 8.59 ± 0.35 U, respectively).
FIG 1.
Aminopeptidase type N (PepN; EC 3.4.11.11) (A), proline iminopeptidase (PepI; EC 3.4.11.9) (B), and prolyl endopeptidyl peptidase (PEP; EC 3.4.21.26) (C) activities of Lactobacillus strains for the Leu-p-nitroanilide (Leu-p-NA), Pro-p-NA, and Z-Gly-Pro-Arg-p-NA substrates, respectively. One unit of activity was defined as the amount of enzyme required to liberate 1 μmol of p-NA per min under the assay conditions. The data are represented as means ± standard deviations from three independent assays. Box plots are also shown, in which the tops and bottoms of the boxes represent the 75th and 25th percentiles of the data, respectively. The tops and bottoms of the error bars represent the 5th and 95th percentiles of the data, respectively. The rhombus in each box plot extends to the outliers (♢). *, selected strains.
Compared to their PepN activity, these strains showed lower proline iminopeptidase (PepI; EC 3.4.11.9) activity (median value, 1.35 U) (Fig. 1B). The strains with the highest PepI activity were L. paracasei BGP2 and LPC01 and especially L. delbrueckii subsp. bulgaricus SP5 (5.73 ± 0.11, 8.05 ± 0.37, and 9.81 ± 0.46 U, respectively). The median value for PEP activity was 1.81 U, and L. paracasei BGP12 and BGP2, L. plantarum LP33, and L. rhamnosus SP1 were the strains with the highest PEP activity (Fig. 1C).
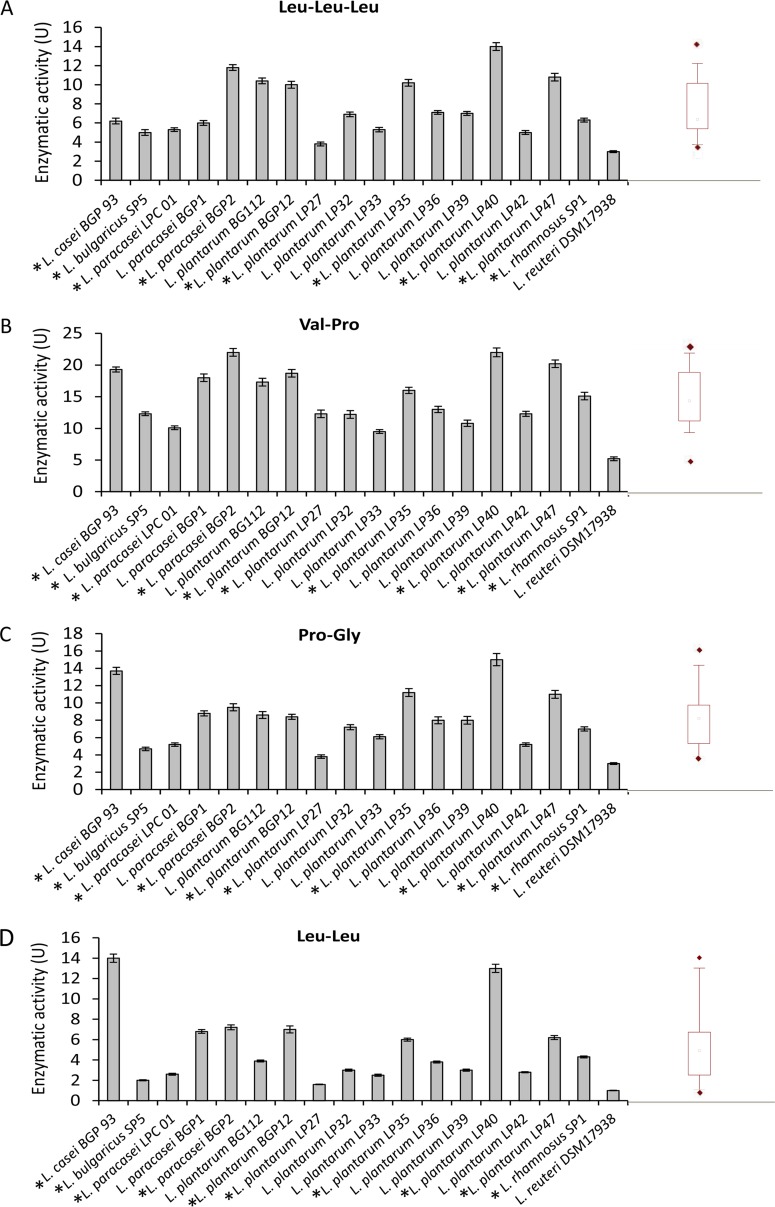
All strains also showed tripeptidase (PepT; EC 3.4.11.4; median value, 6.57 U; Fig. 2A), PepQ (median value, 14.08 U; Fig. 2B), prolinase (PepR; EC 3.4.13.8; median value, 7.99 U; Fig. 2C), and dipeptidase (PepV; EC 3.4.13.11; median value, 3.75 U; Fig. 2D) activities. L. plantarum LP40 was the strain with the highest PepT, PepQ, and PepR activities, whereas L. casei BGP93 had the highest PepV activity.
FIG 2.
Tripeptidase (PepT; EC 3.4.11.4) (A), prolidase (PepQ; EC 3.4.13.9) (B), prolinase (PepR; EC 3.4.13.8) (C), and dipeptidase (PepV; EC 3.4.13.11) (D) activities of Lactobacillus (cell density of 109 CFU/ml) strains for Leu-Leu-Leu, Val-Pro, Pro-Gly, and Leu-Leu substrates, respectively. One unit of activity was defined as the amount of enzyme required to liberate 1 μmol amino acid per min under the assay conditions. The data are represented as the means ± standard deviations from three independent assays. Box plots are also shown, in which the tops and bottoms of the boxes represent the 75th and 25th percentiles of the data, respectively. The tops and bottoms of the error bars represent the 5th and 95th percentiles of the data, respectively. The rhombus in each box plot extends to the outliers (♢). *, selected strains.
The strains with the highest complementary peptidase activities (L. casei BGP93; L. delbrueckii subsp. bulgaricus SP5; L. paracasei LPC01 and BGP2; and L. plantarum BGP12, LP27, LP35, LP40, LP47, and SP1) were selected for further analysis.
Hydrolysis of Pro-rich synthetic peptides.
