Abstract
The papillomavirus E2 protein functions in viral transcriptional regulation, DNA replication, and episomal genome maintenance. Viral genomes are maintained in dividing cells by attachment to mitotic chromosomes by means of the E2 protein. To investigate the chromosomal tethering function of E2, plasmid stability assays were developed in Saccharomyces cerevisiae to determine whether the E2 protein could maintain plasmids containing the yeast autonomous replication sequence replication element but with the centromeric element replaced by E2-binding sites. E2 expression was not sufficient to maintain such plasmids, but plasmid stability could be rescued by expression of the mammalian protein Brd4. In the presence of both Brd4 and E2 proteins, plasmids with multiple E2-binding sites were stable without selection. S. cerevisiae encodes a homolog of Brd4 named Bdf1 that does not contain the C-terminal domain that interacts with the E2 protein. A fusion protein of Bdf1 and the Brd4 C-terminal “tail” could support E2-mediated plasmid maintenance in yeast. Using a panel of mutated E2 proteins, we determined that plasmid stability required the ability of E2 to bind DNA and to interact with Brd4 and mammalian mitotic chromosomes but did not require its replication initiation and transactivation functions. The S. cerevisiae-based plasmid maintenance assays described here are invaluable tools for dissecting mechanisms of episomal viral genome replication and screening for additional host protein factors involved in plasmid maintenance.
Keywords: replication, yeast, segregation, bovine papillomavirus, human papillomavirus
Papillomavirus infections are often long lived and persistent. Viral DNA replicates as a stable episome in dividing infected cells. The viral E2 protein prevents loss of the genome and segregates it to daughter cells by attaching it to host chromosomes during mitosis (1–3). Several viruses have similar strategies for episomal genome maintenance. The Epstein–Barr virus-encoded nuclear antigen 1 (EBNA1) protein of Epstein–Barr virus (EBV) interacts with the cellular mitotic chromosomal protein p40 (hEBP2) to partition the EBV genome (4). The LANA protein of human herpes virus 8 (HHV-8) binds to histone H1, MeCP2, DEK, and Ring3 to tether the genome to chromosomes (5–7).
Papillomavirus genome maintenance is best characterized for bovine papillomavirus type 1 (BPV1). Genome maintenance is mediated by the viral E2 protein, which is composed of two functional domains: an N-terminal transactivation domain and a C-terminal DNA-binding domain separated by a nonconserved hinge. The transactivation domain is sufficient for association of E2 with mitotic chromosomes, which occurs by means of interaction with a host chromosomal protein (8). Point mutations in this domain disrupt chromosomal binding (9, 10). This domain is also critical for viral DNA replication and transcriptional regulation (11, 12). The DNA-binding domain tethers genomes to mitotic chromosomes by means of multiple E2 DNA-binding sites (3).
To investigate the tethering function of E2, plasmid stability assays were established in S. cerevisiae. These assays are based on plasmids that stably replicate by means of the yeast autonomous replication sequence (ARS) and centromeric (CEN) maintenance elements. CEN/ARS plasmids initiate replication at the ARS element and are partitioned by the CEN element (13). Without the CEN element, ARS-containing plasmids replicate but are quickly lost from dividing cells (14, 15). This replication strategy is similar to that of papillomavirus genomes, which contain a replication origin and a separable minichromosome maintenance element that can be replaced by tandem arrays of E2-binding sites (16).
The plasmid stability assays were used to determine whether E2 could directly maintain plasmids in S. cerevisiae. We postulated that ARS plasmids in which the CEN element was replaced by E2-binding sites might be faithfully partitioned if E2 associates with mitotic chromosomes in yeast, as it does in mammalian cells. Using these assays, we show that E2 is unable to maintain such plasmids in yeast. It has been of great interest to identify the chromosomal protein with which E2 interacts to enable papillomavirus genome maintenance. The inability of E2 to maintain plasmids in S. cerevisiae provided an opportunity to identify mammalian proteins that could rescue this function. In this study, we find that plasmid stability can be rescued by the mammalian Brd4 gene.
Brd4 is a bromodomain protein that binds acetylated histones throughout the cell cycle (7). It was recently reported that Brd4 interacts with E2 on mammalian mitotic chromosomes and that disruption of this interaction abrogates the chromosomal association of E2 and viral DNA (17). Our findings that Brd4 can reconstitute E2-mediated plasmid maintenance shows that Brd4 is essential for this process. In addition, the S. cerevisiae assays allowed us to define the functions of E2 required for plasmid maintenance. These S. cerevisiae-based plasmid stability assays are extremely useful tools to further dissect the mechanism of viral genome maintenance and to isolate additional protein factors involved in this mechanism.
Materials and Methods
Yeast Strains and Transformation. The S. cerevisiae strains used were W303 (MATa ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1) (18) and YMB1670 (MATa ade2-101 ade3Δ ura3-52 trp1Δ1 leu2Δ1 lys2-801). Plasmid transformations used the Yeastmaker Transformation System 2 (BD Biosciences). Strains were cultured on yeast extract/peptone/dextrose (YPD) or synthetic complete (SC) dropout media (QBiogene, Irvine, CA).
