Abstract
Accumulating evidence has suggested that germline DNA copy number variations (CNVs) affect various disorders, including human malignancies. However, the significance of CNVs in non-muscle invasive bladder cancer (NMIBC) remains unclear. The purpose of the present study was to identify the role of CNVs in NMIBC. Array comparative genomic hybridization (CGH) analysis was performed to search for candidate CNVs associated with NMIBC susceptibility. Quantitative polymerase chain reaction was carried out to evaluate CNVs associated with patient outcome in 189 NMIBC cases. In total, 11 CNVs were associated with NMIBC risk in array CGH analysis. Out of the 189 CNVs examined, family with sequence similarity 81 member A (FAM81A) and proprotein convertase subtilisin/kexin type 6 (PCSK6) CNVs exhibited a significant association with recurrence and disease progression in NMIBC. PCSK6 has been reported to regulate proliferation and tumor progression in breast and prostate malignancies. Notably, patients with pT1 stage had significantly lower PCSK6 relative copy number than those with pTa (P=0.0196). In multivariate analyses, PCSK6 copy number was an independent prognostic factor for progression-free survival (P=0.0456; risk ratio, 2.17; 95% confidence interval, 1.02–4.82). These data suggest that PCSK6 CNV is a potential new tumor marker for estimating disease progression in NMIBC.
Keywords: DNA copy number variations, non-muscle invasive bladder cancer, PCSK6, FAM81A
Introduction
Urothelial carcinoma of the bladder is the second most common malignancy of the genitourinary tract, the ninth most common cancer and the fourteenth leading cause of mortality due to cancer in the world (1). Approximately 70% of all bladder cancers are non-muscle invasive bladder cancer (NMIBC) (2). Long-term follow-up of low-grade Ta tumors shows a progression rate of ~6%, whereas high-grade T1 tumors exhibit a greater rate of progression, namely ~17% (3,4). Disease progression frequently results in an unfavorable clinical outcome following progression to muscle-invasive tumor or development of distant metastases (3,4). Conventional clinicopathological factors predicting disease progression include number of tumors, tumor diameter, stage, concomitant carcinoma in situ and tumor grade (2). However, the prediction of progression remains difficult based on conventional parameters, and to date, no useful biomarkers have been established for follow-up in routine practice (5).
DNA copy number variations (CNVs) involve gains or losses of several to hundreds kb of genomic DNA among phenotypically normal individuals, and ≥291,801 CNV regions have been identified to date (6). Recent studies have described germline CNVs as potential susceptibility loci for a range of diseases, including infectious, autoimmune and neuropsychiatric diseases (7–10). Concerning malignancies, CNVs have recently been reported as markers predisposing individuals to neuroblastoma, prostate cancer, pancreatic cancer, colorectal cancer, ovarian cancer and breast cancer risk (11–16). However, the significance of CNVs in NMIBC remains unclear. Therefore, the present study investigated the association between CNVs and disease progression in NMIBC. To the best of our knowledge, the present report is the first to confirm CNV as a potential biomarker for assessing disease progression in NMBC.
Materials and methods
Patients
The study group comprised 189 patients who underwent transurethral resection of bladder tumor (TURBT) and were histopathologically diagnosed with pTa and pT1 (N0M0) urothelial carcinoma of the bladder based on the tumor-node-metastasis classification of the International Union Against Cancer (2002) at the first documented diagnosis (17). The patients were followed up from April 2004 to December 2012 at Yamaguchi University Hospital (Ube, Japan). Written informed consent was obtained from each patient. The present study was performed according to the Declaration of Helsinki, and the procedures were approved by the Ethics Committee of Yamaguchi University.
Venous blood samples were collected from each patient, and the original medical records were retrospectively reviewed for collecting data. Tumor grade was classified according to the World Health Organization classification system (1973) (18). The patients' characteristics are summarized in Table I. Patients who underwent TURBT were followed up every 3 months by cystoscopic examination and urine cytology, whereas those with high-risk NMIBC were followed up by computerized tomography scan every year, with a median follow-up period of 81.4 months (range, 1.7–301.9 months). Disease progression was defined when either the recurrent tumor had progressed to muscle-invasive tumor or when development of distant metastases had occurred.
