Abstract
The PTEN (phosphatase and tensin homologue) tumor suppressor protein contains a single catalytic domain with both lipid and protein phosphatase activities. The remaining C-terminal half of the PTEN protein plays a role in its stability and is mutated in many clinical cancer samples. Here, we report that the PTEN C-terminal domain physically interacts with the forkhead-associated domain of the oncogenic MSP58 protein and that this interaction requires PTEN Thr-366. We further show that while MSP58 transforms Pten–/– mouse embryo fibroblasts (MEFs), concurrent introduction of wild-type PTEN causes a dramatic reduction in the number of MSP58-induced transformed foci. This PTEN-mediated inhibition of cellular transformation requires physical interaction as evidenced by the failure of PTEN(T366A) point mutation (residing within the MSP58 interaction domain) to suppress MSP-58-driven transformation. These observations, together with the capacity of catalytically inactive PTEN mutant (G129R) to suppress MSP58 oncogenicity, support the view that the C-terminal region of PTEN directly provides a previously uncharacterized biological function in its ability to regulate cellular transformation.
PTEN (phosphatase and tensin homology, deleted on chromosome ten) is a tumor suppressor gene that is frequently somatically deleted or mutated in a variety of human cancers including those of the brain, endometrium, prostate, and lung (1, 2). Germ-line mutations of PTEN are also the cause of Cowden's disease, an autosomal dominant hamartoma syndrome with increased risk for the development of tumors in a variety of tissues (1, 2).
PTEN dephosphorylates the 3′ position of phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] and phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2] and tyrosine-phosphorylated FAK and Shc. These activities inhibit cell growth and cell migration, respectively (3). The PTEN protein consists of an N-terminal phosphatase domain and a C-terminal domain, which is subdivided into C2, phosphorylation, and PDZ (PSD-95, disc-large, Zonula Occludens-1) binding domains (1). Sequencing of the PTEN gene in spontaneous tumors, and those arising in genetically predisposed individuals, has shown that ≈20% of all known PTEN mutations, including a number of frameshift, nonsense, and missense mutations, target the C-terminal region (2). These data suggest that this region of the PTEN protein may have biological functions distinct from those mediated by its catalytic domain.
The C-terminal domain is phosphorylated in vivo, and this phosphorylation regulates PTEN stability and activity (4). However, the physiologically relevant function of PTEN phosphorylation, other than control of protein stability, continues to be an area of active investigation. Within the C-terminal region, the C2 domain has been shown to regulate the activity and stability of the PTEN protein by binding to the plasma membrane and contributing to the proper orientation of its phosphatase domain (5). The C2 domain can also regulate cell migration, independent of the phosphatase domain (6). However, this inhibition of cell migration can be controlled by PTEN autophosphatase activity (7). It has also been reported that the PTEN C2 domain binds p53 and regulates its transcriptional activity, independent of PTEN phosphatase activity (8). To elucidate the unique functions of the C-terminal region of PTEN, the yeast two-hybrid screen was used to identify its interacting proteins, yielding a known transforming protein, MSP58. Physical interaction with the PTEN C-terminal region functions to suppress the transformation potential of MSP58, and this inhibition does not require PTEN to be catalytically active. These findings point to a previously uncharacterized mechanism through which PTEN can regulate cellular transformation.
Methods
Cell Culture, Retrovirus Infection, and Transfection. Human embryonic kidney 293T cells and Pten–/– MEFs were maintained in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) FBS at 37°C in 5% CO2. Cells were transfected by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Pten–/– MEFs were infected with PTEN and MSP58 coding retroviruses, which were produced in 293T cells. Mammalian expression vectors for PTEN and MSP58 were constructed by PCR with Pfu Turbo DNA polymerase. Various PTEN mutants were constructed in the EGFP and murine stem cell virus vectors by PCR with Pfu Turbo DNA polymerase.
Cell Transformation Assay. Pten–/– MEFs were infected with MSP58 and either wild-type, T336A, or G129R PTEN retroviral vectors, cells were cultured for 40 days, and the resulting foci were stained with 0.5% crystal violet in 20% ethanol and counted.
Antibodies and Immunoblotting Analysis. For whole-cell extracts, cells were washed twice in PBS and lysed in Nonidet P-40 buffer (1% Nonidet P-40/50 mM Tris·HCl, pH 7.4/150 mM NaCl/1 mM EDTA), containing protease and phosphatase inhibitors. Protein samples were separated on SDS-polyacrylamide gels and transferred onto nitrocellulose membranes. Antibodies to Flag (Sigma), GFP (Santa Cruz Biotechnology), PTEN (Santa Cruz Biotechnology), and MSP58 (9) were used for detection.