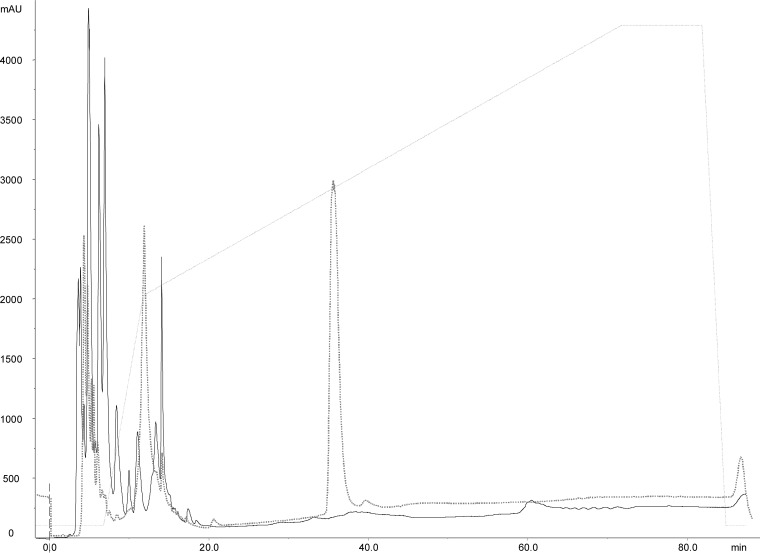
Cells (1 × 109 CFU/ml) were pooled from the following strains: L. casei BGP93; L. delbrueckii subsp. bulgaricus SP5; L. paracasei LPC01 and BGP2; and L. plantarum BGP12, LP27, LP35, LP40, LP47, and SP1. Pooled cells were incubated in a buffer solution with 200 mM either the peptide spanning residues 57 to 68 of the α9-gliadin (α9-gliadin peptide 57-68), A-gliadin peptide 62-75, γ-gliadin peptide 134-153, or the gliadin 33-mer peptide. After 24 h at 37°C, the 33-mer peptide was completely hydrolyzed by the pooled cells (1 × 109 CFU/ml) (Fig. 3). Complete hydrolysis also occurred for α9-gliadin peptide 57-68, A-gliadin peptide 62-75, and γ-gliadin peptide 134-153 (data not shown).
FIG 3.
Hydrolysis of the 33-mer peptide by 10 lactobacilli (109 CFU/ml) (Lactobacillus casei BGP93; Lactobacillus bulgaricus SP5; Lactobacillus paracasei LPC01 and BGP2; and Lactobacillus plantarum BGP12, LP27, LP35, LP40, LP47, and SP1). Reverse-phase high-performance liquid chromatography (RP-HPLC) was used to analyze 750 ppm the 33-mer peptide with probiotic lactobacilli at the beginning of incubation (gray line) and after 24 h of incubation (black line) at 37°C. mAU, milli-absorbance units.
Gluten epitope hydrolysis during simulated gastrointestinal digestion.
The addition of the 10 selected lactobacilli during simulated gastrointestinal digestion significantly decreased the final quantity of gliadin peptides in Triticum aestivum cv. Sagittario compared to that for the control. As estimated by a specific enzyme-linked immunosorbent assay (ELISA), the concentration of gliadin peptides was less than 10 ppm after incubation with the probiotic lactobacilli at 37°C for 360 min. As shown by an R5 antibody-based sandwich and competitive ELISA (R5-ELISA), the concentration of residual gluten in the control (wheat bread digested without probiotics [Ct]) was 1,200 ppm. The probiotic strains hydrolyzed gluten under the simulated gastrointestinal conditions; after 360 min of hydrolysis, the residual gluten levels were less than 20 ppm. Overall, no significant differences (P = 0.584) were found between samples with probiotic strains and skim milk (PB-SM) and samples with probiotic strains without skim milk (PB) (2.94 and 12.45 ppm, respectively). A similar amount of residual gluten was found using the R5-ELISA (data not shown).
Peptides from the hydrolyzed samples were separated by reverse-phase high-performance liquid chromatography (RP-HPLC) and identified by MDLC coupled with nano-ESI–MS/MS. No traces of known gluten epitopes were detected in the samples digested with the addition of the probiotic cells. The lowest concentration detectable by nano-ESI–MS/MS was determined using different concentrations (1 to 100 ppm) of the synthetic immunogenic 33-mer peptide with the digested wheat bread samples (Ct, PB, and PB-SM) (data not shown).
Cytokine expression in duodenal biopsy specimens from patients with CD.
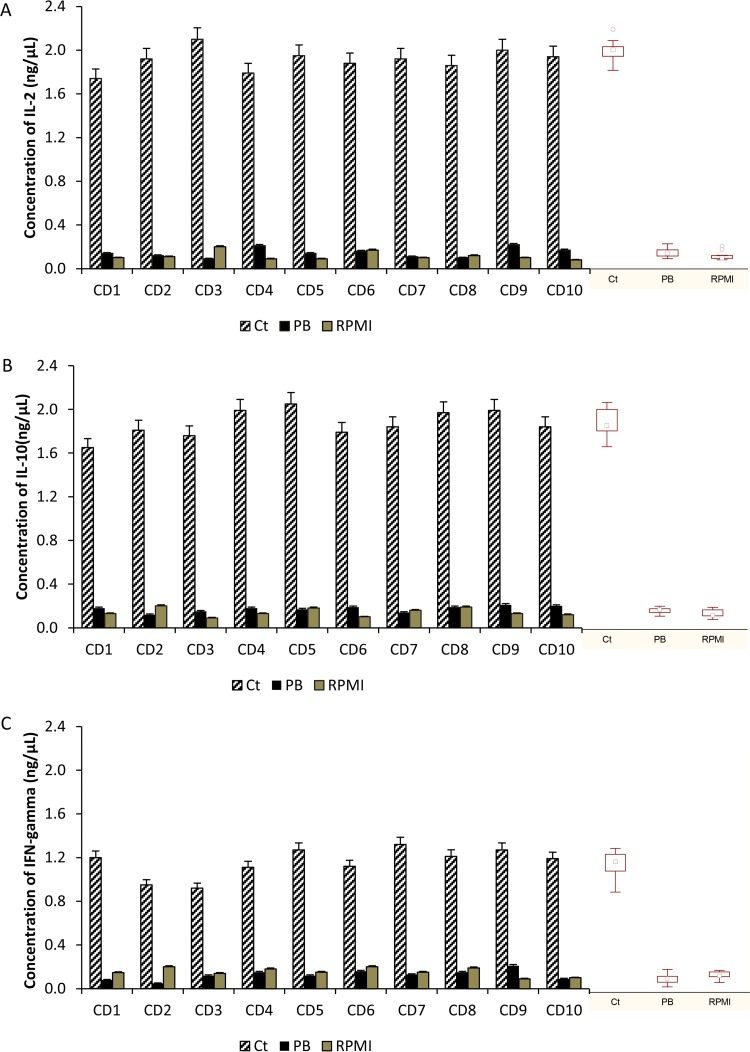
Wheat bread pepsin-trypsin (PT)-digested samples (Ct and PB) were subjected to gliadin and glutenin polypeptide extraction and used (5 mg/ml) to treat duodenal biopsy specimens from CD patients. The stimulated duodenal biopsy specimens produced significantly higher levels of interleukin 2 (IL-2) mRNA in response to PT digestion of wheat bread than in response to the negative control (RPMI 1640) (Fig. 4A). In contrast, no overexpression of IL-2 mRNA was detected in duodenal biopsy specimens treated with PT digestion products, which corresponds to PT digests of wheat bread containing 10 selected probiotic strains. According to the mRNA level, oversynthesis of the IL-2 protein was found only in the supernatant of duodenal biopsy specimens from CD patients treated with the PT digestion products of gliadins from Ct.