Table I.
Patients' demographics and pathological background.
| Factor | No. (%) |
|---|---|
| Sex | |
| Male | 160 (84.7) |
| Female | 29 (15.3) |
| Tumor grade | |
| 1 | 17 (9.0) |
| 2 | 67 (35.4) |
| 3 | 105 (55.6) |
| pT stage | |
| pTa | 84 (44.4) |
| pT1 | 105 (55.6) |
| Concurrent CIS | |
| No | 122 (64.5) |
| Yes | 67 (35.5) |
| Cytology | |
| Negative | 79 (41.8) |
| Positive | 110 (58.2) |
| Recurrence | |
| No | 88 (46.6) |
| Yes | 101 (53.4) |
| Disease progression | |
| No | 157 (83.1) |
| Yes | 32 (16.9) |
| Local | 9 (4.8) |
| Distant | 23 (12.2) |
CIS, carcinoma in situ.
Array comparative genomic hybridization (GCH) assay and data analysis
A total of 67 DNA samples were obtained from the peripheral blood of individuals without a history of human malignancies, including bladder cancer (healthy controls), and 18 DNA samples were obtained from the peripheral blood of patients with a history of NMIBC (6 pTa and 12 pT1). The pathological grades were 1, 2 and 3 for 3, 5 and 10 tumors, respectively. A pool of blood-derived DNA from 30 healthy individuals was used as a reference sample for all the hybridizations performed. The mean age and sex ratio were almost identical in the control and NMIBC patient groups. Assessment of CNVs in the human genome by oligonucleotide array CGH assay (NimbleGen Human CGH 2.1M Whole-Genome Tiling v2.0D Array; Roche Diagnostics, Indianapolis, IN, USA) was performed according to the manufacturer's standard protocol. Array image analysis and data normalization were performed with NimbleScan version 2.5 software (Roche Diagnostics). The normalized data were then processed using Nexus Copy Number version 5.0 software (BioDiscovery, Inc., El Segundo, CA, USA) as previously described (16).
Quantitative polymerase chain reaction (qPCR) and data analysis
qPCR was performed using predesigned TaqMan® Copy Number Assays (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing a primer pair and a fluorescein amidite dye-labeled minor groove binder probe was performed to detect the copy number of the genomic sequence. Copy number assay identity is described in Table II. For the internal control, a predesigned TaqMan® Copy Number Reference Assay RNase P (Applied Biosystems; Thermo Fisher Scientific, Inc.), was used. A total of 189 DNA samples were obtained from the peripheral blood of patients with a history of NMIBC. The calibrator sample for qPCR was the DNA pooled from the 30 healthy volunteers, which was also used as the reference in the array CGH assay, and the copy number of the calibrator sample was assumed to be 2. The 7900HT Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) and the StepOnePlus Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) were used for qPCR analysis. The PCRs were carried out according to the manufacturer's standard protocol using the comparative 2−ΔΔCq method, as previously described (16).
Table II.
CNV markers associated with bladder cancer risk.