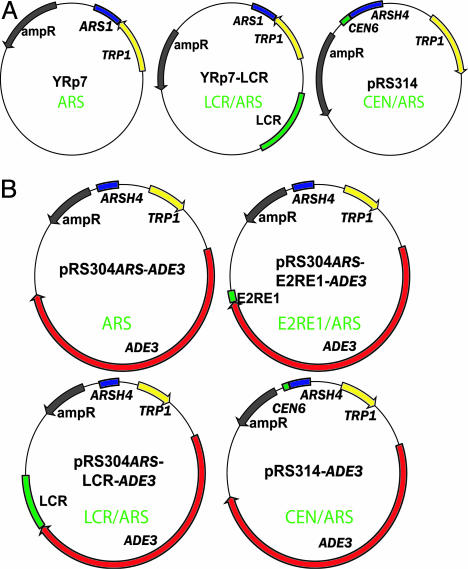
Plasmids. YRp7 and pRS314 are described in refs. 15 and 19. YRp7-LCR contains BPV1 bp 6,946–7,946 in the SalI/BamHI sites of YRp7. The reporter plasmids for the red/white sectoring assay were modified from pRS304 [American Type Culture Collection (ATCC) 77139] and pRS314 (ATCC 77143). pRS304ARS-ADE3 contains the ARSH4 element from pRS314 in the AatII site of pRS304. pRS304ARS-E2RE1-ADE3 contains BPV1 bp 7,611–7,805 in the Asp718I/SalI sites of pRS304ARS. pRS304ARS-LCR-ADE3 contains BPV bp 6,946–7,946 in the pRS304ARS Asp718I/SalI sites. The ADE3 gene from pDK202 (20) was inserted between the BamHI and SalI sites of the plasmids, as indicated.
pADNS-E2 contains a SmaI/HindIII E2 fragment from pTZE2kz (21) in the HindIII/NotI sites of pADNS (22). pRB16-E2 contains a BspMI/BstXI fragment containing the E2 gene and a portion of the ADH promoter from pADNS-E2 in the BspMI/EagI sites of pRB16, which has a crippled ADH promoter (23). p416CYC1-E2 contains E2 SmaI/HindIII fragments from pTZE2kz in the SalI/HindIII sites of p416CYC1 (ATCC 87384) (24). Plasmids expressing mutated E2 proteins are described in refs. 1 and 25. p416CYC1-E2TR was generated by cleaving pTZE2kz with NcoI, religating to remove the transactivation domain, and inserting an XhoI linker into the SmaI site of the vector before inserting into the SalI/HindIII sites of p416CYC1.
pADNS-Brd4 contains an EcoRI fragment from pBSK MCAP WT (26) in the HindIII site of pADNS. pADNS-Brd2 contains an Asp718I/Cfr421 fragment encoding the Brd2 gene from RING3 pBlue Full (26) in the HindIII/Cfr421sites. p40 plasmids (pR425/PGK and p425PGK-hEBP2) are described in ref. 4. An in-frame polylinker was inserted into the HindIII/SfiI sites of pADNS to generate pADNSpl. A NotI/XhoI BDF1 gene fragment from pGEM11Z-yBDF1 (27) was inserted to generate pADNSpl-BDF1. bp 2,149–4,203 of Brd4 were inserted in the NheI/XhoI sites of pGEM11Z-yBDF1. pADNpl-BDF1rcMtail was generated by inserting a NotI/XhoI fragment containing the BDF1rcMtail fusion gene into pADNSpl.
Red/White Sectoring Assay. Transformants were grown in selective medium and then plated on selective and nonselective media containing limiting Ade (21 μg/ml). Plates were incubated at 30°C for 4–7 days and then at 4°C overnight to enhance the red coloration.
Liquid Culture Plasmid-Loss Assay. Transformants were grown to logarithmic phase of growth in selective media. Cultures were diluted to an OD600 of 0.01 Å and grown for 8–11 generations in medium lacking selection for the reporter plasmid. Cultures were diluted to an OD600 of 1.0 Å, with three further 10-fold serial dilutions. Five microliters of each dilution was spotted onto plates selective and nonselective for the reporter plasmid and spread on selective and nonselective media to determine colony counts.