Coimmunoprecipitation. 293T cells were transfected with the appropriate combination of plasmids harvested and extracted in lysis buffer containing 50 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 0.5% Nonidet P-40. Lysates were cleared by centrifugation at 15,000 × g for 20 min at 4°C and analyzed by SDS/PAGE. For immunoprecipitation, lysates were incubated with anti-Flag or anti-GFP antibody-conjugated beads (Sigma and Roche, respectively) for 8 h at 4°C. The immunoprecipitates were washed four times with lysis buffer and then subjected to immunoblotting.
Immunofluorescence. Immunof luorescence with PTEN and MSP58 was performed with mouse monoclonal PTEN (CascadeBioscience, Winchester, MA) and rabbit polyclonal MSP58 antibodies (9). MEFs were fixed for 30 min in 4% paraformaldehyde, incubated for 1 h with anti-PTEN and MSP58 antibodies and then with anti-mouse IgG Alexa Fluor 488 and anti-rabbit IgG 568 secondary antibodies (Molecular Probes), and viewed with a Nikon microscope equipped with a Hamamatsu ORCA-ER camera controlled by metamorph software (Universal Imaging, Downingtown, PA).
Results
The yeast two-hybrid system was used as a means to further elucidate the potential biological functions of the PTEN C-terminal region through the identification of its binding proteins. To that end, yeast two-hybrid library consisting of human and mouse cDNAs were screened with a bait representing the C-terminal region of PTEN (amino acid 186–401, encompassing the C2 and regulatory phosphorylation domains). Among the isolated cDNAs was one encoding MSP58/MCRS1, a 58-kDa microspherule protein (MSP58) also called human microspherule protein 1 (MCRS1) (10, 11). This protein was of particular interest because MSP58 expression has been reported to be induced upon overexpression of the v-jun oncogene in chicken embryo fibroblasts and to confer anchorage-independent growth (12). The physical interaction between PTEN and MSP58 interaction was confirmed on several levels, including coimmunoprecipitation assays in 293T cells, documenting association between endogenous PTEN and MSP58 (Fig. 1 A and B). In addition, GST pull-down assays demonstrated in vitro binding between GST-PTEN and in vitro-translated MSP58 (Fig. 1C). GST-PTEN also immunoprecipitated MSP58 from lysates of 293T cells transfected with a Flag-MSP58 expression plasmid (Fig. 1D). Finally, we also monitored colocalization of PTEN and MSP58 at physiological levels by immunostaining assays performed in Pten+/+ and Pten–/– MEFs with PTEN and MSP58 antibodies (Fig. 1E). These results illustrate that PTEN does colocalize with MSP58 in the nucleus and that this nuclear location of MSP58 was independent of PTEN expression as shown in Pten–/– MEFs. Thus, PTEN directly interacts with MSP58 under in vitro conditions and under endogenous conditions in vivo.
Fig. 1.
PTEN interacts with MSP58. (A) 293T cell lysates were precipitated with anti-PTEN or control IgG antibody followed by immunoblotting for coprecipitated MSP58. (B) Similarly, lysates were precipitated with anti-MSP58 followed by immunoblotting for coprecipitated PTEN. Efficiency of immunoprecipitation was assessed by reprobing each blot with antibodies used for precipitation. (C) 35S-labeled MSP58 protein, produced by TNT T7-coupled reticulocyte lysate, was applied to GST or GST-PTEN prebound to glutathione Sepharose (GST) beads for pull-down assays. (D) Lysates of 293T cells, which were transfected with empty vector or Flag-MSP58 plasmid, were applied to GST beads for pull-down assays. The pull-down product was detected by immunoblotting with anti-Flag tag horseradish peroxidase (HRP)-conjugated antibody. (E) Immunostaining for PTEN and MSP58 MEFs wild-type or null for PTEN.