| Location (GRCh37/hg19) | Cytoband | Gene | CNV assay ID | CNV | P-valuea | Bonferroni |
|---|---|---|---|---|---|---|
| chr2:72,218,053–72,222,026 | p13.3 | CYP26B1 | Hs05873524_cn | Gain | <0.0001 | 0.0149 |
| chr2:101,678,677–101,683,846 | q11.2 | MAP4K4 | Hs02074840_cn | Loss | <0.0001 | 0.0040 |
| chr7:1,237,863–1,240,528 | p22.3 | UNCX | Hs03622829_cn | Loss | <0.0001 | 0.0384 |
| chr8:61,752,169–61,756,537 | q12.2 | CHD7 | Hs02866323_cn | Loss | <0.0001 | 0.0149 |
| chr10:64,949,973–64,952,473 | q21.3 | REEP3 | Hs00735097_cn | Loss | <0.0001 | 0.0002 |
| chr11:256,350–257,345 | p15.5 | Non-coding region of the genome | Hs03791448_cn | Loss | <0.0001 | 0.0331 |
| chr14:50,365,069–50,365,879 | q22.1 | NIN | Hs07054232_cn | Loss | <0.0001 | <0.0001 |
| chr15:57,515,723–57,519,569 | q22.2 | FAM81A | Hs59732286_cn | Loss | <0.0001 | 0.0012 |
| chr15:99,843,944–99,844,859 | q26.3 | PCSK6 | Hs03899300_cn | Loss | <0.0001 | 0.0040 |
| chr16:45,732,422–45,736,896 | q12.1 | NETO2 | Hs02817425_cn | Loss | <0.0001 | 0.0012 |
| chr19:35,123,405–35,132,171 | q12 | C19orf2 | Hs07125447_cn | Loss | <0.0001 | <0.0001 |
Fisher's exact test. CNV, copy number variation; ID, identity; chr, chromosome; Hs, Homo sapiens; CYP26B1, cytochrome P450 family 26 subfamily B member 1; MAP4K4, mitogen-activated protein kinase kinase kinase kinase 4; UNCX, UNC homeobox; CHD7, chromodomain-helicase-DNA-binding protein 7; REEP3, receptor expression-enhancing protein 3; NIN, ninein; FAM81A, family with sequence similarity 81 member A; PCSK6, proprotein convertase subtilisin/kexin type 6; NETO2, neuropilin and tolloid like 2; C19orf2, chromosome 19 open reading frame 2.
Statistical analysis
Statistical analysis was performed using JMP (version 13) statistical software (SAS Institute, Inc., Cary, NC, USA). The Fisher's exact test (Tables II and III) and an unpaired t-test (Fig. 1) were applied to compare variables. CNV markers detected by array CGH assay were calculated using Fisher's exact test and the Bonferroni correction. The probability of survival was calculated by the Kaplan-Meier estimator method, and statistical differences were evaluated by the log-rank test. Categorical variables influencing progression-free survival were compared using Cox proportional hazards regression models. Variables with P<0.05 in univariate analysis were also assessed for their association with progression-free survival in multivariate analysis. For all of the statistical tests, P<0.05 was considered to indicate a statistically significant difference.
Table III.
Correlation of several variables with FAM81A or PCSK6 CNV.
| FAM81A CNV | PCSK6 CNV | |||||
|---|---|---|---|---|---|---|
| Factor | <1.77 | ≥1.77a | P-value | <1.33 | >1.33a | P-value |
| Tumor grade | ||||||
| 1 & 2 | 41 | 32 | 0.1380 | 19 | 54 | 0.0606 |
| 3 | 52 | 64 | 46 | 70 | ||
| pT stage | ||||||
| pTa | 46 | 38 | 0.1896 | 22 | 62 | 0.0449 |
| pT1 | 47 | 58 | 43 | 62 | ||
| Recurrence | ||||||
| No | 37 | 51 | 0.0803 | 26 | 62 | 0.2206 |
| Yes | 56 | 45 | 39 | 62 | ||
| Disease progression | ||||||
| No | 70 | 87 | 0.0062 | 45 | 112 | 0.0004 |
| Yes | 23 | 9 | 20 | 12 | ||
CNV threshold. CNV, copy number variation; FAM81A, family with sequence similarity 81 member A; PCSK6, proprotein convertase subtilisin/kexin type 6.
Figure 1.
