Abstract
Homeodomain-interacting protein kinase-2 (HIPK2) is a serine/threonine kinase involved in transcriptional regulation and apoptosis. The transcriptional corepressor CtBP (carboxyl-terminal binding protein) also plays a fundamental role in these processes. Our previous studies indicate that HIPK2 participates in a pathway of UV-triggered CtBP clearance that results in cell death. HIPK2 phosphorylates CtBP at Ser-422 in vitro. We developed a Ser-422 phospho-specific antibody to demonstrate that CtBP is phosphorylated on this residue in response to UV irradiation. HIPK2 knock-down blocked the UV-induced Ser-422 phosphorylation and degradation. The proteasomal inhibitor MG-132 treatment increased levels of ubiquitinated CtBP, which was induced by UV. Interference with HIPK2 function via the kinase-dead mutant decreased CtBP ubiquitination. Furthermore, a phosphopeptide spanning Ser-422 blocked UV-triggered CtBP degradation, confirming that Ser-422 phosphorylation marks CtBP for clearance. Consequently, interference with HIPK2 action in H1299 cells rescued UV-triggered apoptosis.
Carboxyl-terminal binding protein (CtBP) is a transcriptional corepressor that interacts with a wide variety of cellular factors and carries out important functions in development, cell-cycle regulation, and transformation (1). Microarray assays of mouse embryo fibroblasts (MEFs) derived from animals engineered to contain mutations in the two mammalian CtBP isoforms supported the idea that CtBP is critically involved in gene regulation in mammalian systems (2). Interestingly, several of the genes up-regulated in the knockout MEFs had been linked previously to apoptosis, including PERP (p53-effector related to pmp-22), p21, Noxa, and Bax. Despite the fact that these genes are known p53 targets, CtBP and p53 appear to act through distinct mechanisms. Like cells containing activated p53, the CtBP-null MEFs had an increased sensitivity to proapoptotic stimuli. Thus, CtBP can be considered an antiapoptotic factor. Disruption of the two CtBP isoforms in mice causes lethality at day 8 of development because of a variety of defects (3).
The profound effects of CtBP ablation, both in cell culture and intact animals, suggest that CtBP influences a wide range of biological processes. Relatively little is known about the regulation of transcriptional coactivators and corepressors by intracellular signaling pathways, however. As opposed to modifications of specific DNA-binding factors, phosphorylation of coregulators has the potential to influence a large number of transcriptional programs. Such pathways are just beginning to be characterized. Our previous studies have identified a novel pathway of CtBP regulation (4). In this process, a kinase called homeodomain-interacting protein kinase-2 (HIPK2) is activated by UV irradiation. HIPK2, in turn, targets CtBP for degradation through the proteasomal pathway. By directing CtBP toward degradation, HIPK2 can induce cell death through a p53-independent mechanism. Consistent with this model, short interfering RNA (siRNA) knockdown of CtBP induces apoptosis in cells that are deficient for p53. This pathway may induce apoptosis in p53-deficient tumor cells. The connection between HIPK2 action and CtBP degradation remained unknown, however. In particular, it was not clear how HIPK2 caused CtBP modifications that lead toward proteasomal clearance.
Growth arrest and apoptosis are crucial mechanisms to counteract cellular transformation and carcinogenesis. HIPK2 is a nuclear serine/threonine kinase participating in transcriptional regulation (5, 6), growth control (7), and apoptosis (8, 9). Previously, HIPK2 had been shown to mediate the phosphorylation of p53 in response to UV irradiation, thereby activating p53 function and promoting apoptosis (8, 9). Nevertheless, as we have shown, HIPK2 also promotes apoptosis through its effects on CtBP. We hypothesized that phosphorylation by HIPK2 marks CtBP for clearance. The resultant decrease in CtBP levels, like the HIPK2 activation of p53, sensitizes cells to apoptosis. This model predicts that interference with HIPK2 function might rescue UV-triggered apoptosis in p53-null cells.
To monitor CtBP modification in vivo, we raised a phospho-specific antibody (Ab) and demonstrated that CtBP is indeed phosphorylated on Ser-422 in response to UV irradiation. We also showed that CtBP becomes ubiquitinated after exposure to UV. These modifications can be prevented by blocking the HIPK2 pathway, suggesting that HIPK2 is the main mediator connecting UV irradiation and CtBP regulation. Consequently, interference with the HIPK2 action in H1299 cells rescues their apoptotic response to UV irradiation.
Materials and Methods
Plasmids and Proteins. EGFP-HIPK2 and Myc-HIPK2 (both wild type and the K221R kinase-inactive form) were from Yongsok Kim (National Institutes of Health, Bethesda). FLAG-CtBP and PET24bCtBP are described in ref. 10. CtBP1 was expressed as a His-6 fusion in BL21(DE3) and purified by Ni-nitrilotriacetic acid affinity chromatography (Qiagen, Valencia, CA). HeLa nuclear extract was prepared as described in ref. 11. Phospho-Ser-422 peptide (HPPHAPphosSPGQTVKPE) and the control peptide (HPPHAPSPGQTVKPE) were synthesized by Invitrogen.