Results
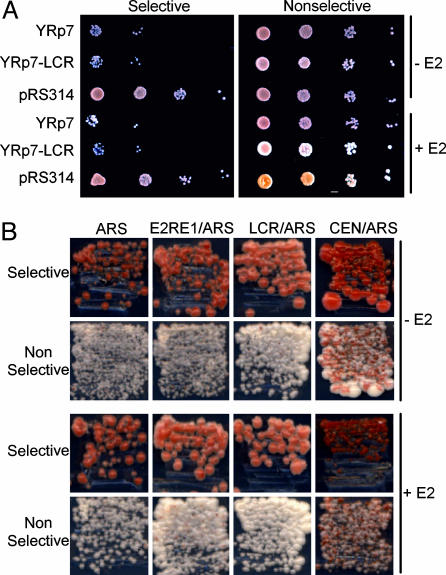
The E2 Protein Cannot Maintain Plasmids with E2-Binding Sites in S. cerevisiae. A plasmid stability assay was developed to investigate the tethering function of the E2 protein and determine whether it could promote plasmid maintenance in S. cerevisiae. A BPV1 fragment (LCR) containing 11 E2-binding sites was inserted into the ARS plasmid Yrp7. The yeast strain W303a was transformed with reporter plasmids YRp7 or YRp7-LCR or CEN/ARS control plasmid pRS314 and selected on SC medium lacking tryptophan (SC-Trp). Plasmid details are given in Fig. 1A and Table 1, which is published as supporting information on the PNAS web site. The resulting strains were transformed with pRB16-E2, which expresses E2 from a crippled ADH promoter, necessary because plasmids expressing higher levels of E2 were growth-inhibitory. Multiple transformants were assayed to determine the relative stability of the reporter plasmids. The strains were grown to logarithmic phase in medium selective for both the reporter and the E2 plasmids (SC-His-Trp) and then transferred to medium selective only for pRB16-E2 (SC-His). Cultures were grown for 11 generations, and the percentage of plasmid retention was determined by comparing colony counts of serial dilutions of the cultures on selective and nonselective media (Fig. 2A). The ARS-containing YRp7 plasmid was retained under nonselective conditions at 2% of that retained in selective conditions, regardless of the presence of E2. Similarly, YRp7-LCR, which contains 11 E2-binding sites in addition to the ARS element, was only retained in 2–3% yeast cells, even in the presence of E2. The positive control CEN/ARS plasmid, pRS314, was retained by 82% of cells. Therefore, the E2 protein is unable to mediate plasmid partitioning in S. cerevisiae, which might indicate the absence or limited conservation of the mitotic chromosomal protein with which E2 normally associates.
Fig. 1.
Reporter liquid culture (A) and red/white sectoring (B) assay plasmids.
Fig. 2.
E2 does not maintain E2-binding site plasmids in S. cerevisiae. (A) Plasmid-loss assay of W303a transformants with indicated reporter plasmids and the E2 expression plasmid pRB16-E2. Transformants were grown in selective medium and transferred for growth for 11 generations in medium nonselective for the reporter plasmids. Aliquots of 10-fold serial dilutions were plated onto media selective and nonselective for the reporter plasmids. (B)A red/white colony sectoring assay. YMB1670 transformants containing the reporter plasmids shown in Fig. 1B and E2 expression plasmid p416CYC1-E2 (or empty vector p416CYC1) were streaked on media selective and nonselective for the reporter plasmids.
A Red/White Sectoring Assay Enables Visualization of Plasmid Maintenance in Individual Colonies. The liquid plasmid stability assay can be used to isolate cellular proteins that can rescue the plasmid-loss phenotype but is limited in its ability to identify these proteins in a pool of transformants. Therefore, we designed a visual colony color assay to facilitate screening for proteins that can rescue E2-mediated plasmid retention. This assay used yeast strains with mutations in the de novo Ade biosynthetic pathway that enable us to visualize plasmid loss on an individual colony basis. ade2 ade3 mutants are cream-colored, because the pathway is blocked at early steps in synthesis. However, expression of a plasmid-based ADE3 gene allows the synthesis of 5-aminoimidazole ribonucleotide (AIR), which is aerobically oxidized to produce a red color. Therefore, ade2 ade3 strains that stably maintain ADE3 plasmids produce red colonies because of the accumulation of AIR. Complete loss of the ADE3 plasmid results in white colonies, and partial loss is seen as white sectors in a red colony. The use of this pathway to visualize yeast markers was first described by Koshland et al. (20).
To develop this assay to study E2-mediated maintenance, the ADE3 gene was cloned into reporter plasmids similar to those described above. pRS304ARS-ADE3 contains only the ARS element; pRS304ARS-E2RE1-ADE3 and pRS304ARS-LCR-ADE3 have, in addition, the BPV1 E2RE1 and LCR elements with 4 or 11 E2-binding sites, respectively. pRS314 ADE3 is the CEN/ARS control plasmid (Fig. 1B and Table 1). We derived the YMB1670 strain (ade2-101 ade3Δ) with mutations in the ADE2 and ADE3 genes for subsequent analyses. This strain was transformed with p416CYC1-E2 and the individual reporter plasmids and selected on SC-Ura-Trp. Individual transformants were streaked on medium either selective or nonselective for the reporter plasmids. As expected, a large percentage of transformants retained the pRS314-ADE3 CEN/ARS plasmid, resulting in red colonies on nonselective medium. Transformants containing pRS304ARS-ADE3, pRS304ARS-E2RE1-ADE3, or pRS304ARS-LCR-ADE3 resulted in white colonies because of loss of these plasmids (Fig. 2B). Therefore, as shown for the liquid culture plasmid-loss assay, E2 was unable to rescue loss of plasmids containing E2-binding sites and an ARS element.