FIG 4.
Effects of pepsin-trypsin (PT) digestion on baker's yeast wheat bread under simulated gastrointestinal conditions in terms of interleukin 2 (IL-2) (A), interleukin 10 (IL-10) (B), and interferon gamma (IFN-γ) (C) mRNA expression. Ct, baker's yeast wheat bread hydrolyzed by gastrointestinal enzymes; PB, baker's yeast wheat bread hydrolyzed by gastrointestinal enzymes and 10 strains of lactobacilli (109 CFU/ml) (Lactobacillus casei BGP93; Lactobacillus delbrueckii subsp. bulgaricus SP5; Lactobacillus paracasei LPC01 and BGP2; and Lactobacillus plantarum BGP12, LP27, LP35, LP40, LP47, and SP1); RPMI, negative control (medium alone); GAPDH, glyceraldehyde-3-phosphate dehydrogenase (endogenous control); CD1 to CD10, celiac disease patients. Box plots are also shown.
Similar trends were also found for IL-10 and interferon gamma (IFN-γ). These genes were overexpressed only in response to the PT digestion products of wheat bread that were not treated with the 10 probiotic strains (Fig. 4B and C). The levels of IL-10 and IFN-γ mRNA in biopsy specimens treated with a PT digest of PB were not significantly (P > 0.05) different from those in the samples treated with RPMI 1640. ELISA analysis confirmed the quantitative real-time PCR (RT-PCR) data for IL-10 and IFN-γ. The highest concentrations of IL-10 and IFN-γ proteins (P < 0.05) were found in biopsy specimens treated with the PT digest of wheat bread without the 10 probiotic strains.
DISCUSSION
It is difficult for many CD patients to strictly adhere to a GFD because traces of gluten are found in the majority of processed foods. In addition, gluten-free products are generally more expensive than their counterparts, and their availability varies around the world, especially in developing countries (28, 29). It is estimated that gluten contamination of GFDs occurs for 32 to 55% of CD patients (30). Probiotics could play a key role in degrading and/or modifying immunogenic epitope contaminants in gluten-free products during gastrointestinal digestion (25, 31). This study demonstrated that 10 strains of probiotic lactobacilli hydrolyze gluten under gastrointestinal conditions. Eighteen commercial strains of probiotic lactobacilli were characterized for peptidase activity (also toward Pro-rich peptides), and all showed a high variability. Previously, it was shown that the probiotic preparation VSL#3, which contains Streptococcus thermophilus, Lactobacillus plantarum, L. acidophilus, L. casei, L. delbrueckii subsp. bulgaricus, Bifidobacterium breve, B. infantis, and B. longum, decreased the toxicity of wheat flour during extended sourdough fermentation (32). However, the ability of VSL#3 or sourdough lactobacilli to hydrolyze Pro-rich peptides and gliadin polypeptides was lost when the individual strains were tested. This occurred because no single lactic acid bacterial or bifidobacterial strain possesses all the peptidases required to degrade the peptides involved in CD (9, 20, 33). According to these findings, 10 strains were selected on the basis of their specific enzymatic activities. When pooled, the 10 strains provided the complete portfolio of peptidases required to degrade the gliadins and glutenin polypeptides involved in CD. It is well-known that several gliadins and glutenin peptides (e.g., α9-gliadin peptide 57-68 [34], A-gliadin peptide 62-75 [35], γ-gliadin peptide 134-153 [36], and the gliadin 33-mer peptide [17]) are recognized by human leukocyte antigen (HLA)-DQ2 (or HLA-DQ8) molecules. HLA molecules bind gliadin and glutenin peptides to CD4+ T cells, which initiate the CD inflammatory processes. Indeed, α9-gliadin peptide 57-68, A-gliadin peptide 62-75, γ-gliadin peptide 134-153, and especially the gliadin 33-mer peptide are not hydrolyzed by gastric and pancreatic proteases, and their small brush border membrane enzymes remain intact for a long time (more than 20 h) in the human intestine (17). The pool of 10 probiotic strains completely hydrolyzed α9-gliadin peptide 57-68, A-gliadin peptide 62-75, γ-gliadin peptide 134-153, and the gliadin 33-mer peptide in vitro.