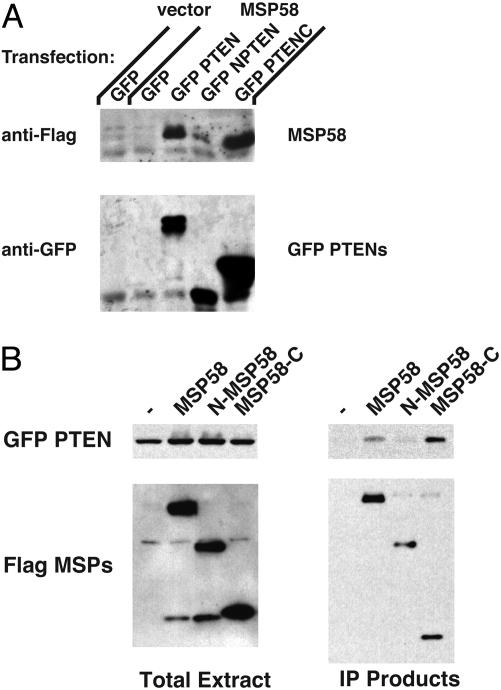
Because we used the PTEN C-terminal region as the bait in the two-hybrid analyses, we infer that MSP58 binds to this region of PTEN. To further define the MSP58-binding domain of PTEN and the PTEN-binding domain of MSP58, we performed a series of experiments in which proteins were coimmunoprecipitated from 293T cells transfected with Flag-MSP58 and various PTEN mutants (Fig. 2A). Full-length PTEN and its C-terminal region bound MSP58, whereas the PTEN phosphatase domain alone could not (Fig. 2 A). We then tested whether the FHA (forkhead-associated) or leucine zipper-like domains contained within MSP58 were required for its interaction with PTEN. The MSP58 FHA domain is necessary and sufficient for interaction with PTEN (Fig. 2B). Therefore, the C-terminal half (the C2 and phosphorylation domains) of PTEN binds MSP58, and the FHA domain of MSP58 binds PTEN.
Fig. 2.
MSP58 binds to the C terminus of PTEN through the MSP58 FHA domain. Lysates of 293T cells, transfected with various combinations of GFP-tagged (PTEN, full-length, amino acids 1–403; NPTEN, amino acids 1–202; PTENC, amino acids 186–403) and Flag-tagged (MSP58, full-length, amino acids 1–462; N-MSP58, amino acids 1–290; MSP58-C, amino acids 290–462) expression plasmids, were used in a coimmunoprecipitation (IP) assay. (A) Lysates of 293T cells were subjected to coimmunoprecipitation with anti-GFP antibody-conjugated agarose beads. Coimmunoprecipitated products were detected by immunoblotting with anti-Flag antibody. For confirmation of IP efficiency, the membrane was stripped and reprobed with anti-GFP antibody. (B) Lysates were incubated with anti-Flag antibody-conjugated agarose. Coimmunoprecipitated products were detected by immunoblotting with anti-GFP horseradish peroxidase (HRP)-conjugated antibody. For confirmation of IP efficiency, the membrane was stripped and reprobed with anti-Flag HRP-conjugated antibody.
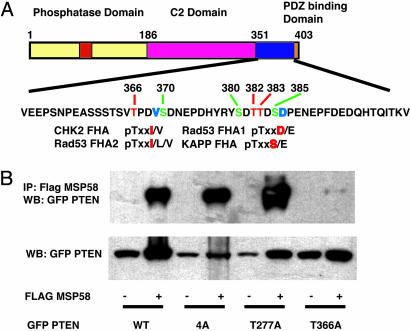
FHA domains recognize phosphothreonine residues with the consensus sequences pTxxI/L/V, pTxxD/V, and pTxxS/E (13, 14). Because the MSP58 FHA domain binds to the C-terminal domain of PTEN, we tested whether MSP58 recognizes a phosphorylated form of PTEN. PTEN can be phosphorylated on Thr-366, Ser-370, Ser-380, Thr-382, Thr-383, and Ser-385 (4, 15). We mutated these serines and threonines to alanine and assessed their interactions with MSP58 (Fig. 3A). We also mutated Thr-277 to alanine, because this site is a perfect match for an FHA domain consensus recognition sequence in the PTEN C-terminal region, although it has not been reported to be phosphorylated. A PTEN 4A mutant (S380A, T382A, T383A, and S385A) and the T277A mutant were still able to bind MSP58; however, the PTEN T366A mutant was deficient for MSP58 binding (Fig. 3B). Therefore, Thr-366 of PTEN is critical for interaction with the MSP58 FHA domain.
Fig. 3.
Thr-366 of PTEN is critical for binding to MSP58. (A) Schematic diagram of phosphorylation sites in the PTEN C-terminal region and the FHA domains of various proteins displaying similarity to PTEN. (B) Lysates of 293T cells, cotransfected with MSP58 and various PTEN mutant expression plasmids, were used in a coimmunoprecipitation (IP) assay. Lysates were incubated with anti-Flag antibody conjugated agarose. Coimmunoprecipitated products were detected by immunoblotting with anti-GFP horseradish peroxidase-conjugated antibody. (Lower) Immunoblotting of GFP PTEN mutants for expression control.