Antibodies (Abs). Anti-CtBP Abs were generated by immunizing rabbits with either the full-length protein or the carboxyl-terminal peptide (12). Anti-ubiquitin Ab was purchased from Pharmingen. Ab to α-tubulin was from Sigma. Polyclonal anti-phospho Ser-422 Ab was generated by immunizing rabbits with phospho-Ser-422 peptide (HPPHAPphosSPGQTVKPE), followed by cross-absorption by a nonphosphorylated form of this peptide (HPPHAPSPGQTVKPE) and affinity purification by using the phosphopeptide column. The affinity-purified Ab was used in this study.
Cell Culture, Treatment, and Transfection. COS-7, H1299, and U2OS cells and CtBP-null MEFs (3) were cultured in DMEM plus 10% FBS and 1% penicillin/streptomycin. The spectral characterization of the UV-B source and the mode of the irradiation of cells with UV-B are described in ref. 13. Proteasome inhibitor MG-132 was from Calbiochem. COS-7 cells were transfected by FuGene 6 (Roche) or Lipofectamine 2000 (Invitrogen). siRNA duplexes were introduced to cells by Lipofectamine 2000. CtBP immunoprecipitated from UV-irradiated COS-7 cells was treated with λ-phosphatase for 30 min at 30°C. Cellular CtBP and DNA staining or the expression of GFP-HIPK2 were examined by fluorescence microscopy.
Dot Blotting and Western Blotting Assays. Serial dilutions of peptides were spotted on a nitrocellulose membrane and blotted by using the Abs purified over the nonphosphorylated or phosphopeptide column. Cell lysate and protein samples were electrophoresed on a SDS/8% polyacrylamide gel and assayed after transfer to a poly(vinylidene difluoride) membrane by using specific Abs.
Immunoprecipitations and in Vivo Ubiquitination. COS-7 cells expressing FLAG-CtBP were pretreated with MG-132 at a concentration of 2.5 μM overnight and irradiated with UV. Anti-FLAG M2-matrix was blocked with BSA and used to precipitate tagged proteins from COS-7 cell extracts in RIPA buffer (0.15 M NaCl/0.05 M Tris·HCl, pH 7.6/1% Nonidet P-40/1% sodium deoxycholate/0.1% SDS) plus protease inhibitors (Complete, Boehringer Mannheim) for 1 h at 4°C. The beads were washed three times with RIPA buffer, boiled in 15 μl of 5× sample buffer (pH 6.8), and electrophoresed on an SDS/8% polyacrylamide gel. After transfer to a poly(vinylidene difluoride) membrane, the bound fraction was assayed for CtBP or ubiquitin by Western blotting using anti-CtBP or -ubiquitin Abs, respectively.
siRNA and RT-PCR. The siRNA vector for reduction of HIPK2 levels is described in ref. 4. FLAG-CtBP, GFP-HIPK2, and HIPK2 siRNA pH1 vectors were cotransfected into COS-7 cells. GFP-HIPK2 was used as a marker. siRNA oligonucleotides for HIPK2 were from Dharmacon Research (Lafayette, CO). Scramble II Duplex was used as control. Cells were transfected with 100 nM oligonucleotides, and RT-PCR was performed to measure the decrease of HIPK2 mRNA. RNA was extracted with an RNeasy kit (Qiagen). DNase I (Ambion, Austin, TX)-treated RNA was used for reverse transcription. RNA was incubated with oligo(dT) primers (Invitrogen) and 1 mM dNTPs at 65°C for 5 min, cooled on ice, then incubated with 10 mM DTT, 5× buffer, and RNasin (Promega) RNase inhibitor at 42°C for 2 min. Reverse transcription was performed by using Super Transcript III (Invitrogen) at 42°C for 50 min. Reverse transcriptase was inactivated at 70°C for 15 min, and the product was treated with RNase A at 37°C for 30 min before PCR amplification.
Chariot Delivery of Peptide. The Ser-422 phosphopeptide or the control (nonphosphorylated) peptide was delivered by Chariot (Active Motif) into COS-7 cells according to the manufacturer's instructions. Briefly, the peptide/Chariot mix was loaded onto cells without other proteins for 1 h before the addition of serum-containing medium. β-gal staining showed that >90% of the cells were transfected.
Apoptosis Measurements. H1299 cells transfected with the kinase-dead HIPK2 or the control plasmid were UV-irradiated. Apoptosis was assayed by Hoechst staining for DNA condensation.
Results
Generation of a Ser-422 Phospho-Specific Ab of CtBP. Previously, we mapped the HIPK2 phosphorylation site on CtBP to Ser-422, which resides at its carboxyl terminus (4). Mutation of this critical serine to alanine (S422A) prevented CtBP degradation in response to either UV irradiation or HIPK2 overexpression, suggesting that phosphorylation of this residue is required for CtBP degradation (4). Consistent with the role of CtBP as an antiapoptotic factor, the S422A mutant rescued the UV-triggered apoptosis better than wild-type CtBP (4). These studies did not examine CtBP phosphorylation in the context of the native protein, however. To further explore the modification of CtBP protein in vivo, we generated polyclonal antisera by immunizing rabbits with a phosphopeptide corresponding to the HIPK2-modification region of CtBP-1.