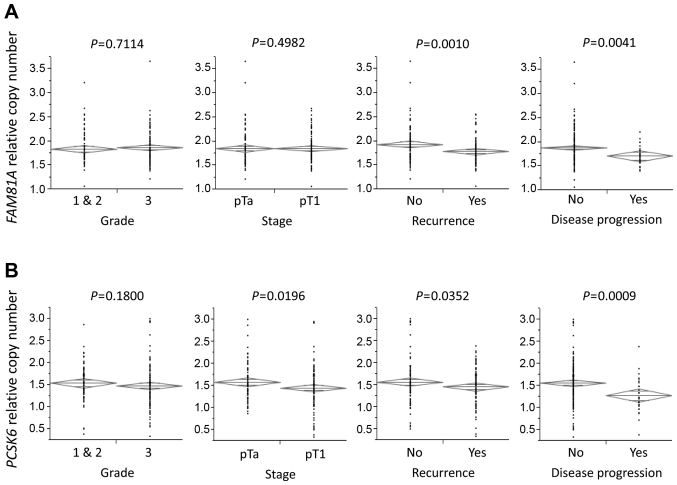
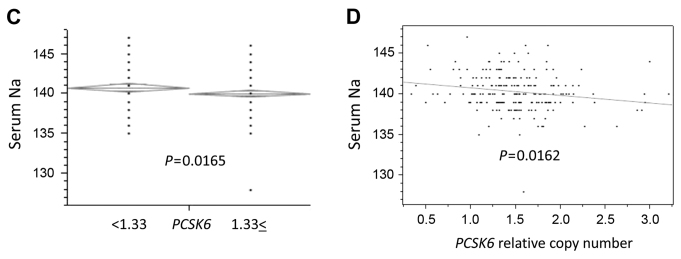
Association of tumor grade, stage, recurrence, disease progression and serum sodium levels with FAM81A or PCSK6 relative copy numbers. (A) Patients with recurrence and disease progression had significantly lower FAM81A relative copy number than those without recurrence and disease progression (P=0.0010 and P=0.0041, respectively). (B) Patients with pT1 stage, recurrence and disease progression had significantly lower PCSK6 relative copy number than those without pT1 stage, recurrence and disease progression (P=0.0196, P=0.0352 and P=0.0009, respectively). The diamonds show the mean (long horizontal line) and 95% confidence interval of the relative copy number. (C) Patients with lower PCSK6 copy number had significantly higher sodium levels in blood than those with higher PCSK6 copy number (P=0.0165). (D) A positive correlation between PCSK6 relative copy number and serum sodium levels was identified in linear regression analysis (P=0.0162). FAM81A, family with sequence similarity 81 member A; PCSK6, proprotein convertase subtilisin/kexin type 6.
Results
CNV markers are associated with bladder cancer risk
Using array CGH assay, 11 CNV regions with significant differences in the frequency of copy number changes between the NMIBC patient group and the control group were identified. The CNV regions reached significance by Bonferroni correction. Therefore, it can be speculated that these regions may involve candidate genes associated with NMIBC risk (Table II).
Association of CNVs with several variables
A case-case study was carried out by qPCR to evaluate the association of the above 11 CNVs with recurrence and disease progression in the present 189 NMIBC cases. Notably, family with sequence similarity 81 member A (FAM81A) and proprotein convertase subtilisin/kexin type 6 (PCSK6) copy numbers, according to these 11 CNVs, exhibited a significant association with recurrence and disease progression in NMIBC (Fig. 1). Another CNV lesion (CYP26B1, MAP4K4, UNCX, CHD7, REEP3, NIN, NETO2 and C19orf2) exhibited a non-significant association with recurrence and disease progression. Therefore, the association between FAM81A and PCSK6 copy numbers and clinicopathological parameters was reviewed. No significant differences were observed in FAM81A copy number between patients with pT1 stage and those with pTa (P=0.4982; Fig. 1A); however, patients with pT1 stage had a significantly lower PCSK6 relative copy number than those with pTa (P=0.0196; Fig. 1B).