Because MSP58 has been shown to transform chicken embryonic fibroblast cells (12), we sought to determine whether interaction with PTEN could regulate the transformation potential of MSP58. To test this, we used Pten–/– MEFs because this allowed us to introduce PTEN or various mutant forms of it in a cellular background devoid of endogenous PTEN. Exogenous expression of MSP58 did transform Pten–/– MEFs (Fig. 4), and when such transformed foci were picked, expanded, and injected s.c. into immunodeficient mice, they formed tumors (data not shown). The coexpression of wild-type PTEN with MSP58 suppressed both the number and size of the resultant foci (Fig. 4). However, coexpression of the PTEN T366A mutant, which does not interact with MSP58, was unable to effect such suppression (Fig. 4). Interestingly, the G129R phosphatase-inactive mutant of PTEN also had suppressive effects, suggesting that PTEN′s phosphatase activity is not required for suppression of MSP58-mediated transformation.
Fig. 4.
PTEN interaction inhibits focus formation by MSP58. Pten–/– MEFs were infected with viruses expressing the indicated proteins. The cells were incubated for 40 days and then fixed and stained with crystal violet.
Discussion
We report here that the C-terminal domain of the PTEN tumor suppressor protein interacts with the oncogenic MSP58/MCRS1, a 58-kDa microspherule protein (MSP58), and human microspherule protein 1 (MCRS1). MSP58/MCRS1 is a nucleolar protein that directly interacts with ICP22, p120, and Daxx. ICP22 is a regulatory protein from herpes simplex virus 1 (10); p120 is a proliferation-related protein expressed at high levels in most human malignant tumor cells (11); and Daxx is a transcriptional repressor and signal transducer for Fas (16). MCRS2, a splice variant of MSP58/MCRS1, binds the Pin2/TRF1 telomere protein and the catalytic hTERT subunit of telomerase (9). Toj3, an MSP58 orthologue, functions downstream of v-jun to transform quail embryo fibroblast (QEF) and chicken embryo fibroblast (CEF) cells (11). TOJ3 is immediately and specifically activated after v-jun induction with kinetics similar to the induction of well characterized direct AP-1 target genes.
We show that MSP58 and PTEN interact through the FHA domain of MSP58 and the C-terminal region of PTEN. The MSP58 FHA domain is well conserved (12). In eukaryotes, FHA domains are found almost exclusively in nuclear proteins linked to the control of transcription, DNA repair, and cell-cycle progression (13). FHA domains mediate protein–protein interactions and bind to phosphopeptides or to peptides in a phosphorylation-dependent manner (13). We show that one PTEN phosphorylation site (T366) is recognized by the FHA domain of MSP58. Phosphorylation of PTEN Thr-366 has been detected in vivo (15), although the PTEN T366 kinase has not yet been identified. Correspondingly, PTEN and MSP58 colocalize in the nucleus in normal MEFs.
Expression of MSP58 can induce transformation of Pten–/– MEFs. Wild-type PTEN can inhibit MSP58-induced transformation, suggesting a synergy between v-Jun activity and loss of PTEN expression in cell transformation. PTEN likely has functions other than inhibition of the PI3K pathway with its lipid-phosphatase activity. There are several reports that the PTEN C2 domain can regulate cell migration (6, 7) and p53-dependent gene expression (8). We add to this evidence by showing that the phosphatase defective PTEN G129R mutant also suppresses MSP58-mediated transformation. These data are consistent with the report that v-jun-derived transformation is independent of the PI3K pathway in QEFs or CEFs (17).
Although the physiological function of MSP58 is still unclear, it might control gene expression by regulating the stability of the Daxx transcriptional repressor protein (16) and telomerase activity (9). MSP58's ability to transform cells seems contrary to a previous report that MSP58 and MCRS2 inhibit telomerase activity (9). However, dephosphorylation of Akt significantly reduces telomerase activity and induces apoptosis (18). Because PTEN inhibits AKT phosphorylation, it likely reduces AKT-dependent telomerase activity. PTEN may also enhance MSP58 inhibition of telomerase activity by interacting with MSP58.
PTEN is an established tumor suppressor protein that down-regulates the PI3K pathway and p53 regulation through its catalytic phosphatase activity. We propose that PTEN suppression of MSP58 transformation through their protein–protein interaction may represent an addition to the ability of PTEN to interfere with processes of tumorigenesis.