Rescue of E2-Mediated Plasmid Stability in S. cerevisiae by Brd4. We used the red/white colony sectoring assay to test the ability of individual proteins to rescue the loss of E2-binding site plasmids. The red/white colony assay can be used to screen libraries or test characterized proteins for their ability to rescue plasmid loss. We selected two candidate proteins to test in this assay because they have been implicated as chromosomal tethering proteins for the episomal viruses EBV and HHV-8. p40 (or hEBP2) interacts with EBNA1 to maintain EBV genomes, and expression of p40 and EBNA1 can reconstitute plasmid partitioning in a liquid assay yeast system (4). Brd2 (or Ring3) colocalizes with the LANA protein of HHV-8 on mitotic chromosomes. Brd2 is a member of the double bromodomain-containing BET family of proteins. This family contains short members and long members that have an additional 650-aa C-terminal “tail.” In addition to Brd2, a short family member, we tested Brd4, a long family member. During the course of this study, You et al. (17) isolated Brd4 as a binding partner of the BPV1 E2 protein.
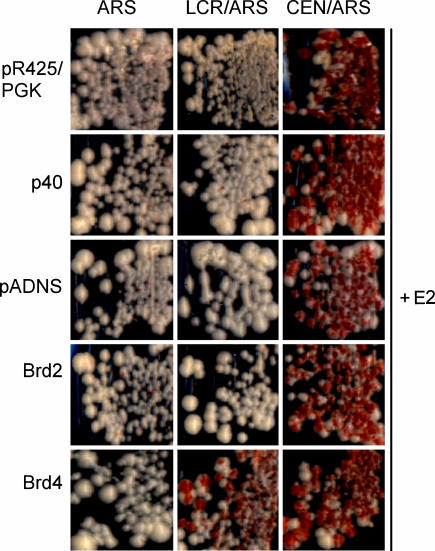
To test whether these mammalian proteins could rescue E2-mediated plasmid stability, the YMB1670 strain was transformed with p416CYC1-E2 plasmid, the reporter plasmids, and the p425PGK-hEBP2 (p40), pADNS-Brd2, or pADNS-Brd4 expression plasmids. Individual colonies were streaked onto medium selective or nonselective for the reporter plasmid. In the case of p40 and Brd2, the colony color on the nonselective plates was indistinguishable from that of transformants containing an empty expression vector, and so these proteins cannot reconstitute E2-mediated plasmid maintenance. However, transformants expressing the Brd4 and E2 proteins and containing pRS304ARS-LCR-ADE3 consistently gave predominantly red colonies under nonselective conditions for the reporter plasmid (Fig. 3). Thus, Brd4 can reconstitute E2-mediated plasmid stability in S. cerevisiae.
Fig. 3.
Mammalian Brd4 protein rescues E2-mediated plasmid maintenance. Red/white sectoring assay of YMB1670 transformants containing p416CYC1-E2, indicated reporter plasmids (see Fig. 1B), and empty vectors (p425/PGK and pADNS) or expression vectors for mammalian candidate proteins p425PGK-hEBP2 (p40), pADNS-Brd2, or pADNS-Brd4. Transformants were streaked onto media nonselective for the reporter plasmids.
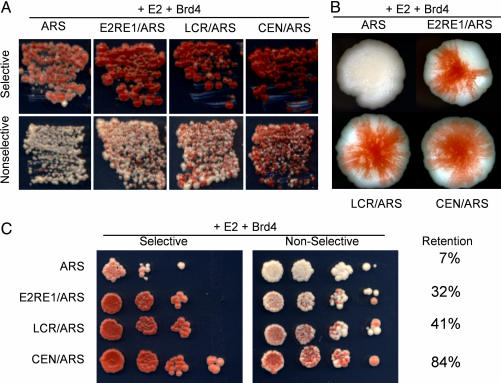
To confirm these results, the assay was performed by using the pRS304ARS-E2RE1-ADE3 plasmid, which contains four E2-binding sites. As shown in Fig. 4A, transformants containing either pRS304ARS-E2RE1-ADE3 or pRS304ARS-LCR-ADE3 reporter plasmids were stable, giving rise to red colonies in the presence of E2 and Brd4. Transformants containing the pRS304ARS-ADE3 plasmid without the BPV1 elements did not produce red colonies. A magnified image of individual colonies is shown in Fig. 4B. Even on selective medium, the colony morphology and coloration of transformants containing pRS304 ARS-E2RE1-ADE3 or pRS304ARS-LCR-ADE3 plasmids was similar to those containing the CEN/ARS plasmid, indicating that these plasmids were stably maintained.