An effective probiotic treatment removes gliadin and glutenin polypeptide contaminants from GFDs by degrading all gluten-derived immunogenic peptides during gastrointestinal digestion. A microbial enzyme treatment for CD patients has been proposed; however, several problems remain regarding its resistance and activity during gastrointestinal digestion (37, 38). Probiotic strains should be able to resist gastric digestion and adhere to the intestinal epithelium (39). It has also been shown that bacteria have a specific transport system to intake immunogenic oligopeptides (19). Under simulated gastrointestinal conditions, the pool of probiotic lactobacilli strongly hydrolyzed gluten during wheat bread digestion. After 360 min, the resultant gluten concentration was less than 10 ppm. As determined by MDLC coupled to nano-ESI–MS/MS, no known immunogenic peptides were detected in the digested wheat bread that was treated with the 10 probiotic strains. Despite the results obtained by chemical and R5-based immunological analyses, the detection of all immunogenic epitopes may be incomplete. Because the possibility of the presence of unknown immunogenic peptides cannot be excluded, cytokine expression in duodenal biopsy specimens from CD patients (38, 40) was studied. In particular, biopsy specimens were treated with wheat bread that was digested in either the presence or absence of probiotics (PB and Ct, respectively). Previously, it was shown that gliadin challenge of untreated and treated CD patients increased cytokine expression (e.g., IFN-γ, IL-2) in the lamina propria, which was associated with significant histological changes in the small intestinal mucosa of these patients (41). IL-10 was also induced in gliadin-stimulated in vitro or in vivo models (42). The amount of IL-10 induced positively correlated with the activity of gliadin-reactive T cells (43, 44). Indeed, IL-10 is an anti-inflammatory cytokine that can be induced under gluten stimulation to suppress inflammatory cytokine secretion from TH1 cells. However, a pilot study with CD patients showed that there were no positive effects (45). After wheat bread digestion, the remaining gluten polypeptides, which are recognized by HLA molecules, activated the T cells from duodenal biopsy specimens, resulting in high levels of cytokine secretion (46, 47). According to these previous studies, the levels of IL-2 and IL-10 were higher in the duodenal biopsy specimens from CD patients treated with digested wheat bread (Ct) than in the untreated duodenal biopsy specimens (RPMI 1640). However, the addition of the 10 probiotic lactobacilli to the simulated gastrointestinal digestion of wheat bread inhibited the induction of IL-2 and IL-10 in the duodenal biopsy specimens from CD patients.
IFN-γ drives the innate and adaptive immune responses that produce strong inflammatory effects leading to mucosal injury and the atrophy of villi (37). It was shown that the use of antibodies to block IFN-γ not only inhibits the activation of metalloproteinases and the influx of gluten peptides through the intestinal barrier but also deters mucosal injury (48, 49). Previously, baked wheat products containing gluten that was hydrolyzed by lactic acid bacteria and fungal proteases during sourdough fermentation did not induce IFN-γ (19) in vitro, thus yielding products that were safe for CD patients (18). As expected, the gastrointestinal digestion products from wheat bread strongly induced IFN-γ synthesis. Interestingly, the level of IFN-γ produced by the duodenal biopsy specimens from CD patients treated with digested wheat bread and the 10 probiotic lactobacilli was similar to the baseline value (RPMI 1640). Previously, it was shown that B. longum CECT 7347 together with gluten in an animal model of gliadin-induced enteropathy reduced the induction of IFN-γ and improved villus width and enterocyte height (50). Other bifidobacterial strains (e.g., Bifidobacterium lactis, B. bifidum) also reduced the level of IFN-γ in an in vitro model (e.g., peripheral blood mononuclear cells) stimulated by gliadin polypeptides (51). Probiotics could be useful for relieving symptoms and reducing molecular mucosal inflammation by downregulating the cytokines involved in CD pathogenesis (52) and hydrolyzing the gluten polypeptides that contaminate food. Probiotic administration could reduce the risk of mucosal inflammation due to the unintentional ingestion of gluten.
The findings of this study provide evidence that the selected probiotic lactobacillus strains have the potential to hydrolyze immunogenic peptides during gastrointestinal digestion, which decreases gluten toxicity for CD patients. Further studies will be performed to reduce the number of strains required without affecting the hydrolytic efficacy toward the immunogenic peptides. The application of probiotics specifically selected for their hydrolytic activity on gluten polypeptides provides a new and safe adjunctive therapy for a GFD.
MATERIALS AND METHODS
Microorganisms and culture conditions.
The following strains were obtained from the probiotic Culture Collection of Sacco Srl (Cadorago, Italy) and selected because of their protease and peptidase activities, including toward Pro-rich peptides: L. casei BGP93; L. delbrueckii subsp. bulgaricus SP5; L. paracasei LPC01, BGP1, and BGP2; L. plantarum BG112, BGP12, LP27, LP33, LP35, LP36, LP39, LP40, LP42, LP47, and LP32; L. rhamnosus SP1; and L. reuteri DSM17938 (33). Strains were propagated for 24 h at 30°C in MRS broth (Oxoid, Basingstoke, Hampshire, United Kingdom). When used for fermentation and enzymatic assays, Lactobacillus cells were cultivated until the late exponential phase of growth was reached (approximately 12 h).
Enzyme assays.
PepN, PepI, and PEP activities were determined as described by De Angelis et al. (33) and Gallo et al. (53) using Leu-p-NA, Pro-p-NA, and Z-Gly-Pro-Arg-p-NA, respectively. The assay mixture contained 900 μl of 2.0 mM substrate in 0.05 M potassium phosphate buffer and 100 μl of cellular suspension (109 CFU) at pH 7.0. The mixture was incubated at 30°C for 1 h, and the absorbance was measured at 410 nm. The data were compared to the values on standard curves generated using p-NA (54). One unit of activity was defined as the amount of enzyme required to liberate 1 μmol of p-NA/min under the assay conditions.
PepT, PepQ, PepR, and PepV activities were determined using Leu-Leu-Leu, Pro-Gly, Val-Pro, and Leu-Leu substrates, respectively. Activities for tri- and dipeptides were determined using the Cd-ninhydrin method (54). The same assay conditions used for the p-NA substrates were used. One unit of activity was defined as the amount of enzyme required to liberate 1 μM amino acid per min under the assay conditions. The data were compared to standard curves created using Leu (54). All synthetic substrates were obtained from Sigma Chemical Co.
Hydrolysis of Pro-rich synthetic peptides.
The following peptides were chemically synthesized and used in this study: α9-gliadin peptide 57-68 (Q-L-Q-P-F-P-Q-P-Q-L-P-Y) (34), A-gliadin peptide 62-75 (P-Q-P-Q-L-P-Y-P-Q-P-Q-S-F-P) (35), γ-gliadin peptide 134-153 (Q-Q-L-P-Q-P-Q-Q-P-Q-Q-S-F-P-Q-Q-Q-R-P-F) (36), and the gliadin 33-mer peptide (L-Q-L-Q-P-F-P-Q-P-Q-L-P-Y-P-Q-P-Q-L-P-Y-P-Q-P-Q-L-P-Y-P-Q-P-Q-P-F) (17). A mixture containing 50 μl of each cellular suspension (109 CFU), 200 mM peptide, and 0.05% (wt/vol) NaN3 in 1 ml of 50 mM phosphate buffer at pH 7.5 was incubated for 24 h at 37°C with stirring (150 rpm). The peptides were evaluated by RP-HPLC using an Äkta purifier system (peptide analysis) and a Biochrom 30 series amino acid analyzer (FAA analysis) as described above.
PT digest.