The low stoichiometry of the phosphorylation of recombinant CtBP by HIPK2 in vitro complicates Ab screening. Consequently, we switched to dot blotting by using the control (HPPHAP-SPGQTVKPE) and the phosphopeptide (HPPHAPphos-SPGQTVKPE) spotted onto nitrocellulose membranes. A serial dilution of both peptides was subjected to Western-blotting using a 1:10,000 dilution of the antisera. A strong immune response was observed for one of the rabbits, and several test bleeds showed >100-fold preference for the phosphopeptide (data not shown).
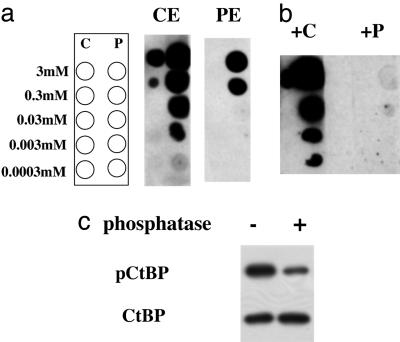
We immobilized the peptides on columns and chose the most promising test bleeds for further purification. The antisera were passed through the control peptide column and then applied to the phosphopeptide column. After extensive washes, the bound material was eluted at low pH, and the input and elutions were subjected to dot blotting. The eluates from the control peptide column still recognized the phosphopeptide, and the fold difference was similar for both eluates (Fig. 1a). This result indicated the presence of a single population of Ab that cross-reacts with the nonphosphorylated peptide. Next, we performed competition assays with either the control or the phosphopeptide during the Ab-binding step. At 0.1 mM concentration, the phosphopeptide effectively blocked binding of the Ab to both the phosphopeptide and the control peptide (Fig. 1b), further suggesting that there is one population of Ab with preference toward the phosphopeptide. The control peptide was ineffective in the competition, demonstrating the low affinity of the Ab toward the nonphosphorylated form. Treatment of CtBP immunoprecipitated from UV-irradiated cells with a Ser/Thr-specific phosphatase eliminated ≈75% the signal detected by the antiphospho-Ser-422 Ab (Fig. 1c).
Fig. 1.
Generation of a Ser-422 phospho-specific Ab. (a) Ab purification. Antisera were passed through the control peptide column and then applied to the phosphopeptide column. After extensive washes, the bound material was eluted at low pH, and the elutions from the control (CE) or the phosphopeptide (PE) column were subjected to dot blotting. The diagram on the left depicts the dot blot layout. Used were 1:10 serial dilutions of the control HPPHAPSPGQTVKPE (C) or the phosphopeptide HPPHAPphosSPGQTVKPE (P). (b) Competition assays. Dot blotting by using the Ser-422 phospho-specific Ab was performed in the presence of either the control (+C) or the phosphopeptide (+P) at 0.1 mM concentration during the Ab-binding step. (c) Phosphatase treatment. CtBP immunoprecipitated from UV-irradiated COS-7 cells was treated with λ-phosphatase and analyzed by using Abs to phospho-Ser-422 (pCtBP) or CtBP.
Phosphorylation on Ser-422 of CtBP in Response to UV Irradiation and HIPK2 Overexpression. HIPK2 has been identified as a stress-activated nuclear Ser/Thr kinase able to phosphorylate p53 family members (8, 9). Previously, we showed that either UV treatment or overexpression of HIPK2 leads to CtBP degradation and that the S422A mutation stabilized the CtBP protein in both settings (4). This finding suggests that phosphorylation of Ser-422 is the cause of CtBP clearance. With a phospho-Ser-422-specific Ab in hand, we set out to study the phosphorylation of CtBP in vivo.
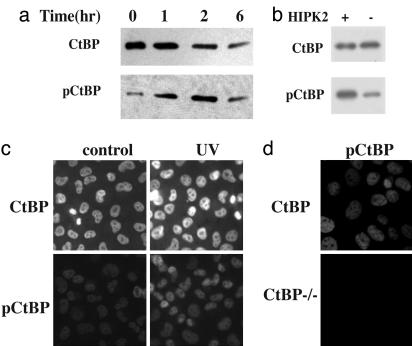
HIPK2 can be activated by UV irradiation. We followed the phosphorylation of CtBP after UV treatment, taking care to account for the crossreactivity of the Ab with the nonphosphorylated form of CtBP. We used a CtBP overexpressing system for some of our studies to take advantage of the quantitative reflection of CtBP levels shown by a previously developed Ab with lower sensitivity. COS-7 cells were transfected with CtBP-expressing vector and irradiated with UV-B. Samples were harvested at different time points. Phosphatase inhibitors were included in the lysis buffer to help preserve the phosphorylation of Ser-422. With the CtBP protein level gradually declining over the time course, the signal from the phospho-specific Ab peaked at 1–2 h after UV irradiation (Fig. 2a), proving that Ser-422 of CtBP was indeed phosphorylated in response to UV irradiation.
Fig. 2.