Fig. 4.
Brd4 maintains plasmids containing E2RE1 or LCR elements. (A) Red/white sectoring assay of YMB1670 transformants containing p416CYC1E2, pADNS-Brd4, and individual reporter plasmids as indicated (see Fig. 1B). Transformants were streaked onto media selective and nonselective for the reporter plasmids. (B) Individual colonies of transformants shown in A on nonselective medium. (C) Plasmid-loss assay of YMB1670 transformants shown in A. After eight generations of growth in nonselective media, 10-fold serial dilutions were plated on media selective and nonselective for the reporter plasmids.
To quantitate E2/Brd4-mediated plasmid stability, we used a liquid culture plasmid-loss assay of transformants containing p416CYC1-E2, a reporter plasmid, and pADNS-Brd4 (Fig. 4C). In parallel to the dilution assay shown in Fig. 4C, aliquots of each dilution were plated to count individual colonies. After growth in nonselective conditions, 41% of transformants retained pRS304ARS-LCR-ADE3 plasmids, and 32% retained pRS304ARS-E2RE1-ADE3 plasmids. The absence of any maintenance element (pRS304ARS-ADE3) resulted in 7% plasmid retention, whereas the authentic yeast CEN element (pRS314-ADE3) conferred 84% retention. Therefore, coexpression of the viral E2 and mammalian Brd4 proteins results in stability of plasmids containing multiple E2-binding sites that is comparable to that conferred by the yeast CEN element.
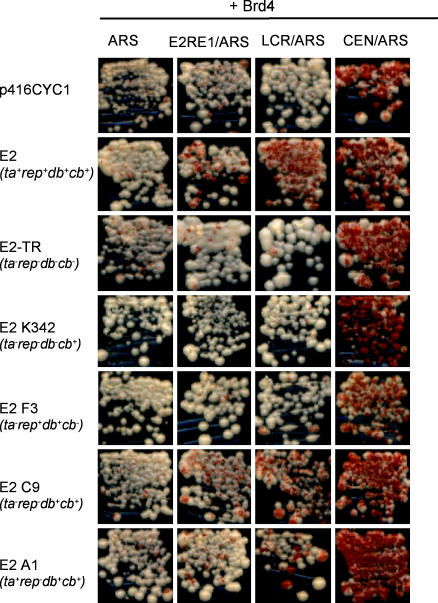
Properties of the E2 Protein Required for Plasmid Stability. Previously, we have shown that only the transactivation domain is necessary for the E2 protein to bind mitotic chromosomes (8). However, E2-binding sites are required to tether a plasmid to mitotic chromosomes (3), and so the E2 DNA-binding domain is likely required for this process. To show that plasmid stabilization is mediated by interaction of E2 and Brd4 and to determine which E2 functions are required, we tested a series of mutated E2 proteins for their ability to maintain plasmids. The YMB1670 strain was transformed with pADNS-Brd4, the reporter plasmids, and expression plasmids for either E2-TR, the shorter repressor form of E2, or E2-K342, an E2 protein defective for DNA binding. Transformants were assayed by using the red/white sectoring assay. In the presence of either E2-TR or E2 K342, transformants containing the LCR plasmid gave white colonies on nonselective media, showing that the reporter plasmid was not retained (Fig. 5). Therefore, both the N-terminal domain, which is absent in E2-TR, and the DNA-binding function of E2 are necessary for plasmid retention in yeast.
Fig. 5.
Requirements for E2-mediated plasmid maintenance. Red/white sectoring assay of YMB1670 transformants with pADNS-Brd4, individual reporter plasmids (see Fig. 1B), and p416CYC1 plasmids expressing WT or mutated E2 proteins. Transformants were streaked on media nonselective for the reporter plasmids. The phenotypes of the E2 proteins are indicated as transactivation (ta), replication (rep), DNA binding (db), and chromosome binding (cb) and are listed in more detail in Table 2.
We recently characterized a series of E2 proteins mutated in the transactivation domain for their ability to regulate transcription, support DNA replication, bind mitotic chromosomes, and interact with Brd4 in vitro (ref. 25 and Table 2, which is published as supporting information on the PNAS web site). E2 F3 (R37A,I73A) is unable to interact with Brd4, bind mitotic chromosomes, or efficiently activate transcription but retains replication and E1-binding functions. In the red/white colony assay, E2 F3 was unable to support Brd4-dependent maintenance of the LCR reporter plasmid (Fig. 5). E2 C9 (Q12N,R68Q) binds mitotic chromosomes and interacts with Brd4 but is defective for transactivation in S. cerevisiae (data not shown) and mammalian cells (25). In the sectoring assay, E2 C9 transformants maintained the LCR reporter plasmid, as indicated by red colonies (Fig. 5). Finally, E2 A1 (E2A,E6A,E13A,E20A), defective only in replication, could support Brd4-mediated retention of pRS304ARS-ADE3-LCR. Immunoprecipitation analysis showed that the mutated proteins were expressed at variable levels; however, the levels of those that did not support plasmid maintenance (F3 and K342) were similar to that of WT E2 (Fig. 7, which is published as supporting information on the PNAS web site). Thus, the E2 transactivation and replication functions are dispensable for plasmid maintenance in S. cerevisiae. Only the abilities of E2 to bind to DNA, interact with Brd4, and associate with mitotic chromosomes are required for plasmid maintenance.