The threshold values of CNVs were set based on the area under the curve (AUC) from the receiver operating characteristic curve for disease progression. The AUC of FAM81A and PCSK6 copy numbers at 1.77 and 1.33 threshold values was 0.65 and 0.70, respectively, with a sensitivity of 71.9 and 65.6%, and a specificity of 55.4 and 73.9%, respectively. In those copy number thresholds, the FAM81A and PCSK6 copy numbers were significantly associated with disease progression (P=0.0062 and P=0.0004, respectively; Table III). PCSK6 copy number was also significantly associated with pT stage (P=0.0449; Table III).
PCSK6 has been reported to regulate sodium homeostasis (19). Therefore, the present study investigated the association between PSCK6 and sodium concentration. Patients with lower PCSK6 copy number had significantly higher serum sodium levels than those with higher PCSK6 copy number (P=0.0330; Fig. 1C). A positive correlation between PCSK6 copy number and serum sodium concentration was identified in linear regression analysis (P=0.0162; Fig. 1D).
Univariate and multivariate analyses of disease progression
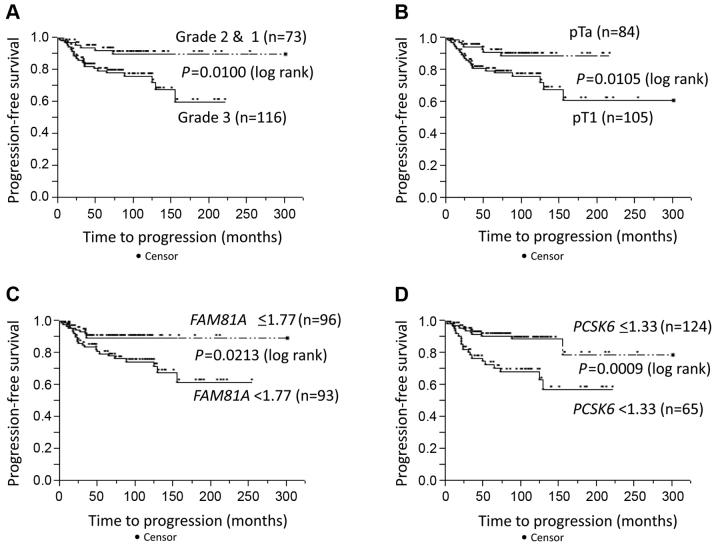
Progression-free survival was evaluated in the context of pathological data and CNVs. Univariate Cox proportional hazards regression analysis revealed that tumor grade (P=0.0067), pT stage (P=0.0079), urine cytology (P=0.0041), and FAM81A (P=0.0190) and PCSK6 (P=0.0013) copy numbers had a significant effect on progression-free survival (Table IV). Progression-free survival rates were plotted using Kaplan-Meier survival curves (Fig. 2). Tumor grade and stage, and FAM81A and PCSK6 copy numbers were significant prognostic factors for disease-specific survival (P=0.0100, P=0.0105, P=0.0213 and P=0.0009, respectively; log-rank test; Fig. 2A-D). In multivariate analyses, PCSK6 copy number was an independent prognostic factor for progression-free survival (P=0.0456; risk ratio, 2.17; 95% confidence interval, 1.02–4.82; Table IV).
Table IV.
Cox proportional hazard model for progression-free survival in non-muscle invasive bladder cancer.
| Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|
| Factor | Category | Risk ratio (95% CI) | P-value | Risk ratio (95% CI) | P-value |
| Tumor grade | Grade 3 vs. 1 & 2 | 3.04 (1.34–8.16) | 0.0067 | 1.62 (0.56–5.33) | 0.3866 |
| pT stage | pT1 vs. pTa | 2.85 (1.30–7.14) | 0.0079 | 1.67 (0.63–5.09) | 0.3168 |
| Concurrent CIS | Yes vs. no | 1.51 (0.74–3.03) | 0.2489 | ||
| Cytology | Negative vs. positive | 3.22 (1.42–8.65) | 0.0041 | 2.05 (0.85–5.77) | 0.1126 |
| FAM81A CNV | <1.77 vs. ≥1.77 | 2.42 (1.15–5.53) | 0.0190 | 1.93 (0.86–4.66) | 0.1110 |
| PCSK6 CNV | <1.33 vs. ≥1.33 | 3.15 (1.56–6.64) | 0.0013 | 2.17 (1.02–4.82) | 0.0456 |
CI, confidence interval; CIS, carcinoma in situ; FAM81A, family with sequence similarity 81 member A; PCSK6, proprotein convertase subtilisin/kexin type 6; CNV, copy number variation.