Phosphorylation of CtBP Ser-422 in response to UV irradiation and HIPK2 expression. (a) Time course of Ser-422 phosphorylation. COS-7 cells were transfected with CtBP-expressing vector and irradiated with UV-B (4,000 J/m2). Samples were harvested at different times and subjected to Western blotting with the Ser-422 phospho-specific (pCtBP) or CtBP Abs. (b) Ser-422 phosphorylation induced by HIPK2 expression. COS-7 cells were transfected with expressing vectors for both CtBP and HIPK2 (+) or the empty vector (–). Samples were subjected to Western blotting with the Ser-422 phospho-specific (pCtBP) or CtBP Ab as indicated. (c) Immunostaining of Ser-422 phosphorylation in response to UV irradiation. One hour after UV irradiation, H1299 cells were fixed and stained with Abs against CtBP or the phospho-Ser-422-specific Ab. Increased staining for pCtBP is evident after UV irradiation. (d) CtBP-null cells with (CtBP) or without (CtBP–/–) CtBP expression were UV-treated and immunostained with the anti-phospho-Ser-422 Ab (pCtBP).
Next, we asked whether the coexpression of HIPK2 also leads to CtBP phosphorylation at Ser-422. Overexpressing HIPK2 causes CtBP clearance, so we collected the samples at an earlier time point. As shown in Fig. 2b, the phospho-signal was stronger in the HIPK2-expressing cells when samples with similar protein levels were assayed.
Ser-422 Phosphorylation of CtBP in the Nucleus. Ser-158 phosphorylation and Lys-428 sumoylation have been linked to CtBP redistribution from the nucleus to the cytoplasma (14, 15). To study the subcellular localization of CtBP after Ser-422 phosphorylation by immunohistochemistry, we fixed H1299 cells at 1 h after UV-irradiation and stained with Abs against CtBP or the phospho-Ser-422-specific Ab (Fig. 2c). Whereas the CtBP protein level remained constant, the phospho-Ser-422 signal was stronger in the cells after UV treatment. No cytoplasmic staining was observed. In a control experiment performed in CtBP-null MEFs, the phospho-Ser-422-specific Ab did not recognize other cellular proteins (Fig. 2d).
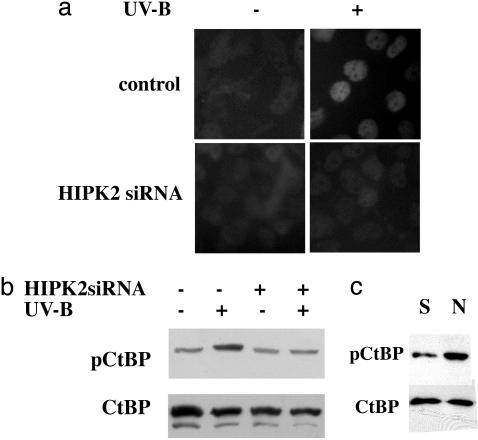
HIPK2 Requirement in Ser-422 Phosphorylation in Response to UV Irradiation. To assess the involvement of HIPK2 in CtBP phosphorylation, we perturbed HIPK2 expression in cells by using siRNA. The DNA sequences of the functional HIPK2 domains are highly conserved in mammals. A siRNA targeting the central region of HIPK2 has been cloned into the pH1 vector and used successfully to block HIPK2 expression (4). First, we introduced these constructs into COS-7 cells. To reveal the siRNA effect, we cotransfected the cells with GFP-HIPK2 and observed the decline of the green fluorescence signal with the siRNA treatment (ref. 4 and data not shown). UV-stimulated CtBP Ser-422 phosphorylation was inhibited by HIPK2 interference (Fig. 3 a and b), demonstrating that HIPK2 is required for the UV-triggered phosphorylation of Ser-422. We obtained similar results with an independent siRNA also directed against HIPK2 (data not shown).
Fig. 3.
HIPK2 involvement in UV-mediated Ser-422 phosphorylation. (a) siRNA to HIPK2 (HIPK2 siRNA) attenuates Ser-422 phosphorylation of endogenous CtBP in H1299 cells in response to UV irradiation. The empty vector, pH1, served as control. Cells were stained with the anti-phospho-Ser-422 Ab. (b) UV-triggered Ser-422 phosphorylation blocked by HIPK2 siRNA. HIPK2 expression in COS-7 cells was perturbed by siRNA (+). The empty vector, pH1, served as control (–). CtBP expressed in COS-7 cells with (+) or without (–) UV irradiation was monitored by Western blotting using the phospho-specific (pCtBP) or CtBP Abs. (c) Basal phosphorylation of CtBP at Ser-422. CtBP from HeLa nuclear extract (N) was compared with recombinant CtBP (S). Bacterially purified CtBP served as titration standard for the nonphosphorylated protein. Samples were subjected to Western blotting with the phospho-specific (pCtBP) or CtBP Abs.