To quantitate the results, we performed a liquid culture plasmid-loss assay. Transformants containing the reporter plasmids, E2 expression plasmids, and Brd4 expression plasmid were used for this assay. Yeast containing Brd4 and E2 K342, E2-TR, or E2 F3 retained the E2-binding site plasmids at levels comparable to pRS304ARS-ADE3 (5–9%; Table 3, which is published as supporting information on the PNAS web site). In contrast, the levels of retention of the E2RE1 and LCR plasmids in yeast expressing E2 C9 or E2 A1 were 18–30% (even although they had reduced expression levels and affinity for Brd4; Fig. 7 and Table 2) compared with 32–42% retention for the WT E2 protein. Therefore, the mammalian Brd4 protein can support E2-mediated plasmid stability in yeast, and the E2 functions required are identical to those that are essential for plasmid maintenance in mammalian cells.
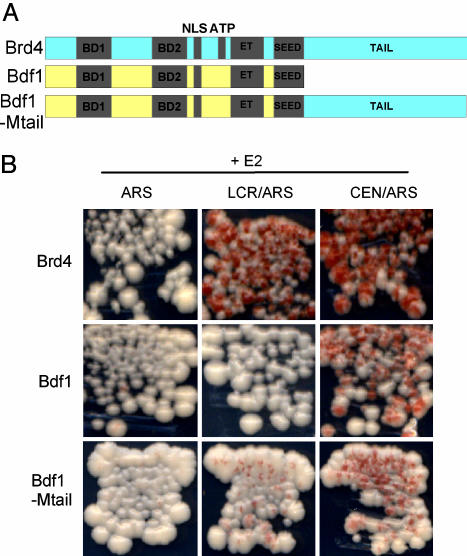
The C-Terminal Region of Brd4 Is Required for E2-Mediated Plasmid Maintenance. The BET protein family contains the yeast member Bdf1. Bdf1 binds yeast mitotic chromosomes (28) and is homologous to Brd4. However, Bdf1 lacks C-terminal sequences found in Brd4 that are important for interaction with the E2 protein (17). Thus, it is likely that the inability of E2 to mediate plasmid maintenance in yeast is due to the absence of this tail region on the Bdf1 protein.
To determine whether the tail region of Brd4 is the missing component for E2-mediated plasmid stability in S. cerevisiae, a fusion protein was generated between Bdf1 and the C-terminal 685 aa of Brd4 (Fig. 6A). The red/white sectoring assay was used to determine whether this fusion protein, Bdf1Mtail, could support E2-mediated plasmid retention. Strain YMB1670 was transformed with p416CYC1-E2, the reporter plasmids, and either Bdf1 or Bdf1Mtail expression plasmids. Individual colonies were streaked onto medium nonselective for the reporter plasmid. As shown in Fig. 6B, ectopically expressed Bdf1 was unable to cooperate with E2 to rescue the loss of pRS304ARS-LCR-ADE3; however, expression of Bdf1Mtail resulted in transformants that retained the reporter plasmid.
Fig. 6.
Bdf1–BRD4 fusion protein supports E2-mediated plasmid maintenance. (A) Diagram of BET family members S. cerevisiae Bdf1 and mouse Brd4. Regions indicated are bromodomains (BD1 and BD2), nuclear localization signal (NLS), ATP-binding motif (ATP), extraterminal domain (ET), and SEED motif. The Brd4 C-terminal region was fused to Bdf1 to generate Bdf1Mtail. (B) Red/white sectoring assay of YMB1670 transformants with p416CYC-E2, individual reporter plasmids (see Fig. 1B), and pADNS-Brd4, pADNSpl-Bdf1, and pADNSpl-Bdf1rcMtail expression plasmids. Transformants were streaked on medium nonselective for the reporter plasmids.
To confirm that the Bdf1Mtail protein could maintain plasmids, plasmid retention was quantitated with the liquid culture plasmid-loss assay (Table 4, which is published as supporting information on the PNAS web site). After eight generations of growth in medium nonselective for the reporter plasmids, only 1.9% of cells contained the negative control pRS304ARS-ADE3, and 49.3% contained the positive control pRS314-ADE3. In comparison, the plasmids containing the E2 DNA-binding sites, pRS304ARS-LCR-ADE3 and pRS304ARS-E2RE1-ADE3, were retained at 11.3% and 5.0%, respectively. Retention was not as robust as that observed with the full-length Brd4 protein (possibly because of growth-inhibitory effects of this protein; note that retention of the control plasmids is also impaired), but the Bdf1Mtail protein could partially support E2-mediated plasmid maintenance. Therefore, the tail region of Brd4 is important for E2-mediated plasmid maintenance in S. cerevisiae, and the lack of this region in Bdf1 likely explains the inability of E2 to maintain plasmids in the absence of Brd4.