Figure 2.
Kaplan-Meier plots of disease-specific survival stratified by (A) tumor grade, (B) stage, (C) FAM81A copy number and (D) PCSK6 copy number. FAM81A, family with sequence similarity 81 member A; PCSK6, proprotein convertase subtilisin/kexin type 6.
Discussion
Since treatment modalities for NMIBC patients vary from simple transurethral resection in low-risk NMIBC patients to recommendation of radical cystectomy in high-risk NMIBC patients, the prediction of biological characteristics of disease progression in individual patients, including invasiveness and metastatic potential, are markedly important for selecting the most appropriate treatment (5). Due to the limited prediction abilities of conventional markers such as tumor grade or stage (2,5), the identification of reliable genetic markers predicting disease progression in NMIBC is urgently required.
Previous reports demonstrated the association between CNVs and the risk of several human malignancies (11–16). PCSK6 is a member of the protease family of proprotein convertases, which activate precursor proteins by cleaving at the specific recognition sequence RXK/RR (20). PCSK6 is important for maintaining sodium homeostasis and normal blood pressure (19). Although PCSK6 has been reported to regulate cell proliferation and tumor progression in breast and prostate cancer (21–23), the exact association between PCSK6 expression and carcinogenesis is controversial. In an experimental model, the overexpression of PCSK6 in immortalized non-tumorigenic or papilloma-derived keratinocytes increased their invasiveness (24), whereas absent or reduced PCSK6 expression was linked to ovarian cancer (25). Reduced germline copy number of PCSK6 was also reported to be associated with breast cancer risk (16). These reports may support the present data that reduced copy number of germline PCSK6 may confer tumor aggressiveness, thus leading to poor disease progression in NMIBC. In addition, a significant reverse correlation between PCSK6 copy number and serum sodium levels was identified in the present study. Since PCSK6 regulates sodium homeostasis, this correlation implies a functional relevance due to gene dosage. The function of a reduced copy number of PCSK6 in normal human cells has not been investigated yet. PCSK6 affects sodium homeostasis; however, this mechanism may be different from tumor progression through PCSK6. The function of FAM81A remains unknown, and there are no reports about FAM81A CNVs. Further studies may be required on the function and significance of PCSK6 and FAM81A CNVs in human malignancies.
In the present study, FAM81A and PCSK6 copy numbers in NMIBC were lower than those in healthy human volunteers, as shown by qPCR (Fig. 1). These data support and validate the data detected by array CGH. Our hypothesis proposed that several candidate genes linked to bladder cancer risk may also affect tumor aggressiveness, thus leading to tumor progression. The present study aimed to determine the prognostic value of CNVs for NMIBC rather than to detect CNVs associated with bladder cancer susceptibility. However, further studies on the association between FAM81A and PCSK6 copy numbers with NMIBC risk may be required to compare a larger sample size of NMIBC cases with healthy volunteers in order to determine CNVs associated with bladder cancer susceptibility.
The present study is constrained by several limitations. First, it is a retrospective study with a limited number of patients. Second, the patients in the present study underwent TURBT by several surgeons over a long time period. Thus, prospective studies including a larger sample size are required to confirm the predictive significance of CNVs in NMIBC. Third, there is a possibility of false positive results in array CGH analysis. CNVs detected by array CGH assay harbor false positives due to a poor signal-to-noise ratio of hybridizations, which leads to considerable variation in the reported CGH ratio (16). Therefore, qPCR with a larger cohort should be carried out to confirm the CNVs that appeared to be associated with bladder cancer susceptibility.