Basal Phosphorylation of CtBP at Ser-422. Next, we asked whether Ser-422 phosphorylation participates in CtBP homeostasis. Recombinant CtBP purified from bacterial extracts is free of phosphorylation and can serve as a titration standard for non-phosphorylated protein. HeLa nuclear extract was assayed by Western blotting using the phospho-specific and CtBP Abs (Fig. 3c). Signals from the samples were within the linear ranges for both Abs (data not shown). At equivalent protein levels, Ser-422 phospho-CtBP from the nuclear extract gave a stronger signal than the recombinant CtBP (Fig. 3c), supporting the idea that Ser-422 is phosphorylated to some degree under basal conditions in HeLa cells. We suggest, therefore, that phosphorylation may contribute to CtBP homeostasis.
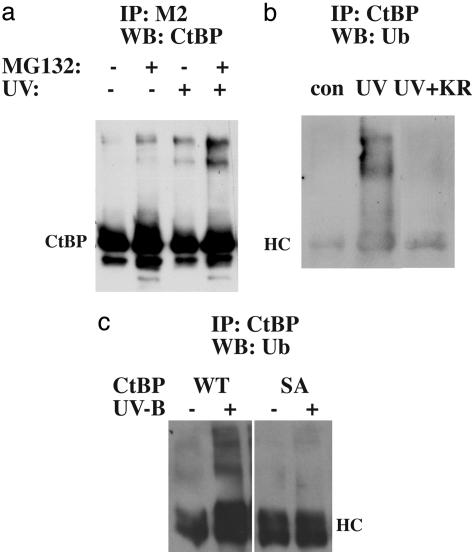
UV Induction of CtBP Ubiquitination. Previously, we showed that UV-triggered CtBP degradation is prevented by proteasomal inhibitor treatment (4). Polyubiquitination has been recognized as a marker for proteasomal clearance. To test this modification of CtBP in vivo, we pretreated the CtBP-expressing cells with MG-132 before irradiation with UV-B. FLAG-tagged CtBP was immunoprecipitated and assayed by Western blotting. A ladder of CtBP signal appeared, most prominently in the UV plus MG-132 treatment sample (Fig. 4a). Some basal modification was observed for CtBP with MG-132 blockage alone, consistent with our data that Ser-422 of CtBP is basally phosphorylated. Next, we pulled down CtBP from MG-132-treated cells and probed with anti-ubiquitin Ab. The high molecular weight ladder of CtBP was positive for ubiquitin (Fig. 4b). Of note, the ubiquitin signal was stronger in the presence of UV irradiation.
Fig. 4.
CtBP ubiquitination. (a) FLAG-CtBP expressed in COS-7 cells was immunoprecipitated and assayed by Western blotting with the CtBP Ab. Cells were treated with MG-132 and UV irradiated, as indicated. (b) Comparison of CtBP ubiquitination in cells with (+KR) or without the expression of the kinase-dead HIPK2. FLAG-CtBP expressed in MG-132 pretreated COS-7 cells before (con) or after (UV) UV irradiation was immunoprecipitated by a rabbit anti-CtBP Ab and assayed by Western blotting using the anti-ubiquitin Ab. HC, heavy chain. (c) CtBP ubiquitination of the wild-type (WT) or the S422A mutant (SA). CtBP expressed in CtBP-null MEFs before (–) or after (+) UV irradiation was immunoprecipitated by a rabbit anti-CtBP Ab and assayed by Western blotting by using the anti-ubiquitin Ab.
To study the contribution of HIPK2 to CtBP ubiquitination, we compared the ubiquitination profile of CtBP in the absence or presence of the kinase-dead HIPK2 KR. Previous studies indicate that this mutant functioned as a dominant negative to stabilize CtBP protein in response to UV irradiation (4). Expression of HIPK2 KR prevented UV-induced Ser-422 phosphorylation (data not shown). Ubiquitination of CtBP was decreased by HIPK2-KR expression in the presence of UV irradiation (Fig. 4b), demonstrating that HIPK2 is required for UV-induced CtBP ubiquitination. Furthermore, ubiquitination of the S422A mutant was not observed (Fig. 4c).
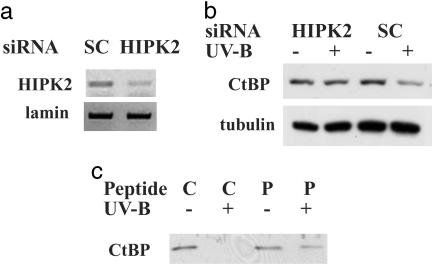
HIPK2-Mediated Ser-422 Phosphorylation in CtBP Degradation. To further investigate the role of HIPK2 in UV-triggered degradation of endogenous CtBP, we introduced a siRNA duplex to interfere with HIPK2 in U2OS cells. This duplex has been shown to efficiently block HIPK2 expression in human cells (16). RT-PCR demonstrated that HIPK2 mRNA was significantly down-regulated by siRNA treatment (Fig. 5a). UV-induced CtBP decrease was prevented by HIPK2 siRNA treatment (Fig. 5b). This information is critical for establishing our model that CtBP is regulated by UV irradiation through the activation of HIPK2.
Fig. 5.