Discussion
The BPV1 E2 protein attaches the viral genome to mitotic chromosomes to ensure its efficient maintenance in dividing cells (1, 2). To further investigate this function of E2, we developed two plasmid-loss assays in S. cerevisiae. In both assays, yeast plasmids were constructed in which the CEN element was replaced by a putative BPV1 segregation element. The liquid culture plasmid-loss assay involves growth of the strains to be tested in the presence or absence of selection for the reporter plasmid, allowing the rate of plasmid loss to be determined. In the visual red/white colony sectoring assay, the reporter plasmid also contained the ADE3 gene, which allows an immediate visual assessment of plasmid maintenance at the individual colony level. The sectoring assay should prove to be extremely useful for screening cDNA libraries for other proteins that can rescue plasmid loss and for further investigation of the mechanism of plasmid maintenance.
Both assays showed that the BPV1 E2 protein could not mediate efficient maintenance of yeast ARS plasmids that contain multiple E2-binding sites in place of the yeast CEN element. Thus, the chromosomal factor with which E2 interacts is either absent in S. cerevisiae or does not have enough homology to interact with E2. Therefore, the plasmid-loss assays were used to test mammalian chromosomal proteins for their ability to rescue plasmid loss. Notably, the EBV EBNA1 protein was also unable to mediate stability of plasmids containing EBNA1-binding sites in S. cerevisiae, but plasmid stability could be rescued by ectopic expression of an EBNA1-associated cellular protein, hEBP2 or p40 (4). S. cerevisiae contains a homolog of hEBP2, but yEBP2 is unable to interact with EBNA1 (4). The HHV-8 LANA protein tethers its genome to mitotic chromosomes in a similar manner. LANA associates with several cellular chromosomal proteins that have been postulated to be important for genome maintenance. One of these proteins, Brd2, is a bromodomain protein that interacts with acetylated histones on mitotic chromosomes (7). In this study, p40 (hEBP2) and Brd2 were tested for their ability to support E2-mediated plasmid maintenance in yeast. A protein from the same family as Brd2, Brd4, also was tested in these assays. We find that Brd4, but not Brd2 or p40, can reconstitute E2-mediated plasmid retention in S. cerevisiae. Consistent with our results, You et al. (17) isolated Brd4 as an E2-binding partner and demonstrated that it was important for maintenance of BPV1 genomes.
The yeast system described here showed that Brd4 could directly mediate maintenance of plasmids in an E2-dependent manner. By using our visual red/white colony assay, we can dissect the role of the Brd4–E2 interaction directly in a plasmid stability assay. You et al. (17) demonstrated that the E2 protein associated with the C-terminal portion of Brd4 and that ectopic expression of this portion of Brd4 could interfere with chromosomal binding of E2 and BPV1-mediated transformation of mouse cells. However, Brd4 is an essential gene that is also involved in the transcriptional regulatory functions of E2 (M. McPhillips, K. Ozato, and A.A.M., unpublished work), and so the effect of the C-terminal portion of Brd4 could be indirect in the latter assay.
The BET family of proteins contains one or more bromodomains, which bind acetylated histones. Certain family members, such as Brd2 and Brd4, remain associated with chromatin throughout mitosis (7). “Long” BET proteins have an additional C-terminal tail region, and it is to this region of Brd4 that E2 binds (17). Notably, the S. cerevisiae Bdf1 protein is a “short” family member, which explains why E2 is unable to mediate plasmid retention in yeast. However, a protein consisting of Bdf1 fused to the C-terminal tail of Brd4 was able to rescue E2-mediated plasmid maintenance in yeast. Attempts to further map the region of Brd4 responsible for rescue of E2 plasmid retention was not feasible because of growth-inhibitory effects of truncated Brd4 proteins in S. cerevisiae.
The yeast plasmid stability assays were used to determine the E2 protein requirements for plasmid retention in yeast. In the presence of Brd4, the E2 K342 protein and the transactivation-defective E2-TR protein were unable to mediate plasmid retention. The E2-TR repressor protein is unable to bind to Brd4 (17) or mammalian mitotic chromosomes (1). E2 K342 can bind mitotic chromosomes by means of the transactivation domain but is defective in DNA binding and should be unable to tether reporter plasmids to the chromosomes. The E2 F3 protein, which is defective in transactivation, Brd4 interaction, and mitotic chromosomal binding but is still functional in replication, was also unable to retain plasmids containing E2-binding sites. However, both the E2 C9 and A1 proteins, which bind Brd4 and mammalian mitotic chromosomes, maintained the reporter plasmids. Plasmids containing the LCR, with 11 binding sites, and plasmids containing E2RE1, with 4 binding sites, were retained in the presence of E2 and Brd4.