To simulate in vivo digestion, gliadins extracted from T. aestivum cv. Sagittario were subjected to sequential PT hydrolysis steps (32) in suspensions containing cells of the 10 pooled Lactobacillus strains (a total of 109 CFU/ml). PT digestion of T. aestivum cv. Sagittario (without Lactobacillus cells) was used as a positive control (19). After digestion, the PT digests were heated at 100°C for 30 min to inactivate the enzymes and then freeze-dried for further analysis.
Hydrolysis of baker's yeast wheat bread.
Simulated gastric and intestinal fluids were added to 5 g of baker's yeast bread, chewed for 30 s, and collected in a beaker with 10 ml of NaK phosphate (0.05 M, pH 6.9). Chewed baker's yeast bread (Ct) was suspended in a simulated gastric juice that contained NaCl (125 mM/liter), KCl (7 mM/liter), NaHCO3 (45 mM/liter), and pepsin (3 g/liter) (Sigma-Aldrich Co., St. Louis, MO, USA). The final pH was adjusted to 2.0 with HCl. To mimic hydrolysis by probiotics colonizing the gastrointestinal tract, baker's yeast bread was further incubated with a final cell density of approximately 9.0 log CFU/ml of the pooled lactobacillus culture (PB). To simulate the presence of other proteins during gastrointestinal digestion, an additional experiment was performed in which reconstituted skim milk (SM; 11% [wt/vol] solids) was added prior to inoculation with the simulated gastric juices at pH 2.0 (PB-SM). The suspension was incubated at 37°C under anaerobic conditions and stirred to simulate peristalsis. After 120 min of gastric digestion, the mixture was collected and suspended in simulated intestinal fluid, which contained 0.1% (wt/vol) pancreatin and 0.15% (wt/vol) Oxgall bile salt (Sigma-Aldrich Co.) at pH 8.0. The suspension was incubated at 37°C under agitation for 360 min. After incubation, the samples were put on ice and immediately analyzed.
Immunological analysis.
An immunological analysis was performed using an R5-ELISA (55, 56). The R5-ELISA was performed using a Transia plate detection kit, and the manufacturer's instructions were followed (Diffchamb, Västra Frölunda, Sweden).
MDLC and nano-ESI-MS/MS.
MDLC coupled with nano-ESI–MS/MS was used to analyze the hydrolyzed samples. The HPLC apparatus consisted of an Ettan MDLC machine (GE Healthcare) equipped with a Zorbax 300 SD C18 precolumn (5 by 0.3 mm) and a Thermo Electron BioBasic-8 column (150 by 0.18 mm). The MDLC was connected to a Finnigan LCQ Deca XP Max ion trap mass spectrometer (Thermo Electron) through its nano-ESI interface. Ten-microliter aliquots of each sample were injected. The HPLC separations were performed at a flow rate of 75 μl/min using a gradient elution of water (eluent A) and 84% acetonitrile (eluent B), both of which contained 0.1% (vol/vol) formic acid. The following program was used: 0% eluent B for 30 min, 0 to 100% (vol/vol) eluent B for 100 min, isocratic elution with 100% eluent B for 100 min, 0% eluent B for 5 min, and column reconditioning for 30 min. The flow rate at the nano-ESI source was 2.5 μl/min. The LCQ spectrometer, which was completely controlled by Xcalibur software (Thermo Electron), was operated in the positive ion mode. MS chromatograms in total ion current-monitoring (m/z range, 50 to 2,000) and select ion-monitoring modes were recorded for each sample.
Mucosal biopsy specimens and organ culture.
The study adhered to the Declaration of Helsinki and was approved by the ethical committee of the Department of Interdisciplinary Medicine, University of Bari Aldo Moro. Duodenal biopsy specimens were obtained from 10 CD patients (age range, 19 to 30 years) following a GFD. All CD patients expressed the HLA-DQ2 phenotype. CD was diagnosed according to European Society for Pediatric Gastroenterology, Hepatology, and Nutrition criteria (57). Immediately after excision, all biopsy specimens were placed in ice-chilled culture medium (RPMI 1640; Gibco-Invitrogen, UK) and transported to the laboratory within 30 min.
Duodenal biopsy specimens were cultured for 4 h using the organ tissue culture method originally described by Browning and Trier (58). Briefly, the biopsy specimens were oriented villous side up on a stainless steel mesh and positioned over the central well of an organ tissue culture dish (Falcon, USA). The well contained RPMI 1640 (Gibco-Invitrogen, UK) supplemented with 15% fetal calf serum (Gibco-Invitrogen, UK) and 1% penicillin-streptomycin (Gibco-Invitrogen, UK). The dishes were placed in an anaerobic jar, which was supplied with 95% O2 and 5% CO2, before the jar was sealed and incubated at 37°C. Four biopsy specimens from each CD patient were cultured with digested baker's yeast wheat bread either in the absence of the 10 probiotic lactobacilli (Ct), in the presence of the 10 probiotic lactobacilli (PB), or in culture medium (RPMI 1640).
RNA extraction and cDNA synthesis.
Biopsy specimens from each patient were rinsed and stored in RNAlater (Qiagen GmbH, Germany) at −80°C to preserve the RNA. Total RNA was extracted from the tissues using an RNeasy minikit (Qiagen GmbH) according to the manufacturer's instructions. The concentration of mRNA was estimated by determination of the UV absorbance at 260 nm. Aliquots of total RNA (500 ng) were reverse transcribed using random hexamers, TaqMan reverse transcription reagents (Applied Biosystems, Monza, Italy), and 3.125 U/μl of MultiScribe reverse transcriptase to a final volume of 50 μl. The cDNA samples were stored at −20°C.
RT-PCR for IFN-γ, IL-2, and IL-10 genes.
RT-PCR was performed in 96-well plates using an ABI Prism 7500HT fast sequence detection system (Applied Biosystems). Data collection and analyses were performed using the machine software. PCR primers and fluorogenic probes for the target genes (IFN-γ, IL-2, and IL-10 genes) and the endogenous control (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) were purchased as a TaqMan gene expression assay and a predeveloped TaqMan assay (Applied Biosystems), respectively. The assays were supplied as a 20× mix of PCR primers and TaqMan Minor Groove Binder 6-carboxyfluorescein dye-labeled probes with a nonfluorescent quencher at the 3′ end of the probe.