CtBP regulation by HIPK2. (a) Efficacy of HIPK2 siRNA. A siRNA duplex targeting HIPK2 or the scrambled duplex (SC) was introduced to cells. RT-PCR was performed to reveal the knockdown of HIPK2 mRNA. Lamin mRNA levels served as control. (b) The role of HIPK2 in UV-triggered degradation of endogenous CtBP. A siRNA duplex was introduced to interfere with HIPK2 expression in U2OS cells. The scrambled duplex (SC) served as control. UV-induced decrease of endogenous CtBP was assayed by Western blotting. Tubulin control remained constant. (c) Phospho-Ser 422 peptide attenuates UV-triggered CtBP degradation. A phosphopeptide (P) or the nonphosphorylated peptide (C) spanning Ser-422 of CtBP was delivered to cells by Chariot. UV-induced decrease of CtBP was assayed by Western blotting.
Our previous results suggest that Ser-422 phosphorylation marks CtBP for clearance (4). This modification on Ser-422 must be read and interpreted by cellular factor(s), and we propose that a specific domain of CtBP containing this critical information should compete with CtBP for the degradation machinery. We addressed this issue by introducing a peptide containing the Ser-422 phosphorylation site. “Chariot” is a delivery reagent that quickly and efficiently transports biologically active proteins, peptides, and Abs directly into cells with an efficiency of 60–95% (17). The Chariot peptide forms a noncovalent bond with the macromolecule of interest, which stabilizes the protein, protecting it from degradation and preserving its natural characteristics during the transfection process. After delivery, the complex dissociates, leaving the macromolecule biologically active and free to proceed to its target organelle.
The Ser-422 phosphopeptide or the control nonphosphorylated peptide were efficiently delivered by Chariot into COS-7 cells. Staining the indicator β-gal showed that >90% of the cells were positive (data not shown). By delivering a specific phosphopeptide encompassing CtBP Ser-422 into cells, we showed that CtBP degradation was attenuated in response to UV irradiation (Fig. 5c). Ser-422 phosphorylation of CtBP was not affected (data not shown), suggesting that the phosphopeptide functions downstream from HIPK2-mediated phosphorylation.
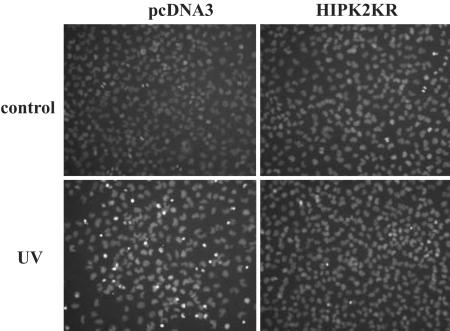
HIPK2 Functional Blockage Rescues the Apoptotic Response. CtBP governs the expression of a panel of proapoptotic genes and CtBP-null cells are prone to apoptosis (2). Because HIPK2 critically down-regulates CtBP levels, blocking the HIPK2 pathway should relieve the stress response of cells. The introduction of the HIPK2 siRNA (data not shown) or kinase-dead HIPK2 KR mutant (Fig. 6) rescued the cell death response of the p53-null H1299 cells after UV irradiation.
Fig. 6.
HIPK2 block rescues the apoptotic response. H1299 cells were transfected with the kinase-dead HIPK2 (HIPK2KR) or the empty vector (pcDNA3). After UV irradiation, the cells were stained with Hoechst to monitor apoptosis-associated chromatin condensation. Two hundred cells from three random fields were counted. The numbers of condensed nuclei are 3.3 ± 1.5 (control) vs. 56.3 ± 7.0 (UV irradiation) for the pcDNA3-transfected cells and 2.3 ± 1.1 (control) vs. 5.3 ± 1.5 (UV irradiation) for the HIPK2KR-transfected cells.
Discussion
The regulation of transcriptional coactivators and corepressors by intracellular signaling pathways coordinates a large number of transcriptional programs and has been under extensive investigation. As one of the central players in transcriptional repression, CtBP regulation has received much attention. One pathway for regulation involves the metabolic intermediates, NAD and NADH. It has been demonstrated that NAD/NADH-induced conformational change in CtBP family members results in an increase in their affinity for PXDLS motif-containing proteins, such as adenoviral E1A and ZEB (12, 18, 19). Another mechanism for regulation is by posttranslational modification. CtBP was reported to be phosphorylated on Ser-158 by Pak1 (14) and sumoylated on Lys-428 by means of the actions of PC2, a polycomb group protein (15, 20). Both modifications have been linked to the regulation of CtBP localization. Furthermore, adenoviral E1A, Epstein–Barr virus oncoprotein EBNA3A, and the nuclear speckle-associated protein Pnn/DRS have been shown to sequester CtBP and thereby relieve CtBP-mediated repression (21–24).
Our previous studies indicate that HIPK2 can phosphorylate CtBP at Ser-422. The S422A mutation is more stable than the wild-type protein and protects better against UV-induced apoptosis (4). In this study, we developed a Ser-422 phospho-specific Ab and demonstrated that CtBP is indeed phosphorylated on this residue in vivo in response to UV treatment. Interference with HIPK2 function blocked the UV-induced Ser-422 phosphorylation and CtBP ubiquitination, thus stabilizing endogenous CtBP after UV irradiation. These studies demonstrate that HIPK2 is the main pathway that connects UV irradiation and CtBP regulation.