The yeast system that we have developed is a powerful yet simple assay that can be used to study many aspects of plasmid and viral genome maintenance and to identify proteins important for these processes. In this study, it was shown that the cellular Brd4 protein could reconstitute E2-mediated plasmid maintenance in yeast. Brd4 belongs to the same family as the proposed cellular tethering protein of HHV-8, Brd2. This strategy of two divergent viruses targeting similar cellular proteins for their tethering function might suggest that the role of this interaction is more involved than a simple attachment to a convenient chromosomal complex.
Supplementary Material
Acknowledgments
We thank Jaquelline Oliveira and Emiko Soeda for comments on the manuscript.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ARS, autonomous replication sequence; CEN, centromeric; EBV, Epstein–Barr virus; EBNA1, EBV-encoded nuclear antigen 1; HHV-8, human herpes virus 8; BPV1, bovine papillomavirus type 1; SC, synthetic complete.
References
- 1.Skiadopoulos, M. H. & McBride, A. A. (1998) J. Virol. 72, 2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehman, C. W. & Botchan, M. R. (1998) Proc. Natl. Acad. Sci. USA 95, 4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ilves, I., Kivi, S. & Ustav, M. (1999) J. Virol. 73, 4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapoor, P., Shire, K. & Frappier, L. (2001) EMBO J. 20, 222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter, M. A. & Robertson, E. S. (1999) Virology 264, 254-264. [DOI] [PubMed] [Google Scholar]
- 6.Platt, G. M., Simpson, G. R., Mittnacht, S. & Schulz, T. F. (1999) J. Virol. 73, 9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dey, A., Chitsaz, F., Abbasi, A., Misteli, T. & Ozato, K. (2003) Proc. Natl. Acad. Sci. USA 100, 8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastien, N. & McBride, A. A. (2000) Virology 270, 124-134. [DOI] [PubMed] [Google Scholar]
- 9.Abroi, A., Ilves, I., Kivi, S. & Ustav, M. (2004) J. Virol. 78, 2100-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng, P.-S., Brokaw, J. L. & McBride, A. A. (2005) J. Virol. 79, 1500-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBride, A. A., Byrne, J. C. & Howley, P. M. (1989) Proc. Natl. Acad. Sci. USA 86, 510-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winokur, P. L. & McBride, A. A. (1996) Virology 221, 44-53. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, L. & Carbon, J. (1980) Nature 287, 504-509. [DOI] [PubMed] [Google Scholar]
- 14.Hsiao, C. L. & Carbon, J. (1979) Proc. Natl. Acad. Sci. USA 76, 3829-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stinchcomb, D. T., Struhl, K. & Davis, R. W. (1979) Nature 282, 39-43. [DOI] [PubMed] [Google Scholar]
- 16.Piirsoo, M., Ustav, E., Mandel, T., Stenlund, A. & Ustav, M. (1996) EMBO J. 15, 1-11. [PMC free article] [PubMed] [Google Scholar]
- 17.You, J., Croyle, J. L., Nishimura, A., Ozato, K. & Howley, P. M. (2004) Cell 117, 349-360. [DOI] [PubMed] [Google Scholar]
- 18.Thomas, B. J. & Rothstein, R. (1989) Cell 56, 619-630. [DOI] [PubMed] [Google Scholar]
- 19.Sikorski, R. S. & Hieter, P. (1989) Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koshland, D., Kent, J. C. & Hartwell, L. H. (1985) Cell 40, 393-403. [DOI] [PubMed] [Google Scholar]
- 21.McBride, A. A., Bolen, J. B. & Howley, P. M. (1989) J. Virol. 63, 5076-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colicelli, J., Birchmeier, C., Michaeli, T., O'Neill, K., Riggs, M. & Wigler, M. (1989) Proc. Natl. Acad. Sci. USA 86, 3599-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brachmann, R. K., Vidal, M. & Boeke, J. D. (1996) Proc. Natl. Acad. Sci. USA 93, 4091-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mumberg, D., Muller, R. & Funk, M. (1995) Gene 156, 119-122. [DOI] [PubMed] [Google Scholar]
- 25.Baxter, M. K., McPhillips, M. G., Ozato, K. & McBride, A. A. (2005) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 26.Dey, A., Ellenberg, J., Farina, A., Coleman, A. E., Maruyama, T., Sciortino, S., Lippincott-Schwartz, J. & Ozato, K. (2000) Mol. Cell. Biol. 20, 6537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ladurner, A. G., Inouye, C., Jain, R. & Tjian, R. (2003) Mol. Cell 11, 365-376. [DOI] [PubMed] [Google Scholar]
- 28.Chua, P. & Roeder, G. S. (1995) Mol. Cell. Biol. 15, 3685-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.