Two-step reverse transcription-PCR was performed using first-strand cDNA with a final concentration of 1× TaqMan gene expression assay mix and 1× TaqMan universal PCR master mix. The final reaction volume was 25 μl. Each sample was analyzed in triplicate, and all experiments were repeated twice. A nontemplate control (RNase-free water) was included with every plate. The following thermal cycler conditions were used: 2 min at 50°C (uracil DNA glycosylase activation), 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. Initially, a standard curve and a validation experiment were performed for each primer/probe set. Six serial dilutions (20 to 0.1 ng/μl) of IFN-γ, IL-2, or IL-10 cDNA were used as a template for each primer/probe set. A standard curve was generated by plotting the threshold cycle (CT) values against the log of the amount of input cDNA. The CT value is the PCR cycle at which an increase in reporter fluorescence above the baseline level is first detected. The average value for the target gene was normalized using an endogenous reference gene (the GAPDH gene). A healthy duodenal biopsy specimen was used to calibrate all of the experiments.
The levels of IFN-γ, IL-2, and IL-10 proteins secreted into the supernatant were quantified by ELISA in 96-well round-bottom plates (Tema Ricerca, Milan, Italy) according to the manufacturer's recommendations.
Statistical analysis.
Experimental data were subjected to analysis of variance (ANOVA), and pairwise comparisons of the treatment mean values were conducted using Tukey's test with a P value of <0.05 and the statistical software Statistica (version 8.0; StatSoft Inc., Tulsa, OK, USA).
ACKNOWLEDGMENTS
We thank Ilario Losito (Department of Chemistry, Centro Interdipartimentale di Ricerca SMART, University of Bari) for his scientific assistance with the nano-ESI–MS/MS analysis and for contributing to critical discussions of the results. We also thank Angela Cassone (Department of Soil, Plant and Food Science, University of Bari) and Francesca Gagliardi (Department of Pediatrics, University of Bari) for their technical assistance.
REFERENCES
- 1.Fasano A, Catassi C. 2001. Current approaches to diagnosis and treatment of celiac disease: an evolving spectrum. Gastroenterology 120:636–651. doi: 10.1053/gast.2001.22123. [DOI] [PubMed] [Google Scholar]
- 2.Mustalahti K, Catassi C, Reunanen A, Fabiani E, Heier M, McMillan S, Murray L, Metzger MH, Gasparin M, Bravi E, Mäki M, Coeliac EU Cluster, Project Epidemiology. 2010. The prevalence of celiac disease in Europe: results of a centralized, international mass screening project. Ann Med 42:587–595. doi: 10.3109/07853890.2010.505931. [DOI] [PubMed] [Google Scholar]
- 3.Cenit MC, Olivares M, Codoñer-Franch P, Sanz Y. 2015. Intestinal microbiota and celiac disease: cause, consequence or co-evolution? Nutrients 7:6900–6923. doi: 10.3390/nu7085314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pagliari D, Urgesi R, Frosali S, Riccioni ME, Newton EE, Landolfi R, Pandolfi F, Cianci R. 2015. The interaction among microbiota, immunity, and genetic and dietary factors is the condicio sine qua non celiac disease can develop. J Immunol Res 2015:123653. doi: 10.1155/2015/123653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanz Y. 2015. Microbiome and gluten. Ann Nutr Metab 67:28–41. doi: 10.1159/000440991. [DOI] [PubMed] [Google Scholar]
- 6.Nagao-Kitamoto H, Kitamoto S, Kuffa P, Kamada N. 2016. Pathogenic role of the gut microbiota in gastrointestinal diseases. Intest Res 14:127–138. doi: 10.5217/ir.2016.14.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ercolini D, Francavilla R, Vannini L, De Filippis F, Capriati T, Di Cagno R, Iacono G, De Angelis M, Gobbetti M. 2015. From an imbalance to a new imbalance: Italian-style gluten-free diet alters the salivary microbiota and metabolome of African celiac children. Sci Rep 5:18571. doi: 10.1038/srep18571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Francavilla R, Ercolini D, Piccolo M, Vannini L, Siragusa S, De Filippis F, De Pasquale I, Di Cagno R, Di Toma M, Gozzi G, Serrazanetti D, De Angelis M, Gobbetti M. 2014. Salivary microbiota and metabolome associated with celiac disease. Appl Environ Microbiol 80:3416–3425. doi: 10.1128/AEM.00362-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Angelis M, Vannini L, Di Cagno R, Cavallo N, Minervini F, Francavilla R, Ercolini D, Gobbetti M. 2016. Salivary and fecal microbiota and metabolome of celiac children under gluten-free diet. Int J Food Microbiol 239:125–132. doi: 10.1016/j.ijfoodmicro.2016.07.025. [DOI] [PubMed] [Google Scholar]
- 10.De Palma G, Nadal I, Collado MC, Sanz Y. 2009. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. Br J Nutr 102:1154–1160. doi: 10.1017/S0007114509371767. [DOI] [PubMed] [Google Scholar]
- 11.Kaukinen K, Lindfors K, Mäki M. 2014. Advances in the treatment of coeliac disease: an immunopathogenic perspective. Nat Rev Gastroenterol Hepatol 11:36–44. doi: 10.1038/nrgastro.2013.141. [DOI] [PubMed] [Google Scholar]
- 12.Sollid LM, Khosla C. 2005. Future therapeutic options for celiac disease. Nat Clin Pract Gastroenterol Hepatol 2:140–147. doi: 10.1038/ncpgasthep0111. [DOI] [PubMed] [Google Scholar]
- 13.Pyle GG, Paaso B, Anderson BE, Allen DD, Marti T, Li Q, Siegel M, Khosla C, Gray GM. 2005. Effect of pretreatment of food gluten with prolyl endopeptidase on gluten-induced malabsorption in celiac sprue. Clin Gastroenterol Hepatol 3:687–694. doi: 10.1016/S1542-3565(05)00366-6. [DOI] [PubMed] [Google Scholar]