HIPK2 is a Ser/Thr kinase that has been shown previously to be activated by UV irradiation (9), TGF-β treatment (16), Wnt signaling, and TAK1 action (25). It plays important roles in transcriptional regulation and promotes cell growth arrest and apoptosis. HIPK2 activation triggers Ser-46 phosphorylation on p53 and prevents MDM2-mediated ubiquitination and degradation of p53 (8, 9, 26), resulting in the apoptotic response. A recent report also demonstrated that HIPK2 controls sensory neuron survival by suppressing the Brn3a-dependent transcription of Brn3a, TrkA, and Bcl-xL (27). Trigeminal sensory neurons, which are especially susceptible to HIPK2-induced apoptosis, express the highest levels of HIPK2 during the peak of apoptosis in vivo, supporting the idea that HIPK2 participates in programmed cell death in the developing peripheral nervous system (28). Our study confirmed that the transcriptional corepressor CtBP is a HIPK2 target. In addition to stabilizing p53, HIPK2 phosphorylates Ser-422 on the antiapoptotic factor CtBP. This action destabilizes CtBP, thus providing an alternative pathway for programmed cell death. Of note, this pathway should be functional in p53-deficient cells, which includes most human tumors.
Precisely how Ser-422 phosphorylation marks CtBP for clearance is uncertain. This modification on Ser-422 must be read and interpreted by the cellular machinery. It is likely that CtBP phosphorylation causes the recruitment of a particular E3 ligase that triggers ubiquitination. Alternatively, novel enzyme(s) and bridging factor(s) might play critical roles in CtBP clearance. Elucidating the mechanism of the phosphorylation-dependent degradation of CtBP is essential for understanding CtBP regulation.
It was interesting that Ser-422 phosphorylation and ubiquitination was observed even in the absence of UV stimulation, suggesting that these modifications are a part of CtBP homeostasis. This finding is in agreement with our previous finding that the Ser-422 mutant of CtBP was detected at a higher level than the wild-type protein when both were expressed in a CtBP-null background (4). Whether basal activity of HIPK2 or other kinases are responsible for this action is unknown.
Posttranslational modifications of protein have been widely used by cells to modulate crucial responses and are usually coordinated events. For example, tight regulation of p53 activation is imperative for preventing tumorigenesis and maintaining normal cell growth. Phosphorylation, acetylation, and ubiquitination of p53 dictate its overall appearance and activity (29). c-Myc, another potent regulator of cellular proliferation and cell fate decision, is also subject to sophisticated control, in that phosphorylation of Ser-62 stabilizes c-Myc, whereas the subsequent phosphorylation at Thr-58 is required for its degradation (30). CtBP has been reported to be phosphorylated on Ser-158 and sumoylated on Lys-428. Whether these modifications are interconnected is currently under investigation.
Whether HIPK2 directly or indirectly phosphorylates CtBP also remains unclear. The c-Jun-amino-terminal kinase has been reported to be activated by ectopic expression of HIPK2 (14) and also can mediate Ser-422 phosphorylation in vitro (Q.Z., unpublished observation). Other nuclear kinases might also phosphorylate Ser-422 and trigger the decrease in CtBP levels, thus sensitizing cells to apoptosis. If so, this hypothesis would suggest that CtBP responds to an even wider-ranging pattern of signaling pathways than is currently imagined. Conceivably, activation of one of these signaling pathways could be used therapeutically to down-regulate CtBP and block the ability of factors such as Evi-1 to mediate cell transformation. Alternatively, because cell adhesion genes like E-cadherin, and proapoptotic genes such as Noxa, Bax, and PERP, are up-regulated by CtBP ablation, it is possible that manipulation of CtBP levels could be used to induce apoptosis and decrease the propensity of certain tumors to metastasize.
Acknowledgments
We thank Y. Kim for providing the HIPK2 cDNA, Y. Liu for imaging, and M. Thayer (Oregon Health and Science University) for the siRNA vector. This work was supported by National Institutes of Health Grants P01DK044239 (to R.H.G.) and K01CA96561 (to Q.Z.).
Abbreviations: CtBP, carboxyl-terminal binding protein; HIPK2, homeodomain-interacting protein kinase-2; MEF, mouse embryonic fibroblast; siRNA, short interfering RNA.