In conclusion, PCSK6 copy number is an independent predictor of progression-free survival in NMIBC. PCSK6 copy number may have significant potential as a biomarker for estimating disease progression in NMIBC patients treated by TURBT, and may help to select patients with NMIBC who may benefit from more aggressive treatment, including radical cystectomy.
Acknowledgements
The authors thank Mrs. Kiyomi Fujita (Department of Urology, Graduate School of Medicine, Yamaguchi University) for her technical assistance.
References
- 1.Mahdavifar N, Ghoncheh M, Pakzad R, Momenimovahed Z, Salehiniya H. Epidemiology, incidence and mortality of bladder cancer and their relationship with the development index in the world. Asian Pac J Cancer Prev. 2016;17:381–386. doi: 10.7314/APJCP.2016.17.1.381. [DOI] [PubMed] [Google Scholar]
- 2.van Rhijn BW, Burger M, Lotan Y, Solsona E, Stief CG, Sylvester RJ, Witjes JA, Zlotta AR. Recurrence and progression of disease in non-muscle-invasive bladder cancer: From epidemiology to treatment strategy. Eur Urol. 2009;56:430–442. doi: 10.1016/j.eururo.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Leblanc B, Duclos AJ, Bénard F, Côté J, Valiquette L, Paquin JM, Mauffette F, Faucher R, Perreault JP. Long-term followup of initial Ta grade 1 transitional cell carcinoma of the bladder. J Urol. 1999;162:1946–1950. doi: 10.1016/S0022-5347(05)68075-5. [DOI] [PubMed] [Google Scholar]
- 4.Palou J, Sylvester RJ, Faba OR, Parada R, Peña JA, Algaba F, Villavicencio H. Female gender and carcinoma in situ in the prostatic urethra are prognostic factors for recurrence, progression, and disease-specific mortality in T1G3 bladder cancer patients treated with bacillus Calmette-Guerin. Eur Urol. 2012;62:118–125. doi: 10.1016/j.eururo.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 5.Babjuk M, Böhle A, Burger M, Capoun O, Cohen D, Compérat EM, Hernández V, Kaasinen E, Palou J, Rouprêt M, et al. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder: Update 2016. Eur Uro. 2017;71:447–461. doi: 10.1016/j.eururo.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 6.Suehiro Y, Furuya T, Sasaki K, Hinota Y. DNA polymorphisms. Rinsho Byori. 2013;61:1001–1007. (In Japanese) [PubMed] [Google Scholar]
- 7.Shlien A, Malkin D. Copy number variations and cancer. Genome Med. 2009;1:62. doi: 10.1186/gm62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wain LV, Armour JA, Tobin MD. Genomic copy number variation, human health, and disease. Lancet. 2009;374:340–350. doi: 10.1016/S0140-6736(09)60249-X. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fanciulli M, Petretto E, Aitman TJ. Gene copy number variation and common human disease. Clin Genet. 2010;77:201–213. doi: 10.1111/j.1399-0004.2009.01342.x. [DOI] [PubMed] [Google Scholar]
- 11.Diskin SJ, Hou C, Glessner JT, Attiyeh EF, Laudenslager M, Bosse K, Cole K, Mossé YP, Wood A, Lynch JE, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–991. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W, Sun J, Li G, Zhu Y, Zhang S, Kim ST, Sun J, Wiklund F, Wiley K, Isaacs SD, et al. Association of a germ-line copy number variation at 2p24.3 and risk for aggressive prostate cancer. Cancer Res. 2009;69:2176–2179. doi: 10.1158/0008-5472.CAN-08-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lucito R, Suresh S, Walter K, Pandey A, Lakshmi B, Krasnitz A, Sebat J, Wigler M, Klein AP, Brune K, et al. Copy-number variants in patients with a strong family history of pancreatic cancer. Cancer Biol Ther. 2007;6:1592–1599. doi: 10.4161/cbt.6.10.4725. [DOI] [PubMed] [Google Scholar]