- 14.Shan L, Qiao SW, Arentz-Hansen H, Molberg Ø, Gray GM, Sollid LM, Khosla C. 2005. Identification and analysis of multivalent proteolytically resistant peptides from gluten: implication for celiac sprue. J Protein Res 4:1732–1741. doi: 10.1021/pr050173t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rauhavirta T, Qiao SW, Jiang Z, Myrsky E, Loponen J, Korponay-Szabo IR, Salovaara H, Garcia-Horsman JA, Venalainen J, Mannisto PT, Collighan R, Mongeot A, Griffin M, Maki M, Kaukinen K, Lindfors K. 2011. Epithelial transport and deamidation of gliadin peptides: a role for coeliac disease patient immunoglobulin A. Clin Exp Immunol 164:127–136. doi: 10.1111/j.1365-2249.2010.04317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caputo I, Lepretti M, Martucciello S, Esposito C. 2010. Enzymatic strategies to detoxify gluten: implications for celiac disease. Enzyme Res 2010:174354. doi: 10.4061/2010/174354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shan L, Molberg Ø, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C. 2002. Structural basis for gluten intolerance in celiac sprue. Science 297:2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 18.Greco L, Gobbetti M, Auricchio R, Di Mase R, Paparo F, Di Cagno R, De Angelis M, Rizzello CG, Cassone A, Terrone G, Timpone L, D'Aniello M, Troncone R, Auricchio S. 2011. Safety for celiac patients of baked goods made of wheat flour hydrolyzed during food processing. Clin Gastroenterol Hepatol 9:24–29. doi: 10.1016/j.cgh.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 19.Rizzello CG, De Angelis M, Di Cagno R, Camarca A, Silano M, Losito I, De Vincenzi M, De Bari MD, Palmisano F, Maurano F, Gianfrani C, Gobbetti M. 2007. Highly efficient gluten degradation by lactobacilli and fungal proteases during food processing: new perspectives for celiac disease. Appl Environ Microbiol 73:4499–4507. doi: 10.1128/AEM.00260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Angelis M, Cassone A, Rizzello CG, Gagliardi F, Minervini F, Calasso M, Di Cagno R, Francavilla R, Gobbetti M. 2010. Mechanism of degradation of immunogenic gluten epitopes from Triticum turgidum L. var. durum by sourdough lactobacilli and fungal proteases. Appl Environ Microbiol 76:508–518. doi: 10.1128/AEM.01630-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duar RM, Clark KJ, Patil PB, Hernández C, Brüning S, Burkey TE, Madayiputhiya N, Taylor SL, Walter J. 2015. Identification and characterization of intestinal lactobacilli strains capable of degrading immunotoxic peptides present in gluten. J Appl Microbiol 118:515–527. doi: 10.1111/jam.12687. [DOI] [PubMed] [Google Scholar]
- 22.Caminero A, Herran AR, Nistal E, Perez-Andres J, Vaquero L, Vivas S, Ruiz de Morales JMG, Albillos SM, Casqueiro J. 2014. Diversity of the cultivable human gut microbiome involved in gluten metabolism: isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol Ecol 88:309–319. doi: 10.1111/1574-6941.12295. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Feo M, Wei G, Blumenkranz G, Dewhirst FE, Schuppan D, Oppenheim FG, Helmerhorst EJ. 2013. The cultivable human oral gluten-degrading microbiome and its potential implications in coeliac disease and gluten sensitivity. Clin Microbiol Infect 19:E386–E394. doi: 10.1111/1469-0691.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zamakhchari M, Wei G, Dewhirst F, Lee J, Schuppan D, Oppenheim FG, Helmerhorst EJ. 2011. Identification of Rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PLoS One 6:e24455. doi: 10.1371/journal.pone.0024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laparra JM, Sanz Y. 2010. Bifidobacteria inhibit the inflammatory response induced by gliadins in intestinal epithelial cells via modifications of toxic peptide generation during digestion. J Cell Biochem 109:801–807. doi: 10.1002/jcb.22459. [DOI] [PubMed] [Google Scholar]
- 26.Orlando A, Linsalata M, Notarnicola M, Tutino V, Russo F. 2014. Lactobacillus GG restoration of the gliadin induced epithelial barrier disruption: the role of cellular polyamines. BMC Microbiol 14:19. doi: 10.1186/1471-2180-14-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cinova J, De Palma G, Stepankova R, Kofronova O, Kverka M, Sanz Y, Tuckova L. 2011. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: study in germ-free rats. PLoS One 6:e16169. doi: 10.1371/journal.pone.0016169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barada K, Bitar A, Mokadem MA, Hashash JG, Green P. 2010. Celiac disease in Middle Eastern and North African countries: a new burden? World J Gastroenterol 16:1449–1457. doi: 10.3748/wjg.v16.i12.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee AR, Ng DL, Zivin J, Green PHR. 2007. Economic burden of a gluten-free diet. J Hum Nutr Diet 20:423–430. doi: 10.1111/j.1365-277X.2007.00763.x. [DOI] [PubMed] [Google Scholar]
- 30.Silvester JA, Rashid M. 2007. Long-term follow-up of individuals with celiac disease: an evaluation of current practice guidelines. Can J Gastroenterol 21:557–564. doi: 10.1155/2007/342685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindfors K, Blomqvist T, Juuti-Uusitalo K, Stenman S, Venäläinen J, Mäki M, Kaukinen K. 2008. Live probiotic Bifidobacterium lactis bacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin Exp Immunol 152:552–558. doi: 10.1111/j.1365-2249.2008.03635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Angelis M, Rizzello CG, Fasano A, Clemente MG, De Simone C, Silano M, De Vincenzi M, Losito I, Gobbetti M. 2006. VSL#3 probiotic preparation has the capacity to hydrolyze gliadin polypeptides responsible for celiac sprue. Biochim Biophys Acta 1762:80–93. doi: 10.1016/j.bbadis.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 33.De Angelis M, Di Cagno R, Gallo G, Curci M, Siragusa S, Crecchio C, Parente E, Gobbetti M. 2007. Molecular and functional characterization of Lactobacillus sanfranciscensis strains isolated from sourdoughs. Int J Food Microbiol 114:69–82. doi: 10.1016/j.ijfoodmicro.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 34.Arentz-Hansen H, Körner R, Molberg O, Quarsten H, Vader W, Kooy YM, Lundin KEA, Koning F, Roepstorff P, Sollid LM, McAdam SN. 2000. The intestinal T cell response to alpha-gliadin in adult coeliac disease is focused on a single deamidated glutamine targeted by tissue transglutaminase. J Exp Med 191:603–612. doi: 10.1084/jem.191.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silano M, De Vincenzi M. 1999. Bioactive antinutritional peptides derived from cereal prolamins: a review. Nahrung 43:175–184. doi:. [DOI] [PubMed] [Google Scholar]