References
- 1.Chinnadurai, G. (2002) Mol. Cell 9, 213–224. [DOI] [PubMed] [Google Scholar]
- 2.Grooteclaes, M., Deveraux, Q., Hildebrand, J., Zhang, Q., Goodman, R. H. & Frisch, S. M. (2003) Proc. Natl. Acad. Sci. USA 100, 4568–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hildebrand, J. D. & Soriano, P. (2002) Mol. Cell. Biol. 22, 5296–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang, Q., Yoshimatsu, Y., Hildebrand, J., Frisch, S. M. & Goodman, R. H. (2003) Cell 115, 177–186. [DOI] [PubMed] [Google Scholar]
- 5.Kim, Y. H., Choi, C. Y., Lee, S. J., Conti, M. A. & Kim, Y. (1998) J. Biol. Chem. 273, 25875–25879. [DOI] [PubMed] [Google Scholar]
- 6.Choi, C. Y., Kim, Y. H., Kwon, H. J. & Kim, Y. (1999) J. Biol. Chem. 274, 33194–33197. [DOI] [PubMed] [Google Scholar]
- 7.Pierantoni, G. M., Fedele, M., Pentimalli, F., Benvenuto, G., Pero, R., Viglietto, G., Santoro, M., Chiariotti, L. & Fusco, A. (2001) Oncogene 20, 6132–6141. [DOI] [PubMed] [Google Scholar]
- 8.D'Orazi, G., Cecchinelli, B., Bruno, T., Manni, I., Higashimoto, Y., Saito, S., Gostissa, M., Coen, S., Marchetti, A., Del Sal, G., et al. (2002) Nat. Cell Biol. 4, 11–19. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann, T. G., Moller, A., Sirma, H., Zentgraf, H., Taya, Y., Droge, W., Will, H. & Schmitz, M. L. (2002) Nat. Cell Biol. 4, 1–10. [DOI] [PubMed] [Google Scholar]
- 10.Zhang, Q., Yao, H., Vo, N. & Goodman, R. H. (2000) Proc. Natl. Acad. Sci. USA 97, 14323–14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, Q., Vo, N. & Goodman, R. H. (2000) Mol. Cell. Biol. 20, 4970–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, Q., Piston, D. W. & Goodman, R. H. (2002) Science 295, 1895–1897. [DOI] [PubMed] [Google Scholar]
- 13.Iordanov, M. S., Choi, R. J., Ryabinina, O. P., Dinh, T. H., Bright, R. K. & Magun, B. E. (2002) Mol. Cell. Biol. 22, 5380–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnes, C. J., Vadlamudi, R. K., Mishra, S. K., Jacobson, R. H., Li, F. & Kumar, R. (2003) Nat. Struct. Biol 10, 622–628. [DOI] [PubMed] [Google Scholar]
- 15.Kagey, M. H., Melhuish, T. A. & Wotton, D. (2003) Cell 113, 127–137. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, T. G., Stollberg, N., Schmitz, M. L. & Will, H. (2003) Cancer Res. 63, 8271–8277. [PubMed] [Google Scholar]
- 17.Morris, M. C., Depollier, J., Mery, J., Heitz, F. & Divita, G. (2001) Nat. Biotechnol. 19, 1173–1176. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, V., Carlson, J. E., Ohgi, K. A., Edwards, T. A., Rose, D. W., Escalante, C. R., Rosenfeld, M. G. & Aggarwal, A. K. (2002) Mol. Cell 10, 857–869. [DOI] [PubMed] [Google Scholar]
- 19.Fjeld, C. C., Birdsong, W. T. & Goodman, R. H. (2003) Proc. Natl. Acad. Sci. USA 100, 9202–9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, X., Sun, B., Liang, M., Liang, Y. Y., Gast, A., Hildebrand, J., Brunicardi, F. C., Melchior, F. & Feng, X. H. (2003) Mol. Cell 11, 1389–1396. [DOI] [PubMed] [Google Scholar]
- 21.Schaeper, U., Boyd, J. M., Verma, S., Uhlmann, E., Subramanian, T. & Chinnadurai, G. (1995) Proc. Natl. Acad. Sci. USA 92, 10467–10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grooteclaes, M. L. & Frisch, S. M. (2000) Oncogene 19, 3823–3828. [DOI] [PubMed] [Google Scholar]
- 23.Hickabottom, M., Parker, G. A., Freemont, P., Crook, T. & Allday, M. J. (2002) J. Biol. Chem. 277, 47197–47204. [DOI] [PubMed] [Google Scholar]
- 24.Alpatov, R., Munguba, G. C., Caton, P., Joo, J. H., Shi, Y., Hunt, M. E. & Sugrue, S. P. (2004) Mol. Cell. Biol. 24, 10223–10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanei-Ishii, C., Ninomiya-Tsuji, J., Tanikawa, J., Nomura, T., Ishitani, T., Kishida, S., Kokura, K., Kurahashi, T., Ichikawa-Iwata, E., Kim, Y., et al. (2004) Genes Dev. 18, 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Stefano, V., Blandino, G., Sacchi, A., Soddu, S. & D'Orazi, G. (2004) Oncogene 23, 5185–5192. [DOI] [PubMed] [Google Scholar]
- 27.Wiggins, A. K., Wei, G., Doxakis, E., Wong, C., Tang, A. A., Zang, K., Luo, E. J., Neve, R. L., Reichardt, L. F. & Huang, E. J. (2004) J. Cell Biol. 167, 257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doxakis, E., Huang, E. J. & Davies, A. M. (2004) Curr. Biol. 14, 1761–1765. [DOI] [PubMed] [Google Scholar]
- 29.Brooks, C. L. & Gu, W. (2003) Curr. Opin. Cell Biol. 15, 164–171. [DOI] [PubMed] [Google Scholar]
- 30.Sears, R., Nuckolls, F., Haura, E., Taya, Y., Tamai, K. & Nevins, J. R. (2000) Genes Dev. 14, 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]