Acknowledgments
We gratefully acknowledge discussions, especially those with Dr. Robert Bachoo (Dana–Farber Cancer Institute) and Michelle Mendoza (Biomedical Sciences Graduate Program, University of California at San Diego), as well as those with Drs. Elizabeth Maher, Cameron Brennan, Lynda Chin, David Louis, and David Rowitch of our program project consortium. This work was supported, in part, by Scholar Awards for cancer research from the Kimmel Foundation and the V Foundation (to F.B.F.), National Cancer Institute Grant CA95616 (to W.K.C., F.B.F., and R.A.D.P.), and a National Foundation for Cancer Research Fellow Award (to W.K.C.).
Abbreviations: FHA, forkhead-associated; MEF, mouse embryo fibroblast.
See Commentary on page 2677.
References
- 1.Maehama, T., Taylor, G. S. & Dixon, J. E. (2001) Annu. Rev. Biochem. 70, 247–279. [DOI] [PubMed] [Google Scholar]
- 2.Eng, C. (2003) Hum. Mutat. 22, 183–198. [DOI] [PubMed] [Google Scholar]
- 3.Larsen, M., Tremblay, M. L. & Yamada, K. M. (2003) Nat. Rev. Mol. Cell Biol. 4, 700–711. [DOI] [PubMed] [Google Scholar]
- 4.Vazquez, F., Ramaswamy, S., Nakamura, N. & Sellers, W. R. (2000) Mol. Cell. Biol. 20, 5010–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee, J. O., Yang, H., Georgescu, M. M., Di Cristofano, A., Maehama, T., Shi, Y., Dixon, J. E., Pandolfi, P. & Pavletich, N. P. (1999) Cell 99, 323–334. [DOI] [PubMed] [Google Scholar]
- 6.Maier, D., Jones, G., Li, X., Schonthal, A. H., Gratzl, O., Van Meir, E. G. & Merlo, A. (1999) Cancer Res. 59, 5479–5482. [PubMed] [Google Scholar]
- 7.Raftopoulou, M., Etienne-Manneville, S., Self, A., Nicholls, S. & Hall, A. (2004) Science 303, 1179–1181. [DOI] [PubMed] [Google Scholar]
- 8.Freeman, D. J., Li, A. G., Wei, G., Li, H. H., Kertesz, N., Lesche, R., Whale, A. D., Martinez-Diaz, H., Rozengurt, N., Cardiff, R. D., et al. (2003) Cancer Cell 3, 117–130. [DOI] [PubMed] [Google Scholar]
- 9.Song, H., Li, Y., Chen, G., Xing, Z., Zhao, J., Yokoyama, K. K., Li, T. & Zhao, M. (2004) Biochem. Biophys. Res. Commun. 316, 1116–1123. [DOI] [PubMed] [Google Scholar]
- 10.Ren, Y., Busch, R. K., Perlaky, L. & Busch, H. (1998) Eur. J. Biochem. 253, 734–742. [DOI] [PubMed] [Google Scholar]
- 11.Bruni, R. & Roizman, B. (1998) J. Virol. 72, 8525–8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bader, A. G., Schneider, M. L., Bister, K. & Hartl, M. (2001) Oncogene 20, 7524–7535. [DOI] [PubMed] [Google Scholar]
- 13.Durocher, D. & Jackson, S. P. (2002) FEBS Lett. 513, 58–66. [DOI] [PubMed] [Google Scholar]
- 14.Li, J., Lee, G. I., Van Doren, S. R. & Walker, J. C. (2000) J. Cell Sci. 23, 4143–4149. [DOI] [PubMed] [Google Scholar]
- 15.Miller, S. J., Lou, D. Y., Seldin, D. C., Lane, W. S. & Neel, B. G. (2002) FEBS Lett. 528, 145–153. [DOI] [PubMed] [Google Scholar]
- 16.Lin, D. Y. & Shih, H. M. (2002) J. Biol. Chem. 277, 25446–25456. [DOI] [PubMed] [Google Scholar]
- 17.Aoki, M., Blazek, E. & Vogt, P. K. (2001) Proc. Natl. Acad. Sci. USA 98, 136–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haendeler, J., Hoffmann, J., Rahman, S., Zeiher, A. M. & Dimmeler, S. (2003) FEBS Lett. 536, 180–186. [DOI] [PubMed] [Google Scholar]