- 36.Aleanzi M, Demonte AM, Esper C, Garcilazo S, Waggener M. 2001. Celiac disease: antibody recognition against native and selectively deaminated gliadin peptides. Clin Chem 47:2023–2028. [PubMed] [Google Scholar]
- 37.Makharia GK. 2014. Current and emerging therapy for celiac disease. Front Med (Lausanne) 1:6. doi: 10.3389/fmed.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shan L, Marti T, Sollid LM, Gray GM, Khosla C. 2004. Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for celiac sprue. Biochem J 383:311–318. doi: 10.1042/BJ20040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campieri M, Gionchetti P. 1999. Probiotics in inflammatory bowel disease: new insight to pathogenesis or a possible therapeutic alternative? Gastroenterology 116:1246–1249. doi: 10.1016/S0016-5085(99)70029-6. [DOI] [PubMed] [Google Scholar]
- 40.Kilmartin C, Lynch S, Abuzakouk M, Wieser H, Feighery C. 2003. Avenin fails to induce a Th1 response in coeliac tissue following in vitro culture. Gut 52:47–52. doi: 10.1136/gut.52.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kontakou M, Przemioslo RT, Sturgess RP, Limb GA, Ellis HJ, Day P, Ciclitira PJ. 1995. Cytokine mRNA expression in the mucosa of treated coeliac patients after wheat peptide challenge. Gut 37:52–57. doi: 10.1136/gut.37.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsberg G, Hernell O, Melgar S, Israelsson A, Hammarstrom S, Hammarstrom ML. 2002. Paradoxical coexpression of proinflammatory and down-regulatory cytokines in intestinal T cells in childhood celiac disease. Gastroenterology 123:667–678. doi: 10.1053/gast.2002.35355. [DOI] [PubMed] [Google Scholar]
- 43.Gianfrani C, Levings MK, Sartirana C, Mazzarella G, Barba G, Zanzi D, Camarca A, Iaquinto G, Giardullo N, Auricchio S, Troncone R, Roncarolo MG. 2006. Gliadin-specific type 1 regulatory T cells from the intestinal mucosa of treated celiac patients inhibit pathogenic T cells. J Immunol 177:4178–4186. doi: 10.4049/jimmunol.177.6.4178. [DOI] [PubMed] [Google Scholar]
- 44.Salvati VM, Mazzarella G, Gianfrani C, Levings MK, Stefanile R, De Giulio B, Iaquinto G, Giardullo N, Auricchio S, Roncarolo MG, Troncone R. 2005. Recombinant human interleukin 10 suppresses gliadin dependent T cell activation in ex vivo cultured coeliac intestinal mucosa. Gut 54:46–53. doi: 10.1136/gut.2003.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulder CJ, Wahab PJ, Meijer JW, Metselaar E. 2001. A pilot study of recombinant human interleukin-10 in adults with refractory coeliac disease. Eur J Gastroenterol Hepatol 13:1183–1188. doi: 10.1097/00042737-200110000-00010. [DOI] [PubMed] [Google Scholar]
- 46.van de Wal Y, Kooy YM, van Veelen P, Vader W, August SA, Drijfhout JW, Peña SA, Koning F. 1999. Glutenin is involved in the gluten-driven mucosal T cell response. Eur J Immunol 29:3133–3139. doi:. [DOI] [PubMed] [Google Scholar]
- 47.Søndergaard I, Jensen K, Krath B. 1994. Classification of wheat varieties by isoelectric focusing patterns of gliadins and neural network. Electrophoresis 15:584–588. doi: 10.1002/elps.1150150181. [DOI] [PubMed] [Google Scholar]
- 48.Bethune MT, Siegel M, Howles-Banerji S, Khosla C. 2009. Interferon-gamma released by gluten-stimulated celiac disease-specific intestinal T cells enhances the transepithelial flux of gluten peptides. J Pharmacol Exp Ther 329:657–668. doi: 10.1124/jpet.108.148007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Przemioslo RT, Lundin KE, Sollid LM, Nelufer J, Ciclitira PJ. 1995. Histological changes in small bowel mucosa induced by gliadin sensitive T lymphocytes can be blocked by anti-interferon gamma antibody. Gut 36:874–879. doi: 10.1136/gut.36.6.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laparra JM, Olivares M, Gallina O, Sanz Y. 2012. Bifidobacterium longum CECT 7347 modulates immune responses in a gliadin-induced enteropathy animal model. PLoS One 7:e30744. doi: 10.1371/journal.pone.0030744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Sousa Moraes LF, Grzeskowiak LM, de Sales Teixeira TF, do Carmo Gouveia Peluzio M. 2014. Intestinal microbiota and probiotics in celiac disease. Clin Microbiol Rev 27:482–489. doi: 10.1128/CMR.00106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Losurdo G, Principi M, Iannone A, Ierardi E, Di Leo A. 2016. The interaction between celiac disease and intestinal microbiota. J Clin Gastroenterol 2015:S145–S147. [DOI] [PubMed] [Google Scholar]
- 53.Gallo G, De Angelis M, McSweeney PLH, Corbo MR, Gobbetti M. 2005. Partial purification and characterization of an X-prolyl dipeptidyl aminopeptidase from Lactobacillus sanfranciscensis CB1. Food Chem 91:535–544. doi: 10.1016/j.foodchem.2004.08.047. [DOI] [Google Scholar]
- 54.Gobbetti M, Lanciotti R, De Angelis M, Corbo MR, Massini R, Fox PF. 1999. Study of the effects of temperature, pH, NaCl, and aw on the proteolytic and lipolytic activities of cheese-related lactic acid bacteria by quadratic response surface methodology. Enzyme Microb Technol 25:795–809. doi: 10.1016/S0141-0229(99)00110-6. [DOI] [Google Scholar]
- 55.Ferre S, Garcìa E, Mendez E. 2004. Measurement of hydrolysed gliadins by a competitive ELISA based on monoclonal antibody R5: analysis of syrups and beers. In Stern M. (ed), Proceedings of the 18th Meeting of the Working Group on Prolamin Analysis and Toxicity. Verlag Wissenschaftliche Scripten, Zwickau, Germany. [Google Scholar]
- 56.Valdés I, Garcìa E, Lorente M, Mèndez E. 2003. Innovative approach to low-level gluten determination in foods using a novel sandwich enzyme-linked immunosorbent assay protocol. Eur J Gastroenterol Hepatol 15:465–474. [DOI] [PubMed] [Google Scholar]
- 57.European Society of Paediatric Gastroenterology and Nutrition. 1990. Revised criteria for diagnosis of coeliac disease. Report of working group of European Society of Paediatric Gastroenterology and Nutrition. Arch Dis Child 65:909–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Browning TH, Trier JS. 1969. Organ culture of mucosal biopsies of small intestine. J Clin Invest 48:1423–1432. doi: 10.1172/JCI106108. [DOI] [PMC free article] [PubMed] [Google Scholar]