- 14.Thean LF, Loi C, Ho KS, Koh PK, Eu KW, Cheah PY. Genome-wide scan identifies a copy number variable region at 3q26 that regulates PPM1L in APC mutation-negative familial colorectal cancer patients. Genes Chromosomes Cancer. 2010;49:99–106. doi: 10.1002/gcc.20724. [DOI] [PubMed] [Google Scholar]
- 15.Yoshihara K, Tajima A, Adachi S, Quan J, Sekine M, Kase H, Yahata T, Inoue I, Tanaka K. Germline copy number variations in BRCA1-associated ovarian cancer patients. Genes Chromosomes Cancer. 2011;50:167–177. doi: 10.1002/gcc.20841. [DOI] [PubMed] [Google Scholar]
- 16.Suehiro Y, Okada T, Shikamoto N, Zhan Y, Sakai K, Okayama N, Nishioka M, Furuya T, Oga A, Kawauchi S, et al. Germline copy number variations associated with breast cancer susceptibility in a Japanese population. Tumour Biol. 2013;34:947–952. doi: 10.1007/s13277-012-0630-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobin LH, Gospodarowicz M, Wittekind C, editors. TNM classification of malignant tumors. 7th. UICC International Union Against Cancer; 2009. pp. 262–265. [Google Scholar]
- 18.Mostofi FK, Sobin LH, Torloni H. International Histological Classification of Tumors. No. 10. World Health Organization; Geneva: 1973. Histologic typing of urinary bladder tumors. [Google Scholar]
- 19.Chen S, Cao P, Dong N, Peng J, Zhang C, Wang H, Zhou T, Yang J, Zhang Y, Martelli EE, et al. PCSK6-mediated corin activation is essential for normal blood pressure. Nat Med. 2015;21:1048–1053. doi: 10.1038/nm.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molloy SS, Anderson ED, Jean F, Thomas G. Bi-cycling the furin pathway: From TGN localization to pathogen activation and embryogenesis. Trends Cell Biol. 1999;9:28–35. doi: 10.1016/S0962-8924(98)01382-8. [DOI] [PubMed] [Google Scholar]
- 21.Lapierre M, Siegfried G, Scamuffa N, Bontemps Y, Calvo F, Seidah NG, Khatib AM. Opposing function of the proprotein convertases furin and PACE4 on breast cancer cells' malignant phenotypes: Role of tissue inhibitors of metalloproteinase-1. Cancer Res. 2007;67:9030–9034. doi: 10.1158/0008-5472.CAN-07-0807. [DOI] [PubMed] [Google Scholar]
- 22.Wang F, Wang L, Pan J. PACE4 regulates proliferation, migration and invasion in human breast cancer MDA-MB-231 cells. Mol Med Rep. 2015;11:698–704. doi: 10.3892/mmr.2014.2691. [DOI] [PubMed] [Google Scholar]
- 23.Kang S, Zhao Y, Hu K, Xu C, Wang L, Liu J, Yao A, Zhang H, Cao F. miR-124 exhibits antiproliferative and antiaggressive effects on prostate cancer cells through PACE4 pathway. Prostate. 2014;74:1095–1106. doi: 10.1002/pros.22822. [DOI] [PubMed] [Google Scholar]
- 24.Mahloogi H, Bassi DE, Klein-Szanto AJ. Malignant conversion of non-tumorigenic murine skin keratinocytes overexpressing PACE4. Carcinogenesis. 2002;23:565–572. doi: 10.1093/carcin/23.4.565. [DOI] [PubMed] [Google Scholar]
- 25.Fu Y, Campbell EJ, Shepherd TG, Nachtigal MW. Epigenetic regulation of proprotein convertase PACE4 gene expression in human ovarian cancer cells. Mol Cancer Res. 2003;1:569–576. [PubMed] [Google Scholar]